Abstract
The transforming growth factor-β (TGFβ) family plays a critical regulatory role in repair and coordination of remodeling after cutaneous wounding. TGFβ1-mediated chemotaxis promotes the recruitment of fibroblasts to the wound site and their resultant myofibroblastic transdifferentiation that is responsible for elastic fiber deposition and wound closure. TGFβ3 has been implicated in an antagonistic role regulating overt wound closure and promoting ordered dermal remodeling. We generated a mutant form of TGFβ3 (mutTGFβ3) by ablating its binding site for the latency-associated TGFβ binding protein (LTBP-1) in order to improve bioavailability and activity. The mutated cytokine is secreted as the stable latency-associated peptide (LAP)-associated form and is activated by normal intracellular and extracellular mechanisms including integrin-mediated activation but is not sequestered. We show localized intradermal transduction using a lentiviral vector expressing the mutTGFβ3 in a mouse skin wounding model reduced re-epithelialization density and fibroblast/myofibroblast transdifferentiation within the wound area, both indicative of reduced scar tissue formation.
Introduction
The application of gene therapy for scar-free repair of dermal wounds is a relatively untapped area of regenerative medicine. Elective and emergency surgeries involve incision wounds that under most instances will leave a permanent scar that can have significant psychological consequences to the patient. In addition, the treatment of keloids and diabetic ulcers are major therapeutic targets. Emerging protocols seek to combine either gene-,1,2,3 cellular-,4,5,6 or molecular-based approaches7,8,9,10 to facilitate scar-free wound closure. The modulation of growth factors, most notably the transforming growth factor-β (TGFβ) family, at the site of cutaneous wounding has been a major area of investigation.7,11,12
The role of TGFβ1 in mediating rapid wound closure and fibrotic scars is well documented.13,14,15,16 Local TGFβ1 activity promotes fibroblast migration to the dermal wound site followed by transdifferentiation of these cells to myofibroblasts, which secrete elastic fibers resulting in rapid contraction and wound closure followed by scar tissue formation.17,18 We have sought to modulate this profibrotic response using a gene therapy approach to overexpress the TGFβ1 antagonist TGFβ3 in order to reduce scarring. The relative ratios of the two family members have been shown to be critical in regulating ordered dermal regeneration or disordered repair.19,20,21,22
The application of recombinant active TGFβ3 to the site of dermal wounds is already in human trials and has proved to be safe and efficacious.23 However, active TGFβ is known to have a half-life of only 2–3 minutes in plasma compared to ~100 minutes for the latency-associated peptide (LAP) bound inactive form.24 Furthermore, the TGFβ3 LAP contains an integrin binding RGD domain that is now known to activate the LAP-associated cytokine.25,26,27
We present data describing the development of a lentiviral vector system to deliver maximal quantities of TGFβ3 in a form that will have the greatest therapeutic effect. We have designed a mutant TGFβ3 (mutTGFβ3) variant ablating the binding site of the sequestering protein, latency-associated TGFβ binding protein (LTBP-1), but retaining the activating RGD integrin-binding domain. Overexpression of this mutant resulted in greater bioavailability of active cytokine assayed by reporter gene assays and scrape assays in vitro. Furthermore, we show that the gene therapy approach was able to mediate a reduction in markers of scar tissue formation in a mouse skin wounding model possibly by a mechanism mediated by integrin-LAP activation of TGFβ3.
Results
Vector construction
Lentivectors expressing either wild-type LAP-TGFβ3 (Lnt-TGFβ3) or containing a C25G mutation within the LAP protein (Lnt-mutTGFβ3) were generated. The transgenes were expressed from a bicistronic foot-and-mouth disease virus–derived 2A cassette co-expressing green fluorescent protein (GFP) to mark transduced cells.28,29
Ablation of LTBP-1 binding to the mutTGFβ3
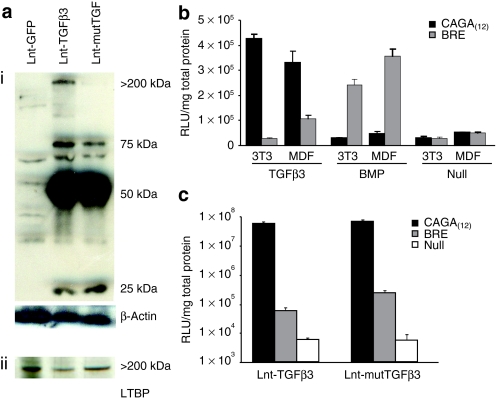
To first confirm expression of TGFβ3 in transduced cells, we performed a western blot using a TGFβ3 antibody on transduced murine dermal fibroblast (MDF) cell lysates under nonreducing conditions to retain noncovalent associations [Figure 1a(i)]. A strong 50-kDa band was present corresponding to heterodimers of LAP-TGF-β3 in TGFβ3- and mutTGFβ3-expressing cell lysates. A weaker band was observed at 25 kDa corresponding to the activated homodimer and a still weaker band at 75 kDa, which is consistent with uncleaved heterodimers of GFP.2A.TGF-β3. This was confirmed by the presence of this band on reprobing the blot with a GFP antibody (data not shown). Importantly, blots consistently showed a band present at ~200–250 kDa for the TGFβ3 but not the mutTGFβ3-transduced cells, the correct size range for the LAP-TGFβ3/LTBP-1 complex. A separate western blot for LTBP-1 on the same cell lysates showed the same >200-kDa band indicating that this band represents LTBP-1 [Figure 1a(ii)]. These data confirm that the C25G mutation ablates or significantly reduces mutTGFβ3 binding to LTBP-1.
Figure 1.
Transduction of murine fibroblasts with lentiviruses encoding TGFβ3 results in efficient transgene expression and functional activation of specific bioresponders. (a) To confirm the presence of transgenic protein in transduced murine dermal fibroblast (MDF) cells, lysates were subjected to western blot under nonreducing conditions with a murine TGFβ3-specific antibody. A strong 50-kDa band was evident in MDF cells transduced with the TGFβ3 containing lentivectors and not in control-transduced cells. This band corresponds to heterodimers of latency-associated peptide-TGFβ3. (i) There were also bands at 25 kDa corresponding to the active homodimer of TGFβ3 and a weaker band at 75 kDa, which corresponds to the predicted size of the uncleaved bicistronic precursor protein GFP.2A.TGFβ3 or GFP.2A.mutTGFβ3. (ii) A >200-kDa band corresponding to the size expected for the TGFβ3/LTBP-1 complex is evident in the wild-type TGFβ3-transduced cells but not the mutant. To confirm this to be the TGFβ3/LTBP-1 complex lysates were blotted with LTBP-1 antibody. (b) Transgenic cell lines were generated from NIH-3T3 cells and primary adult MDFs that contain a single stably integrated copy of either the TGFβ-specific enhancer elements controlling a murine minimal promoter (CAGA(12)) or a bone morphogenic protein–specific Id1 enhancer elements (BRE) both driving the luciferase reporter gene. These cells were validated by exogenous application of either TGFβ3 or BMP4. Cells were lysed 6 hours after cytokine application and assayed for luciferase activity (n = 4 ± SEM). (c) MDF cells, MDF/CAGA(12)-Luc cells, or MDF/BRE-Luc cells were transduced with either Lnt-TGFβ3, Lnt-mutTGFβ3, or Lnt-GFP at 10 MOI. Cells were lysed after 48 hours and luciferase activity assayed (n = 4 ± SEM). LTBP, latency-associated TGFβ binding protein; RLU, relative light units; TGF, transforming growth factor.
TGFβ3/mutTGFβ3 expression after lentiviral transduction of murine fibroblasts provide Smad-specific responses
In order to quantitatively analyze transgene functional activity, we generated novel cell lines containing Smad-specific bioresponsive transgenic elements. Enhancer/promoter elements specific and responsive to TGFβ [CAGA(12)] or alternatively bone morphogenic protein (BMP) responsive element (BRE) as a control have been described in the literature.30,31 We engineered these elements driving the luciferase reporter gene into a lentiviral backbone and generated viral preps for each. Adult primary MDFs and mouse fibroblast 3T3 cells were transduced and clonal populations, containing on average a single viral integration (data not shown), were amplified. The CAGA(12)-Luc and BRE-Luc MDF cell lines were validated by the application of exogenous TGFβ3 and BMP4 followed by luciferase assay after 6 hours (Figure 1b). Both elements responded specifically to the appropriate cytokine and were broadly nonresponsive in all other conditions although there was a low but unexpected response above background from the BRE-Luc MDF cells exposed to TGFβ3. Next, we used these cells as an assay platform to quantify TGFβ3 secreted into conditioned medium from MDFs transduced with either Lnt-TGFβ3 or Lnt-mutTGFβ3. MDFs were transduced at a multiplicity of infection of 10 and grown for a further 72 hours. These cells were then cocultured at a ratio of 1:1 with our CAGA(12)-Luc and BRE-Luc MDF cell lines. Cells were lysed after 48 hours and luciferase expression measured (Figure 1c). In coculture experiments, cells transduced with TGFβ3- and mutTGFβ3-expressing constructs generated a TGFβ-specific luciferase response >30,000- and >40,000-fold higher than background whereas the BMP-specific response, although significantly above background, were only 13- and 71-fold, respectively. There was no substantial difference in CAGA(12)-Luc stimulation between TGFβ3- or mutTGFβ3-expressing cells.
mutTGFβ3 retards fibroblast migration and facilitates Smad2/3 intracellular signaling in an integrin-specific manner
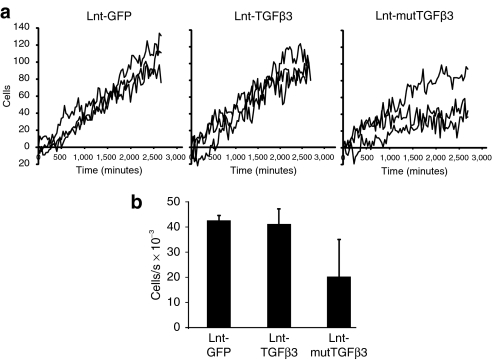
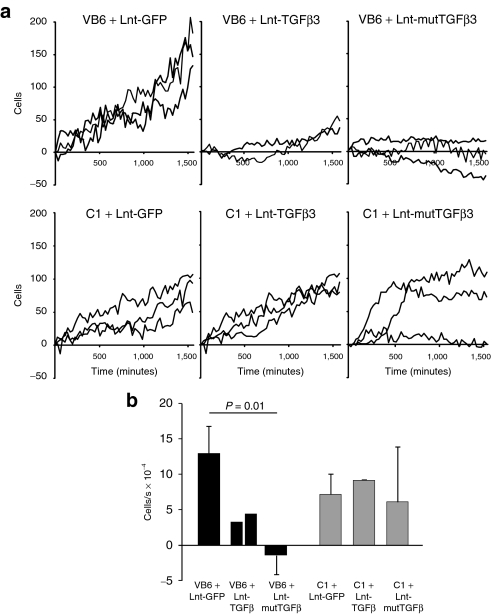
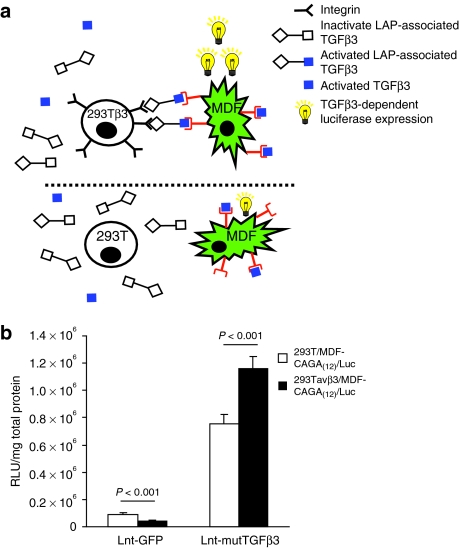
To assess whether transgenic expression of TGFβ3 has an effect on fibroblast migration, we carried out scrape assays on MDF cells transduced with lentivector expressing either wild-type or mutTGFβ3 or GFP at multiplicity of infection = 10 (Figure 2a and Supplementary Movie S1A). Over a period of 48 hours, there was no significant difference between cells transduced with a control GFP virus and those expressing wild-type TGFβ3. However, there was a significant retardation of migration between cells transduced with Lnt-GFP, Lnt-TGFβ3, and Lnt-mutTGFβ3 [Figure 2b; one-way general linear model (GLIM), GFP versus TGFβ3 versus mutTGFβ3: P = 0.045]. We hypothesized that this retardation of migration was mediated through increased bioavailability of mutTGFβ3 and its interaction with integrin. The main integrin heterodimer by which TGFβ is known to interact is αvβ6 so we carried out scrape assays with the β6-transfected VB6 cell line that overexpresses β6 and its parental cell line C1 when transduced with our expression constructs at multiplicity of infection = 10. VB6 cells have been modified to express high levels of αvβ6 and have been shown to migrate toward LAP using αvβ6 alone.32 The differential effect upon cell migration of TGFβ3 and mutTGFβ3 was statistically significant and this was fundamentally different between VB6 and C1 cell types (two-way GLIM; first factor: control versus TGFβ3 versus mutTGFβ3: P = 0.034; second factor: VB6 cells versus C1 cells: P = 0.055; factor interaction: P = 0.014). Migration of VB6 cells expressing mutTGFβ3 was significantly retarded compared with the control group (Bonferroni simultaneous test: P = 0.01; Figure 3 and Supplementary Movie S1B (VB6),GLIM (C1)). To test whether an interaction with integrin increased intracellular signaling, we designed an experiment where CAGA(12)-Luc MDF cells transduced with Lnt-mutTGFβ3 or control Lnt-GFP (multiplicity of infection = 10) were cocultured 1:1 with 293T cells or 293T cells stably expressing β3 integrin (293Tβ3). There was a significant increase in luciferase activity from mutTGFβ3-expressing CAGA(12)-Luc MDF cells when cocultured with β3 integrin overexpressing cells (Figure 4). This effect was particularly impressive given that, in contrast, in the control group of lnt-GFP-transduced CAGA(12)-Luc MDF cells, coculture with 293Tβ3 cells reduced luciferase expression compared with 293T parental cells. Collectively, these data suggest that mutTGFβ3 interacts with integrins and mediates intracellular Smad2/3 signaling.
Figure 2.
Expression of mutTGFβ3 retards wound closure of in vitro scrape assays in murine dermal fibroblasts (MDFs). (a) MDF cells were transduced with lentivectors expressing either wild-type or mutant (C25G LAP) TGFβ3 or GFP control (10 multiplicity of infection) then seeded on plastic and grown to 90% confluence. A single midline scrape was made in each well and real-time imaging carried out every 10 minutes over 48 hours full data stream is shown in Supplementary Movie S1A. (b) Migration rates for each group were derived by performing linear regression on the data set from each well (n = 3 ± SEM). mutTGFβ3, mutant transforming growth factor β3.
Figure 3.
mutTGFβ3 retardation of wound closure is mediated by integrin. (a) Scrape assays were set up with the VB6 cell line that overexpresses β6 VB6 integrin compared to their parental line C1. Cells were transduced with lentivectors expressing wild-type or mutant TGFβ3 or GFP control. Imaging was carried out as described over 48 hours (Supplementary Movie S1B). (b) Migration rates for each group were derived by performing linear regression on the data set from each well (n = 2–3 ± SEM). GFP, green fluorescent protein; mutTGFβ3, mutant transforming growth factor β3.
Figure 4.
mutTGFβ3-activated intracellular Smad2/3 signaling is increased in the presence of integrin. (a) Murine dermal fibroblast (MDF) cells stably expressing a single copy of the Smad2/3 responsive element CAGA(12) and minimal promoter driving luciferase were transduced with Lnt-mutTGFβ3 or Lnt-GFP (multiplicity of infection = 10). These cells were cocultured with 293T cells overexpressing β3 integrin 293Tβ3 or 293T parental cells were. Overexpression of integrin could increase activation of mutTGFβ3 in an RGD-specific manner. The green jagged cells represent the reporter MDF/CAGA(12)-Luc cells, the circular cells represent integrin overexpressing (top panel) or their parental line (bottom panel). The blue squares represent activated TGFβ3, the open squares represent inactive latency-associated peptide (LAP)-associated TGFβ3. LAP association with integrin activates a tethered form of TGFβ3. (b) Cocultures were grown for 48 hours then lysed and luciferase activity quantified. There was a significant increase in luciferase activity in the presence β3 integrin (n = 4 ± SEM). mutTGFβ3, mutant transforming growth factor β3.
Intradermal injection of Lnt-mutTGFβ3 at the site of cutaneous wounding in vivo results in a reduction of re-epithelialization density
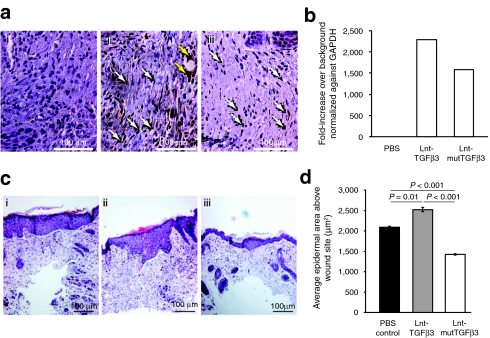
In order to assess therapeutic potential, a single dose of Lnt-mutTGFβ3, Lnt-TGFβ3 (1 × 109 virus particles), or vehicle control was applied intradermally to opposing flanks of adult CD1 mice (n = 6). Four hours later a single full-thickness cutaneous incision 2 cm in length was made across the site of injection. Mice were killed after 14 days and the site of wounding assessed by histological, immunohistochemical, and molecular analyses. Successful transduction was confirmed by staining sections from the center of the wound area with an anti-GFP antibody (Figure 5a). There was no staining apparent in the vehicle control [Figure 5a(i)] but widespread GFP staining in the GFP.2A.TGFβ3- [Figure 5a(ii)] and GFP.2A.mTGFβ3 [Figure 5a(iii)]-transduced wounds. The majority of cells transduced were fibroblastic in morphology although there was also evidence of transduction of other cell types such endothelial cells and follicular cell types (as identified by histology). Quantitative PCR analysis of tissue dissected from the wound site revealed that there was a higher level of GFP expression in animals transduced with the TGFβ3 vector compared to mutTGFβ3 consistent within data sets suggesting a slight difference in transduction efficiency (Figure 5b).
Figure 5.
Mice were treated with a single intradermal injection of 1 × 109 vp of lentivirus expressing either TGFβ3, mutTGFβ3, or a sham PBS injection. This was followed by a full-thickness midline incision of the skin over the injection site 4 hours later. After 14 days, the animals were sacrificed and the incision site was dissected out and analyzed by immunohistochemistry and real-time qPCR. (a) Sections were stained for GFP (i) PBS, (ii) Lnt-TGFβ3, (iii) Lnt-mutTGFβ3 which showed GFP positive fibroblasts (white arrow) and other cell types such as endothelium (yellow arrows). Images are representative for each treatment (n = 6). (b) Relative levels of GFP were quantified using qPCR with an n of three samples carried out duplicate. (c) H&E stained sections were analyzed for re-epithelialization area at the site of incision (i) PBS, (ii) Lnt-TGFβ3, and Lnt-mutTGFβ3. (d) Quantification of re-epithelialization using Image J analysis software of at least 10 sections cut at 50 micron intervals across the wound site for each treatment group and expressed in microns sq. (n = 4, 3, 4, respectively). mutTGFβ3, mutant transforming growth factor β3; PBS, phosphate-buffered saline; qPCR, quantitative PCR; vp, virus particles.
We analyzed wound re-epithelialization area on hematoxylin and eosin stained sections taken at 50-µm intervals from the center of each wound site [Figure 5c(i–iii)] and quantified using image analysis software (Image J; NIH, Bethesda, MD) (Figure 5d). Vector treatment had a significant effect on wound healing [one-way GLIM, phosphate-buffered saline (PBS) versus Lnt-TGFβ3 versus Lnt-mutTGFβ3: P < 0.001]. Vehicle- and Lnt-TGFβ3-treated wound closure resulted in a thickened epidermal layer but Lnt-mutTGFβ3-treated animals showed a substantial reduction in epidermal thickness at the wound site. In many instances, the wound site was indiscernible from the peripheral continuum. The epidermal area of Lnt-mutTGFβ3-treated wounds was significantly smaller than either Lnt-TGFβ3- or vehicle-treated wounds (Bonferroni simultaneous test: P < 0.001 for both).
Lnt-mutTGFβ3 transduction results in a reduction of fibroblast to myofibroblast differentiation
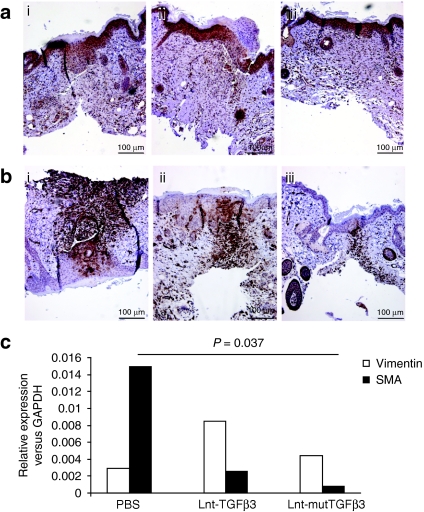
We further investigated the migration of fibroblasts at the wound site and their subsequent transdifferentiation to myofibroblasts. Sections were immunostained for the presence of vimentin, a fibroblast-specific marker (Figure 6a) and smooth muscle actin (SMA), a myofibroblast marker (Figure 6b). Furthermore, representative wound site tissues were analyzed by semiquantitative reverse transcription-PCR for the presence of mRNA for the same markers (Figure 6c). There was no significant difference between the treatment groups for expression of vimentin. This would suggest that TGFβ3 does not act on the recruitment of fibroblasts to the dermal wound site. However, there was a significant effect of vector injection on SMA expression (one-way GLIM, PBS versus Lnt-TGFβ3 versus Lnt-mutTGFβ3: P = 0.023). There was a significant decrease in the quantity of myofibroblasts evident from SMA staining at the wound site in mutTGFβ3 compared with PBS injection (Bonferroni simultaneous test: P = 0.037). Collectively, the in vivo data show that transgenic expression of the mutant LAP(C25G)-TGFβ3 at the site of cutaneous wounding results in a significant reduction in re-epithelialization density and fibroblast to myofibroblast differentiation, both markers of scarring.
Figure 6.
Transgenic overexpression of TGFβ3 decreases fibroblast to myofibroblast differentiation at the site of cutaneous wounding in vivo. (a,b) Wound sections were stained immunohistochemically for fibroblast (a: vimentin) and myofibroblast (b: SMA) markers after treatment with [a and b(i)] PBS, [a and b(ii)] Lnt-TGFβ3, or [a and b(iii)] Lnt-mutTGFβ3. Representative images from at least three sections from each animal (n = 6). (c) Real-time reverse transcription-PCR showed that there was a small but nonsignificant increase in vimentin expression but a significant decrease within both TGFβ3 application groups and the PBS control (n = 4) as well as a significant decrease between the Lnt-mutTGFβ3 and Lnt-TGFβ3 treatment groups. PBS, phosphate-buffered saline; SMA, smooth muscle actin; TGF, transforming growth factor.
Discussion
We have designed a proof-of-principle gene therapy strategy based on TGFβ3 transgenic expression for the reduction of scar tissue formation after cutaneous wounding. There are currently Phase II/III clinical trials underway using application of exogenous active TGFβ3 but the cytokine is unstable in this form used in these trials and as such the therapeutic window is limited. Such treatment is viable for incisional wounds in elective surgery but less so for trauma, keloids, or diabetic ulcers. We hypothesized that gene delivery of the complete latency-associated TGFβ3 could provide a stable supply of endogenously activated cytokine at the site of cutaneous wounding if LTBP-mediated sequestering could be ablated. Furthermore, the LAP is known to contain an RGD motif that associates with integrin subunits β3/β6 that activates the tethered form of TGFβ3.25,26,27 We believe that this component of TGFβ3 activation is critical in cell:cell and cell:extracellular matrix communication that mediates coordinated wound closure.
We generated two lentiviral expression vectors containing either the murine LAP-TGFβ3 or a mutant version with a C25G amino acid substitution in the LAP protein designed to remove the Cys residue that forms a disulfide bridge between LAP and LTBP-1. Primary MDFs transduced with these lentivectors were analyzed for expression of TGFβ3 by western blot under nonreducing conditions. The expected expression profile of active and inactive heterodimers was observed, but importantly a band was evident at >200 kDa for the TGFβ3-expressing cells but not mutTGFβ3 which was consistent in size with the LAP-TGFβ3/LTBP-1 complex. We further confirmed this by reprobing the blot with an LTBP-1 antibody and the same sized band was apparent.
To test the functional activity of these vectors, we generated novel bioresponsive MDF cells containing a single stably expressing copy of the firefly luciferase gene under the control of either a TGFβ (Smad2/3) or BMP (Smad1/5/8) responsive promoter/enhancer element. These were compared to the 3T3 mouse fibroblast cell line. Previous analyses with such responsive elements have been conducted by transient transfection on permissive cell lines33,34,35,36 or using a mink lung epithelial cell line, which may bear little phenotypic resemblance to clinically relevant cell types.37 All four of our cell lines were strongly responsive to specific growth factor activation. The primary MDF-CAGA(12)-Luc (Smad2/3) and MDF-BRE-Luc (Smad1/5/8) cells were used to assay the biological activity of our TGFβ3-expressing lentivectors. Both vectors efficiently activated the Smad2/3 responsive reporter in MDFs. The Smad1/5/8 responsive reporter was also activated above background but many thousand-fold less. This low level of activation could be due to diverse crosstalk between the two signaling pathways or nonspecific activation at high concentrations of TGFβ3 but would require further investigation.
Having shown evidence that LTBP-1 sequestering is ablated in our mutant, we designed experiments to show that the LAP and mutated LAP (C25G) of TGFβ3 interacted with and were activated by integrins. Our initial analysis was to assess cell migration using scrape assays as integrins are known to be involved in cell motility.38 We initially used primary MDFs transduced with the mutant and wild-type TGFβ3 constructs compared to GFP control-transduced cells. Interestingly, there was no significant difference in scrape closure for wild-type TGFβ3-expressing cells versus controls but a significant retardation in cell migration was observed in the mutTGFβ3-expressing cells. Thus, we conclude that the mutTGFβ3 is interacting with MDFs via a mechanism that retards migration and is biologically more available than the wild-type protein. We carried out further scrape assays with the VB6 cell line that overexpresses β6 integrin, which has been shown to interact with TGFβ family members.25,26,27 In VB6 cells but not the parental cell line, C1 migration was significantly perturbed in cells expressing the mutant when compared to the wild-type form of TGFβ3 or control cells. This retardation in cell migration would suggest that TGFβ3 does interact with integrin but that TGFβ3 interaction with LTBP-1 masks any phenotypic effect possibly by reducing bioavailability. We would have ideally liked to corroborate our data by using integrin inhibitors; however, the real-time imaging is a closed system and inhibitors would have to be added multiply over the imaging time frame to be effective.
The scrape assays show that mutTGFβ3 significantly decreases cell migration in integrin overexpressing cells, which is indicative of direct interaction between cytokine and integrin but does not provide evidence of the activation of downstream intracellular signaling. In an attempt to resolve this, we designed a coculture experiment where MDF-CAGA(12)-Luc Smad2/3 reporter cells were transduced with Lnt-mutTGFβ3 or control vector and mixed with 293T cells or 293T cells overexpressing β3 integrin (293Tβ3). We observed a significant increase in Smad2/3-mediated luciferase reporter expression 293Tβ3 cocultures compared to controls. In combination, these data provide strong evidence that mutTGFβ3 is more bioavailable than the wild-type; it interacts with and is activated in the presence of integrin.
In order to evaluate the efficacy of lentiviral delivery of mutTGFβ3 as a gene therapy for cutaneous wounding, we administered a single dose of 1 × 109 virus particles to the flanks of mice before a full-thickness cutaneous incisional wound. Mice were killed after 14 days and the wound site assessed for markers of scar tissue formation. We chose this time frame as full-thickness dermal wounds in mice are known to repair quicker than in humans. A 14-day period has previously been shown to encompass wound closure and re-epithelialization.39,40 Transduction efficiency was assessed by GFP expression using GFP antibody staining of sections and quantitative reverse transcription-PCR on mRNA prepared from tissue surrounding the wound site. We observed consistently higher levels of transduction in the GFP.2A.TGFβ3-treated group than the GFP.2A.mutTGFβ3 group even though both viral preparations had been normalized by physical titer before application. A possible explanation for this anomaly is that there was a higher comparative biological titer (infectious units) for the LNT-TGFβ3 than the Lnt-mutTGFβ3 despite the physical titers (viral particles) being the same. However, despite this difference, it was starkly apparent on analysis of hematoxylin and eosin stained histological sections that the re-epithelialized epidermis was significantly smaller in area above the wound site in mutTGFβ3-treated skin compared to TGFβ3- or PBS control-treated wounds. There was no significant difference between TGFβ3- and PBS-treated wounds. It was apparent from multiple sections that re-epithelialization in the mutTGFβ3-treated wounds was similar and in some cases indiscernible to that of the surrounding skin. We conclude that coordinate wound re-epithelialization is enhanced by the overexpression of mutTGFβ3.
Having observed a more coordinated re-epithelialization at the wound site in mutTGFβ3-transduced skin, we analyzed other aspects of wound healing. Wound closure is characterized by fibroblast migration to the site of injury followed by fibroblast to myofibroblast transdifferentiation. To assess whether TGFβ3 overexpression negatively affects this process, we stained treatment and control group wound sections and quantified vimentin and αSMA mRNA from tissue: markers of fibroblasts and myofibroblasts, respectively. There was no significant difference in the amount of vimentin in each sample group, which leads us to believe that the overexpression of either mutant or wild-type TGFβ3 at 14 days does not lead to a reduction in fibroblast number. However, of great significance is the pronounced reduction in levels of the myofibroblast marker αSMA the mutant but not wild-type TGFβ3 treatment compared to PBS controls. This provides strong evidence that the presence of mutTGFβ3 at the wound site significantly reduces the amount of myofibroblasts present.
In conclusion, we have shown proof-of-principle for a gene therapy treatment strategy for coordinated wound closure and reduction of scar tissue formation after cutaneous wounding. Our data corroborates the clinical efficacy shown by Ferguson's team that TGFβ3 is a viable antiscarring treatment.8 We have shown that the TGFβ3 LAP has therapeutic significance by controlling cytokine release and integrin-mediated activation. By mutating the TGFβ3 LAP's LTBP-1 binding domain while retaining the integrin-binding RGD domain, we have retained the activation component of LAP while increasing bioavailability at the wound site. We believe that this activity is instrumental in controlling fibroblast to myofibroblast transdifferentiation. Significantly, this component is lacking in current therapeutic strategies. This study was designed using lentiviral vectors as they have been shown to elicit comparatively low immune and localized inflammatory responses.41,42 An ideal preclinical strategy would be to combine these attributes with transient expression as therapeutic strategies for chronic wound closure would involve a treatment window of weeks and not months. Although we have used integrating lentivirus vectors in these proof-of principle studies, in future studies we aim to utilize novel integration defective lentivectors43,44 to effect transient expression and avoid adverse events associated with genome integration.45
Materials and Methods
Construction, production, and validation of lentiviral constructs. For the production of pseudotyped lentivector, third-generation human immunodeficiency virus-1 cassettes based on those originally described by Dull et al. were used.46 Latency-associated TGFβ3 plasmid pTGFβ3 (GenBank: NM_009368) was obtained from ATCC (Manassas, VA). TGFβ3 was PCR amplified from pTGFβ3 including flanking EcoRI and SalI sites using the following primers 5′-gaattctatgcacttgcaaagggctctgg-3′, 5′-gtcgacttattatcagctgcacttacac-3′ and cloned downstream (and in-frame) of the 2A sequence in pSP72-2A. Subsequently, enhanced GFP was PCR amplified with NotI/XhoI flanking sites using the following primers 5′-gcggccgcatggtgagcaagg-3′, 5′-ctcgagccttgtacagctcgtccatgcc-3′ and cloned upstream of and in-frame with the 2A sequence by a NotI/XhoI digest creating the bicistronic vector pSP72-GFP-2A-TGFβ3. GFP-2A-TGFβ3 was subsequently excised using a NotI/SalI digest and inserted into our K2 lentivector to provide K2cmvGFP-2A-TGFβ3 (Lnt-TGFβ3). mutTGFβ3 was produced from pSP72-2A-TGFβ3 by site-directed mutagenesis (Stratagene, Amsterdam, Netherlands) of the 25th residue of the TGFβ3 LAP mutating a Cys→Gly by a single T→G base change with the following mutagenesis primers; 5′-ccactggcaccacgttggac ttcggccacatcaagaagaagag-3′ 5′-ctcttcttcttgatgtggccgaagtccaacgtggtgccagtgg-3′ to produce the vector Lnt-mutTGFβ3.
Lentiviral vector expressing GFP under the cytomegalovirus promoter and the plasmid expressing the vesicular stomatitis G protein have been previously described.47,48 Lentivectors expressing Smad-specific promoter driven luciferase vectors were constructed as follows. Both the pCAGA(12)-Luc and the pBRE2-Luc inserts (plasmids were a kind gift from Louise Reynolds, QMUL and based on a pGL3 background) were excised by cutting with MluI/XbaI and ligated directly into MluI/NheI sites of the K2 lentivector multicloning site. All positive clones were validated by sequencing. Lentivectors were prepared as previously described.47 Viral titers were determined by transduction and flow cytometry of GFP expression in 293T cells, or by p25 enzyme-linked immunosorbent assay (Beckman Coulter, High Wycome, UK) or colorimetric Reverse Transcriptase assay (Roche Diagnostics, Burgess Hill, UK).49
Evaluation of TGFβ3 transgene expression and function
Smad-activated luciferase assay: MDF were isolated as previously described by DiPersio et al.50 MDF or NIH-3T3 cells were expanded to ~60% confluence and transduced with an human immunodeficiency virus-1 based third-generation lentivirus expressing a luciferase reporter gene under the control of either a Smad 2/3-specific CAGA(12) enhancer30 or a Smad 1/5-specific BRE2 promoter31 combined with a minimal promoter. Clones that contained, on average, a single integration of transgenic reporter were isolated. Integrations were analyzed by quantitative PCR on an ABI Prism 7000 real-time PCR machine using the following primers for viral integrations (WPRE: 5′-tggattctgcgcggga-3′ 5′-gaaggaaggtccgctggatt-3′) and the mouse-specific primers for estimating cell number (Titin M ex5: 5′-aaaacgagcagtgacgtgagc-3′ 5′-ttcagtcatgctgctagcgc-3′ primers and plasmid standard were a kind gift from Anne Galy, Genethon, France). Cells were then replated 72 hours later in the absence of serum and either subjected to exogenous TGFβ3, BMP4 (both R&D Systems, Abingdon, UK) and lysed after 6 hours or cocultured with MDFs transduced with LntTGFβ3/Lnt-GFP and lysed 48 hours later. Cell lysate was assayed for luciferase expression using the Promega luciferase assay kit and a Berthold Flash'n'Glow LB955 (Berthold, Herts, UK) luminometer. Relative luciferase activity was expressed in arbitrary units with respect to total protein measured by standard Bradford assay as by manufacturer's instructions (Bio-Rad, Herts, UK).
Western blot: Rabbit anti-TGFβ3 antibody and sc-83 rabbit polyclonal IgG were supplied by Santa Cruz Biotechnology (Santa Cruz, CA) rabbit anti-GFP polyclonal antibody was supplied by AbCam (Cambridge, UK). LTBP-1 antibody was a kind gift from Sarah Dallas, University of Missouri, USA. Nondenaturing western blots were carried out using standard protocols.
In vitro scrape assays: MDF, VB6, C1 cells were transduced with lentivector either expressing TGF-β3, mutTGFβ3, or a GFP control and allowed to reach 80–90% confluence. Twenty-four hours later, cells were serially washed in PBS to remove serum and grown in Advanced Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with ITS+1 (Sigma-Aldrich, St Louis, MO). Scratches (1 mm) were made across the diameter of each well using CytoSelect inserts (Cell Biolabs, San Diego, CA). Scratch closure was measured using a Cell-IQ (Chipman Technologies, Tampere, Finland) real-time imaging platform and data managed using Chipman Technologies software.
In vitro coculture assays: 293Tβ3 and the parental 293T cells were transduced with lentivirus either expressing TGF-β3, mutTGFβ3, or a GFP control were mixed 1:1 with MDF-CAGA(12)-Luc cells. Cells were cocultured for 48 hours in serum-depleted Advanced Dulbecco's modified Eagle's medium (Invitrogen) medium supplemented with ITS+1 (Sigma-Aldrich). Cells were subsequently lysed and luciferase expression quantified.
Intradermal injection and full-thickness dermal wounding in vivo: Male CD1 mice (Harlan UK, Wyton, UK) were used in this study. Mice were anesthetized by inhalation of isofluorane (Abbot Laboratories, Maidenhead, UK). For full-thickness cutaneous incision, a 2-cm line was drawn on the underside midline of each animal and at the midpoint of that line a single, intradermal injection of high titer lentivirus (25 µl volume) was administered. The resultant bleb was softly massaged into the skin and the animals allowed to recover. Thirty minutes later a single 2-cm, full-thickness dermal incision was performed along the midline and a single 5/0 Mersilk suture (Ethicon, Brussels, Belgium) placed ~3 mm from each end to secure the wound. Animals were kept in a warmed cage in an undisturbed environment until awake and active. Animals were killed at defined time points, the wound site excised, dissected, and halved. Half was frozen at −80 °C for isolation of mRNA, the other half fixed in 4% paraformaldehyde and analyzed histologically. All animal work was carried out under United Kingdom Home Office regulations and was compliant with the guidelines of the Imperial College London ethical review committee.
Real-time semiquantitative PCR: Total RNA was extracted from frozen dermal tissue using the RNeasy kit (Qiagen, Crawley, UK) as per manufacturer's instructions. RNA preparations were subjected to DNase I treatment (1 U/µg total RNA) (Invitrogen) for 30 minutes at 37 °C before complete denturation of the enzyme by heating to 70 °C for 15 minutes. First strand complementary DNA was synthesized from 2 µg total RNA using MoMLV Reverse Transcriptase (Roche Applied Science) and random hexamer primer as per manufacturer's instructions. Real-time PCR was carried out using a SYBR Green qPCR kit (Eurogentec, Liege, Belgium) and ABI Prism (Applied Biosystems, Warrington, UK). Primers were as follows; GFP 5′-taaacggccacaagttcagcgtgt-3′, 5′-ttctcgttggggtctttgctcag-3′, vimentin 5′-aagagaactttgccgttgaa-3′, 5′-gtgatgctgagaagtttcgt-3′, SMA 5′-ag attgtccgtgacatcaagg-3′, ttgtgtctagaggcagagc-3′, GAPDH 5′-tgcaccaccaactgc ttag-3′, 5′-ggatgcagggatgatgttc-3′.
Histochemistry and immunohistochemistry: Excised skin tissue sections were fixed in 4% paraformaldeyhde for 16 hours at 4 °C and 4-µm paraffin sections cut at 50-µm intervals. Sections were stained for the following polyclonal antibody markers, all raised in rabbit, and counterstained with hematoxylin and eosin: anti-GFP (AbCam), anti-mouse SMA, anti-mouse vimentin, and anti-mouse pancytokeratin (all Santa Cruz Biotechnology). Antibody was detected using the Envision+ system-HRP kit (Dako, Ely, UK) and slides analyzed by differential interference contrast imaging on an Olympus BX51 microscope (Olympus, Southend-on-Sea, UK). When comparing sections, positively stained cells were counted and confirmed by the presence of a single localized nucleus. All animal experiments were carried out on cohorts of six mice and for every mouse at least four independent sections were counted with at least 100 µm between any serial sections to avoid counting cells twice.
Statistical analyses. For analysis of scrape closure (Figures 2 and 3) each cell migration data set was normalized to zero starting point and a linear regression performed to derive a rate of migration (line gradient) for each well. This provided a single data point for each well. For comparison between multiple groups with either one or two levels, a GLIM was used. For pairwise comparisons, Bonferroni simultaneous tests were subsequently performed (Figures 2 and 3). All analysis was performed using Minitab software (Minitab, Myerstown, PA).
SUPPLEMENTARY MATERIAL Movie S1. Images were captured every 10 minutes over a period of 48 hours using a x10 objective and digital zoom on a Cell-IQ (Chipman Technologies) imaging platform.
Acknowledgments
The plasmid encoding the foot-and-mouth disease virus 2A sequence (pSP72-2A) was a kind gift from Michael Milsom, Cincinnati Harvard Medical School, Children's Hospital Boston, Boston, MA, USA. The plasmids encoding the Smad-responsive elements were a kind gift from Louise Reynolds, Cancer Research UK, Barts & The London, Queen Mary's School of Medicine & Dentistry, London, UK. The β3 integrin overexpressing 293T cell line was a kind gift from John Olsen, CF Pulmonary Research & Treatment Center, UNC, NC, USA. The β6 overexpressing cell line VB6 and its parental line C1 was a kind gift from Gareth Thomas, School of Medicine & Cancer Studies, University of Southampton. Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element and murine titin real-time PCR standards and plasmids were a kind gift from Anne Galy, Genethon, Paris. Real-time imaging was carried out using a Cell-IQ platform (Chipman Technologies, Tampere, Finland). We are grateful for the continued technical assistance of Geraint Wilde and David Bolton of Chipman Technologies.
Supplementary Material
Images were captured every 10 minutes over a period of 48 hours using a x10 objective and digital zoom on a Cell-IQ (Chipman Technologies) imaging platform.
References
- Lee JA, Conejero JA, Mason JM, Parrett BM, Wear-Maggitti KD, Grant RT, et al. Lentiviral transfection with the PDGF-B gene improves diabetic wound healing. Plast Reconstr Surg. 2005;116:532–538. doi: 10.1097/01.prs.0000172892.78964.49. [DOI] [PubMed] [Google Scholar]
- Stoff A, Rivera AA, Mathis JM, Moore ST, Banerjee NS, Everts M, et al. Effect of adenoviral mediated overexpression of fibromodulin on human dermal fibroblasts and scar formation in full-thickness incisional wounds. J Mol Med. 2007;85:481–496. doi: 10.1007/s00109-006-0148-z. [DOI] [PubMed] [Google Scholar]
- Tyrone JW, Mogford JE, Chandler LA, Ma C, Xia Y, Pierce GF, et al. Collagen-embedded platelet-derived growth factor DNA plasmid promotes wound healing in a dermal ulcer model. J Surg Res. 2000;93:230–236. doi: 10.1006/jsre.2000.5912. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J., and, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Woodley DT, Remington J, Huang Y, Hou Y, Li W, Keene DR, et al. Intravenously injected human fibroblasts home to skin wounds, deliver type VII collagen, and promote wound healing. Mol Ther. 2007;15:628–635. doi: 10.1038/sj.mt.6300041. [DOI] [PubMed] [Google Scholar]
- Wu Y, Chen L, Scott PG., and, Tredget EE. Mesenchymal stem cells enhance wound healing through differentiation and angiogenesis. Stem Cells. 2007;25:2648–2659. doi: 10.1634/stemcells.2007-0226. [DOI] [PubMed] [Google Scholar]
- Chu Y, Guo F, Li Y, Li X, Zhou T., and, Guo Y. A novel truncated TGF-beta receptor II downregulates collagen synthesis and TGF-beta I secretion of keloid fibroblasts. Connect Tissue Res. 2008;49:92–98. doi: 10.1080/03008200801913924. [DOI] [PubMed] [Google Scholar]
- Occleston NL, O'Kane S, Goldspink N., and, Ferguson MW. New therapeutics for the prevention and reduction of scarring. Drug Discov Today. 2008;13:973–981. doi: 10.1016/j.drudis.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Rodgers KE, Espinoza T, Felix J, Roda N, Maldonado S., and, diZerega G. Acceleration of healing, reduction of fibrotic scar, and normalization of tissue architecture by an angiotensin analogue, NorLeu3-A(1-7) Plast Reconstr Surg. 2003;111:1195–1206. doi: 10.1097/01.PRS.0000047403.23105.66. [DOI] [PubMed] [Google Scholar]
- Sun W, Lin H, Xie H, Chen B, Zhao W, Han Q, et al. Collagen membranes loaded with collagen-binding human PDGF-BB accelerate wound healing in a rabbit dermal ischemic ulcer model. Growth Factors. 2007;25:309–318. doi: 10.1080/08977190701803885. [DOI] [PubMed] [Google Scholar]
- Liu W, Wang DR., and, Cao YL. TGF-beta: a fibrotic factor in wound scarring and a potential target for anti-scarring gene therapy. Curr Gene Ther. 2004;4:123–136. doi: 10.2174/1566523044578004. [DOI] [PubMed] [Google Scholar]
- Sadick H, Herberger A, Riedel K, Bran G, Goessler U, Hoermann K, et al. TGF-beta1 antisense therapy modulates expression of matrix metalloproteinases in keloid-derived fibroblasts. Int J Mol Med. 2008;22:55–60. doi: 10.3892/ijmm.22.1.55. [DOI] [PubMed] [Google Scholar]
- Arany PR, Flanders KC, Kobayashi T, Kuo CK, Stuelten C, Desai KV, et al. Smad3 deficiency alters key structural elements of the extracellular matrix and mechanotransduction of wound closure. Proc Natl Acad Sci USA. 2006;103:9250–9255. doi: 10.1073/pnas.0602473103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft GS, Yang X, Glick AB, Weinstein M, Letterio JL, Mizel DE, et al. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat Cell Biol. 1999;1:260–266. doi: 10.1038/12971. [DOI] [PubMed] [Google Scholar]
- Flanders KC. Smad3 as a mediator of the fibrotic response. Int J Exp Pathol. 2004;85:47–64. doi: 10.1111/j.0959-9673.2004.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., and, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Grinnell F. Fibroblasts, myofibroblasts, and wound contraction. J Cell Biol. 1994;124:401–404. doi: 10.1083/jcb.124.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., and, Parkhurst SM. Parallels between tissue repair and embryo morphogenesis. Development. 2004;131:3021–3034. doi: 10.1242/dev.01253. [DOI] [PubMed] [Google Scholar]
- Cowin AJ, Brosnan MP, Holmes TM., and, Ferguson MW. Endogenous inflammatory response to dermal wound healing in the fetal and adult mouse. Dev Dyn. 1998;212:385–393. doi: 10.1002/(SICI)1097-0177(199807)212:3<385::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Whitby DJ., and, Ferguson MW. The extracellular matrix of lip wounds in fetal, neonatal and adult mice. Development. 1991;112:651–668. doi: 10.1242/dev.112.2.651. [DOI] [PubMed] [Google Scholar]
- Lu L, Saulis AS, Liu WR, Roy NK, Chao JD, Ledbetter S, et al. The temporal effects of anti-TGF-beta1, 2, and 3 monoclonal antibody on wound healing and hypertrophic scar formation. J Am Coll Surg. 2005;201:391–397. doi: 10.1016/j.jamcollsurg.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Soo C, Beanes SR, Hu FY, Zhang X, Dang C, Chang G, et al. Ontogenetic transition in fetal wound transforming growth factor-beta regulation correlates with collagen organization. Am J Pathol. 2003;163:2459–2476. doi: 10.1016/s0002-9440(10)63601-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occleston NL, Laverty HG, O'Kane S., and, Ferguson MW. Prevention and reduction of scarring in the skin by Transforming Growth Factor beta 3 (TGFbeta3): from laboratory discovery to clinical pharmaceutical. J Biomater Sci Polym Ed. 2008;19:1047–1063. doi: 10.1163/156856208784909345. [DOI] [PubMed] [Google Scholar]
- Wakefield LM, Winokur TS, Hollands RS, Christopherson K, Levinson AD., and, Sporn MB. Recombinant latent transforming growth factor beta 1 has a longer plasma half-life in rats than active transforming growth factor beta 1, and a different tissue distribution. J Clin Invest. 1990;86:1976–1984. doi: 10.1172/JCI114932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annes JP, Chen Y, Munger JS., and, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165:723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y, Ihn H, Jinnin M, Mimura Y., and, Tamaki K. Involvement of alphavbeta5 integrin in the establishment of autocrine TGF-beta signaling in dermal fibroblasts derived from localized scleroderma. J Invest Dermatol. 2006;126:1761–1769. doi: 10.1038/sj.jid.5700331. [DOI] [PubMed] [Google Scholar]
- Asano Y, Ihn H, Yamane K, Jinnin M, Mimura Y., and, Tamaki K. Increased expression of integrin alpha(v)beta3 contributes to the establishment of autocrine TGF-beta signaling in scleroderma fibroblasts. J Immunol. 2005;175:7708–7718. doi: 10.4049/jimmunol.175.11.7708. [DOI] [PubMed] [Google Scholar]
- de Felipe P, Hughes LE, Ryan MD., and, Brown JD. Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J Biol Chem. 2003;278:11441–11448. doi: 10.1074/jbc.M211644200. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, et al. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S., and, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynskyi O., and, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- Thomas GJ, Poomsawat S, Lewis MP, Hart IR, Speight PM., and, Marshall JF. alpha v beta 6 Integrin upregulates matrix metalloproteinase 9 and promotes migration of normal oral keratinocytes. J Invest Dermatol. 2001;116:898–904. doi: 10.1046/j.1523-1747.2001.01352.x. [DOI] [PubMed] [Google Scholar]
- Filyak Y, Filyak O, Souchelnytskyi S., and, Stoika R. Doxorubicin inhibits TGF-beta signaling in human lung carcinoma A549 cells. Eur J Pharmacol. 2008;590:67–73. doi: 10.1016/j.ejphar.2008.05.030. [DOI] [PubMed] [Google Scholar]
- Kaivo-Oja N, Mottershead DG, Mazerbourg S, Myllymaa S, Duprat S, Gilchrist RB, et al. Adenoviral gene transfer allows Smad-responsive gene promoter analyses and delineation of type I receptor usage of transforming growth factor-beta family ligands in cultured human granulosa luteal cells. J Clin Endocrinol Metab. 2005;90:271–278. doi: 10.1210/jc.2004-1288. [DOI] [PubMed] [Google Scholar]
- Kopp J, Preis E, Said H, Hafemann B, Wickert L, Gressner AM, et al. Abrogation of transforming growth factor-beta signaling by SMAD7 inhibits collagen gel contraction of human dermal fibroblasts. J Biol Chem. 2005;280:21570–21576. doi: 10.1074/jbc.M502071200. [DOI] [PubMed] [Google Scholar]
- von Bubnoff A, Peiffer DA, Blitz IL, Hayata T, Ogata S, Zeng Q, et al. Phylogenetic footprinting and genome scanning identify vertebrate BMP response elements and new target genes. Dev Biol. 2005;281:210–226. doi: 10.1016/j.ydbio.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Abe M, Harpel JG, Metz CN, Nunes I, Loskutoff DJ., and, Rifkin DB. An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal Biochem. 1994;216:276–284. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
- Thomas GJ, Hart IR, Speight PM., and, Marshall JF. Binding of TGF-beta1 latency-associated peptide (LAP) to alpha(v)beta6 integrin modulates behaviour of squamous carcinoma cells. Br J Cancer. 2002;87:859–867. doi: 10.1038/sj.bjc.6600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ferrer M, Afshar-Sherif AR, Uwamariya C, de Crombrugghe B, Davidson JM., and, Bhowmick NA. Dermal transforming growth factor-beta responsiveness mediates wound contraction and epithelial closure. Am J Pathol. 2010;176:98–107. doi: 10.2353/ajpath.2010.090283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg H, Schulz T, Krueger C., and, Vollmar B. Consequences of surgical stress on the kinetics of skin wound healing: partial hepatectomy delays and functionally alters dermal repair. Wound Repair Regen. 2009;17:367–377. doi: 10.1111/j.1524-475X.2009.00490.x. [DOI] [PubMed] [Google Scholar]
- Gjertsson I, Laurie KL, Devitt J, Howe SJ, Thrasher AJ, Holmdahl R, et al. Tolerance induction using lentiviral gene delivery delays onset and severity of collagen II arthritis. Mol Ther. 2009;17:632–640. doi: 10.1038/mt.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Gunther R, Duan W, Wendell S, Kaemmerer W, Kafri T, et al. Biodistribution and toxicity studies of VSVG-pseudotyped lentiviral vector after intravenous administration in mice with the observation of in vivo transduction of bone marrow. Mol Ther. 2002;6:19–29. doi: 10.1006/mthe.2002.0630. [DOI] [PubMed] [Google Scholar]
- Rahim AA, Wong AM, Howe SJ, Buckley SM, Acosta-Saltos AD, Elston KE, et al. Efficient gene delivery to the adult and fetal CNS using pseudotyped non-integrating lentiviral vectors. Gene Ther. 2009;16:509–520. doi: 10.1038/gt.2008.186. [DOI] [PubMed] [Google Scholar]
- Apolonia L, Waddington SN, Fernandes C, Ward NJ, Bouma G, Blundell MP, et al. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol Ther. 2007;15:1947–1954. doi: 10.1038/sj.mt.6300281. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley SM, Howe SJ, Sheard V, Ward NJ, Coutelle C, Thrasher AJ, et al. Lentiviral transduction of the murine lung provides efficient pseudotype and developmental stage-dependent cell-specific transgene expression. Gene Ther. 2008;15:1167–1175. doi: 10.1038/gt.2008.74. [DOI] [PubMed] [Google Scholar]
- Johnson LG, Mewshaw JP, Ni H, Friedmann T, Boucher RC., and, Olsen JC. Effect of host modification and age on airway epithelial gene transfer mediated by a murine leukemia virus-derived vector. J Virol. 1998;72:8861–8872. doi: 10.1128/jvi.72.11.8861-8872.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- DiPersio CM, Shah S., and, Hynes RO. alpha 3A beta 1 integrin localizes to focal contacts in response to diverse extracellular matrix proteins. J Cell Sci. 1995;108 (Pt 6):2321–2336. doi: 10.1242/jcs.108.6.2321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Images were captured every 10 minutes over a period of 48 hours using a x10 objective and digital zoom on a Cell-IQ (Chipman Technologies) imaging platform.