Abstract
Retroviral vectors remain the most efficient and widely applied system for induction of pluripotency. However, mutagenic effects have been documented in both laboratory and clinical gene therapy studies, principally as a result of dysregulated host gene expression in the proximity of defined integration sites. Here, we report that cells with characteristics of pluripotent stem cells can be produced from normal human fibroblasts in the absence of reprogramming transcription factors (TFs) during lentiviral (LV) vector–mediated gene transfer. This occurred via induced alterations in host gene and microRNA (miRNA) expression and detrimental changes in karyotype. These findings demonstrate that vector-induced genotoxicity may alone play a role in somatic cell reprogramming derivation and urges caution when using integrating vectors in this setting. Clearer understanding of this process may additionally reveal novel insights into reprogramming pathways.
Introduction
Induced pluripotent stem cell (iPS) generation was first described in an elegant set of experiments detailing γ-retroviral-mediated gene overexpression of pluripotency-associated, and oncogenic, transcription factors (TFs; Sox2, c-Myc, Oct3/4, and KLF4) in adult mouse dermal fibroblasts.1 This landmark finding has been confirmed in many research laboratories, using both mouse and human fibroblasts, with Nanog and LIN28 (oncogene) capable of replacing Klf4 and cMYC.2,3 The finding that some of the original TFs could be replaced highlighted the potential for novel TFs to be identified in a scenario where reprogramming is not reliant upon overexpression of the aforementioned factors.
Retroviral vectors [based on γ-retroviral or lentiviral (LV) backbones] have been widely applied to this system due to their high gene-transfer efficiency (reviewed in ref. 4). Although the reprogramming procedure is relatively robust and validated in several laboratories, the reprogramming efficiency remains very low indicating that other factors may be involved as a consequence of integration events with integrating systems. Indeed, circumstantially this would appear to be confirmed with integration-free iPS cell generation [using a variety of methods including adenoviruses, plasmids, transposons, and recombinant proteins (reviewed in refs. 4,5,6,7)] proving difficult in human cells and the reprogramming efficiency decreasing substantially as compared with their integrating counterparts.8,9 Several issues regarding the use of integrating vectors (retroviral or LV) include: alteration of gene function as a result of genomic insertion;10 clonal evolution of cells due to a vector integration event activating proto-oncogenic activity; and viral transgene reactivation.11 However, owing to reprogramming efficiency, these systems (especially integrating LV) remain the “gold standard” in the laboratory setting. Mutagenic effects of retroviral vectors are well documented in both laboratory and clinical gene therapy studies, principally as a result of dysregulated host gene expression in the proximity of specific integration sites.10,12 It is, therefore, conceivable that genotoxicity can in itself contribute to reprogramming of somatic cells, either alone or in combination with expression of relevant TFs. Thus, the effect of the LV gene delivery per se on the recipient cells deserves careful analysis for both laboratory and clinical applications of reprogramming.
Results
Colony appearance
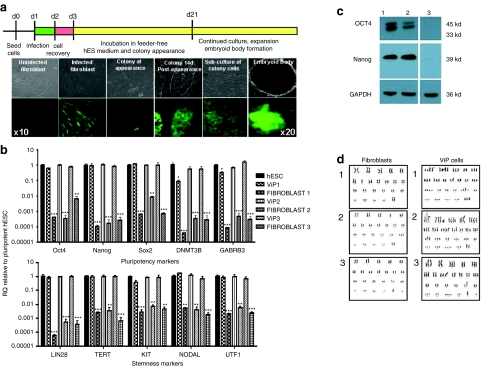
We transduced 2 × 104 cells/well (in 12-well plates) of primary human fibroblast cultures (n = 7 subjects; aged 27–55 years) with a self-inactivating (SIN) LV expressing green fluorescent protein (GFP) under regulatory control of the spleen focus-forming virus (SFFV) long terminal repeat sequence. Total multiplicities of infection (MOIs) of 0, 10, 50, and 200 were used. Subsequently, cells were incubated in feeder-free human embryonic stem cell (hESC) culture media.13 At 14–21 days post-infection, colonies of cells, hereby termed LV-induced pluripotent-like ViP cells, with typical pluripotent morphology appeared in 200 MOI-infected fibroblast cultures derived from 37- and 29-year-old female and 33-year-old male (Figure 1a, Supplementary Figure S1) but not in any other culture (overall efficiency = 0.000004%. see Supplementary Materials and Methods for individual efficiency values). Cells within colonies displayed high-level GFP expression that was maintained for the entire duration of culture (Figure 1a). DNA fragment analysis using microsatellite markers confirmed that cells were derived from the parental fibroblasts (Supplementary Table S1). Karyotyping revealed pseudohypotriploid cell clones with a similar broad range of genetic abnormalities including trisomies and tetrasomies in every cell (n = 21 cells) (Figure 1d; Supplementary Table S2). Identical chromosome aberrations were observed in chromosome 3, 4, 9, and 13 in every ViP line and in chromosome 1, 3, 4, 8, 13, 22, and X in 2 out of 3 ViP lines (Supplementary Table S2). Parental cells were 46XX or 46XY karyotype (Figure 1d).
Figure 1.
Colony appearance and characterization of pluripotency. (a) Schematic and images documenting timeline of colony appearance. See also Supplementary Figure S1 and Table S1. (b) Expression of pluripotency and stemness genes. Data are mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. hESC. In all cases, ViP cells acquire expression of pluripotency and stemness genes. (c) Western blot analysis of pluripotency markers (Oct4, Nanog, and Sox2) protein expression. hESC (lane 1) and ViP (lane 2) cells express pluripotency genes. No expression observed in parental fibroblasts (lane 3). (d) Karyotype analysis of ViP cells and parental fibroblasts. Cells are termed pseudohypotriploid, with a broad range of genetic abnormalities present on every cell analyzed, including a plethora of translocations, deletions, trisomies, and tetrasomies on every chromosome. Representative images are shown for each ViP line (n = 21 cells/clone). See also Supplementary Table S2. hESC, human embryonic stem cell; ViP, LV-induced pluripotent-like cells.
Characterization of pluripotency
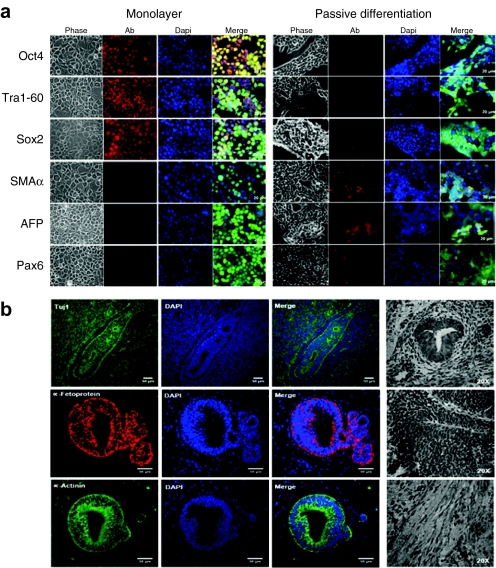
Upon subculture in feeder-free hESC conditions, colony-derived cells demonstrated the ability to self-renew, as determined by >20 passages, and formed embryoid bodies (Figure 1a). Gene expression analysis revealed a profile similar to pluripotent hESCs, with induction of pluripotency markers (Oct4, Nanog, Sox2, DNMT3B, and GABRB3; Figure 1b,c) and stemness markers (Lin28, TERT, Kit, Nodal, and UTF1; Figure 1b). No expression of gene markers from the three germ layers was detected (data not shown). Parental fibroblasts did not express any pluripotency or stemness markers (Figure 1b) and p53 expression was normal (Supplementary Figure S2). ViP cells maintained in hESC culture media exhibited morphological characteristics of pluripotent stem cells and stained positive for the classic pluripotency markers (Oct4, Tra1-60, and Sox2), and negative for germ layer markers: ectoderm (Paired box gene 6), endoderm (α-fetoprotein), and mesoderm (smooth muscle actin-α Figure 2a). Conversely, ViP cells passively differentiated to embryoid bodies lost expression of pluripotency markers and acquired characteristic markers for the three germ layers (Figure 2a). Additionally, ViP cells were capable of forming teratomas in vivo, containing cells from all germ layers (Figure 2b); ectoderm (β-III tubulin), endoderm (α-fetoprotein), and mesoderm (α-actinin). No induction of tumour-associated markers (data not shown) or altered p53 expression14 (Supplementary Figure S2) was observed.
Figure 2.
In vitro and in vivo differentiation of ViP cells. (a) Immunocytofluorescent analysis of ViP cells in pluripotent monolayer conditions (expressing pluripotency markers but not differentiation markers, with typical pluripotent morphology) and after EB passive differentiation (lack of pluripotency markers, induction of germ layer markers and associated morphological changes). (b) Teratoma formation in vivo. Expression of germ layer markers and phase images. See also Supplementary Figure S2. ViP, LV-induced pluripotent-like cells.
Genotoxic event characterization
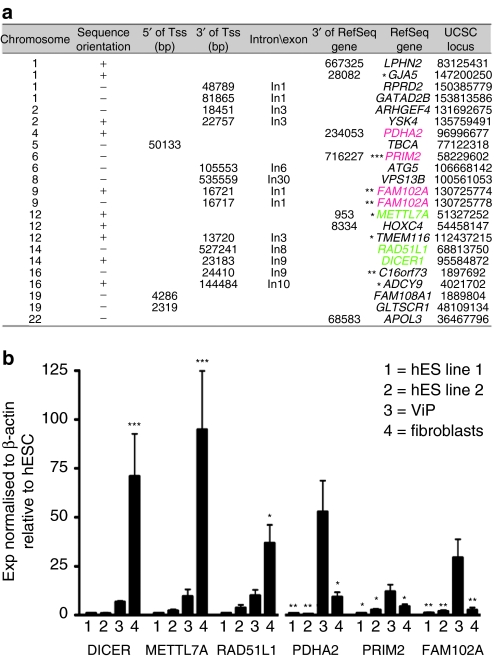
Using comprehensive genome-wide integration site analysis and sequencing, we identified 23 unique vector insertion sites in one ViP cell clone (Figure 3a). Vector target site distribution showed typical LV-specific preferences for insertions in RefSeq genes.15 We found seven LV-tagged genes recently described as iPS signature genes (Figure 3a).16 Furthermore, we identified two insertions in opposite orientation spaced by 4 bp in FAM102A (Figure 3a), recently identified as a late iPS signature gene.17 We screened for potential dysregulation of genes located next to vector integrants in one ViP clone. mRNA expression analysis revealed dysregulation in 6/10 analyzed genes in one of the clones, including integrations located in or near: DICER; methyltransferase-like 7A (METTL7A); RAD51L1; pyruvate dehydrogenase (lipoamide) α2 (PDHA2); the DNA primase polypeptide 2 (PRIM2); HOXC4; and FAM102A (Figure 3b). Transcript expression analysis of genes located next to vector integrants revealed induction of expression of FAM102A, PDHA2, and PRIM2 compared to pluripotent hESC or donor fibroblasts (Figure 3b). Furthermore, reduced expression of METTL7A; RAD51L1, thought to be essential for DNA repair by homologous recombination; and the microRNA (miRNA) biogenesis regulator, DICER, was observed resulting in levels similar to those observed in hESC (Figure 3b).
Figure 3.
Proviral insertion site characterization. (a) Chromosomal location of LV insertion sites. The distance to transcription start sites (TSS) and the nearest Refseq genes are listed. Some of the genes tagged by LV insertion sites were analyzed for their gene expression status revealing genes showing increased expression compared to hESC or parental fibroblasts (PDHA2, PRIM2, FAM102A), whereas others showed reduced expression (METTL7A, RAD51L1, DICER1). LV targeted genes recently described as early, late, and common signatures17 are marked. In total four genes (GJA5, PRIM2, METTL7A, TMEM116, ADCY9) described as early (*) and three genes (PRIM2, FAM102A, C16orf73) resembling late (**) iPSC signatures were identified. The PRIM2 gene is also part of common (***) iPSC signatures. (b) Expression of genes located next to vector insertion sites. Data are mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. hESC (DICER; METTL7a; RAD51L1) and vs. ViP (PDHA2; PRIM2; FAM102A). hES, human embryonic stem; LV, lentiviral; ViP, LV-induced pluripotent-like cells.
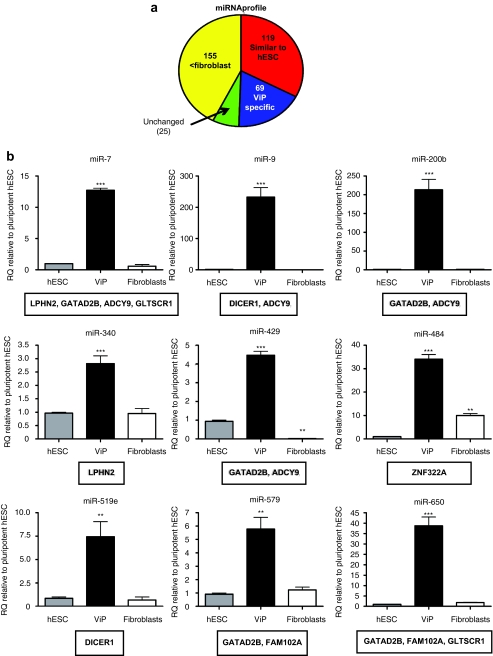
We asked whether reduced expression levels of the DICER gene revealed any distinct miRNA expression profile in the ViP clone. Analyses of miRNA profiles by TaqMan low-density array identified 119 miRNAs with similar levels to hESC (Figure 4a), and reduced expression of 155 miRNAs compared to parental fibroblasts (Figure 4a) including the Let7 family and miR-145 (targets include pluripotency-associated genes; Supplementary Figure S3). An additional subset of 69 ViP-specific miRNAs (Figure 4a), with predictive target genes in the vicinity of vector integrants, e.g. DICER and FAM102A (Figure 4b), was observed.
Figure 4.
MicroRNA expression profile. (a) Chart depicting proportional changes of miRNA expression profile. (b) Expression of (subset of) ViP-specific miRNAs with predictive target genes located near to sites of vector integrants. Data are mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. hESC. hESC (gray bars), ViP (black bars), parental fibroblasts (white bars). Target genes are listed in box below graph. See also Supplementary Figure S3. hESC, human embryonic stem cell; miRNA, microRNA; ViP, LV-induced pluripotent-like cells.
Discussion
This study reports, for the first time, independent reprogramming of human diploid fibroblasts from three individuals in the absence of transgenic reprogramming factors after LV-mediated gene transfer. This appeared to occur via induction of DNA damage associated with insertional mutagenesis and substantial chromosomal and transcriptional changes in recipient cells. Pluripotency of human stem cells is conventionally assessed by their ability to differentiate into all three germ layers both in vitro and in vivo, while losing expression of pluripotency-associated genes.4 ViP cells did not express genes indicative of specific germ cell layers when maintained under hESC culture conditions. In addition, we demonstrated that ViP cells possessed the capability to form embryoid bodies in vitro and teratomas in vivo containing cells from all three germ layers. Induction of pluripotency was not associated with altered p53 expression, recently identified as a gate-keeper of reprogramming.18,19,20 No pluripotency-associated genes (including TERT, also associated with tumorigenesis21) were expressed in the parental fibroblast samples, and we did not detect any difference in p53 expression between the parental fibroblasts, derived pluripotent-like cells and hESC samples. Together these findings strongly suggest that the driving force behind the appearance of the pluripotent-like cells in our transduced cultures was the vector itself.
To date, no association between retroviral integration site selection and direct reprogramming has been documented,22,23 suggesting that vector-mediated mutagenesis is noncontributory.24,25,26 Although there is no evidence of shared integration site selection between the selected iPS cell lines previously analyzed,26 the biological relevance of potential vector-induced effects remain unclear, owing in part to technological limitations in identification of all insertion sites.27 Thus, speculation regarding an existence of vector integration contribution to reprogramming continues.27 Using comprehensive genome-wide integration site analysis and sequencing,28 we have identified unique vector insertion sites in ViP cells. Interestingly, we identified seven vector insertions within RefSeq genes,15 which have recently been described as iPS signature genes,16 and two insertions in opposite orientation spaced by 4 bp in FAM102A, (recently identified as a late iPS signature gene)17 similar to recently described double insertions in human iPS cells.26 ViP cells demonstrated altered expression of LV-tagged genes associated with cell cycle and miRNA biogenesis as compared to their parental fibroblasts, implying that cell cycle and miRNA regulation may be implicated in the genotoxic events inducing pluripotency. miR-302, which is highly expressed in hESCs, has been implicated in reprogramming of tumour cells.29 Alternatively, downregulation of specific miRNAs may be beneficial for inducing pluripotency. For instance, the reprogramming gene Lin28 inhibits the biogenesis of the let7 family of miRNAs.30 Recent reports have confirmed that suppression of let7 activation promotes reprogramming efficiency when combined with expression of Oct4, Sox2, and KLF4.31 It has also been recently demonstrated that miR-145, which is induced during differentiation, silences the self-renewal and pluripotency mechanism in ESC by directly suppressing Oct4, Sox2, and KLF4.32 Consistent with this finding, ViP cells have reduced expression of mature miR-145 and let-7e as compared to their parental fibroblasts, but similar expression levels to pluripotent stem cells. Indeed the miRNA profile in ViP was largely akin to that observed in hESCs. Additionally, we identified a subset of ViP-specific miRNAs. Within this subset, we observed high expression of mature miR-9 and miR-519e, both of which target DICER, expression of which was augmented as compared with parental fibroblasts. Interestingly, ViP-specific miR-579 and miR-650 are predicted to target FAM102A, but in ViP cells FAM102A mRNA expression is induced when compared with pluripotent hESC and parental fibroblasts.
Unlike most conventional iPS, all ViP lines generated in this study were aneuploidic. However, they exhibit identical karyotypic abnormalities in chromosomes 3, 4, 9 and in chromosomes 1, 3, 4, 8, 13, 22, and X in 2 out of 3 independently derived lines. It is not yet clear why LV-mediated gene transfer was associated with aneuploidy. We did not observe cells with aneuploidy in parental fibroblasts; therefore, we can suggest that these karyotypic changes are restricted to the pluripotent-like ViP cells and occurred as a consequence of vector-mediated integration and/or DNA damage. Whether these cells are truly pluripotent or transformed is not clear; however, they display a number of characteristics that are akin to those utilized to identify human iPS cells (reviewed in ref. 4). We observed 23 insertions in ViP cells, which is within a similar range to that reported in a comparison of a selection of iPS lines where the number of insertion sites ranged from 5 to 15 per individual iPS clone.26
Current research in the reprogramming field is endeavouring to generate transgene-free methods of deriving iPS cells but with the caveat that reprogramming efficiency of nonintegrating systems remains very low, so the majority of research on iPS biology as compared to hESC cells and their subsequent differentiation for analyzing disease modeling is performed on iPS cells derived from integrating systems of reprogramming. These data demonstrate for the first time that a pluripotent-like cell can be derived by integrating LV constructs in the absence of exogenous TFs and suggests that other mechanisms in addition to the expression of reprogramming factors could be involved in the higher incidence of integration-vector-induced pluripotency. This is an important consideration when analyzing data from iPS cells obtained from integrating systems even in circumstances where the majority of the proviral construct is excised, and has implications for interpretation of studies using iPS cells.
The potential therapeutic applications of iPS cells are extensive and without limitation; however, understanding of the interactions that govern iPS cell generation at the molecular level is still inadequate, with better knowledge and identification of novel reprogramming factors a prerequisite for progression toward the clinic. More broadly, integrating vectors require careful safety evaluation relating to their application to gene therapy and cell reprogramming. We have reported the generation of pluripotent-like cells from a high dose of integrating LV in the absence of exogenous reprogramming TFs, which suggests that there is a potential for an integration-related phenomenon in cellular reprogramming, a possibility that should not be discounted when analysing experiments performed on cells generated from an integrating system.
Materials and Methods
Cell lines and culture conditions. Informed patient consent was obtained in writing from each patient before tissue collection after local ethical committee approval for the study was granted. Primary human dermal fibroblasts were initiated from explants obtained from punch biopsy and were used between passages 4 and 10. All cultures were routinely cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), penicillin (50 µg/ml; Invitrogen), streptomycin (50 µg/ml; Invitrogen), ℓ-glutamate (2 mmol/l; Invitrogen) and sodium pyruvate (1 mmol/l; Invitrogen), denoted “complete media”. HEK293T cells (ATCC, Teddington, UK) were maintained in complete media. Cell lines were cultivated at 37 °C in a humidified atmosphere containing 5% CO2.
Before LV infection, cells were seeded at a density of 2 × 104/well (12-well plate) on 1 mg/ml fibronectin (Merck Chemicals, Nottingham, UK) and infected in complete media. After LV infection and cell recovery, fibroblasts were cultured in a chemically defined media,33 supplemented with 50% human Umbilical Wharton Inner Layer (Cellartis, Göteborg, Sweden) fibroblast-conditioned VitrohES media (VitroLife, Gothenburg, Sweden) and 10 ng/ml basic fibroblast growth factor (Invitrogen).
Embryoid bodies were formed by forced aggregation as previously described34 and permitted to reattach to 1 mg/ml fibronectin-coated dishes prior to staining.
LV vectors and hESC infection. LV vectors were produced by triple transient transfection of HEK293T cells with a packaging plasmid (pCMVΔ8.74), a plasmid encoding the envelope of vesicular stomatitis virus (Plasmid Factory, Bielefeld, Germany) and pHR-SIN-SFFV-GFP,35 employing polyethylenimine (Sigma-Aldrich, St Louis, MO) as previously described.35 For infection, 2 × 104 cells were transduced with pHR-SIN-SFFV-GFP lentivirus with an MOI of 0, 10, 50, or 200. Cells were incubated in complete media containing LV particles and 4 µg/ml Polybrene (Sigma-Aldrich) for 18 hours at 37 °C in a humidified atmosphere containing 5% CO2. LV particles were removed and media was replaced with fresh complete media for an additional 24 hours to permit cell recovery.
mRNA and miRNA quantitative-reverse transcription-PCR analysis. Total cellular RNA was isolated from ViP cells, parental fibroblasts or pluripotent hESCs with RNAeasy Mini Kit (Qiagen, Crawley, UK). mRNA expression of 92 validated genes associated with stem cell pluripotency and differentiation to all three germ layers were analyzed using the Human Stem Cell Pluripotency Taqman Low-Density Array fluidic card (Applied Biosystems, Warrington, UK). Human miRNAs were analyzed using the Human MicroRNA Array v1.0 (Applied Biosystems). p53 expression was analyzed using an inventoried p53-specific gene expression assay (Applied Biosystems). The samples were run on an ABI Prism 7900Ht sequence Detection System (Applied Biosystems) in triplicate, and mean values used in subsequent analyses.
miRNA predictive targets. miRNA predictive targets were identified using miRbase v14.0 and Targetscan.
Immunocytofluorescence. Cultured cells were incubated at 4 °C overnight with the following antibodies: mouse IgG monoclonal antihuman OCT4 (1:200; Santa Cruz Biotechnology, Heidelberg, Germany); mouse IgG monoclonal antihuman Sox2 (1:200; Abcam, Cambridge, UK); monoclonal antihuman Tra1-60 (1:200; Santa Cruz Biotechnology); mouse IgG monoclonal antihuman smooth muscle actin-α (1:200; Dako UK, Ely, UK); mouse IgG monoclonal antihuman α-fetoprotein (1:100; Abcam); rabbit polyclonal antihuman Paired box gene 6 (1:1,000; Abcam); rabbit polyclonal antihuman vimentin (1:200; Abcam) and detected with a 1:500 dilution of TRITC (543)-conjugated goat anti-mouse/goat anti-rabbit/antibody (Molecular Probes, Invitrogen) in phosphate-buffered saline for 60 minutes at room temperature. Cells were counterstained and mounted under a glass coverslip using ProLong Gold (Invitrogen). Images were captured on the Axiovert 200M microscope (Zeiss, Hertfordshire, UK).
Teratoma generation. A volume of 1 × 106 iPS cells in matrigel (per site per injection) were injected subcutaneously into the dorsal flank of common γ-chain−/−, RAG2−/− C5−/− mice. Mice were killed between 2 and 8 weeks after injection and teratoma was dissected and processed for hematoxylin/eosin staining and immunostaining with suitable antibodies. The following primary antibodies along with suitable secondary conjugated antibodies were used in this study: anti-tubulin β-III isoform (ectodermal derivatives), anti-α-actinin (mesodermal derivatives), and anti-α-fetoprotein (endodermal derivatives) (all from Millipore, Watford, UK). All images were taken with a Zeiss Axiovert 135 inverted microscope and edited with Adobe Photoshop CS4 software.
Comprehensive genome-wide integration site analysis. LV insertion sites were cloned by linear amplification-mediated-PCR36 and nonrestrictive linear amplification-mediated-PCR28 from 100 to 300 ng genomic DNA and sequenced by massive parallel pyrosequencing (GS FLX/454; Roche, Mannheim, Germany). Vector–cellular junctions were bioinformaticaly identified using Blast2Seq and the Smith–Waterman algorithm (Jaligner). Vector insertion loci were mapped to the human genome via BLAT (Assembly February 2009).
Chromosome analysis. Chromosome harvesting and analysis was performed by the West of Scotland Regional Cytogenetics Service. ViP and fibroblast cultures at identical passage numbers were manually harvested using routine techniques following 3-hour incubation with 1 µg/ml colcemid (Invitrogen). Slides were G-banded using the Thermo Scientific Varistain Gemini (Trypsin Sigma, Leishman's Sigma) and were scanned on the Genetix GSL 120 (Genetix, New Milton, UK). Chromosome analysis and checking was performed, by HPC registered Clinical Scientists, on screen using Genetix Cytovision software (Genetix Europe, Gateshead, UK).
Statistical analysis. Before any statistical analysis data were tested for and shown to exhibit Gaussian distribution. Gaussian distribution was determined by applying the Shapiro–Wilk normality test to the data. Where appropriate, values were presented as means ± SEM. Comparison was determined by repeated measures analysis of variance. Significant differences were assigned using Dunnett's post hoc test. The criterion for significance for all tests was set at P < 0.05. Specific software was used to assist in the data analysis (GraphPad Prism v4.0b for Macintosh; GraphPad Software, San Diego, CA).
SUPPLEMENTARY MATERIAL Figure S1. Morphology of pluripotent-like cell colony appearance. Top panel shows parental fibroblasts from 3 patients, bottom panel the corresponding pluripotent-like cells with typical pluripotent morphology. Fibroblast morphology-like cells in bottom panel are non-reprogrammed cells (related to Figure 1). Figure S2. p53 expression in hESC, ViP and parental fibroblast cells. No significant difference was observed between cells (related to Figure 2). Figure S3. Expression of miRNAs with pluripotency-associated predictive target genes. Data are mean ± S.E.M; *P < 0.05, ***P < 0.001 vs. hESC. hESC (white bars), ViP (red bars), parental fibroblasts (green bars). Target genes are listed in box below graph (related to Figure 4). Table S1. DNA fragment analysis. Table S2. List of chromosome aberrations. Chromosome analysis of the cell lines was carried out using standard G-banding techniques at passage 7. Chromosome analysis shows pseudohypotriploid cell lines. This means that there are between 58-68 chromosomes per cell but the chromosome complement is not normal. Of 20 cells examined, a range of trisomies and tetrasomies are present in each cell. Both clonal and single cell structural abnormalities are also present. Abnormalities are coloured according to the numbers of cells they have been seen in: Pink = 15/20 cells or more; Blue = 10-14/20 cells; Green = 2-9/20 cells; Black = 1/20 cells. * denote chromosome aberrations that are common to two or more ViP lines (related to Figure 1). Materials and Methods.
Acknowledgments
We thank Nicola Britton at the British Heart Foundation Glasgow Cardiovascular Research Centre (BHF GCRC) for technical assistance and Stuart Nicklin (BHF GCRC) for assistance in editing the manuscript. This work was supported by the British Heart Foundation. Confocal images of teratoma were taken at the Confocal facility, UCL Institute of Child Health with the help of Dr Bertrand Vernay.
Supplementary Material
Morphology of pluripotent-like cell colony appearance. Top panel shows parental fibroblasts from 3 patients, bottom panel the corresponding pluripotent-like cells with typical pluripotent morphology. Fibroblast morphology-like cells in bottom panel are non-reprogrammed cells (related to Figure 1).
p53 expression in hESC, ViP and parental fibroblast cells. No significant difference was observed between cells (related to Figure 2).
Expression of miRNAs with pluripotency-associated predictive target genes. Data are mean ± S.E.M; *P < 0.05, ***P < 0.001 vs. hESC. hESC (white bars), ViP (red bars), parental fibroblasts (green bars). Target genes are listed in box below graph (related to Figure 4).
DNA fragment analysis.
List of chromosome aberrations. Chromosome analysis of the cell lines was carried out using standard G-banding techniques at passage 7. Chromosome analysis shows pseudohypotriploid cell lines. This means that there are between 58-68 chromosomes per cell but the chromosome complement is not normal. Of 20 cells examined, a range of trisomies and tetrasomies are present in each cell. Both clonal and single cell structural abnormalities are also present. Abnormalities are coloured according to the numbers of cells they have been seen in: Pink = 15/20 cells or more; Blue = 10-14/20 cells; Green = 2-9/20 cells; Black = 1/20 cells. * denote chromosome aberrations that are common to two or more ViP lines (related to Figure 1).
REFERENCES
- Takahashi K., and, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Maherali N., and, Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Okita K., and, Yamanaka S. Induction of pluripotency by defined factors. Exp Cell Res. 2010;316:2565–2570. doi: 10.1016/j.yexcr.2010.04.023. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K., and, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E., and, Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J Clin Invest. 2010;120:51–59. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Araúzo-Bravo MJ., and, Schöler HR. Generation of induced pluripotent stem cells from neural stem cells. Nat Protoc. 2009;4:1464–1470. doi: 10.1038/nprot.2009.173. [DOI] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC., and, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T., and, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Baum C. Insertional mutagenesis in gene therapy and stem cell biology. Curr Opin Hematol. 2007;14:337–342. doi: 10.1097/MOH.0b013e3281900f01. [DOI] [PubMed] [Google Scholar]
- Kane NM, Meloni M, Spencer HL, Craig MA, Strehl R, Milligan G, et al. Derivation of endothelial cells from human embryonic stem cells by directed differentiation: analysis of microRNA and angiogenesis in vitro and in vivo. Arterioscler Thromb Vasc Biol. 2010;30:1389–1397. doi: 10.1161/ATVBAHA.110.204800. [DOI] [PubMed] [Google Scholar]
- Zhao T., and, Xu Y. p53 and stem cells: new developments and new concerns. Trends Cell Biol. 2010;20:170–175. doi: 10.1016/j.tcb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KD, Seong SK, Park YM, Choi Y, Park JH, Lee SH, et al. Telomerase reverse transcriptase (TERT) Related with Telomerase Activity Regulates Tumorigenic Potential of Mouse Embryonic Stem Cells. Stem Cells Dev. 2010. [DOI] [PubMed]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN., and, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Varas F, Stadtfeld M, de Andres-Aguayo L, Maherali N, di Tullio A, Pantano L, et al. Fibroblast-derived induced pluripotent stem cells show no common retroviral vector insertions. Stem Cells. 2009;27:300–306. doi: 10.1634/stemcells.2008-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler T, Cantilena A, Métais JY, Xu X, Nguyen AD, Borate B, et al. No evidence for clonal selection due to lentiviral integration sites in human induced pluripotent stem cells. Stem Cells. 2010;28:687–694. doi: 10.1002/stem.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RG. Does retroviral insertional mutagenesis play a role in the generation of induced pluripotent stem cell. Mol Ther. 2008;16:1354–1355. doi: 10.1038/mt.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15:1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ., and, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL., and, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA., and, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Liu Y, Song Z, Zhao Y, Qin H, Cai J, Zhang H, et al. A novel chemical-defined medium with bFGF and N2B27 supplements supports undifferentiated growth in human embryonic stem cells. Biochem Biophys Res Commun. 2006;346:131–139. doi: 10.1016/j.bbrc.2006.05.086. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Anderson D, Priddle H, Barbadillo Muñoz MD, Chamberlain S, Allegrucci C, et al. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphology of pluripotent-like cell colony appearance. Top panel shows parental fibroblasts from 3 patients, bottom panel the corresponding pluripotent-like cells with typical pluripotent morphology. Fibroblast morphology-like cells in bottom panel are non-reprogrammed cells (related to Figure 1).
p53 expression in hESC, ViP and parental fibroblast cells. No significant difference was observed between cells (related to Figure 2).
Expression of miRNAs with pluripotency-associated predictive target genes. Data are mean ± S.E.M; *P < 0.05, ***P < 0.001 vs. hESC. hESC (white bars), ViP (red bars), parental fibroblasts (green bars). Target genes are listed in box below graph (related to Figure 4).
DNA fragment analysis.
List of chromosome aberrations. Chromosome analysis of the cell lines was carried out using standard G-banding techniques at passage 7. Chromosome analysis shows pseudohypotriploid cell lines. This means that there are between 58-68 chromosomes per cell but the chromosome complement is not normal. Of 20 cells examined, a range of trisomies and tetrasomies are present in each cell. Both clonal and single cell structural abnormalities are also present. Abnormalities are coloured according to the numbers of cells they have been seen in: Pink = 15/20 cells or more; Blue = 10-14/20 cells; Green = 2-9/20 cells; Black = 1/20 cells. * denote chromosome aberrations that are common to two or more ViP lines (related to Figure 1).