Abstract
Reversible posttranslational modification of internal lysines in many cellular or viral proteins is now emerging as part of critical signalling processes controlling a variety of cellular functions beyond chromatin and transcription. This paper aims at demonstrating the role of lysine acetylation in the cytoplasm driving and coordinating key events such as cytoskeleton dynamics, intracellular trafficking, vesicle fusion, metabolism, and stress response.
1. Introduction
The story of cytoplasmic protein lysine acetylation begins long after the discovery of posttranslational acetylation of histones in the sixties [1]. In fact only in 1985 tubulin was described as the first acetylated cytoplasmic protein [2, 3]. Since then numerous other cytoplasmic proteins have been found acetylated. The first global proteomic study on acetylated proteins describes 37 acetylated proteins in the cytoplasmic fraction of Hela cells and 133 in mouse liver mitochondria [4]. In another study about 250 acetylated proteins, presumably localized in the cytoplasm, have been identified [5].
Lysine (K) acetylation is catalysed by a lysine acetyltransferase (KAT) formerly called histone acetyltransferase (HAT) (for new nomenclature see Allis et al. [6]), which transfers the acetyl-group of acetyl-CoA to the epsilon-amino group of an internal lysine residue. The reverse reaction is accomplished by deacetylases, which can be divided into several classes. The class I, IIa, IIb, and IV enzymes are zinc dependant, whereas members of the class III family (also called sirtuins) use NAD+ as a cofactor for the deacetylation reaction.
The high number of acetylated proteins present in the cytoplasm points to a critical role for this posttranslational modification in the regulation of cytoplasmic events. In this paper we will focus on selected examples illustrating the role of reversible acetylation in the cytoplasm and we will mention some proteins, identified by proteomic approaches as being acetylated, when it could be important in the context of the discussed cellular processes. We will also provide an overview on what is known about the cytoplasmic localisation of the enzymes implicated in lysine (de)acetylation.
2. Cytoplasmic Localisation of KATs and HDACs
2.1. KATs
Most of the extensively characterised acetyltransferases are known as nuclear enzymes (see Table 1 for overview). Even Hat1, the first identified acetyltransferase, is predominantly localized in the nucleus, although it has been characterized as a type B acetyltransferase which refers to its role in the cytoplasm where it acetylates newly synthesised histones [7–9]. Under some circumstances, like early during development or in colorectal tumors, the cytoplasmic fraction of Hat1 increases [10, 11]. In addition, it has recently been shown that two different isoforms of Hat1 are expressed in keratinocytes, which differ in their cellular localisation [12].
Table 1.
Lysine (K) acetyltransferases.
| New Name | Former name human | Former name D. melanogaster | Former name S. cerevisiae | Former name S. pombe | HAT complexes |
|---|---|---|---|---|---|
| GNAT family | |||||
| KAT1 | HAT1 | CG2051 | Hat1 | Hat1/Hag603 | HAT-B |
| KAT2 | dGCN5/PCAF | Gcn5 | Gcn5 | SAGA, ADA, ATAC | |
| KAT2A | hGCN5 | STAGA, TFTC | |||
| KAT2B | PCAF | PCAF complex | |||
| KAT9 | ELP3 | dELP3/CG15433 | Elp3 | Elp3 | Elongator for RNA polymerase II |
| HsMEC-17 | |||||
| p300/CBP family | |||||
| KAT3 | dCBP/NEJ | ||||
| KAT3A | CBP | ||||
| KAT3B | P300 | ||||
| MYST family | |||||
| KAT5 | TIP60/PLIP | dTIP60 | Esa1 | Mst1 | NuA4 |
| KAT6 | (CG1894) | Sas3 | (Mst2) | NuA3 | |
| KAT6A | MOZ/MYST3 | ||||
| KAT6B | MORF/MYST4 | ||||
| KAT7 | HBO1/MYST2 | (Mst2) | |||
| KAT8 | hMOF/MYST1 | dMOF (CG1894) | Sas2 | (Mst2) | MSL complex |
| nuclear receptor coactivators | |||||
| KAT13A | SRC-1 | ||||
| KAT13B | ACTR | ||||
| SRC-3 | |||||
| TIF-2 | |||||
| GRIP1 | |||||
| ATF-2 | |||||
| Divers | |||||
| KAT4 | TAF1 (TAFII250) | dTAF1 | Taf1 | Taf1 | TFIID |
| KAT10 | Hap2 | ||||
| KAT11 | Rtt109 | ||||
| KAT12 | TFIIIC90 | ||||
| KAT13C | P160 | ||||
| KAT13D | CLOCK | ||||
Although acetyltransferases are considered mostly nuclear, an increasing number of studies reports on their nucleocytoplasmic transport. For instance PCAF and Gcn5 become phosphorylated following growth factor receptor signalling, which induces their translocation to the nucleus [13]. The cellular localisation of PCAF is not only regulated by phosphorylation. In fact PCAF can autoacetylate lysine residues within its nuclear localisation signal (NLS) and deacetylation of these lysine residues leads to cytoplasmic accumulation of PCAF [14]. CBP and p300 behave almost like Hat1 since, during oocyte maturation, they are first found in the cytoplasm before being imported into the nucleus [15]. Furthermore, similar to Hat1, p300 is found in the cytoplasm in breast carcinomas but not in the adjacent normal mammary gland [16]. Both nuclear localisation and nuclear export signals have been found in Tip60, a member of the MYST family of acetyltransferases. Tip60 can be recruited to the plasma membrane by the amyloid precursor protein, which induces its phosphorylation and subsequent translocation to the nucleus [17]. In addition Tip60 appears in two splice variants. Whereas the longer isoform is essentially found in the nucleus, the shorter form, Tip60 beta (also called PLIP), missing exon 5, is located in both the cytoplasm and the nucleus and interacts with cytosolic phospholipase A2 [18, 19]. The acetyltransferase ATF2 also has nuclear localisation and export signals and is able to shuttle between the nucleus and the cytoplasm. Heterodimerisation with c-Jun in the nucleus is necessary to retain ATF2 in the nuclear compartment [20].
The most astonishing fact is that, although tubulin was the first acetylated protein described in the cytoplasm [2, 3] and a tubulin acetyltransferase activity had already been purified and characterised in 1986 [21], the scientific community had to wait until 2009 to put a name on an enzyme able to acetylate α-tubulin. In fact the acetyltransferase Elp3, which is the catalytic subunit of the transcriptional elongator complex, was found able to acetylate tubulin, which is essential for the maturation of cortical neurons [22]. The role of Elp3 as a tubulin acetyltransferase important for neuronal development has been confirmed in a genetic RNAi suppression screen for regulators of α-tubulin acetylation using the nematode Caenorhabditis elegans [23]. Besides its role in tubulin acetylation, Elp3 has been implicated in the regulation of other processes outside the nucleus such as stress signalling, tRNA modification, exocytosis and actin dynamics [24–28]. A study by Esberg et al., however, questions the role of Elp3 as an acetyltransferase since various effects observed in Elp3 deficient cells could be attributed to its role in tRNA modifications and more specifically to the absence of wobble uridine modified lysine and glutamine tRNA species [29, 30]. A recently published paper describes Gcn5 as another acetyltransferase, capable of acetylating α-tubulin after its recruitment to microtubules via a cytoplasmic proteolytic fragment of myc [31]. Importantly, another recent paper identifies Mec-17, a Gcn5- related protein, as a major alpha tubulin acetyltransferase [32]. Finally, two reports suggest that enzymes having N-acetyltransferase activity could also acetylate internal lysine residues and contribute to tubulin acetylation [33, 34].
2.2. HDACs
Compared to the Hat families there are by far more examples of deacetylases located in the cytoplasm (see Table 2 for list of HDACs). Even members of the type I family of deacetylases, which in the past have been considered as strictly nuclear proteins, can be found in the cytoplasm. For instance HDAC1 is a nuclear enzyme which, under pathological situations, is exported via CRM1 into the cytoplasm where it binds to kinesin motors, hindering cargo transport [35]. HDAC3, another type I deacetylase, is found in the nucleus and the cytoplasm and has both nuclear export as well as nuclear localisation signals [36]. It is retained in the cytoplasm via its interaction with IkappaBalpha [37] until TNF-alpha signalling leads to the degradation of this binding partner [38]. Furthermore HDAC3 can associate with the plasma membrane where it is phosphorylated by src thereby increasing its activity [39]. Surprisingly, in contrast to the other class I enzymes which are more nuclear, HDAC8 is essentially in the cytoplasm where it associates with smooth muscle alpha actin to regulate cell contractility [40, 41].
Table 2.
Lysine (K) deacetylases.
| Name | Localisation | Cytoplasmic substrates |
|---|---|---|
| Class I | ||
| HDAC1 | mainly nucleus | |
| HDAC2 | nucleus | |
| HDAC3 | mainly nucleus/cytoplasm | |
| HDAC8 | mainly cytoplasm | |
| class IIa | ||
| HDAC4 | cytoplasm/nucleus | MLP, DNAJB8 |
| HDAC5 | cytoplasm/nucleus | |
| HDAC7 | cytoplasm/mitochondria/nucleus | |
| HDAC9 | cytoplasm/nucleus | |
| Class IIb | ||
| HDAC6 | cytoplasm | tubulin, cortactin, Hsp90, β-catenin, peroxiredoxin |
| HDAC10 | cytoplasm/nucleus | |
| Class IV | ||
| HDAC11 | mainly nucleus | |
| Class III(sirtuins) | ||
| sirt1 | cytoplasm/nucleus | cortactin, Atg5, Atg7, Atg8 |
| sirt2 | cytoplasm | tubulin, cortactin? |
| sirt3 | mitochondria/long form cytoplasm | |
| sirt4 | mitochondria | |
| sirt5 | mitochondria | |
| sirt6 | nucleus | |
| sirt7 | nucleus | |
Class IIa deacetylases (4, 5, 7 and 9) [42] are known to shuttle between the nucleus and the cytoplasm [43, 44]. HDAC7 is the only member of this family reported to be present also in mitochondria; it leaves mitochondria and the nucleus to accumulate in the cytoplasm after initiation of apoptosis [45]. All class IIa members have nuclear localisation signals and binding sites for proteins of the 14-3-3 family. An important regulatory mechanism relies on the phosphorylation of these 14-3-3 binding sites inducing the interaction with 14-3-3 proteins and subsequent accumulation in the cytoplasm. Cytoplasmic accumulation has been attributed to 14-3-3-mediated CRM1- dependant nuclear export but two publications have shown more recently that phosphorylation can target the NLS and that the deacetylases are retained in the cytoplasm after binding of 14-3-3 to the phosphorylated NLS and disruption of the nuclear import [46, 47]. Independent of its phosphorylation status, the nuclear export of HDAC4 is regulated by oxidation of two of its cystein residues resulting in an intramolecular disulfide bridge, whose reduction inhibits nuclear export [48]. HDAC7 is another example of a class IIa enzyme, which can be exported from the nucleus independently of its phosphorylation status and 14-3-3 binding [49].
The translocation of type I and IIa HDACs into the cytoplasm in all the examples mentioned above (except HDAC8) is generally considered as a regulatory mechanism, which serves to reduce their action in the nucleus and so far cytoplasmic substrates have only been found for HDAC4 (as discussed below).
The most extensively studied cytoplasmic deacetylase is the type IIb enzyme HDAC6, which has several cytoplasmic substrates, including tubulin [50–52], cortactin [53], Hsp90 [54–57], β-catenin [58], and peroxiredoxin [59]. Although it bears intrinsic nuclear import and export signals, HDAC6 is almost exclusively localised in the cytoplasm [60, 61], where its actions underlie multiple regulatory processes. One way to regulate its action is to change its localisation within the cytoplasm. As mentioned below HDAC6 is translocated together with Hsp90 and Rac1 to membrane ruffles after PDGF stimulation where it influences actin dynamics [62]. Tubulin deacetylation by HDAC6 can be prevented by Cbl, due to competitive binding to β-tubulin [63]. Another way influencing its activity is the binding to various partner proteins either directly or indirectly. An example of an indirect interaction essential for HDAC6 activity is the formation of a tripartite complex composed of the farnesyltransferase, HDAC6, and microtubules. Disruption of the complex by KO of the alpha subunit of the farnesyltransferase or by an inhibitor of this enzyme leads to enhanced tubulin acetylation [64]. BBIP10, a protein essential for primary cilia formation also binds to and inhibits HDAC6, but so far it is not clear whether this interaction is direct or not [65]. An example of a direct interaction is the binding of Dia2 to one of the two deacetylase domains of HDAC6, which leads to activation of HDAC6 in osteoclasts [66]. Two other examples are the microtubule-associated protein tau and the tubulin polymerisation promoting protein TPPP/p25, which can bind to HDAC6 and inhibit its activity leading to a subsequent increase in microtubule acetylation [67–69]. Ilp45 (a newly discovered protein) as well as CYLD, a deubiquitinating enzyme, can both inhibit HDAC6 by direct binding to the deacetylase domains of HDAC6, which in the case of Ilp45 will also lead to HDAC6 degradation [70, 71]. A third way to regulate HDAC6's activity is by its posttranslational modification. It has been shown for instance that HDAC6 interacts with EGFR and is phosphorylated by EGFR at Tyr570 after ligand-induced receptor activation [72]. This reduces HDAC6 activity leading to tubulin acetylation and accelerated delivery of endocytosed EGFR to lysosomes. In contrast Aurora A, a mitotic ser/thr kinase, phosphorylates HDAC6 leading to its activation resulting in tubulin deacetylation and resorption of primary cilia in human retinal cells [73]. In this context it is worth mentioning that HDAC6 (as well as other HDACs) can be found in complexes together with phosphatases and both enzymes are active under these conditions [74]. HDAC6 activity, besides being regulated by phosphorylation, can also be reduced by p300-mediated acetylation [75]. In addition to its deacetylase function, HDAC6 has other important cellular tasks, such as cargo transport by binding to the dynein and kinesin motor complexes, or binding to mono and polyubiquitin, which is necessary for aggresome formation and induction of quality control autophagy (QC-autophagy) (for review see [76, 77]).
HDAC10, the other class IIb enzyme, is present in the cytoplasm and the nucleus and, in contrast to the class IIa family members, is not exported through a CRM1-mediated transport mechanism [78–81]. Finally HDAC11, the only type IV deacetylase, is essentially a nuclear enzyme but can be found in the cytoplasm in latent HIV infected cells [82, 83].
The sirtuin or class III family of deacetylases are a functionally distinct family for two reasons. First they depend on the cofactor NAD+ for their activity, and second they do not release free acetate after hydrolyses of the acetyl group but rather transfer it onto ADP-ribose. In mammals, seven sirtuins have been described (Sirt1-7) [84]. Only Sirt1, Sirt2, and Sirt3 have a true deacetylase activity all the others have only ADP-ribosyltransferase activity. At least, this was considered the truth until it was observed that Sirt6 could deacetylate lysine 9 of histone H3 [85]. No cytoplasmic localisation, however, has been observed for Sirt6; we will therefore focus only on Sirt1-3 [86].
Sirt1 has both a nuclear localisation and a CRM1-dependent export signal and, in most cells, it is present both in the nucleus and the cytoplasm [87]. Upon induction of neural differentiation it is transiently transported into the nucleus but for NGF-induced neurite outgrowth it has to be present in the cytoplasm [88, 89]. As discussed below, Sirt1 has important roles in the cytoplasm linked to autophagy and cell migration via deacetylation of cortactin [90, 91].
Sirt2 is almost only localised in the cytoplasm due to an active CRM1-dependent export mechanism [92]. It has been implicated in tubulin deacetylation [93] and seems to deacetylate tubulin especially under certain circumstances, for instance during mitosis to regulate mitotic microtubule dynamics [94] or during oligodendrocyte differentiation [95]. It was suggested that Sirt2 inhibition could reduce alpha synuclein toxicity in Parkinson disease, but the molecular interplay has not been identified [96, 97].
Finally, Sirt3 is known as a mitochondrial enzyme (for a detailed review see [84]), but a long form of Sirt3 partially localised in the cytoplasm has been recently described [98].
At last, it is worth mentioning that quite a few nuclear and cytoplasmic acetylated proteins are targeted by both, class I/II as well as class III, deacetylases as for instance histones [99], tubulin [50–52, 93], Ku70 [98, 100], p53 [101, 102], and cortactin [53, 91]. For most of these examples it is not clear whether HDACs of different classes show a preference for particular acetylated lysine residues in these proteins. Actually, motif preferences for lysine acetylation only start to emerge. The local sequence context around acetylated lysine residues in cytoplasmic proteins appears to be different from that of histones but similar to that of other nuclear proteins. Furthermore mitochondrial acetylation motifs differ from acetylation sites found in both histones and other nuclear/cytoplasmic proteins [4, 5]. In spite of these advances a precise consensus sequence for the substrate specificity of different Kats or HDACs has yet to be defined.
3. Functional Implications of Cytoplasmic Lysine Acetylation
3.1. Regulation of the Cytoskeleton
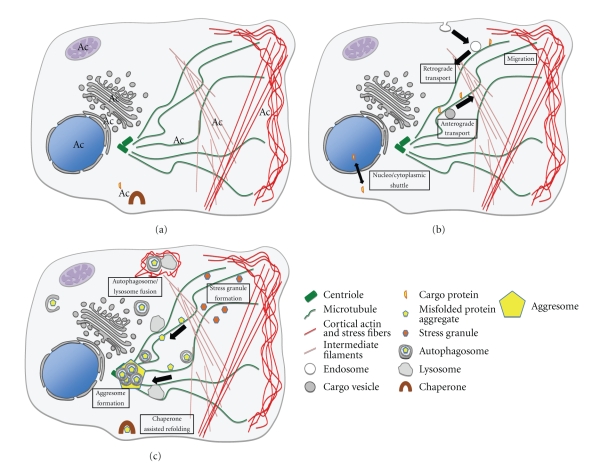
Since the cytoskeleton is involved in many cytoplasmic processes, we will start by describing what is known about the regulatory effect of acetylation on the three main types of cytoskeletal components, namely, actin filaments, microtubules, and intermediate filaments (Figures 1(a) and 1(b)).
Figure 1.
Schematic overview of cytoplasmic mechanisms regulated by acetylation. (a) Localisation of acetylation events within a cell. Indicated are individual acetylated proteins and cytoskeletal structures (cortical actin and actin stress fibers, microtubules, intermediate filaments) as well as organelles (nucleus, mitochondria, ER, ERGIC, golgi) in which acetylation and deacetylation events take place. (b) Involvement of reversible acetylation in the regulation of various cytoplasmic processes, including cell migration following cytoskeleton remodelling, individual protein transport as well as vesicular transport and nucleocytoplasmic shuttling of proteins. (c) Regulation of stress response and cytoplasmic cleanup systems by various acetylation/deacetylation events.
3.1.1. Actin Filaments
The globular monomeric form of actin (G-actin) polymerises to form a double helix (filamentous or F-actin), which can then be bundled into stress fibers. There are three major isoforms of actin. Beta and gamma actin form the stress fibers are important for cell shape and cell movement. Alpha actin constitutes the microfilaments in muscle cells, which together with myosin assures the traction forces necessary not only for muscle contraction but also for cytoplasmic streaming in nonmuscle cells. Using proteomic approaches it has been shown that all three actin isoforms can be acetylated [4, 5] and acetylation of lysine 61 in gamma actin may result in stabilisation of actin stress fibers [4]. Not only actin itself but also several regulatory proteins of the actin cytoskeleton are modified by acetylation. For instance six out of the seven subunits of the Arp2/3 complex, which is important for actin nucleation, are acetylated [5]. Cortactin binds to F-actin in the cell cortex and can recruit the Arp2/3 complex to the cortical actin cytoskeleton when it has been activated by phosphorylation. But phosphorylation is not the only regulatory modification of cortactin. It has been shown that cortactin can be acetylated by p300 or PCAF on nine different lysine residues present in its central repeat domain [53, 91]. When acetylated its translocation to the cell periphery is inhibited and its actin binding capacity reduced. This leads to diminished actin dynamics at the cell periphery and thus to altered cell motility. Deacetylation of cortactin is mediated via HDAC6 and Sirt1 (maybe also Sirt2), which may act in cooperation to induce cell migration [53, 91]. In fact the actin cytoskeleton is essential for cell migration since it is necessary for the formation of membrane ruffles or lamellipodia, filopodia, and actin stress fibers, which reflect different dynamic states of the actin cytoskeleton. Actin dynamics is also regulated by small GTPases of the Rho family. A simplified general scheme implicates RhoA in stress fiber formation, Rac1 in lamellipodia and cdc42 in filopodia formation. The activation of these G-proteins in turn depends on the action of GDIs (GDP dissociation inhibitors), GAPs (GTPase-activating proteins), and GEFs (GDP/GTP exchange factors). It has been shown that acetylation of RhoGDI alpha prevents its inhibitory action on Rho family members leading to enhanced stress fiber and filopodia formation [4]. RhoA can also be inhibited by the E-cadherin-binding protein p120 catenin. Acetylation of p120 changes its subcellular localisation thereby relieving its inhibitory action on RhoA [4]. Rac1 activation, leading to membrane ruffle formation, associated macropinocytosis, and cell migration, depends on deacetylation of the chaperon Hsp90 by HDAC6, but the precise sequence of events has not been elucidated [62].
Muscle contraction can also be regulated by acetylation as observed for actomyosin filament activity in cardiac muscle cells. In fact the muscle LIM protein (MLP), which colocalizes with myofilaments at the Z-disc of sarcomeres, is a mechanical stretch sensor. Acetylation of MLP by PCAF enhances the calcium sensitivity of myofilaments. HDAC4 seems to be the deacetylase responsible for MLP deacetylation [103].
3.1.2. Microtubules
As already mentioned, α-tubulin, which together with β-tubulin forms the heterodimeric building block of microtubules, was the first cytoplasmic protein described to be acetylated [2, 3]. α-tubulin is acetylated on lysine 40 which, until recently, was considered to be the only acetylated lysine residue in tubulin isoforms. Interestingly, in a recent systematic proteomic identification approach for acetylated proteins, lysine 40 of α-tubulin was not identified. This is most probably due to the fact that an immunoprecipitation step using a pan acetylated lysine antibody has been used in this approach, which most probably has a low affinity for this particular residue. However, several other acetylated lysine residues in different tubulin isoforms (α as well as β subunits) were detected, but their role and significance have still to be elucidated [5]. Even our understanding of the functional role of the acetylation at lysine 40 of α-tubulin is not clear cut. The reason for this may come from the fact that the expression of nonacetylatable α-tubulin does not result in any obvious phenotype [104–106], nor does hyperacetylation following KO of HDAC6 [107]. In general, acetylated microtubules are considered to represent the subpopulation of more stable microtubules in the cell. There has been however a long debate in the literature as to whether acetylation is the cause or a consequence of microtubule stabilisation. While it has been suggested that acetylation just accumulates with time in microtubules with a long half-life [108], others have proposed that acetylation could directly stabilise microtubules [50, 51]. Yet, hyperacetylation in neuronal cells does not lead to microtubule stabilisation [109]. Also, in cells KO for the tubulin deacetylase HDAC6, hyperacetylation of microtubules is not accompanied by detyrosination, which is the other hallmark of stable microtubules [62]. Thus we are proposing an alternative hypothesis for the role of tubulin acetylation. In fact, acetylated microtubules are not very abundant in most of the cells cultured in vitro. There are, however, a few exceptions. At least three cell types have heavily acetylated microtubules in their cytoplasm; these are neurons, platelets, and megacaryocytes, the platelet progenitors. In addition there are cellular substructures formed by microtubules, which are also heavily acetylated like primary cilia, flagella, mitotic spindles and midbodies. What all these structures and cell types have in common is the need of microtubule bundles. Thus acetylation may allow for more efficient bundling of microtubules, which in turn may lead to enhanced stability of these microtubule bundles. In support of this hypothesis are several observations described in the literature. Naranatt et al. have shown that, 30 min after infection of cells by HHV-8, a transient increase of microtubule acetylation is seen which is accompanied by a thickening of microtubule bundles. Both, acetylation as well as the thickening of microtubules, come back to basal levels about 2-3 hours after infection [110]. Similarly, pneumolysin, a virulence factor of streptococcus pneumonia, can induce microtubule acetylation and concomitant microtubule bundling [111]. Also, overexpression of calpain 6, a catalytically inactive calpain isoform, induces hyperacetylation and bundling of microtubules [112]. Another example is the tubulin polymerisation promoting protein TPPP/p25, which induces not only tubulin polymerisation but also acetylation and subsequent bundling and stabilisation of microtubules [68]. Microtubule bundles are also promoted by p180, a rough ER associated protein, after induction of tubulin acetylation [113]. In addition, early during neuronal commitment an acetylated array of microtubules is formed which is arranged in a bundle of parallel microtubules [114].
Besides its contribution in stabilisation and/or bundling of microtubules, tubulin acetylation plays an important role in cellular transport events as discussed below. It should be mentioned here that there is a coordinated interplay between the actin cytoskeleton and the microtubular network of which one coordinator is the RhoA effector Dia [115, 116], which can bind to microtubules and, as mentioned above, also binds to actin filaments. In fact Dia has been shown to influence microtubule orientation, stability and acetylation states. Some of the studies conducted so far describe enhanced microtubule acetylation following RhoA and Dia activation [110, 117–119]. It is not known, however, whether in these cases enhanced acetylation is due to direct Dia mediated HDAC6 inhibition or to activation of a tubulin acetyltransferase. In another study using osteoclasts, Dia activation leads to the opposite effect namely deacetylation of microtubules due to the interaction of Dia with, and concomitant activation of, HDAC6 [66]. The outcome of Dia activation on microtubule acetylation/stabilisation may thus be cell type dependent. Further complexity might be added to the finetuning of these regulatory events by the fact that a Dia-related isoform seems to be acetylated itself [5].
3.1.3. Intermediate Filaments
At least some of the constituents of the third class of cytoskeletal elements, the intermediate filaments, have also been found acetylated. One example is vimentin, which is acetylated on several lysine residues [5], another is cytokeratin 8, which is acetylated on three lysine residues. In contrast to tubulin and actin however, in this case, acetylation seems to destabilize the polymer [120, 121].
3.2. Transport
Cellular transport along microtubule tracks is particularly important in cells, which have long extensions like axons and dendrites of neuronal cells. This may be the reason why the important role of microtubule acetylation for cargo transport has first been observed in neurons. The microtubule motor kinesin is implicated in anterograde transport, whereas dynein motors are involved in retrograde transfer of cellular materials (Figure 1(b)). It has been shown that binding of kinesin-1 to and mobility on microtubules is enhanced by tubulin acetylation and thus delivery of Jip-1 (a cargo protein of kinesin-1) to neurite tips is accelerated [122]. Another example of more efficient kinesin-1-mediated transport along acetylated microtubules is the vesicular transport of the neurotrophic factor BDNF. In neurons of Huntington disease patients, BDNF is not transported and secreted efficiently due to a polyglutamine expansion in the htt protein, which is unable to stimulate microtubule-based transport. Hyperacetylation of microtubules can rescue this deficiency. In addition the authors show that both kinesin as well as dynein-dependent microtubular transport is enhanced by tubulin acetylation [109]. Regulation of receptor trafficking by enhancing the speed of endosome transport is also augmented by microtubule acetylation [72, 123]. EGFR vesicle recycling however is not influenced under these conditions [123]. Collectively, these observations indicate that the transport of individual cargo proteins, as well as some but not all vesicular transport processes, are regulated by microtubule acetylation (Figure 1(b)).
Viruses have developed strategies to profit from the host cellular transport machinery for efficient infection. The herpes virus HHV-8 for instance is able to enhance microtubule acetylation by activation of RhoA and its effector Dia2, which speeds the dynein dependent delivery of viral DNA to the nucleus [110]. The adenovirus acts in a similar way [119]. The vaccinia virus instead inhibits RhoA and, in turn, Dia2 leading to diminished tubulin acetylation but increased tubulin dynamics in the cell periphery, which in this case could help viral release [117]. Efficient invasion of bladder cells by uropathogenic E. coli bacteria also requires HDAC6-mediated deacetylation of microtubules [124].
In the context of cellular transport mechanisms it is worth mentioning that several motor proteins have themselves been identified as being acetylated and future studies will have to tell whether transport regulation attributed to microtubule acetylation could have been influenced also by motor protein acetylation [5].
3.3. Translation, Quality Control, and Cytoplasmic Cleanup
Acetylation has been widely studied for its role in transcriptional regulation in the nucleus and it is almost astonishing that, to our knowledge, no observation on translational regulation by acetylation has been described so far. It is worth mentioning, however, that in global proteomic studies a huge number of translation initiation factors or ribosomal proteins have been found acetylated [4, 5]. Thus a possible role of acetylation in translational control mechanisms remains to be discovered. The amount of expressed proteins is not only controlled at the transcriptional and translational levels but also by regulation of their half-life. Acetylation is widely used to influence protein stability in both directions but, since this topic has been reviewed recently [125], it will not be discussed here.
Besides quantity control, the cell is also equipped with quality control and cleanup systems for cellular proteins to avoid the accumulation of misfolded, aggregated proteins in the cytoplasm, which would have a deleterious effect on coordinated normal cellular functions (Figure 1(c)). Following translation many newly synthesised proteins have to be assisted for correct folding or assembly into multisubunit complexes. This is ensured by chaperones. Chaperones are also implicated in the management of misfolded proteins which arise following different environmental stress conditions like heat shock, oxidative stress and aging, or pathological expression of proteins due to mutations or overexpression. In case of an unsuccessful management by chaperones, misfolded proteins are ubiquitinated and degraded by the proteasome. After a prolonged or extensive stress period or under experimental overexpression of proteins, the chaperone/proteasome pathway cannot cope anymore with the massive conformational defects of proteins, which accumulate and aggregate in the cytoplasm. This gives rise to a more global stress response, leading to stress granules and aggresome formation and induction of QC-autophagy. The important role of HDAC6 in all of these different cellular defence mechanisms has been well described (for review see [76, 77]) and will not be discussed in detail in this paper. We will rather emphasize the critical importance of some acetylated lysine residues within other key actors involved in these processes.
3.3.1. Chaperones/Proteasome
Chaperones are well known as proteins assisting nucleosome formation. There are however also several chaperones important for the folding or assembly of cytoplasmic protein complexes and many chaperones have been discovered following heat shock treatment, which leads not only to their transcriptional induction but also to their immediate activation. Hsp90 can be acetylated on several lysine residues and its activation state depends on its acetylation status. Its immediate activation is mediated by HDAC6-dependent deacetylation, which is essential for ATP binding of Hsp90 [54] and for its binding to its cochaperone p23 as well as to various client proteins like AhR (aryl hydrocarbon receptor) [126], Bcr-Abl, AKT, c-Raf [54], or the glucocorticoid receptor [57, 127]. Besides other acetylated lysine residues deacetylation of lysine 294 in Hsp90 is particularly important for cochaperone and client binding [56] but, as mentioned above, HDAC6 mediated deacetylation of this site is also responsible for Hsp90 mediated Rac1 activation resulting in actin reorganisation and cell migration [62]. The different functional roles of Hsp90 could be dictated by its cellular localisation since Hsp90 is translocated to membrane ruffles to mediate its role in actin remodelling [62]. By deacetylation of Hsp90, HDAC6 may regulate its own survival since HDAC6 has been shown to be itself an Hsp90 client protein [128].
Alpha A-crystallin, a chaperone in the soluble fraction of eye lenses, becomes acetylated on lysine 70, a region supposed to be important for its activity, and alphaB-crystallin appears to be acetylated on lysine 92 [129, 130]. Crystallins may thus be another example of chaperones whose activity is regulated by acetylation. The chaperone DNAJB8 can be acetylated on two conserved lysine residues situated in its carboxyterminal part. Although these lysine residues are not implicated in substrate binding, deacetylation mediated by HDAC4 activates the chaperone and suppresses cytotoxic protein aggregation [131].
As mentioned above, in case chaperones are unable to assist correct folding of their client proteins, these misfolded proteins are eliminated by proteasomal degradation. One example is again Hsp90. In its inactive ADP-bound state it associates with the cochaperone Hsp70, which prepares the misfolded client protein for degradation by its association with a ubiquitin ligase. Hsp70 is also hyperacetylated following either class I HDAC or HDAC6 inhibition. In contrast to Hsp90, hyperacetylation of Hsp70 promotes its interaction with client proteins leading to their ubiquitination and degradation by the proteasome [132].
3.3.2. Stress Granules/Aggresome/Autophagy
When the load of misfolded proteins is too high the cells suspend their normal function and respond with an immediate block of mRNA translation (except for stress response factors like heat shock proteins/chaperones). Accumulation of the halted transcripts gives rise to the appearance of stress granules. In parallel, the accumulating misfolded proteins become ubiquitinated and transported along microtubules to the centrosome where they form the so-called aggresome. HDAC6 is essential for both stress granule and aggresome formation [133, 134]. It has been suggested that HDAC6 could act in the transport process as an adaptor by binding to the dynein motor complex on the one hand and binding via its ubiquitin binding domain to ubiquitinated proteins on the other [133]. Parkin, a ubiquitin E3 ligase (often mutated in Parkinson disease), tightly binds to HDAC6 and is also transported by HDAC6 in a microtubule motor dependent manner to the forming aggresome. There may be ongoing parkin-mediated ubiquitination of misfolded proteins during transport along the microtubules [135, 136]. Given that microtubule motor-dependent transport is enhanced when microtubules are acetylated one would expect that the deacetylase activity of HDAC6 is inhibited during these transport processes and that deacetylase deficient forms of HDAC6 should be able to rescue stress granule and aggresome formation in HDAC6 KO cells. Since this is not the case and in addition, HDAC6 and parkin are both active during the transport to the aggresome, it seems that a deacetylation event is essential for aggresome and stress granule formation. Whether tubuline is the relevant substrate or not remains to be determined.
Aggresomes or smaller ubiquitinated misfolded protein aggregates are cleared by basal or QC-autophagy. Double membranous vesicular compartments called autophagosomes are formed in order to isolate the cytoplasm containing the material to be eliminated. These vesicles then fuse with lysosomes, and the lysosomal enzymes ensure the proteolytic digestion of their content. HDAC6 is essential for the delivery of autophagosomal constituents and lysosomes to the pericentriolar localised aggresome by transport along microtubule tracts, as observed for the clearing of aggregated huntington [137]. HDAC6 then plays a second role in the fusion between autophagosomes and lysosomes by recruiting and deacetylating cortactin which in turn recruits actin filaments to tether the two vesicle populations [138, 139]. QC-autophagy can be distinguished from starvation-induced autophagy since the latter does not depend on HDAC6. Both, however, depend on the same autophagy-related genes (Atg). Recent studies have shown that starvation-induced autophagy is regulated by the acetylation status of Atg5, Atg7, and Atg8, which are proteins required for the formation of the autophagosome. They are acetylated by p300, which prevents autophagosome formation [140]. Deacetylation is accomplished by Sirt1 and is essential for the induction of autophagy following nutrient deprivation [90]. This observation correlates with the fact that Sirt1 is implicated in metabolic processes and upregulated during fasting conditions [141]. Stimulation of starvation-induced autophagy requires also hyperacetylation of tubulin, which could explain why the absence of HDAC6 does not prevent this type of autophagy [142].
3.4. Plasma Membrane and Organelles
Several ion pumps of the plasma membrane, like the Na+/K+-ATPase and the Ca2± ATPase can associate with acetylated microtubules or acetylated tubulin dimers but not with unacetylated tubulin. Association with acetylated tubulin inhibits the pumps and may serve at the same time as an anchorage site for the microtubular network to the plasma membrane [143]. Acetylated tubulin at the plasma membrane is also found in another context. The viral protein gp120 of HIV binds to its receptor CD4 in the plasma membrane of T cells and induces tubulin acetylation and accumulation at contact sites. These plasma membrane domains rich in acetylated tubulin could be entry sites for HIV, since active HDAC6 diminishes HIV infection [144].
Mitochondria harbour an enormous amount of acetylated proteins, and there are several publications on the important role of acetylation for the regulation of metabolic and age-related processes, which could not be included in this paper.
Interestingly, recent studies describe acetylation and deacetylation events occurring in the lumen of the ER and Golgi apparatus, respectively. Bace1, the protease responsible for amyloid precursor protein cleavage, and the LDL receptor become acetylated in the ERGIC [145, 146]. Acetylation is essential for their stabilisation and progression along the secretory pathway. Both proteins are deacetylated in the Golgi apparatus. This implies not only the presence of Hats and HDACs in these cellular compartments but also an ER import system for acetyl-CoA. While the responsible Hat enzymes have been identified and appear to be type II membrane proteins, the HDACs still remain to be discovered and the acetyl-CoA transporter is currently under investigation [147].
3.5. Nuclear-Cytoplasmic Shuttle
Besides the regulation of protein stability, activity and interactions, acetylation is widely used for regulating the cellular localization of proteins, especially for nuclear import and export (Figure 1(b)). Most proteins, which shuttle between the nucleus and the cytoplasm, are acetylated by p300/CBP, including HNF-4, CIITA, PCNA, SRY, cAbl, CtBP2, p53, PAP, and β-catenin, RECQL4 [148–157]. Two out of those can also be acetylated by PCAF [154, 155], and there is only one report published so far where Gcn5-mediated acetylation plays a role, concerning CDC6 [158]. Until now no general rule for the localisation of the acetylated subpopulation of proteins has been deciphered. For some proteins, acetylation enhances localisation in the cytoplasm [149, 151, 153, 154, 158, 159], whereas for others acetylation will preferentially favour a nuclear localization [148, 150, 152, 155, 156, 160, 161]. The mechanism by which acetylation influences cellular localisation can be either the modification of an interaction with a binding partner leading to retention in a particular compartment, or an altered interaction with nuclear import/export factors. For poly(A) polymerase (PAP) and the DNA helicase RECQL4 it was shown that acetylation disrupts their interaction with the nuclear import factors leading to their accumulation in the cytoplasm [149, 153]. In contrast, SRY acetylation provokes its nuclear import, which also depends on its interaction with a nuclear import factor, but in this case acetylation induces an increased interaction of the acetylated form with importin beta [148]. Interestingly, in order to stay in the nucleus HNF-4 has to be kept in an acetylated state at its NLS. Deacetylation leads to CRM1-dependent export most probably after a conformational change liberating its nuclear export signal [156]. Similarly, cytoplasmic localisation of acetylated p53 depends on CRM1 binding to its nuclear export signal which is accessible only when p53 oligomerization is prevented by lysine acetylation [151]. Another example of CRM1-dependent export regulated by acetylation is NF-kappaB. When present in the nucleus the RelA subunit of NF-kappaB can be acetylated on several lysine residues, which differentially influence its nuclear export mediated by the NF-kappaB inhibitor IkappaBalpha [161, 162].
Interestingly, the nuclear import factor importin alpha is itself targeted by reversible acetylation by p300/CBP, which stimulates heterodimer formation with importin beta [163, 164]. In addition, several other transport factors are among the acetylated proteins identified by Choudhary et al. These include importin beta as well as CAS, the exportin responsible for recycling importin alpha to the cytoplasm, and the more general export receptor CRM1 [5].
4. Concluding Remarks
The overwhelming amount of published work on important aspects of cytoplasmic acetylation precludes an exhaustive overview covering the current knowledge (for additional aspects see [165]). We have therefore focused on selected examples illustrating some important points and wish to apologize for other interesting observations, which have not been included.
Having discussed the importance of acetylation in the cytoplasm, it may be worth insisting on the fact that cells should be considered as entities which cannot be strictly divided into two different compartments. The interpretation of some of the results might have been biased by indirect effects due to transcriptional regulation or leakage of nuclear proteins during biochemical purification steps. Also proteins generally considered as typical cytoplasmic residents do play important roles in the nucleus. Actin, for instance, known as a major cytoskeletal and thus cytoplasmic constituent, also plays important roles in the nucleus as a part of transcription or DNA remodelling complexes. Even tubulin was recently found translocating between the nucleus and the cytoplasm [166]. To circumvent problems related to the presence of the nucleus for the characterisation of cytoplasmic acetylation events, we propose platelets as a good model system. Platelets are devoid of a nucleus but are equipped with all the important players to execute known cytoplasmic processes like exocytosis, adhesion, aggregation, shape change, remodelling of the cytoskeleton, and even apoptosis-related events. Furthermore all these processes have to be tightly regulated and induction has to be extremely rapid to ensure hemostasis and prevent thrombosis.
We would like to conclude with the prediction that, although numerous acetylated proteins have been already identified in the cytoplasm, there are likely more to be discovered. This idea is supported by the fact that acetylation of lysine 40 of alpha tubulin has not been detected in the proteomic survey of acetylated proteins by Choudhary et al. [5] and other acetylation sites may have been missed for the same technical reasons (see above). New strategies including bioinformatic prediction approaches [167, 168] may lead us to new candidates for reversible acetylation. We therefore believe that the story will not end here and future will tell us more about acetylated proteins and exciting new regulatory mechanisms. Finally this paper highlights the importance of reversible cytoplasmic protein acetylation in a wide range of cellular functions and regulatory mechanisms. The functional significance of this modification has been largely recognized in the past for transcriptional control mechanisms in the nucleus and at present, as summarized here, also for cytoplasmic events. Thus the functional role of reversible protein lysine acetylation now definitely approaches the regulatory power of protein phosphorylation as rightly predicted by Kouzarides [169].
Acknowledgments
The authors gratefully acknowledge Dr. Sophie Rousseaux and Dr. Olivier Destaing for critical reading of the paper. Saadi Khochbin's laboratory was supported by INCa-DHOS, ANR blanche “EpiSperm” and “Empreinte”, and ARC research programs.
Abbreviations
- ADP:
Adenosine diphosphate
- AhR:
Aryl hydrocarbon Receptor
- Arp:
Actin-related protein
- ATAC:
Ada Two-A Containing
- ATF2:
Activating Transcription Factor 2
- ATP:
Adenosine triphosphate
- BBIP10:
BBSome Interacting Protein 10
- BDNF:
Brain Derived Neurotrophic Factor
- Cbl:
Casitas B-lineage lymphoma
- CBP:
CREB Binding Protein
- CD:
Cluster of Differentiation
- CDC6:
Cell Division Cycle 6
- CRM1:
Chromosome Region Maintenance 1
- EGFR:
Epidermal Growth Factor Receptor
- Elp3:
Elongator complex protein 3
- ER:
Endoplasmic Reticulum
- ERGIC:
Endoplasmic Reticulum—Golgi Intermediate Compartment
- Gcn5:
General control of amino acid synthesis 5
- Hat:
Histone acetyltransferase
- HDAC:
Histone deacetylase
- HHV-8:
Human Herpes Virus 8
- HIV:
Human Immunodeficiency Virus
- HNF-4:
Hepatocyte Nuclear Factor 4
- HSP90:
Heat Shock Protein 90
- KAT:
lysine (K) acetyltransferase
- LDL:
Low-density Lipoprotein
- MYST:
MOZ, Ybf2/Sas3, Sas2, Tip60
- NAD:
Nicotinamide Adenine Dinucleotide
- NF-kappaB:
Nuclear Factor-kappaB
- NGF:
Nerve Growth Factor
- NLS:
Nuclear Localization Signal
- PAP:
Poly A Polymerase
- PCAF:
p300/CBP-associated Factor
- PCNA:
Proliferating Cell Nuclear Antigen
- PDGF:
Platelet-derived Growth Factor
- QC-autophagy:
Quality Control-autophagy
- Rac1:
Ras-related C3 botulinum toxin substrate 1
- RECQL4:
RecQ protein-like 4
- Rho:
Ras homolog gene
- RNAi:
RNA interference
- SAGA:
Spt-Ada-Gcn5 Acetyltransferase
- Sirt:
Sirtuin
- SRY:
Sex-determining Region Y
- STAGA:
SPT3-TAF9-GCN5/PCAF acetylase
- TFTC:
TBP-free TAF-containing Complex
- Tip60:
Tat interacting protein 60
- TNF:
Tumor Necrosis Factor
- TPPP:
Tubulin Polymerization-Promoting Protein
- tRNA:
transfer RNA.
References
- 1.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.L’Hernault SW, Rosenbaum JL. Chlamydomonas α-tubulin is posttranslationally modified by acetylation on the ε-amino group of a lysine. Biochemistry. 1985;24(2):473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 3.Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. Journal of Cell Biology. 1985;101(6):2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SC, Sprung R, Chen Y, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular Cell. 2006;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 6.Allis CD, Berger SL, Cote J, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 7.Parthun MR. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26(37):5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- 8.Mersfelder EL, Parthun MR. Involvement of Hat1p (Kat1p) catalytic activity and subcellular localization in telomeric silencing. The Journal of Biological Chemistry. 2008;283(43):29060–29068. doi: 10.1074/jbc.M802564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz-García AB, Sendra R, Galiana M, Pamblanco M, Pérez-Ortín JE, Tordera V. Hat1 and Hat2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. The Journal of Biological Chemistry. 1998;273(20):12599–12605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- 10.Imhof A, Wolffe AP. Purification and properties of the Xenopus Hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry. 1999;38(40):13085–13093. doi: 10.1021/bi9912490. [DOI] [PubMed] [Google Scholar]
- 11.Seiden-Long IM, Brown KR, Shih W, et al. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene. 2006;25(1):91–102. doi: 10.1038/sj.onc.1209005. [DOI] [PubMed] [Google Scholar]
- 12.Lebel EA, Boukamp P, Tafrov ST. Irradiation with heavy-ion particles changes the cellular distribution of human histone acetyltransferase HAT1. Molecular and Cellular Biochemistry. 2010;339(1-2):271–284. doi: 10.1007/s11010-010-0390-0. [DOI] [PubMed] [Google Scholar]
- 13.Wong K, Zhang J, Awasthi S, et al. Nerve growth factor receptor signaling induces histone acetyltransferase domain-dependent nuclear translocation of p300/CREB-binding protein-associated factor and hGCN5 acetyltransferases. The Journal of Biological Chemistry. 2004;279(53):55667–55674. doi: 10.1074/jbc.M408174200. [DOI] [PubMed] [Google Scholar]
- 14.Blanco-García N, Asensio-Juan E, De La Cruz X, Martínez-Balbás MA. Autoacetylation regulates P/CAF nuclear localization. The Journal of Biological Chemistry. 2009;284(3):1343–1352. doi: 10.1074/jbc.M806075200. [DOI] [PubMed] [Google Scholar]
- 15.Kwok RPS, Liu X-T, Smith GD. Distribution of co-activators CBP and p300 during mouse oocyte and embryo development. Molecular Reproduction and Development. 2006;73(7):885–894. doi: 10.1002/mrd.20440. [DOI] [PubMed] [Google Scholar]
- 16.Fermento ME, Gandini NA, Lang CA, et al. Intracellular distribution of p300 and its differential recruitment to aggresomes in breast cancer. Experimental and Molecular Pathology. 2010;88(2):256–264. doi: 10.1016/j.yexmp.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Hass MR, Yankner BA. A γ-secretase-independent mechanism of signal transduction by the amyloid precursor protein. The Journal of Biological Chemistry. 2005;280(44):36895–36904. doi: 10.1074/jbc.M502861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ran Q, Pereira-Smith OM. Identification of an alternatively spliced form of the Tat Interactive Protein (Tip60), Tip60(β) Gene. 2000;258(1-2):141–146. doi: 10.1016/s0378-1119(00)00410-8. [DOI] [PubMed] [Google Scholar]
- 19.Sheridan AM, Force T, Yoon H-J, et al. PLIP, a novel splice variant of Tip60, interacts with group iv cytosolic phospholipase A2, induces apoptosis, and potentiates prostaglandin production. Molecular and Cellular Biology. 2001;21(14):4470–4481. doi: 10.1128/MCB.21.14.4470-4481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Deng X, Shyu YJ, Jian JL, Taparowsky EJ, Hu C-D. Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. The EMBO Journal. 2006;25(5):1058–1069. doi: 10.1038/sj.emboj.7601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maruta H, Greer K, Rosenbaum JL. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. Journal of Cell Biology. 1986;103(2):571–579. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creppe C, Malinouskaya L, Volvert M-L, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of α-tubulin. Cell. 2009;136(3):551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 23.Solinger JA, Paolinelli R, Klöß H, et al. The Caenorhabditis elegans elongator complex regulates neuronal α-tubulin acetylation. PLoS Genetics. 2010;6(1) doi: 10.1371/journal.pgen.1000820. Article ID e1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen LD, Naumanen T, Knudsen A, et al. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. Journal of Cell Science. 2008;121(6):854–864. doi: 10.1242/jcs.013722. [DOI] [PubMed] [Google Scholar]
- 25.Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Molecular Cell. 2005;17(6):841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Svejstrup JQ. Elongator complex: how many roles does it play? Current Opinion in Cell Biology. 2007;19(3):331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Holmberg C, Katz S, Lerdrup M, et al. A novel specific role for IκB kinase complex-associated protein in cytosolic stress signaling. The Journal of Biological Chemistry. 2002;277(35):31918–31928. doi: 10.1074/jbc.M200719200. [DOI] [PubMed] [Google Scholar]
- 28.Huang B, Johansson MJO, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11(4):424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esberg A, Huang B, Johansson MJO, Byström AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Molecular Cell. 2006;24(1):139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Tuck S, Byström AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genetics. 2009;5(7) doi: 10.1371/journal.pgen.1000561. Article ID e1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conacci-Sorrell M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes α-tubulin acetylation and cell differentiation. Cell. 2010;142(3):480–493. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akella JS, Wloga D, Kim J, et al. MEC-17 is an α-tubulin acetyltransferase. Nature. 2010;467(7312):218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohkawa N, Sugisaki S, Tokunaga E, et al. N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes to Cells. 2008;13(11):1171–1183. doi: 10.1111/j.1365-2443.2008.01235.x. [DOI] [PubMed] [Google Scholar]
- 34.Shen Q, Zheng X, McNutt MA, et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Experimental Cell Research. 2009;315(10):1653–1667. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Kim JY, Shen S, Dietz K, et al. HDAC1 nuclear export induced by pathological conditions is essential for the onset of axonal damage. Nature Neuroscience. 2010;13(2):180–189. doi: 10.1038/nn.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W-M, Tsai S-C, Wen Y-D, Fejé G, Seto E. Functional domains of histone deacetylase-3. The Journal of Biological Chemistry. 2002;277(11):9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- 37.Viatour P, Legrand-Poels S, Van Lint C, et al. Cytoplasmic IκBα increases NF-κB-independent transcription through binding to histone deacetylase (HDAC) 1 and HDAC3. The Journal of Biological Chemistry. 2003;278(47):46541–46548. doi: 10.1074/jbc.M306381200. [DOI] [PubMed] [Google Scholar]
- 38.Gao Z, He Q, Peng B, Chiao PJ, Ye J. Regulation of nuclear translocation of HDAC3 by IκBα is required for tumor necrosis factor inhibition of peroxisome proliferator- activated receptor γ function. The Journal of Biological Chemistry. 2006;281(7):4540–4547. doi: 10.1074/jbc.M507784200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longworth MS, Laimins LA. Histone deacetylase 3 localizes to the plasma membrane and is a substrate of Src. Oncogene. 2006;25(32):4495–4500. doi: 10.1038/sj.onc.1209473. [DOI] [PubMed] [Google Scholar]
- 40.Waltregny D, Glénisson W, Tran SL, et al. Histone deacetylase HDAC8 associates with smooth muscle α-actin and is essential for smooth muscle cell contractility. The FASEB Journal. 2005;19(8):966–968. doi: 10.1096/fj.04-2303fje. [DOI] [PubMed] [Google Scholar]
- 41.Waltregny D, De Leval L, Glénisson W, et al. Expression of histone deacetylase 8, a class I histone deacetylase, is restricted to cells showing smooth muscle differentiation in normal human tissues. American Journal of Pathology. 2004;165(2):553–564. doi: 10.1016/S0002-9440(10)63320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: conducting development and differentiation. The International Journal of Developmental Biology. 2009;53(2-3):291–301. doi: 10.1387/ijdb.082698mm. [DOI] [PubMed] [Google Scholar]
- 43.Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. The EMBO Journal. 1999;18(18):5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kao H-Y, Verdel A, Tsai C-C, Simon C, Juguilon H, Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. The Journal of Biological Chemistry. 2001;276(50):47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 45.Bakin RE, Jung MO. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. The Journal of Biological Chemistry. 2004;279(49):51218–51225. doi: 10.1074/jbc.M409271200. [DOI] [PubMed] [Google Scholar]
- 46.Nishino TG, Miyazaki M, Hoshino H, Miwa Y, Horinouchi S, Yoshida M. 14-3-3 regulates the nuclear import of class IIa histone deacetylases. Biochemical and Biophysical Research Communications. 2008;377(3):852–856. doi: 10.1016/j.bbrc.2008.10.079. [DOI] [PubMed] [Google Scholar]
- 47.Harrison BC, Huynh K, Lundgaard GL, Helmke SM, Perryman MB, McKinsey TA. Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. FEBS Letters. 2010;584(6):1103–1110. doi: 10.1016/j.febslet.2010.02.057. [DOI] [PubMed] [Google Scholar]
- 48.Ago T, Liu T, Zhai P, et al. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133(6):978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 49.Gao C, Li X, Lam M, Liu Y, Chakraborty S, Kao H-Y. CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14-3-3s. FEBS Letters. 2006;580(21):5096–5104. doi: 10.1016/j.febslet.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 50.Hubbert C, Guardiola A, Shao R, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 51.Matsuyama A, Shimazu T, Sumida Y, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO Journal. 2002;21(24):6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Li N, Caron C, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO Journal. 2003;22(5):1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Yuan Z, Zhang Y, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Molecular Cell. 2007;27(2):197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. The Journal of Biological Chemistry. 2005;280(29):26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 55.Aoyagi S, Archer TK. Modulating molecular chaperone Hsp90 functions through reversible acetylation. Trends in Cell Biology. 2005;15(11):565–567. doi: 10.1016/j.tcb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Scroggins BT, Robzyk K, Wang D, et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Molecular Cell. 2007;25(1):151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kovacs JJ, Murphy PJM, Gaillard S, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Molecular Cell. 2005;18(5):601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Zhang X, Polakiewicz RD, Yao T-P, Comb MJ. HDAC6 is required for epidermal growth factor-induced β-catenin nuclear localization. The Journal of Biological Chemistry. 2008;283(19):12686–12690. doi: 10.1074/jbc.C700185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parmigiani RB, Xu WS, Venta-Perez G, et al. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(28):9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verdel A, Curtet S, Brocard M-P, et al. Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Current Biology. 2000;10(12):747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 61.Bertos NR, Gilquin B, Chan GKT, Yen TJ, Khochbin S, Yang X-J. Role of the tetradecapeptide repeat domain of human histone deacetylase 6 in cytoplasmic retention. The Journal of Biological Chemistry. 2004;279(46):48246–48254. doi: 10.1074/jbc.M408583200. [DOI] [PubMed] [Google Scholar]
- 62.Gao Y-S, Hubbert CC, Lu J, Lee Y-S, Lee J-Y, Yao T-P. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Molecular and Cellular Biology. 2007;27(24):8637–8647. doi: 10.1128/MCB.00393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purev E, Neff L, Horne WC, Baron R. c-Cbl and Cbl-b act redundantly to protect osteoclasts from apoptosis and to displace HDAC6 from β-tubulin, stabilizing microtubules and podosomes. Molecular Biology of the Cell. 2009;20(18):4021–4030. doi: 10.1091/mbc.E09-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou J, Vos CC, Gjyrezi A, et al. The protein farnesyltransferase regulates HDAC6 activity in a microtubule-dependent manner. The Journal of Biological Chemistry. 2009;284(15):9648–9655. doi: 10.1074/jbc.M808708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loktev AV, Zhang Q, Beck JS, et al. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Developmental Cell. 2008;15(6):854–865. doi: 10.1016/j.devcel.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Destaing O, Saltel F, Gilquin B, et al. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. Journal of Cell Science. 2005;118(13):2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- 67.Perez M, Santa-Maria I, De Barreda EG, et al. Tau—an inhibitor of deacetylase HDAC6 function. Journal of Neurochemistry. 2009;109(6):1756–1766. doi: 10.1111/j.1471-4159.2009.06102.x. [DOI] [PubMed] [Google Scholar]
- 68.Tokési N, Lehotzky A, Horváth I, et al. TPPP/p25 promotes tubulin acetylation by inhibiting histone deacetylase 6. The Journal of Biological Chemistry. 2010;285(23):17896–17906. doi: 10.1074/jbc.M109.096578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding H, Dolan PJ, Johnson GVW. Histone deacetylase 6 interacts with the microtubule-associated protein tau. Journal of Neurochemistry. 2008;106(5):2119–2130. doi: 10.1111/j.1471-4159.2008.05564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Y, Song SW, Sun J, Bruner JM, Fuller GN, Zhang W. IIp45 inhibits cell migration through inhibition of HDAC6. The Journal of Biological Chemistry. 2010;285(6):3554–3560. doi: 10.1074/jbc.M109.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wickström SA, Masoumi KC, Khochbin S, Fässler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. The EMBO Journal. 2010;29(1):131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deribe YL, Wild P, Chandrashaker A, et al. Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Science Signaling. 2009;2(102):p. ra84. doi: 10.1126/scisignal.2000576. [DOI] [PubMed] [Google Scholar]
- 73.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent aurora a activation induces disassembly of the primary cilium. Cell. 2007;129(7):1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brush MH, Guardiola A, Connor JH, Yao T-P, Shenolikar S. Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. The Journal of Biological Chemistry. 2004;279(9):7685–7691. doi: 10.1074/jbc.M310997200. [DOI] [PubMed] [Google Scholar]
- 75.Han Y, Jeong HM, Jin Y-H, et al. Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochemical and Biophysical Research Communications. 2009;383(1):88–92. doi: 10.1016/j.bbrc.2009.03.147. [DOI] [PubMed] [Google Scholar]
- 76.Matthias P, Yoshida M, Khochbin S. HDAC6 a new cellular stress surveillance factor. Cell Cycle. 2008;7(1):7–10. doi: 10.4161/cc.7.1.5186. [DOI] [PubMed] [Google Scholar]
- 77.Boyault C, Sadoul K, Pabion M, Khochbin S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene. 2007;26(37):5468–5476. doi: 10.1038/sj.onc.1210614. [DOI] [PubMed] [Google Scholar]
- 78.Tong JJ, Liu J, Bertos NR, Yang X-J. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Research. 2002;30(5):1114–1123. doi: 10.1093/nar/30.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guardiola AR, Yao T-P. Molecular cloning and characterization of a novel histone deacetylase HDAC10. The Journal of Biological Chemistry. 2002;277(5):3350–3356. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- 80.Fischer DD, Cai R, Bhatia U, et al. Isolation and characterization of a novel class II histone deacetylase, HDAC10. The Journal of Biological Chemistry. 2002;277(8):6656–6666. doi: 10.1074/jbc.M108055200. [DOI] [PubMed] [Google Scholar]
- 81.Kao H-Y, Lee C-H, Komarov A, Han CC, Evans RM. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. The Journal of Biological Chemistry. 2002;277(1):187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- 82.Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. Journal of Virology. 2009;83(10):4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. The Journal of Biological Chemistry. 2002;277(28):25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 84.Vaquero A. The conserved role of sirtuins in chromatin regulation. The International Journal of Developmental Biology. 2009;53(2-3):303–322. doi: 10.1387/ijdb.082675av. [DOI] [PubMed] [Google Scholar]
- 85.Kawahara TLA, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009;136(1):62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Molecular Biology of the Cell. 2005;16(10):4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. The Journal of Biological Chemistry. 2007;282(9):6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 88.Sugino T, Maruyama M, Tanno M, Kuno A, Houkin K, Horio Y. Protein deacetylase SIRT1 in the cytoplasm promotes nerve growth factor-induced neurite outgrowth in PC12 cells. FEBS Letters. 2010;584(13):2821–2826. doi: 10.1016/j.febslet.2010.04.063. [DOI] [PubMed] [Google Scholar]
- 89.Hisahara S, Chiba S, Matsumoto H, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(40):15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.In HL, Cao L, Mostoslavsky R, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y, Zhang M, Dong H, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28(3):445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 92.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS ONE. 2007;2(8, article e784) doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Molecular Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 94.Inoue T, Hiratsuka M, Osaki M, et al. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26(7):945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- 95.Tang BL, Chua CEL. SIRT2, tubulin deacetylation, and oligodendroglia differentiation. Cell Motility and the Cytoskeleton. 2008;65(3):179–182. doi: 10.1002/cm.20253. [DOI] [PubMed] [Google Scholar]
- 96.Garske AL, Smith BC, Denu JM. Linking SIRT2 to Parkinson’s disease. ACS Chemical Biology. 2007;2(8):529–532. doi: 10.1021/cb700160d. [DOI] [PubMed] [Google Scholar]
- 97.Outeiro TF, Kontopoulos E, Altmann SM, et al. Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317(5837):516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 98.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Molecular and Cellular Biology. 2008;28(20):6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shahbazian MD, Grunstein M. Functions of Site-Specific histone acetylation and deacetylation. Annual Review of Biochemistry. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 100.Cohen HY, Lavu S, Bitterman KJ, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Molecular Cell. 2004;13(5):627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 101.Juan L-J, Shia W-J, Chen M-H, et al. Histone deacetylases specifically down-regulate p53-dependent gene activation. The Journal of Biological Chemistry. 2000;275(27):20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 102.Vaziri H, Dessain SK, Eaton EN, et al. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 103.Gupta MP, Samant SA, Smith SH, Shroff SG. HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. The Journal of Biological Chemistry. 2008;283(15):10135–10146. doi: 10.1074/jbc.M710277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fukushige T, Siddiqui ZK, Chou M, et al. MEC-12, an α-tubulin required for touch sensitivity in C. elegans. Journal of Cell Science. 1999;112(3):395–403. doi: 10.1242/jcs.112.3.395. [DOI] [PubMed] [Google Scholar]
- 105.Gaertig J, Cruz MA, Bowen J, Gu L, Pennock DG, Gorovsky MA. Acetylation of lysine 40 in α-tubulin is not essential in Tetrahymena thermophila. Journal of Cell Biology. 1995;129(5):1301–1310. doi: 10.1083/jcb.129.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kozminski KG, Diener DR, Rosenbaum JL. High level expression of nonacetylatable α-tubulin in Chlamydomonas reinhardtii. Cell Motility and the Cytoskeleton. 1993;25(2):158–170. doi: 10.1002/cm.970250205. [DOI] [PubMed] [Google Scholar]
- 107.Zhang Y, Kwon S, Yamaguchi T, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Molecular and Cellular Biology. 2008;28(5):1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(8):4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dompierre JP, Godin JD, Charrin BC, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. Journal of Neuroscience. 2007;27(13):3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Naranatt PP, Krishnan HH, Smith MS, Chandran B. Kaposi’s sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-Diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. Journal of Virology. 2005;79(2):1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Iliev AI, Djannatian JR, Opazo F, et al. Rapid microtubule bundling and stabilization by the Streptococcus pneumoniae neurotoxin pneumolysin in a cholesterol-dependent, non-lytic and Src-kinase dependent manner inhibits intracellular trafficking. Molecular Microbiology. 2009;71(2):461–477. doi: 10.1111/j.1365-2958.2008.06538.x. [DOI] [PubMed] [Google Scholar]
- 112.Tonami K, Kurihara Y, Aburatani H, Uchijima Y, Asano T, Kurihara H. Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Molecular and Cellular Biology. 2007;27(7):2548–2561. doi: 10.1128/MCB.00992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ogawa-Goto K, Tanaka K, Ueno T, et al. p180 is involved in the interaction between the endoplasmic reticulum and microtubules through a novel microtubule-binding and bundling domain. Molecular Biology of the Cell. 2007;18(10):3741–3751. doi: 10.1091/mbc.E06-12-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Falconer MM, Vielkind U, Brown DL. Establishment of a stable, acetylated microtubule bundle during neuronal commitment. Cell Motility and the Cytoskeleton. 1989;12(3):169–180. doi: 10.1002/cm.970120306. [DOI] [PubMed] [Google Scholar]
- 115.Ishizaki T, Morishima Y, Okamoto M, Furuyashiki T, Kato T, Narumiya S. Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nature Cell Biology. 2001;3(1):8–14. doi: 10.1038/35050598. [DOI] [PubMed] [Google Scholar]
- 116.Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nature Cell Biology. 2001;3(8):723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- 117.Arakawa Y, Cordeiro JV, Way M. F11L-mediated inhibition of RhoA-mDia signaling stimulates microtubule dynamics during vaccinia virus infection. Cell Host and Microbe. 2007;1(3):213–226. doi: 10.1016/j.chom.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 118.Palazzo AF, Eng CH, Schlaepfer DD, Marcantonio EE, Gundersen GG. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science. 2004;303(5659):836–839. doi: 10.1126/science.1091325. [DOI] [PubMed] [Google Scholar]
- 119.Warren JC, Rutkowski A, Cassimeris L. Infection with replication-deficient adenovirus induces changes in the dynamic instability of host cell microtubules. Molecular Biology of the Cell. 2006;17(8):3557–3568. doi: 10.1091/mbc.E05-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Drake PJM, Griffiths GJ, Shaw L, Benson RP, Corfe BM. Application of high-content analysis to the study of post-translational modifications of the cytoskeleton. Journal of Proteome Research. 2009;8(1):28–34. doi: 10.1021/pr8006396. [DOI] [PubMed] [Google Scholar]
- 121.Leech SH, Evans CA, Shaw L, et al. Proteomic analyses of intermediate filaments reveals cytokeratin8 is highly acetylated—implications for colorectal epithelial homeostasis. Proteomics. 2008;8(2):279–288. doi: 10.1002/pmic.200700404. [DOI] [PubMed] [Google Scholar]
- 122.Reed NA, Cai D, Blasius TL, et al. Microtubule acetylation promotes Kinesin-1 binding and transport. Current Biology. 2006;16(21):2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 123.Gao Y-S, Hubbert CC, Yao T-P. The microtubule-associated histone deacetylase 6 (HDAC6) regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation. The Journal of Biological Chemistry. 2010;285(15):11219–11226. doi: 10.1074/jbc.M109.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dhakal BK, Mulvey MA. Uropathogenic escherichia coli invades host cells via an HDAC6-modulated microtubule-dependent pathway. The Journal of Biological Chemistry. 2009;284(1):446–454. doi: 10.1074/jbc.M805010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 2008;90(2):306–312. doi: 10.1016/j.biochi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 126.Kekatpure VD, Dannenberg AJ, Subbaramaiah K. HDAC6 modulates Hsp90 chaperone activity and regulates activation of aryl hydrocarbon receptor signaling. The Journal of Biological Chemistry. 2009;284(12):7436–7445. doi: 10.1074/jbc.M808999200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 127.Murphy PJM, Morishima Y, Kovacs JJ, Yao T-P, Pratt WB. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. The Journal of Biological Chemistry. 2005;280(40):33792–33799. doi: 10.1074/jbc.M506997200. [DOI] [PubMed] [Google Scholar]
- 128.Rao R, Fiskus W, Yang Y, et al. HDAC6 inhibition enhances 17-AAG mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood. 2008;112(5):1886–1893. doi: 10.1182/blood-2008-03-143644. [DOI] [PubMed] [Google Scholar]
- 129.Lin PP, Barry RC, Smith DL, Smith JB. In vivo acetylation identified at lysine 70 of human lens αA- crystallin. Protein Science. 1998;7(6):1451–1457. doi: 10.1002/pro.5560070622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lapko VN, Smith DL, Smith JB. In vivo carbamylation and acetylation of water-soluble human lens αB-crystallin lysine 92. Protein Science. 2001;10(6):1130–1136. doi: 10.1110/ps.40901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hageman J, Rujano MA, van Waarde MAWH, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Molecular Cell. 2010;37(3):355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 132.Wang Y, Wang S-Y, Zhang X-H, et al. FK228 inhibits Hsp90 chaperone function in K562 cells via hyperacetylation of Hsp70. Biochemical and Biophysical Research Communications. 2007;356(4):998–1003. doi: 10.1016/j.bbrc.2007.03.076. [DOI] [PubMed] [Google Scholar]
- 133.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao T-P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115(6):727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 134.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes and Development. 2007;21(24):3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jiang Q, Ren Y, Feng J. Direct binding with histone deacetylase 6 mediates the reversible recruitment of parkin to the centrosome. Journal of Neuroscience. 2008;28(48):12993–13002. doi: 10.1523/JNEUROSCI.2860-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Olzmann JA, Li A, Chudaev MV, et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. Journal of Cell Biology. 2007;178(6):1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated Huntingtin. The Journal of Biological Chemistry. 2005;280(48):40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 138.Lee J-Y, Yao T-P. Quality control autophagy: a joint effort of ubiquitin, protein deacetylase and actin cytoskeleton. Autophagy. 2010;6(4):555–557. doi: 10.4161/auto.6.4.11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee J-Y, Koga H, Kawaguchi Y, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. The EMBO Journal. 2010;29(5):969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. The Journal of Biological Chemistry. 2009;284(10):6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boily G, Seifert EL, Bevilacqua L, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3(3) doi: 10.1371/journal.pone.0001759. Article ID e1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Geeraert C, Ratier A, Pfisterer SG, et al. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. The Journal of Biological Chemistry. 2010;285(31):24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zampar GG, Chesta ME, Carbajal A, et al. Acetylated tubulin associates with the fifth cytoplasmic domain of Na+/K+-ATPase: possible anchorage site of microtubules to the plasma membrane. Biochemical Journal. 2009;422(1):129–137. doi: 10.1042/BJ20082410. [DOI] [PubMed] [Google Scholar]
- 144.Valenzuela-Fernandez A, Álvarez S, Gordon-Alonso M, et al. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Molecular Biology of the Cell. 2005;16(11):5445–5454. doi: 10.1091/mbc.E05-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Costantini C, Ko MH, Jonas MC, Puglielli L. A reversible form of lysine acetylation in the ER and Golgi lumen controls the molecular stabilization of BACE1. Biochemical Journal. 2007;407(3):383–395. doi: 10.1042/BJ20070040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jonas MC, Costantini C, Puglielli L. PCSK9 is required for the disposal of non-acetylated intermediates of the nascent membrane protein BACE1. EMBO Reports. 2008;9(9):916–922. doi: 10.1038/embor.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mi HK, Puglielli L. Two Endoplasmic Reticulum (ER)/ER golgi intermediate compartment-based lysine acetyltransferases post-translationally regulate BACE1 levels. The Journal of Biological Chemistry. 2009;284(4):2482–2492. doi: 10.1074/jbc.M804901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thevenet L, Méjean C, Moniot B, et al. Regulation of human SRY subcellular distribution by its acetylation/deacetylation. The EMBO Journal. 2004;23(16):3336–3345. doi: 10.1038/sj.emboj.7600352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shimazu T, Horinouchi S, Yoshida M. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. The Journal of Biological Chemistry. 2007;282(7):4470–4478. doi: 10.1074/jbc.M609745200. [DOI] [PubMed] [Google Scholar]
- 150.Naryzhny SN, Lee H. The post-translational modifications of proliferating cell nuclear antigen: acetylation, not phosphorylation, plays an important role in the regulation of its function. The Journal of Biological Chemistry. 2004;279(19):20194–20199. doi: 10.1074/jbc.M312850200. [DOI] [PubMed] [Google Scholar]
- 151.Kawaguchi Y, Ito A, Appella E, Yao T-P. Charge modification at multiple c-terminal lysine residues regulates p53 oligomerization and its nucleus-cytoplasm trafficking. The Journal of Biological Chemistry. 2006;281(3):1394–1400. doi: 10.1074/jbc.M505772200. [DOI] [PubMed] [Google Scholar]
- 152.Zhao L-J, Subramanian T, Zhou Y, Chinnadurai G. Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. The Journal of Biological Chemistry. 2006;281(7):4183–4189. doi: 10.1074/jbc.M509051200. [DOI] [PubMed] [Google Scholar]
- 153.Dietschy T, Shevelev I, Pena-Diaz J, et al. P300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. Journal of Cell Science. 2009;122(8):1258–1267. doi: 10.1242/jcs.037747. [DOI] [PubMed] [Google Scholar]
- 154.di Bari MG, Ciuffini L, Mingardi M, Testi R, Soddu S, Barilà D. c-Abl acetylation by histone acetyltransferases regulates its nuclear-cytoplasmic localization. EMBO Reports. 2006;7(7):727–733. doi: 10.1038/sj.embor.7400700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Spilianakis C, Papamatheakis J, Kretsovali A. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Molecular and Cellular Biology. 2000;20(22):8489–8498. doi: 10.1128/mcb.20.22.8489-8498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Molecular Cell. 2000;5(4):745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 157.Firestein R, Blander G, Michan S, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3(4) doi: 10.1371/journal.pone.0002020. Article ID e2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nature Structural and Molecular Biology. 2009;16(4):412–420. doi: 10.1038/nsmb.1583. [DOI] [PubMed] [Google Scholar]
- 159.Valacco MP, Varone C, Malicet C, Cánepa E, Iovanna JL, Moreno S. Cell growth-dependent subcellular localization of p8. Journal of Cellular Biochemistry. 2006;97(5):1066–1079. doi: 10.1002/jcb.20682. [DOI] [PubMed] [Google Scholar]
- 160.Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1: possible role in AMP-activated protein kinase activation. The Journal of Biological Chemistry. 2008;283(41):27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen L-F, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293(5535):1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 162.Kiernan R, Brès V, Ng RWM, et al. Post-activation turn-off of NF-κB-dependent transcription is regulated by acetylation of p65. The Journal of Biological Chemistry. 2003;278(4):2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 163.Bannister AJ, Miska EA, Görlich D, Kouzarides T. Acetylation of importin-α nuclear import factors by CBP/p300. Current Biology. 2000;10(8):467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- 164.Ryan CM, Harries JC, Kindle KB, Collins HM, Heery DM. Functional interaction of CREB binding protein (CBP) with nuclear transport proteins and modulation by HDAC inhibitors. Cell Cycle. 2006;5(18):2146–2152. doi: 10.4161/cc.5.18.3207. [DOI] [PubMed] [Google Scholar]
- 165.Close P, Creppe C, Gillard M, et al. The emerging role of lysine acetylation of non-nuclear proteins. Cellular and Molecular Life Sciences. 2010;67(8):1255–1264. doi: 10.1007/s00018-009-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Akoumianaki T, Kardassis D, Polioudaki H, Georgattos SD, Theodoropoulos PA. Nucleocytoplasmic shuttling of soluble tubulin in mammalian cells. Journal of Cell Science. 2009;122(8):1111–1118. doi: 10.1242/jcs.043034. [DOI] [PubMed] [Google Scholar]
- 167.Schwartz D, Chou MF, Church GM. Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Molecular and Cellular Proteomics. 2009;8(2):365–379. doi: 10.1074/mcp.M800332-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Basu A, Rose KL, Zhang J, et al. Proteome-wide prediction of acetylation substrates. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation. EMBO Journal. 2000;19(6):1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]