Abstract
RNA polymerase III (Pol III) transcribes small untranslated RNAs, such as tRNAs. To define the Pol III transcriptome in Saccharomyces cerevisiae, we performed genome-wide chromatin immunoprecipitation using subunits of Pol III, TFIIIB and TFIIIC. Virtually all of the predicted targets of Pol III, as well as several novel candidates, were occupied by Pol III machinery. Interestingly, TATA box-binding protein occupancy was greater at Pol III targets than virtually all Pol II targets, and the highly occupied Pol II targets are generally strongly transcribed. The temporal relationships between factor occupancy and gene activity were then investigated at selected targets. Nutrient deprivation rapidly reduced both Pol III transcription and Pol III occupancy of both a tRNA gene and RPR1. In contrast, TFIIIB remained bound, suggesting that TFIIIB release is not a critical aspect of the onset of repression. Remarkably, TFIIIC occupancy increased dramatically during repression. Nutrient addition generally reestablished transcription and initial occupancy levels. Our results are consistent with active Pol III displacing TFIIIC, and with inactivation/release of Pol III enabling TFIIIC to bind, marking targets for later activation. These studies reveal new aspects of the kinetics, dynamics, and targets of the Pol III system.
RNA polymerase III (Pol III) in Saccharomyces cerevisiae transcribes genes for small structural RNAs important for many cellular processes. Among the known Pol III targets are structural RNAs for translation (all tRNAs, 5S ribosomal RNA, 7SL RNA), tRNA processing (RPR1; the RNA of the ribonuclease P complex), and splicing (i.e., U6). The S. cerevisiae genome is predicted to contain 280 targets, including 275 tRNA genes. The Pol III transcription machinery is highly conserved in eukaryotes, and consists of the multisubunit Pol III polymerase, two complexes (TFIIIB and TFIIIC) required for promoter recognition and/or initiation, and an additional factor (TFIIIA) required only for 5S rDNA transcription (1–3).
Several reviews have addressed the function and regulation of the Pol III machinery in detail (2–4), and here we provide a brief summary. Pol III promoters contain two conserved DNA sequence elements, termed A and B boxes, which are located within the transcribed region and direct the Pol III machinery to target genes (5, 6). The A and B boxes are recognized by the TFIIIC complex, which can bind in the absence of TFIIIB and Pol III, establishing TFIIIC as the promoter specificity factor (7–11). Because most Pol III targets are relatively short (<100 bases), TFIIIC can encompass nearly the entire gene (12).
TFIIIC interacts with TFIIIB, enabling recruitment of TFIIIB to Pol III promoters (13, 14). TFIIIB contains the subunits Brf1 (TFIIB-related factor), Bdp1/Tfc5/B″, and the TATA-binding protein (TBP). TBP is the only subunit of the basal factors not dedicated solely to Pol III transcription; TBP is used by all three RNA polymerases and is required for transcription at both TATA-containing and TATA-less promoters (15). Brf1 is similar to the Pol II initiation factor TFIIB, and binds TBP. Thus, the Pol II and Pol III systems share the use of TBP itself and an interacting TFIIB-related factor.
Pol III consists of 17 subunits, and our studies involve Rpc82 (dedicated to Pol III) and Rpc40 (also a member of RNA Pol I). Although TFIIIB and TFIIIC cooperate in the recruitment of Pol III, TFIIIB appears to have more extensive and important interactions with Pol III (2). TFIIIC is required for the recruitment of TFIIIB and Pol III to Pol III targets in vivo. However, TFIIIB is capable of Pol III recruitment and reinitiation in the absence of TFIIIC in vitro at certain loci (16). Taken together, TFIIIC is important for efficient promoter recognition and for the recruitment of TFIIIB and Pol III, whereas TFIIIB and Pol III are essential for transcript initiation and production (16).
Because Pol III gene products are required for rapid cell growth, their transcription rates are tightly coupled to environmental status. Several conditions related to nutrient deprivation and cell stress can lead to a rapid and/or dramatic reduction in Pol III transcription (17–20). Stress conditions are relayed to the Pol III machinery by various signaling pathways, including those mediated by CK2 and Pkc1 (19–21). Recently, Maf1 has also emerged as an important negative regulator of Pol III, likely interfering with TFIIIB function (22, 23).
Several important questions regarding Pol III regulation remain. For example, the true scope of the Pol III transcriptome is based largely on sequence prediction, and has never been tested by occupancy studies. Also, the extent to which factor recruitment/occupancy correlates with activity in vivo has not been addressed in detail. For example, it is not clear whether TFIIIC is removed from the transcribed region (which contains its binding site) during active transcription in vivo, or whether the initiation factor TFIIIB or Pol III itself must release from targets during the onset of repression in vivo. Here, we use occupancy studies to identify the S. cerevisiae Pol III transcriptome. We also explore the temporal relationships between gene activity and factor occupancy, and reveal both expected and unexpected features of Pol III system regulation.
Methods
Strains and Nutrient Deprivation. For a list of strain genotypes, see Table 1, which is published as supporting information on the PNAS web site. Nutrient deprivation and reintroduction regimen was as follows: rich medium [time (T) = 0], yeast peptone (YP) with 2% glucose, OD600 0.8; deprivation (T = 12, 25, 75, and 180 min), 0.15× YP lacking glucose, starts at OD600 0.8; reintroduction (T = +25 and +75 min), 0.5× YP with 1% glucose, starts at OD600 ≈ 0.5.
Chromatin Immunoprecipitation (ChIP) Microarray. For the details of our ChIP procedure, microarray preparation, and data analysis, please refer to Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Quantitative PCR (qPCR) Analysis. Real-time qPCR was performed by using an iCycler machine and iQ Sybr Green Supermix reagent (Bio-Rad). For a list of primers, see Table 2, which is published as supporting information on the PNAS web site. PCR of ChIP DNA was quantified in triplicate by using a standard curve, established with PCRs of serial 10-fold dilutions of a representative input DNA. This quantitation method normalizes for differences in amplification efficiency between primer sets and yields a value that may be considered the relative abundance of a particular target. Occupancy level was determined by dividing the relative abundance of an experimental target by the relative abundance of a control target (CDC2 or TRA1). This ratio represents the enrichment of ChIP DNA over the input DNA for a specific target versus the control target.
Results
Defining the RNA Polymerase III Transcriptome. The distribution of the Pol III machinery in the haploid genome of S. cerevisiae during growth in rich medium was determined through ChIP, fluorescent labeling of enriched DNA fragments, and hybridization to an array bearing segments of the entire yeast genome (see Supporting Materials and Methods). We performed separate ChIP microarray experiments with two members of each of the basal transcription complexes: Pol III (Rpc82 and Rpc40), TFIIIB (Brf1 and TBP), and TFIIIC (Tfc1 and Tfc6). TFIIIA was not included as it is reported to have a single essential target, 5S rDNA (24). Input and ChIP-enriched DNA were labeled (Cy3 for input, Cy5 for ChIP-enriched) and hybridized competitively to glass slides containing the entire yeast genome arrayed in ≈14,000 segments, parsed as ORFs and intergenic regions (see Supporting Materials and Methods).
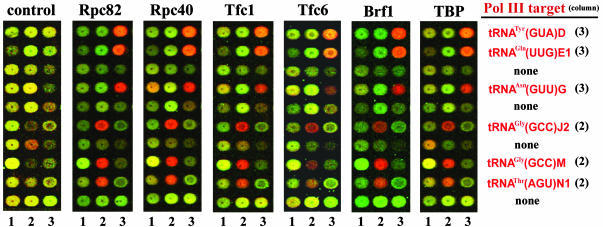
Visual inspection of the scanned arrays revealed robust enrichment of Pol III targets (Fig. 1). Enriched segments generate high Cy5/Cy3 ratios, which appear as red color (Cy5 as red, Cy3 as green). Images of a representative grid of 30 spots/segments from the genomic array that contain both tRNA-linked and unlinked segments are provided for comparative purposes (Fig. 1); we note that arrayed spots/segments are not adjacent on the chromosomal physical map. Whereas segments from the control ChIP (Fig. 1 Left) show essentially no enrichment, those derived from ChIP of the Pol III machinery show strong and consistent enrichment of Pol III targets. For example, in the Rpc82 ChIP experiments, the median tRNA-linked segment is enriched 6.2-fold, whereas the median ratio for the same targets in the control experiment was 1.0. The normalized spot/segment intensity ratios and average percentile ranks for each experiment discussed in this work are available in Data Set 1, which is published as supporting information on the PNAS web site.
Fig. 1.
ChIP of the Pol III machinery enriches segments linked to Pol III genes. Spot/segment color reflects the relative enrichment of DNA homologous to the segment on the array (orange/red = high enrichment, see Results). An identical array grid of 30 spots/segments derived from each ChIP microarray experiment. Rows that contain a tRNA-linked segment are identified at right, along with the respective column number.
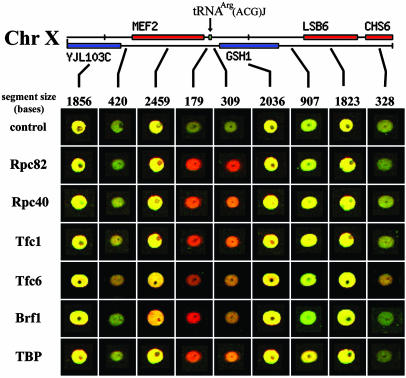
Apparent enrichment peaks at Pol III targets on the chromosomal physical map, as shown at tRNAArg(ACG)J on chromosome X (Fig. 2). Here, spots/segments were culled from either control or Pol III ChIP arrays and arranged according to the physical map. Segments >2 kb from the tRNA are not highly enriched, whereas those flanking the tRNA show strong enrichment. As the average length of the sheared DNA used for ChIP is ≈1–2 kb, modest enrichment of segments surrounding the Pol III target is often observed, but peak enrichment is strongly linked to the tRNA locus (Fig. 2 and data not shown).
Fig. 2.
Alignment of ChIP microarray results with the physical map, centered on a representative tRNA target. A 10-kb segment of chromosome X is depicted. For each member of the Pol III machinery, spots/segments were culled from different locations on the same array and arranged on the grid according to the physical map.
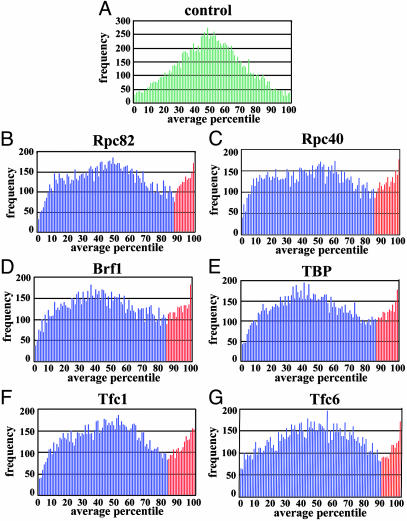
To more rigorously define enriched segments, we applied a method developed by Brown and colleagues (25, 26). First, the Cy3 and Cy5 intensities were determined for every segment and normalized. The Cy5/Cy3 ratio for each segment was then assigned a percentile rank (0–100%) that reflected relative enrichment. Segments were then sorted into bins; the most enriched segment (Cy5/Cy3 ratio »1) was placed in the 99–100% bin, the median segment (Cy5/Cy3 ratio of ≈1) was placed in the 49–50% bin, and the least enriched (Cy5/Cy3 ratio «1) was placed in the 0–1% bin. In a single experiment, each percentile bin receives an equal number of genomic segments. Two separate ChIP microarray experiments (with each subunit) allowed us to generate two independent percentile rank values for each genomic segment. These values were then combined to arrive at an average percentile rank for each segment. Those segments enriched in both independent ChIPs generated very high average percentiles, whereas those not enriched generated average percentiles centering around the 50th percentile. For our control ChIP, a plot that compared the number of segments (frequency) versus average percentile rank provided, as expected, a simple Gaussian distribution curve (Fig. 3A). In contrast, plots derived from ChIPs of Pol III machinery generated bimodal distributions, each with a distinctive trough. Nearly identical bimodal distributions were also obtained for the alternative subunits of each complex. We define those segments that generate average percentile ranks greater than those located at the base of the trough (the cutoff) as occupied by a Pol III member (shown in red, Fig. 3 B–E).
Fig. 3.
Defining Pol III targets by percentile rank analysis. Percentile rank values for each segment were determined as described in Results. Average percentile rank values were plotted versus the number of segments with the same average percentile rank (frequency). (A) Segments from the untagged strain generate a Gaussian distribution curve. (B–G) Segments from tagged strains or using TBP antisera generate bimodal distribution curves. Those segments to the right of the trough (cutoff) are colored red and considered enriched. Cutoffs: Rpc82, 83%; Rpc40, 83%; Brf1, 82%; TBP, 85%; Tfc1, 83%; Tfc6, 81%.
Our criterion for defining a segment as occupied by the Pol III machinery was that one member of each of the three complexes generated an average percentile greater than the cutoff. For tRNA genes, we evaluated occupancy at the 5′ or 3′ proximal segment to avoid the problem of cross-hybridization between similar tRNA genes. By this criterion, 264 of the 275 predicted tRNA genes are occupied [including the pseudo-tRNA gene tRNAAsp(GUC)N], as well as RPR1, SNR6, SCR1, and 5S rDNA. In the vast majority of cases, all six of the tagged members of the Pol III machinery occupied both the 5′ and 3′ segments flanking Pol III targets. However, by a less stringent criterion (occupancy by at least one member of each Pol III complex within 1 kb), all 275 tRNA genes are occupied. Finally, RNA170 (27) was only occupied above the cutoff by TFIIIC. Taken together, all of the predicted Pol III genes except RNA170 are occupied by all three Pol III complexes under exponential growth conditions.
Loci with Unexpected Occupancy by Pol III Machinery. In addition to the expected Pol III targets, several additional loci were clearly occupied by TFIIIC (Table 3, which is published as supporting information on the PNAS web site). Of these, two were additionally occupied by TFIIIB and Pol III: SNR52 and YML089C. We used the program trnascan-se (28), visual inspection, and comparisons to the genomic sequences of these loci in closely related yeasts to identify potential A and B boxes for these seven loci (Table 3). However, of all seven loci, only the SNR52 locus showed a detectable message by Northern analysis (Fig. 6, which is published as supporting information on the PNAS web site). Based on precedence with other Pol III-derived RNAs (non-tRNAs like RPR1), the 5′ region of the RNA containing the A and B box is removed by splicing, generating a mature RNA. In keeping with this idea, probes to the A or B boxes identify a rare 250-base RNA, whereas a probe to the predicted mature form reveals both this rare 250-base RNA and an abundant ≈92-base RNA. Together, these results suggest that SNR52 is a target of the Pol III machinery, although genetic analysis is required for verification.
Genome-Wide Occupancy of TBP. TBP occupancy at Pol III genes was striking, showing occupancy ratios of Pol III targets comparable to other members of the Pol III machinery. In fact, the highest percentile ranks of occupied segments are comprised almost exclusively of Pol III targets. Apart from Pol III targets, several hundred ORF and intergenic segments containing RNA polymerase II targets were occupied above the cutoff values. To designate true Pol II targets, we selected TBP-occupied ORF segments that were both above the cutoff and not occupied by other Pol III members (209 total). These genes include ribosomal protein genes, SNR genes, and translation factors. Others have estimated the transcription rate of most Pol II genes in the genome (29), and the TBP-occupied ORFs correlate with high transcription (see Discussion).
Response of the Pol III System to Nutrient Deprivation. We then investigated the relationships between gene activity and occupancy by the Pol III machinery. Pol III transcription is high in rich medium, and repressed during nutrient deprivation (17). Accordingly, we subjected yeast cultures bearing tagged Pol III machinery to the following regimen: logarithmic growth (nutrient replete), nutrient deprivation, and nutrient reintroduction. Cultures were initially grown to early log phase in rich medium, and an aliquot was removed and processed (see Supporting Materials and Methods). Cultures were then resuspended in low-nutrient medium. Aliquots were removed at 12, 25, 75, and 180 min and processed for RNA and ChIP analysis. We did not observe a significant change in the numbers of unbudded cells at T = 180 min (data not shown), suggesting that a transition to stationary phase had not occurred. Nutrients were then reintroduced, and aliquots were removed after an additional 25 and 75 min.
We selected two tRNA genes and RPR1 for analysis. Monitoring the rate of Pol III transcription by target RNA abundance is complicated by the exceptional stability of the mature forms of many structural RNAs. Most Pol III targets [such as tRNAPhe-(GAA)P2 and RPR1] transcribe RNAs that are rapidly spliced, but their unspliced forms are still detectable by Northern blot analysis at steady-state. Therefore, Pol III transcription rates are considered proportional to the abundance of the unspliced form compared with Pol II-transcribed U4 RNA. Northern analyses established the impact of our regimen on transcription rates at tRNAPhe(GAA)P2 and RPR1 (Figs. 4A and 5A). At both loci, nutrient deprivation rapidly reduced transcript levels, and we termed this period (within 25 min) the acute phase of repression. Transcript levels remained low until nutrient reintroduction restored their levels.
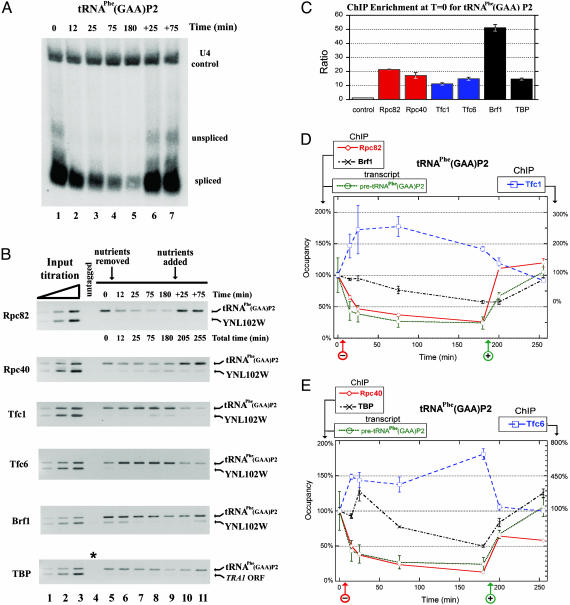
Fig. 4.
Activity–occupancy analysis of the Pol III machinery at tRNAPhe(GAA)P2 in response to nutrient availability. (A) Relative transcript levels of tRNAPhe(GAA)P2 determined by Northern analysis of total RNA from strain YBC1846 (similar results were obtained with untagged wild-type strain FT4; data not shown) using a probe to the mature form of the tRNA to detect both the unspliced and spliced forms. A probe to Pol II-transcribed U4 serves as a control. (B) Multiplex PCR analysis of tRNAPhe(GAA)P2 occupancy (see supporting information for details). Titration of the input DNA establishes that amplification is linear in the range tested, and ChIP from an untagged strain (FT4) provides little or no product (lane 4). The asterisk in lane 4 indicates no antisera control. (C) The initial occupancy levels of members of the Pol III machinery in rich medium (T = 0 in the regimen). The amplicon for tRNAPhe(GAA)P2 encompasses the entire tRNA gene. The control amplicon for all subunits except TBP is within the Pol II-transcribed CDC2 ORF. For TBP, the control amplicon is within the TRA1 ORF to ensure that the distance to a TATA box is >4 kb. PCR product accumulation was quantified by Sybr Green fluorescence (see Methods). Error bars represent ±1 SD for triplicate qPCR quantitations of a representative experiment. These initial occupancy levels (T = 0) are set to 100% for D and E.(D and E) Occupancy of tRNAPhe(GAA)P2 and analysis of the corresponding transcript in response to nutrient availability. One member of each complex is shown in each graph for clarity. Data from a representative ChIP experiment of each subunit is provided. Error bars represent ± 1SD for triplicate qPCR quantitations of a representative experiment. Average quantitation (quantified from blot in A and a replicate, not shown) of the level of unspliced tRNAPhe(GAA)P2 versus U4 from two independent experiments provides the transcript level, where the initial ratio is set to 100% and error bars are ±1 SD for the two independent experiments. Arrows at the bottom indicate nutrient deprivation (red arrow, –) and reintroduction (green arrow, +).
Fig. 5.
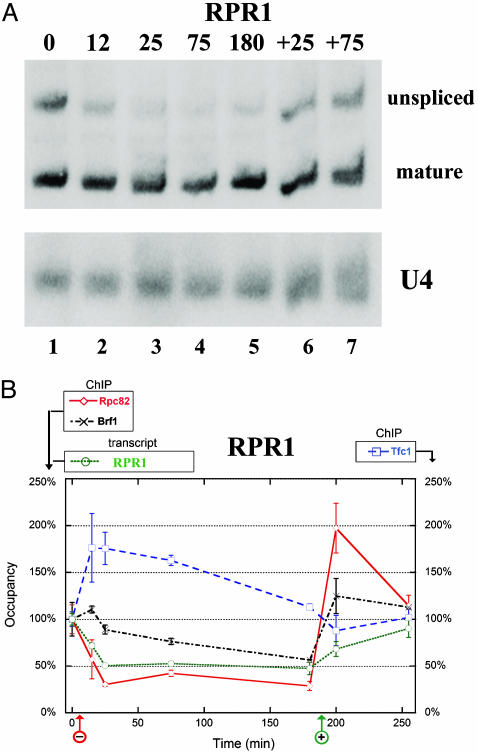
Activity–occupancy analysis of the Pol III machinery at RPR1 in response to nutrient availability. (A) Relative transcript levels of RPR1 determined by Northern analysis of total RNA from strain YBC1846 (similar results were obtained with untagged wild-type strain FT4; data not shown), using a probe to the mature form of RPR1 to detect both the unspliced and spliced forms. A probe to Pol II-transcribed U4 serves as a control. (B) Occupancy of RPR1 and analysis of the corresponding transcript in response to nutrient deprivation. The initial occupancy levels of members of the Pol III machinery in rich medium (T = 0 in the regimen) are as follows: Rpc82, 9.3-fold; Brf1, 18-fold; Tfc1, 7.1-fold. These ratios are set to 100% in the graph. The data are treated similarly to that in Fig. 4D, except the experimental target amplicon is for RPR1, which includes a large part of the transcribed region. The control amplicon is within the Pol II-transcribed CDC2 ORF.
We used multiplex PCR to measure occupancy at tRNAPhe-(GAA)P2 and observed strong enrichment with all Pol III machinery tested (Fig. 4B, lane 5, T = 0 min). The regimen revealed a remarkable set of trends: during acute repression, Pol III occupancy is markedly reduced, TFIIIB remains, and TFIIIC occupancy is increased dramatically. During prolonged repression (75–180 min), TFIIIB occupancy is reduced at tRNAPhe(GAA)P2 (with some variation), whereas TFIIIC occupancy levels remain high. Nutrient addition then largely reestablished the initial occupancy levels. Protein levels of the tagged components did not change appreciably during the regimen (data not shown). In many cases, our enrichment of Pol III targets was so robust that the control locus used (YNL102W or TRA1) was not detectable by this method, and were therefore difficult to quantify (Fig. 4B and data not shown).
To further examine these trends and more accurately quantify enrichment, we used real-time qPCR. With this method, enrichment was reproducibly high at tRNAPhe(GAA)P2 (Fig. 4C). To compare occupancy changes of each Pol III member during the regimen, we set their initial occupancy values (at T = 0) as 100%. A plot of occupancy versus time is provided for a representative ChIP experiment (with PCR analysis performed in triplicate)(Fig. 4 D and E). Components of each Pol III complex were split into separate graphs for clarity (note the alternative scale for TFIIIC components, at the right of each graph). Our qPCR results essentially parallel those obtained from multiplex PCR analysis; during acute repression Pol III largely leaves, TFIIIB remains and TFIIIC occupancy dramatically increases. During prolonged repression, we again observe a reduction in TFIIIB occupancy (with some variation), whereas TFIIIC occupancy levels remain high. Nutrient reintroduction largely restored occupancy levels to the initial state. Virtually identical trends were obtained at an additional tRNA locus, tRNALys(CUU)G1 (Fig. 7, which is published as supporting information on the PNAS web site), suggesting that this observation extends to other tRNAs. A similar trend, although less robust for TFIIIC, was also observed at RPR1 (Fig. 5B), showing that this trend is not limited to tRNAs. Thus, at the loci tested, nutrient deprivation is accompanied by a reduction in transcription and Pol III occupancy and an increase in TFIIIC occupancy. TFIIIB remains during the onset of repression, with some reduction in occupancy observed during prolonged repression.
Discussion
The Pol III Transcriptome. Our genome-wide occupancy assessment of the Pol III machinery establishes its presence at 264 of the 275 tRNA genes, 5S rDNA, SNR6, SCR1, and RPR1. The 12 predicted Pol III loci not occupied by our stringent criteria all display high levels of occupancy by most members within a 1-kb region (RNA170 excepted). Taken together, the Pol III machinery occupies all predicted Pol III targets except RNA170, based on our criteria. However, RNA170 and seven novel loci are strongly occupied by TFIIIC. Others have shown that RNA170 is transcribed by Pol III (27). The low levels of Pol III and TFIIIB present at this locus suggest that RNA170 may be transcribed at very low levels. Two of these loci, SNR52 and YML089C, show robust occupancy by all members of the Pol III machinery tested. SNR52 transcription was previously attributed to Pol II; however, a near-consensus A and B box was identified, and a transcript that includes the predicted A box and B box was detected. Although our data strongly suggest its inclusion as a Pol III target, genetic evidence is required for certainty. For the other loci identified, Northern analysis did not detect a transcript, so their candidacy is based on occupancy by TFIIIC alone. Further studies are required to determine whether Pol III generates transcripts at these loci under certain conditions or whether TFIIIC has an independent function.
A particularly interesting feature of this work was the exceptionally high occupancy of TBP at Pol III genes. In addition to Pol III targets, several hundred Pol II targets were occupied by TBP above the cutoff value. Remarkably, Pol III targets are transcribed at levels of up to 50 RNAs per min (30), suggesting that high levels of transcription might require the constant presence of TBP. Consistent with this idea, the Pol II genes identified as highly occupied are highly transcribed (median of 50 mRNA transcripts per h) in comparison to all Pol II genes (median of 2 mRNAs per h), based on the activity estimates of others (29). It remains possible that TBP is more efficiently precipitated in TFIIIB than in TFIID, which might increase its detection at Pol III targets.
Activity–Occupancy Relationships. We aimed to define correlations between gene activity and factor occupancy at Pol III genes. Our occupancy determinations were performed by using tags on two different members of each complex, and both members displayed similar trends. Thus, we consider enrichment to reflect occupancy and not simply occlusion or availability of the epitope tag under particular conditions. At the tRNA loci tested and RPR1, transcription is strongly reduced, Pol III rapidly leaves, TFIIIB largely remains, and TFIIIC occupancy strongly increases. Therefore, at these loci transcription is directly correlated with Pol III occupancy.
The work of others suggests that inhibition of TFIIIB function is an important aspect of repression (18, 22, 31, 32), and, in higher eukaryotes, TFIIIB may release from certain Pol III targets during the repression process (31, 32). However, TFIIIB appears to remain during acute repression in our studies, suggesting that large changes in TFIIIB occupancy are not correlated with transcriptional activity at the targets tested. During prolonged repression (75–180 min) TFIIIB occupancy trends downward (with some variability in extent), but is still largely present. Among the interesting questions raised is whether the TFIIIB occupancy detected during repression represents retention of the residing TFIIIB or recruitment of a new population, and whether the TFIIIB present is in an inactivated form. Also of interest is whether Pol III repression occurs by the same mechanisms in the acute and prolonged phases and in response to all types of cell stress.
Clearly, the most striking trend observed is the dramatic increase in TFIIIC occupancy at all loci tested during acute repression. TFIIIC is not required for transcriptional reinitiation in vitro, and the presence of its binding sites (A and B boxes) within the transcribed region have led to the proposal that TFIIIC might be removed as the elongating polymerase transcribes through its binding site, leaving a Pol III-TFIIIB reinitiation complex (4, 30, 33, 34). Our occupancy data provides in vivo support for this proposal, and is consistent with the following model (see Fig. 8, which is published as supporting information on the PNAS web site). The occupancy observed by all three members of the Pol III machinery in rich medium reflects an equilibrium between three states: (i) an initiation complex (containing all three complexes), (ii) a reinitiation complex (which recycles, containing only TFIIIB and Pol III), and (iii) a preinitiation complex (containing only TFIIIC and TFIIIB). Our data suggest this equilibrium shifts in response to altered growth conditions. We first consider the initiation complex, whose commitment to RNA synthesis leads to the displacement of TFIIIC from its binding site lowering the apparent occupancy of TFIIIC. TFIIIC may then leave the template or remain tethered to Pol III or TFIIIB. Active transcription of Pol III targets may involve a recycling TFIIIB–Pol III reinitiation complex that our occupancy data suggest dominates the steady state in nutrient replete conditions. However, this recycling reinitiation complex may display a limited half-life, leading to passive loss of Pol III at a slow rate. Nutrient deprivation increases Pol III loss and/or prevents reassociation, driving the equilibrium to the inactive state. Loss of active Pol III allows TFIIIC to reoccupy the template, greatly increasing its apparent occupancy. TFIIIC then either recruits a new TFIIIB or simply joins the resident TFIIIB. We suggest that the nascent TFIIIC–TFIIIB preinitiation complex is either active (competent to recruit Pol III) or inactive (not competent); the former dominates during nutrient replete conditions and the latter during nutrient deprivation/repression. The TFIIIC–TFIIIB preinitiation complex may be unstable during prolonged repression, with some loss of TFIIIB occurring. Similar activity–occupancy relationships may also occur in response to other types of cell stress, but remain to be tested. Other models for repression are also consistent with our data. For example, repression might increase TFIIIC-binding affinity, enabling occupancy of a larger fraction of targets and possible interference with Pol III activity.
Taken together, our work has defined the Pol III transcriptome and provided an activity–occupancy framework for understanding Pol III regulation. Signaling pathways involving Pkc1, casein kinase II, and Maf1 are known to play central roles in the regulation of Pol III activity, and likely function through modification of or interaction with the Pol III machinery (19–23). Further experiments are required to define the precise roles played by the basal machinery and its regulators to arrive at the occupancy relationships described.
Supplementary Material
Acknowledgments
We thank Brian Dalley, Adrienne Tew, and Qian Yang in the Huntsman Cancer Institute microarray facility for preparing and processing genomics resources. We also thank Jason Lieb and Patrick Brown for assistance with the ChIP microarray procedure and analysis, Kevin Struhl for strains, and Roger Kornberg for TBP antisera. This work was supported by the National Institutes of Health Grants 5 T32 DK007115 (to J. Kushner for the support of D.N.R.), and CA24014 (for core facilities), the Huntsman Cancer Institute (for support of A.J.S.), and by the Howard Hughes Medical Institute (for support of J.T.H. and genomics resources).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Pol III, RNA polymerase III; ChIP, chromatin immunoprecipitation; TBP, TATA-binding protein; qPCR, quantitative PCR; T, time.
Note Added in Proof. While this manuscript was in review, Harismendy et al. (35) published a genome-wide occupancy analysis of the Pol III machinery in yeast and arrived at similar conclusions.
References
- 1.Huet, J., Manaud, N., Dieci, G., Peyroche, G., Conesa, C., Lefebvre, O., Ruet, A., Riva, M. & Sentenac, A. (1996) Methods Enzymol. 273, 249–267. [DOI] [PubMed] [Google Scholar]
- 2.Geiduschek, E. P. & Kassavetis, G. A. (2001) J. Mol. Biol. 310, 1–26. [DOI] [PubMed] [Google Scholar]
- 3.Schramm, L. & Hernandez, N. (2002) Genes Dev. 16, 2593–2620. [DOI] [PubMed] [Google Scholar]
- 4.Dieci, G. & Sentenac, A. (2003) Trends Biochem. Sci. 28, 202–209. [DOI] [PubMed] [Google Scholar]
- 5.Galli, G., Hofstetter, H. & Birnstiel, M. L. (1981) Nature 294, 626–631. [DOI] [PubMed] [Google Scholar]
- 6.Sakonju, S., Bogenhagen, D. F. & Brown, D. D. (1980) Cell 19, 13–25. [DOI] [PubMed] [Google Scholar]
- 7.Conesa, C., Swanson, R. N., Schultz, P., Oudet, P. & Sentenac, A. (1993) J. Biol. Chem. 268, 18047–18052. [PubMed] [Google Scholar]
- 8.Parsons, M. C. & Weil, P. A. (1990) J. Biol. Chem. 265, 5095–5103. [PubMed] [Google Scholar]
- 9.Parsons, M. C. & Weil, P. A. (1992) J. Biol. Chem. 267, 2894–2901. [PubMed] [Google Scholar]
- 10.Lefebvre, O., Carles, C., Conesa, C., Swanson, R. N., Bouet, F., Riva, M. & Sentenac, A. (1992) Proc. Natl. Acad. Sci. USA 89, 10512–10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanson, R. N., Conesa, C., Lefebvre, O., Carles, C., Ruet, A., Quemeneur, E., Gagnon, J. & Sentenac, A. (1991) Proc. Natl. Acad. Sci. USA 88, 4887–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartholomew, B., Kassavetis, G. A., Braun, B. R. & Geiduschek, E. P. (1990) EMBO J. 9, 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaussivert, N., Conesa, C., Shaaban, S. & Sentenac, A. (1995) J. Biol. Chem. 270, 15353–15358. [DOI] [PubMed] [Google Scholar]
- 14.Rameau, G., Puglia, K., Crowe, A., Sethy, I. & Willis, I. (1994) Mol. Cell. Biol. 14, 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez, N. (1993) Genes Dev. 7, 1291–1308. [DOI] [PubMed] [Google Scholar]
- 16.Kassavetis, G. A., Braun, B. R., Nguyen, L. H. & Geiduschek, E. P. (1990) Cell 60, 235–245. [DOI] [PubMed] [Google Scholar]
- 17.Clarke, E. M., Peterson, C. L., Brainard, A. V. & Riggs, D. L. (1996) J. Biol. Chem. 271, 22189–22195. [DOI] [PubMed] [Google Scholar]
- 18.Zaragoza, D., Ghavidel, A., Heitman, J. & Schultz, M. C. (1998) Mol. Cell. Biol. 18, 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nierras, C. R. & Warner, J. R. (1999) J. Biol. Chem. 274, 13235–13241. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y., Moir, R. D., Sethy-Coraci, I. K., Warner, J. R. & Willis, I. M. (2000) Mol. Cell. Biol. 20, 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghavidel, A. & Schultz, M. C. (2001) Cell 106, 575–584. [DOI] [PubMed] [Google Scholar]
- 22.Upadhya, R., Lee, J. & Willis, I. M. (2002) Mol. Cell 10, 1489–1494. [DOI] [PubMed] [Google Scholar]
- 23.Pluta, K., Lefebvre, O., Martin, N. C., Smagowicz, W. J., Stanford, D. R., Ellis, S. R., Hopper, A. K., Sentenac, A. & Boguta, M. (2001) Mol. Cell. Biol. 21, 5031–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camier, S., Dechampesme, A. M. & Sentenac, A. (1995) Proc. Natl. Acad. Sci. USA 92, 9338–9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. & Brown, P. O. (2001) Nature 409, 533–538. [DOI] [PubMed] [Google Scholar]
- 26.Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28, 327–334. [DOI] [PubMed] [Google Scholar]
- 27.Olivas, W. M., Muhlrad, D. & Parker, R. (1997) Nucleic Acids Res. 25, 4619–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holstege, F. C., Jennings, E. G., Wyrick, J. J., Lee, T. I., Hengartner, C. J., Green, M. R., Golub, T. R., Lander, E. S. & Young, R. A. (1998) Cell 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 30.Dieci, G., Giuliodori, S., Catellani, M., Percudani, R. & Ottonello, S. (2002) J. Biol. Chem. 277, 6903–6914. [DOI] [PubMed] [Google Scholar]
- 31.Crighton, D., Woiwode, A., Zhang, C., Mandavia, N., Morton, J. P., Warnock, L. J., Milner, J., White, R. J. & Johnson, D. L. (2003) EMBO J. 22, 2810–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felton-Edkins, Z. A., Fairley, J. A., Graham, E. L., Johnston, I. M., White, R. J. & Scott, P. H. (2003) EMBO J. 22, 2422–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huibregtse, J. M. & Engelke, D. R. (1989) Mol. Cell. Biol. 9, 3244–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dieci, G. & Sentenac, A. (1996) Cell 84, 245–252. [DOI] [PubMed] [Google Scholar]
- 35.Harismendy, O., Gendrel, C. G., Soularue, P., Gidrol, X., Sentenac, A., Werner, M. & Lefebvre, O. (2003) EMBO. J. 22, 4738–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.