Abstract
BACKGROUND
High mobility group AT-hook 1 (HMGA1) proteins are architectural transcription factors that are overexpressed by pancreatic adenocarcinomas. The authors hypothesized that tumor HMGA1 status represents a novel prognostic marker in pancreatic adenocarcinoma. They also tested the hypothesis that HMGA1 promotes anchorage-independent cellular proliferation and in vivo tumorigenicity.
METHODS
Tumor HMGA1 expression was examined by immunohistochemical analysis of tissues from 89 consecutive patients who underwent resection for pancreatic adenocarcinoma. Short-hairpin RNA (shRNA)-mediated RNA interference was used to silence HMGA1 expression in MiaPaCa2 and PANC1 pancreatic cancer cells. Anchorage-independent proliferation was assessed by using soft agar assays. The roles of phosphatidylinositol 3-kinase (PI3-K)/Akt and extracellular signal-regulated kinase (ERK) signaling were investigated by using specific inhibitors and adenoviral dominant-negative/active Akt constructs. In vivo tumorigenicity was assessed by using a nude mouse xenograft model.
RESULTS
Tumor HMGA1 expression was detected in 93% of patients with pancreatic adenocarcinoma. Patients with HMGA1-negative tumors had a significantly longer median survival than patients with HMGA1-expressing cancers in univariate analysis (P = .0028) and in multivariate analysis (P<.05). shRNA-mediated HMGA1 silencing resulted in significant reductions in anchorage-independent proliferation in soft agar. Forced HMGA1 overexpression promoted proliferation in soft agar through a process that was dependent on PI3-K/Akt-activited signaling, but not on mitogen-activated protein kinase (MEK)/ERK signaling. Targeted silencing of HMGA1 reduced tumor growth in vivo through reduced proliferation (Ki-67 index) and increased apoptosis (terminal deoxynucleotidyl transferase nick-end labeling).
CONCLUSIONS
The current findings suggested that HMGA1 is an independent predictor of poor postoperative survival in patients with pancreatic adenocarcinoma. Furthermore, HMGA1 promotes tumorigenicity through a PI3-K/Akt-dependent mechanism. HMGA1 warrants further evaluation as a prognostic marker and therapeutic target in pancreatic cancer.
Keywords: high mobility group AT-hook 1, Akt, extracellular signal-regulated kinase, growth, survival, pancreatic adenocarcinoma
Pancreatic adenocarcinoma is the fourth leading cause of cancer-related death in the United States. The overall prognosis for patients who are diagnosed with this malignancy remains dismal, with 5-year survival rates averaging <5%.1 The rational identification of clinically relevant therapeutic targets based on an under-standing of the biologic mechanisms underlying the aggressive behavior of pancreatic cancer is a high-priority goal.
The human high mobility group AT-hook 1 gene HMGA1, which is located on chromosomal locus 6p21, encodes 2 HMGA1 splice variants (HMGA1a and HMGA1b).2 These HMGA1 proteins are architectural transcription factors that regulate gene expression in vivo.3,4 They form stereo-specific, multiprotein complexes termed “enhanceosomes” on the promoter/enhancer regions of genes, where they bind to the minor groove of AT-rich DNA sequences.3,5 HMGA1 proteins are overexpressed in a range of human cancers.6–9 By using a small sample of tumor tissues, Abe and colleagues previously demonstrated that HMGA1 is overexpressed in pancreatic cancers, although the clinical relevance of HMGA1 expression in this tumor type remains uncertain.10 The degree of tumor HMGA1 expression reportedly is correlated inversely with patient survival duration in some human cancers.6,7 These correlative data suggest that HMGA1 may play an important role in cancer progression; however, the mechanisms by which HMGA1 may mediate features of the malignant phenotype are poorly understood. We demonstrated previously that HMGA1 promotes pancreatic cancer cellular invasiveness and that the targeted suppression of HMGA1 reduces metastasis in vivo.11 We also demonstrated that HMGA1 promotes resistance to anoikis (apoptosis caused by the loss of substratum attachment)12 and chemoresistance to gemcitabine in pancreatic cancer cells.13 Although we have demonstrated that HMGA1 affects the metastatic and apoptotic processes, its role in pancreatic tumor growth is unknown, and its potential as an antigrowth therapeutic target remains to be explored.
To establish the clinical relevance of tumor HMGA1 expression in patients with pancreatic cancer, we examined tumor HMGA1 expression in a large cohort of patients using a tissue microarray (TMA) with clinicopathologic correlates. Our findings suggest that tumor HMGA1 expression status represents a biomarker that can be used to predict postoperative survival in patients who have undergone surgical resection for pancreatic adenocarcinoma. In this study, to further evaluate the potential roles of HMGA1 in mediating the malignant phenotype, we tested the hypotheses that HMGA1 promotes anchorage-independent cellular proliferation in pancreatic adenocarcinoma cells and that suppression of HMGA1 expression would impair anchorage-independent proliferation in vitro and tumor growth in vivo. Our observations indicate that HMGA1 indeed promotes pancreatic adenocarcinoma tumorigenesis and that a key effector of HMGA1-induced, anchorage-independent growth is the phosphoinositidyl-3 kinase (PI3-K)/Akt pathway.
MATERIALS AND METHODS
Tissue Microarray Construction and Analysis
Under an Institutional Review Board-approved study protocol, pathology reports were searched to identify patients who underwent curative surgical resection for pancreatic adenocarcinoma between the years 1991 and 2002 at Brigham and Women’s Hospital. A pancreatic adenocarcinoma TMA was constructed from 89 consecutive patients who underwent curative resection for pancreatic adenocarcinoma. Formalin-fixed, paraffin-embedded specimens were used to construct the pancreatic adenocarcinoma TMA. Representative tumor regions were selected from each tissue block, and 2 tissue cores (0.6 mm in greatest dimension) were taken from each region using an automated tissue arrayer (Beecher Instruments, Sun Prairie, Wis). Cores also were taken from normal adjacent pancreas for use as internal controls. To avoid areas of pancreas that may have harbored premalignant changes (eg, desmoplasia), we selected only cores that were normal morphologically as internal controls during the TMA construction. Standard hematoxylin and eosin-stained slides from each tumor and its surrounding normal pancreas were reviewed by a single pathologist (M.R.).
Five-micrometer sections were cut from each recipient block to make the TMA slides. Clinical information, including age, sex, and use of chemotherapy, was gathered retrospectively from patient records. Pathologic findings, including tumor size, stage, lymphovascular invasion (LVI), perineural invasion (PNI), differentiation, surgical resection margin status, and lymph node status, were obtained from original pathology reports. Pathologic staging was updated according to current American Joint Committee on Cancer guidelines.
TMA slides were deparaffinized and processed using a streptavidin-biotin-peroxidase complex method. Antigen retrieval was performed by microwave heating sections in 10 mM sodium citrate buffer, pH 6.0, for 10 minutes. After quenching of endogenous peroxidase activity and blocking nonspecific binding, anti-HMGA1 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif) was added at a 1:50 dilution; then, slides were incubated at 4 °C overnight. The secondary biotinylated rabbit antigoat antibody (DAKO, Carpinteria, Calif) was used at a dilution of 1:200 for 30 minutes at 37 °C. Next, sections were incubated with streptavidin-biotin complex/horseradish peroxidase (1:100 dilution; DAKO) for 30 minutes at 37 °C. Chromogenic immunolocalization was determined by exposure to 0.05% 3,3-diaminobenzidine tetrahydrochloride. Normal serum was used in the place of primary antibody as a negative control. Slides were reviewed by 2 independent observers who were blinded to clinical and pathologic data. HMGA1 was scored according to nuclear staining intensity as follows: 0, no staining or weak-intensity staining in <5% of cells; 1, weak-intensity staining; 2, moderate-intensity staining; 3, strong-intensity staining. For statistical analyses, expression was dichotomized into an HMGA1-negative group (score 0) and an HMGA1-positive group (scores ≥1). In cases of disagreement, a consensus was reached by joint review.
Cells and Cell Culture
MiaPaCa2 and PANC1 human pancreatic ductal adenocarcinoma cells were obtained from American Type Culture Collection (Manassas, Va). Cells were maintained in Dulbecco Modified Eagle Medium containing 10% fetal bovine serum (Gibco Life Technologies Inc., Gaithersburg, Md).
Reagents and dominant-negative Akt adenovirus
The PI3-K inhibitor LY294002 and mitogen-activated protein kinase 1/2 (MEK1/2) inhibitor PD98059 were purchased from Calbiochem (San Diego, Calif). Anti-HMGA1 and antilamin B1 antibodies were obtained from Santa Cruz Biotechnology. Adenovirus carrying dominant-negative (Ad-DN-Akt) and dominant-active (Ad-myr-Akt) Akt1 and control cytomegalovirus (Ad-CMV-null) were obtained from Vector BioLabs (Philadelphia, Pa). Adenoviral infection was performed at a multiplicity of infection of 10 in the presence of 6 μg/mL polybrene for 24 hours. Experiments were performed on cells 48 hours after infection.
Plasmid-mediated HMGA1 RNA interference
Hairpin RNA interference plasmids were obtained from The RNA-mediated Interference (RNAi) Consortium (Mission TRC-Hs 1.0; Sigma Aldrich, St. Louis, Mo). The sequences of short-hairpin RNA (shRNA) targeting the human HMGA1 gene were as follows: shHMGA1-1 plasmid, 5′-CAACTCCAGGAAGGAAACC AA-3′; and shHMGA1-2 plasmid, 5′-CCTTGGCC TCCAAGCAGGAAA-3′. The control plasmid, which has a scrambled, nontargeting shRNA sequence, was obtained from Addgene (Cambridge, Mass). Pooled stable transfectants were established using puromycin (InvivoGen, San Diego, Calif) selection.
Expression vector and transfection
The HMGA1 coding sequence was amplified by polymerase chain reaction (PCR) from IMAGE clone 5399570 by using gene-specific primers that were modified to include the appropriate restriction sites at their 5′ end. The following primers were used: forward, 5′-TTTTGATATCATGAGTGAGTCGAGCTCGAAG-3′ and backward, 5′-TTTTGAATTCTCACTGCTCCTCCTCCGAGGA-3′. Purified PCR products were digested with EcoRV and EcoRI before ligation into an EcoRV/EcoRI-digested pIRES-puro3 vector (Clontech, Palo Alto, Calif). The expression plasmids were named pIRES-HMGA1. MiaPaCa2 cells were transfected with pIRES-HMGA1 or with empty pIRES-puro3, which acted as a control, using Lipofectamine 2000 (Invitrogen). Clones pIRES-HMGA1.1 and pIRES-HMGA1.2 were selected by using puromycin (InvivoGen) were used for further studies, because they expressed the highest levels of HMGA.
Soft agar colony formation assay
Assays were performed by using a cell transformation detection assay according to the manufacturer’s instructions (Chemicon, Temecula, Calif). Briefly, assays were performed in 6-well plates with 5 × 103 cells, resuspended as a single cell suspension in 0.4% agar, and layered on top of 0.8% agar. Plates were incubated for 10 to 12 days. Colonies were stained and counted manually at high-power magnification (×40). The counting was performed for 10 fields in each well, and at least 6 wells per condition were counted in each experiment. Average values from 3 independent experiments were calculated. The relative number of colonies was calculated by dividing each value by the mean value of the control group.
Western blot analysis
Total cell extracts were prepared with Phosphosafe lysis buffer (Novagen, San Diego, Calif). Nuclear extracts were prepared by using NE-PER Nuclear Extraction Reagents (Pierce, Rockford, Ill). Protein concentrations were measured by using a bicinchoninic acid assay kit (Sigma) with bovine serum albumin as a standard. Cell lysates that contained 50 μg protein or nuclear protein that contained 10 μg protein were subjected to 10% sodium dodecyl sulfate/polyacrylamide gel electrophoresis, as described previously.11 Chemiluminescence detection (Amersham Biosciences, NJ) was performed in accordance with the manufacturer’s instructions.
Nude mouse subcutaneous xenograft model
Male athymic nu/nu mice aged 5 weeks were obtained from Harlan Sprague-Dawley (Indianapolis, Ind). Mice housed in a pathogen-free facility were observed for signs of tumor growth, activity, feeding, and pain in accordance with the guidelines of the Harvard Medical Area Standing Committee on Animals. To determine the effect of HMGA1 gene silencing on in vivo growth, 2 × 106 MiaPaCa2 cells and PANC1 stable transfectants that expressed the control or HMGA1 shRNA (shHMGA1.1 plasmid) were implanted subcutaneously in nude mice. Tumor dimensions were measured weekly by using micrometer calipers. Tumor volumes were calculated as follows: volume = 1/2 a × b2, where a and b represented the larger and smaller tumor dimensions, respectively. Eight weeks after implantation, the primary tumor was excised, fixed in formalin, and embedded in paraffin.
Immunohistochemistry
Xenograft tumor sections (5 μm) were deparaffinized and processed by using the streptavidin-biotin-peroxidase complex method described above. Sections were incubated with anti-Ki-67 (DAKO) at 4 °C overnight at 1:200 dilution. The secondary antibody was biotinylated rabbit-antimouse antibody (DAKO), which was used at 1:200 dilution for 30 minutes at 37 °C. Tumor cells were considered positive for the Ki-67 antigen if there was intranuclear staining. Cells with positively stained nuclei were counted at ×40 magnification in 5 random fields from each section.
Apoptosis staining
After preparation of 5-μm tumor sections, apoptosis was quantified by using a commercially available terminal deoxynucleotidyl transferase nick-end labeling (TUNEL) kit (Chemicon). The number of apoptotic cells was counted in 5 random fields from each section.
Statistical Analysis
Differences between groups were analyzed using Student t tests, multifactorial analyses of variance of initial measurements, and Mann-Whitney U tests for nonparametric data, as appropriate, with Statistica software (version 5.5; StatSoft, Inc., Tulsa, Okla). In cases in which averages were normalized to controls, the standard deviations of each nominator and denominator were taken into account in calculating the final standard deviation. P < .05 was considered statistically significant.
RESULTS
Pancreatic Adenocarcinoma TMA: Patient Characteristics
The cohort consisted of 89 patients with pathologically proven pancreatic adenocarcinoma (42 men and 47 women). The mean age at diagnosis was 63 years (median age, 63 years; age range 34–84 years). The median survival was 16.6 months (range, 91–3462 days). The actuarial 1-year survival rate was 70.3%, and the 5-year survival rate was 8.1%. A summary of the clinicopathologic characteristics of the cohort is provided in Table 1.
TABLE 1.
Clinicopathologic Characteristics of the Pancreatic Adenocarcinoma Cohort
| Characteristics | No. of patients (%) |
|---|---|
| Age, y | |
| Median | 63 |
| Range | 34284 |
| Sex | |
| Men | 42 |
| Women | 47 |
| Overall disease stage | |
| I | 13 (15) |
| II | 74 (83) |
| III | 0 (0) |
| IV | 2 (2) |
| Lymph node status | |
| Negative | 34 (38) |
| Positive | 55 (62) |
| Pathologic tumor size, cm | |
| Median | 2.70 |
| Range | 0.10–8.40 |
| Histopathologic differentiation | |
| Well | 8 (9) |
| Moderate | 47 (53) |
| Poor | 34 (38) |
HMGA1 Expression in Normal Tissue and Pancreatic Adenocarcinoma Specimens
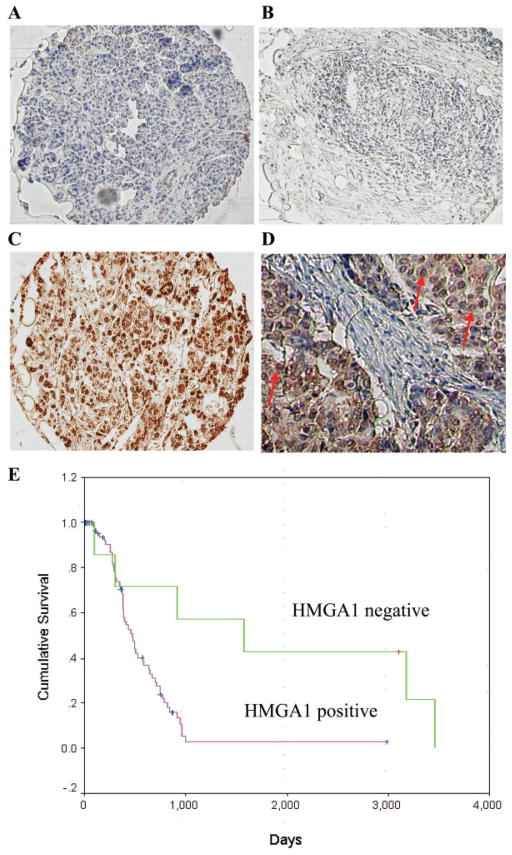
Paraffin-embedded specimens were used to construct a pancreatic adenocarcinoma TMA. For each patient within the TMA, cores also were taken from adjacent normal pancreas to act as internal controls and to assess the expression of HMGA1 in normal pancreas. After immunostaining with anti-HMGA1 antibody, HMGA1 expression was scored according to nuclear intensity. Expression was dichotomized into an HMGA1-negative group (score 0) and an HMGA1-positive group (scores ≥1). On immunohistochemical analysis, we detected the presence of nuclear HMGA1 expression in 83 of 89 (93%) pancreatic adenocarcinoma specimens (Fig. 1C). The majority of normal pancreatic ducts had no detectable nuclear HMGA1 expression (Fig. 1A), whereas some normal pancreatic ducts had very weak expression. In the majority of tumor specimens (52%), the degree of HMGA1 staining was graded with a score ≥2 (score 1, 42% [37 of 89 tumors]; score 2, 35% [31 of 89 tumors]; score 3, 17% [5 of 89 tumors]). HMGA1 staining predominantly was localized to the nucleus of cancerous cells with some staining of the cytoplasm (Fig. 1C). A detailed pathology case review of the 6 specimens with absent tumor HMGA1 expression (Fig. 1B) demonstrated no atypical features for pancreatic adenocarcinoma.
FIGURE 1.
Immunohistochemical staining for high mobility group AT-hook 1 (HMGA1) in normal pancreas and pancreatic adenocarcinoma specimens using a tissue microarray. (A) Normal pancreas (HMGA1 negative). (B) Pancreatic adenocarcinoma (HMGA1 negative). (C) Pancreatic adenocarcinoma (HMGA1 positive). (D) High-power magnification (original magnification, ×400) of an HMGA1-positive section that has intense nuclear staining for HMGA1 (indicated by red arrows) with some staining of cytoplasm. (E) Kaplan-Meier analysis of overall survival for patients with pancreatic adenocarcinoma based on HMGA1 expression. Survival of immunohistochemically HMGA1-negative patients was compared with survival of HMGA1-positive patients by using the log-rank test (P =.0028).
Tumor HMGA1 Expression Status: Association With Clinicopathologic Features
To identify associations of HMGA1 expression (HMGA1-negative expression vs HMGA1-positive-expression) with clinicopathologic variables, the variables were dichotomized as shown in Table 2. There were no significant differences patients with positive or negative tumor HMGA1 expression were compared with respect to patient age, sex, tumor size, differentiation, LVI, PNI, receipt of chemotherapy, margin status, lymph node involvement, and disease stage (Fisher exact test; P >.05) (Table 2). In a Kaplan-Meier analysis, patients who had no HMGA1 expression (HMGA1-negative tumors) had significantly longer overall postoperative survival (mean, 5.7 years; median, 4.3 years) compared with patients who had HMGA1-positive tumors (mean, 1.6 years; median, 1.3 years; P =.0028; log-rank test) (Fig. 1D). For the patients who had positive HMGA1 expression, increasing degree of HMGA1 expression was not associated with worse clinicopathologic features or shorter survival.
TABLE 2.
Associations of High Mobility Group AT-hook 1 Expression With Clinicopathologic Features
| Variable | HMGA1 status |
P | |
|---|---|---|---|
| Negative (N = 6) | Positive (N = 83) | ||
| Age, y | |||
| ≤64 | 3 | 44 | 1.000 |
| >64 | 3 | 39 | |
| Sex | |||
| Men | 2 | 40 | .680 |
| Women | 4 | 43 | |
| Tumor grade (differentiation) | |||
| 1 | 1 | 6 | .713 |
| 2 | 3 | 44 | |
| 3 | 2 | 32 | |
| Tumor size, cm | |||
| <2.5 | 3 | 38 | .416 |
| >2.5 | 3 | 55 | |
| Lymph node metastasis | |||
| No | 4 | 30 | .197 |
| Yes | 2 | 53 | |
| Lymphovascular invasion | |||
| No | 5 | 49 | .397 |
| Yes | 1 | 34 | |
| Perineural invasion | |||
| No | 3 | 40 | 1.000 |
| Yes | 3 | 43 | |
| Microscopic margin status | |||
| Negative | 3 | 49 | .690 |
| Positive | 3 | 34 | |
| Tumor location | |||
| Head | 6 | 77 | 1.000 |
| Tail | 0 | 6 | |
| Tumor classification | |||
| T1/T2 | 2 | 11 | .210 |
| T3/T4 | 4 | 72 | |
| Chemotherapy | |||
| No | 1 | 4 | .272 |
| Yes | 3 | 59 | |
HMGA1 indicates high mobility group AT-hook 1.
HMGA1 Represents an Independent Prognostic Indicator in Pancreatic Adenocarcinoma
To assess whether HMGA1 expression was an independent predictor of overall postoperative survival, a Cox proportional-hazards model was created in a forward fashion that included only the covariates that had a statistically significant correlation (inclusion threshold, P ≤.05) with postoperative survival. Univariate analysis demonstrated that increasing tumor size, poor tumor differentiation, and HMGA1-positive tumors were significant predictors of poorer survival (P < .05) (Table 3). Furthermore, multivariate analysis demonstrated that, after correction for confounding variables, HMGA1 expression remained a significant independent prognosticator for postoperative survival (P =.001) (Table 3).
TABLE 3.
Predictors of Postoperative Survival: High Mobility Group AT-hook 1 as an Independent Prognostic Indicator
| Risk factor | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| Hazard | 95% CI | P | Hazard | 95% CI | P | |
| Increasing age | 1.015 | 0.994–1.037 | .171 | 1.008 | 0.983–1.033 | .545 |
| Women | 0.742 | 0.442–1.244 | .258 | 1.056 | 0.576–1.936 | .861 |
| Increasing tumor size | 1.217 | 1.022–1.450 | .028* | 1.398 | 1.130–1.730 | .002* |
| Presence of lymph node metastasis | 1.733 | 0.995–3.019 | .052 | 1.551 | 0.781–3.082 | .210 |
| Advanced tumor stage (T3/T4 vs T1T/2) | 2.034 | 0.906–4.567 | .085 | |||
| Local invasion | 1.078 | 0.612–1.897 | .796 | |||
| Poor tumor differentiation | 1.859 | 1.183–2.921 | .007* | 3.203 | 1.746–5.877 | .000* |
| Presence of PNI | 1.088 | 0.651–1.819 | .746 | |||
| Presence of LVI | 1.704 | 0.982–2.958 | .058 | 1.710 | 0.944–3.098 | .077 |
| Presence of tumor at microscopic margin | 1.490 | 0.886–2.505 | .133 | |||
| Chemotherapy treatment | 0.440 | 0.132–1.474 | .183 | |||
| HMGA1 expression | 5.473 | 1.614–18.557 | .006* | 12.474 | 2.705–57.520 | .001* |
95% CI indicates 95% confidence interval; LVI, lymphovascular invasion; PNI, perineural invasion; HMGA1, high mobility group AT-hook 1.
P < .05.
Stable RNAi-mediated Suppression of HMGA1 Expression Inhibits Anchorage-independent Growth
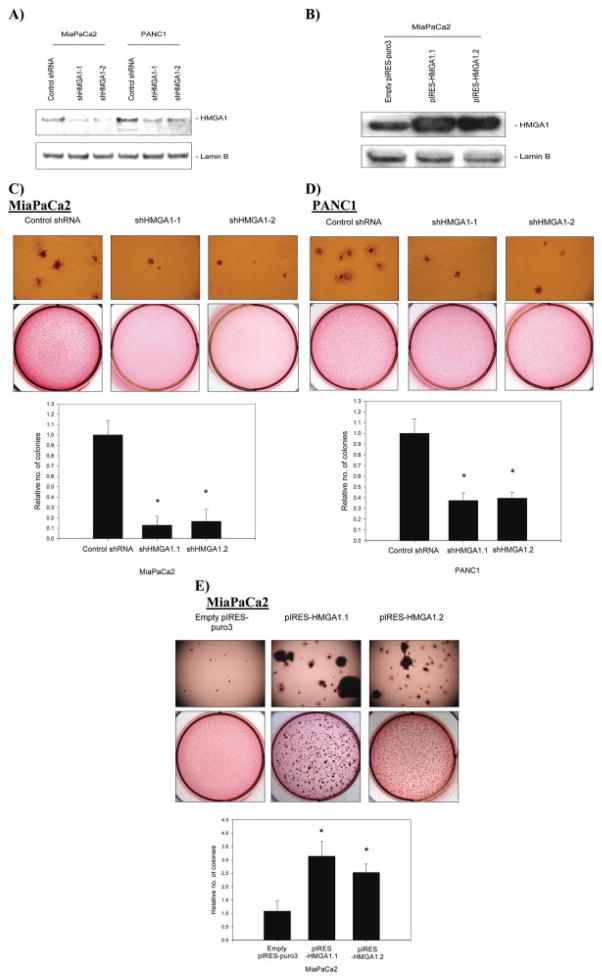
Both MiaPaCa2 and PANC1 pancreatic adenocarcinoma cell lines expressed HMGA1, and MiaPaCa2 cells had a lower expression level of HMGA1 at baseline. We used 2 independent shRNA target sequences (called shHMGA1-1 and shHMGA1-2) to suppress HMGA1 expression. Each of these shRNA sequences was associated with an approximately 80% reduction in HMGA1 expression in MiaPaCa2 cells (Fig. 2A), as confirmed by Western blot analyses of nuclear extracts. These same shRNA sequences were associated with approximately 55% to 60% reductions in HMGA1 expression in PANC1 cells (Fig. 2A).
FIGURE 2.
(A) Stable silencing of high mobility group AT-hook 1 (HMGA1) expression using 2 short-hairpin RNA (shRNA) expression vectors with independent target sequences (shHMGA1-1 and shHMGA1-2) was confirmed on Western blot analysis of nuclear extracts. Controls were shRNA expression vectors with a scrambled, nontargeting sequence. Greater suppression of HMGA1 expression was achieved in MiaPaCa2 cells, in which there was approximately 80% silencing with shRNA sequences. In PANC1 cells, there was 55% to 60% suppression of HMGA1 expression with each shRNA sequence. (B) Results confirmed that 2 clones of MiaPaCa2 cells stably overexpressed HMGA1 (pIRES-HMGA1.1 and pIRES-HMGA1.2) on Western blot analysis of nuclear extracts. Blots shown are representative of 3 independent experiments. (C) The effects of modulating HMGA1 expression on anchorage-independent growth was assessed by using soft agar assays. Stable HMGA1 silencing using each of 2 independent shRNA sequences (shHMGA1-1 and shHMGA1-2) resulted in reductions in soft agar colony formation in both MiaPaCa2 cells and PANC1 cells compared with the scrambled control shRNA-transfected cells. Effects on soft agar growth were greater in Mia-PaCa2 cells, corresponding to greater silencing of HMGA1 in these cells. An asterisk indicates P < .05 versus control shRNA. (D) Overexpression of HMGA1 in MiaPaCa2 clones (pIRES-HMGA1.1 and pIRES-HMGA1.2) resulted in unequivocal increases in anchorage-independent growth in soft agar. The number and size of colonies in soft agar clearly were larger with overexpression of HMGA1. An asterisk indicates P < .05 versus empty pIRES-puro3-transfected cells. Values are means (±standard deviation).
In MiaPaCa2 cells, the high degree of HMGA1 silencing induced by each of the shRNA sequences was associated with marked reductions in growth in soft agar (Fig. 2C). Similar but less marked reductions in growth in soft agar were observed for PANC1 cells (Fig. 2D) corresponding to lower degrees of HMGA1 silencing in this cell line.
Forced HMGA1 Overexpression Promotes Anchorage-independent Growth
Given the relatively low baseline expression level of HMGA1 in MiaPaCa2 cells, we chose this cell line to test the effects of forced HMGA1 overexpression. We transfected this cell line with an overexpression vector that carried the full-length HMGA1 combinational DNA (cDNA). Two clones that stably overexpressed HMGA1 were selected and named pIRES-HMGA1.1 and pIRES-HMGA1.2. The degree of HMGA1 overexpression (2.5-fold and 3-fold overexpression, respectively, over controls) was documented on Western blot analyses of nuclear extracts (Fig. 2B). HMGA1 overexpression was associated with increased colony formation in soft agar (Fig. 2E).
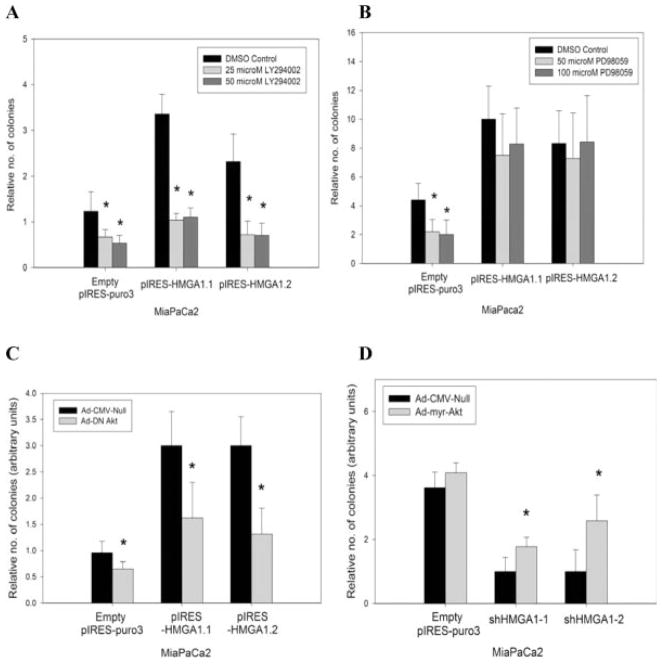
HMGA1-induced Increases in Anchorage-independent Growth Are PI3-K/Akt-dependent but Not MEK/Extracellular Signal-regulated Kinase-dependent
The importance of the PI-3K/Akt and MEK/extracellular signal-regulated kinase (ERK) pathways in tumor growth has been described previously.14,15 Our group also previously demonstrated that Akt and ERK activation depends on HMGA1 expression levels.11,12 Therefore, we sought to determine the dependence of HMGA1-induced increase in soft agar growth on these pathways. Given the effects of HMGA1 on Akt activation, we performed soft agar assays in the presence of the PI-3K inhibitor LY294002. At concentrations of either 25 μM or 50 μM, LY294002 was associated with significant reductions in soft agar growth by the pIRES-HMGA1.1 and pIRES-HMGA1.2 clones at levels similar to those exhibited by parental MiaPaCa2 cells and MiaPaCa2 cells stably transfected with empty pIRES-puro3 vector (Fig. 3A). In contrast, the MEK/ERK inhibitor PD98059 (at concentrations of either 50 μM and 100 μM) had no impact on soft agar growth by the pIRES-HMGA1.1 and pIRES-HMGA1.2 clones (Fig. 3B). In addition, infection of Ad-DN-Akt abrogated soft agar growth in the pIRES-HMGA1.1 and pIRES-HMGA1.2 clones (Fig. 3C). Infection of Ad-myr-Akt significantly increased the soft agar growth in Mia-PaCa2 cells stably transfected with shHMGA1-1 and shHMGA1-2 (Fig. 3D).
FIGURE 3.
(A) Given the effect of high mobility group AT-hook 1 (HMGA1) on Akt activation, soft agar assays were performed in the presence of 25 μM and 50 μM of LY294002 (a specific phosphatidylinositol 3-kinase [PI3-K] inhibitor). Clones pIRES-HMGA1.1 and pIRES-HMGA1.2 exhibited significant inhibition of soft agar growth in the presence of LY294002 at either concentration compared with the dimethyl sulfoxide (DMSO)-treated controls. Although LY294002 also had an effect on soft agar growth in pIRES-empty puro3 controls, the degree of inhibition clearly was less marked compared with that in the pIRES-HMGA1.1 and pIRES-HMGA1.2 clones. An asterisk indicates P < .05 versus DMSO-treated controls. (B) Mitogen-activated protein kinase(MEK)/extracellular signal-regulated kinase (ERK) inhibitor PD98059 had no effects on HMGA1 overexpression-induced increases in soft agar growth, because the pIRES-HMGA1.1 and pIRES-HMGA1.2 clones exhibited no significant reductions in growth even in high concentrations (50 μM and 100 μM) of PD98059. This is in contrast to the significant inhibition of soft agar growth when the pIRES-puro3 controls were exposed to PD98059. An asterisk indicates P < .05 versus DMSO-treated controls. (C) Infection of the pIRES-HMGA1.1 and pIRES-HMGA1.2 clones with dominant-negative Akt adenovirus resulted in significant reductions in HMGA1 overexpression-induced increase in soft agar growth. Dominant-negative Akt adenovirus resulted in a small effect on soft agar growth in the empty pIRES-puro3 control cells. An asterisk indicates P < .05 versus control adenovirus-cytomegalovirus (Ad-CMV-Null). (D) Conversely, infection of adenovirus-expressing, constitutively active Akt (Ad-myr-Akt) rescued the ability to grow under anchorage-independent conditions in shHMGA1-1 and shHMGA1-2 stable transfectants. No effects were observed when empty pIRES-puro3 controls were infected with constitutively active Akt adenovirus. An asterisk indicates P < .05 versus control adenovirus (Ad-CMV-Null). Values are means (±standard deviations).
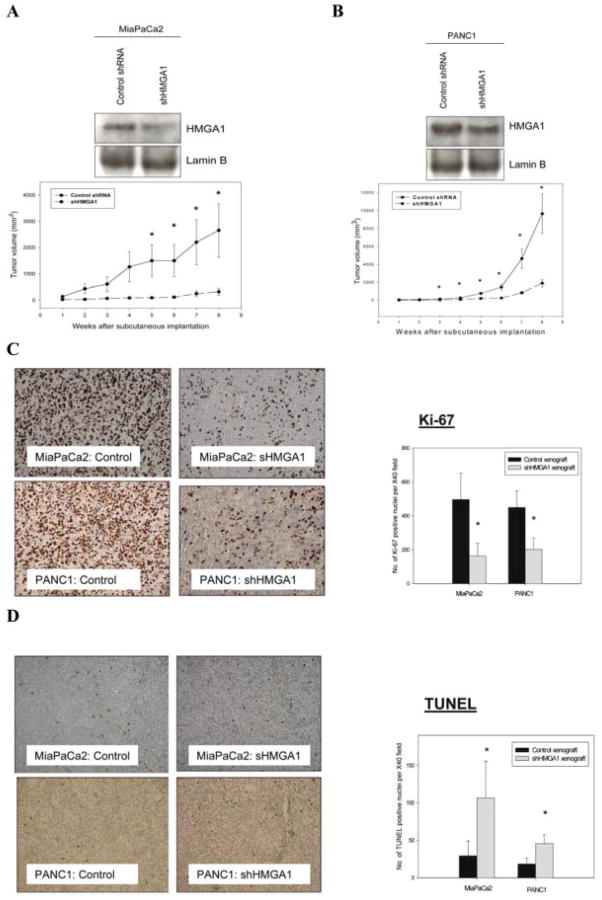
HMGA1 Silencing Resulted in Significant Inhibition of Tumor Growth in Vivo
Tumors derived from the subcutaneous implantation of MiaPaCa2 and PANC1 cells that were stably transfected with HMGA1 shRNA vectors exhibited reduced growth rates in nude mice compared with the growth in corresponding controls (tumors derived from Mia-PaCa2 and PANC1 cells stably transfected with control shRNA vectors) during the 8-week period after implantation (Fig. 4A,B). Stable knockdown of HMGA1 was confirmed by performing Western blot analysis on nuclear extracts of tumor xenografts (Fig. 4A and B). Immunohistochemical analysis of tumors that were harvested at the end of this observation period suggested that HMGA1 silencing was associated with an inhibition of tumor cell proliferation (Ki-67 reactivity) (Fig. 4C) and an increase in tumor apoptosis (TUNEL staining) (Fig. 4D).
FIGURE 4.
(A,B) Stable silencing of high mobility group AT-hook 1 (HMGA1) resulted in significant attenuation in the growth of tumors derived from subcutaneous implantation of MiaPaCa2 cells and PANC1 cells in nude mice. Mice (n = 6 per group) were implanted subcutaneously with stably transfected cells (either a scrambled short-hairpin RNA [shRNA] control or an shHMGA1-1 plasmid). Subcutaneous tumor size was monitored weekly for 8 weeks. Stable HMGA1 silencing was confirmed by Western blot analysis of nuclear extracts from explanted xenograft tumors. Values are means (±standard error of the means). An asterisk indicates P < .05 versus control shRNA xenografts. Suppression of HMGA1 resulted in reduction of Ki-67 immunoreactivity in vivo. Each tumor slide was stained for Ki-67, and the numbers of Ki-67–positive cells were counted in at least 5 randomly selected fields at ×40 magnification. Representative tumor sections stained for Ki-67 immunoreactivity in MiaPaCa2 and PANC1 tumor xenografts are shown. An asterisk indicates P < .05 versus control shRNA trasnfectant-derived xenografts. (D) HMGA1 silencing led to increased apoptosis in tumor xenografts, as demonstrated on terminal deoxynucleotidyl transferase nick-end labeling (TUNEL) staining. TUNEL-positive cells were counted in at least 5 randomly selected fields at ×40 magnification in each xenograft slide. Representative tumor sections stained for TUNEL in MiaPaCa2 and PANC1 tumor xenografts are shown. An asterisk indicates P < .05 versus control shRNA transfectant-derived xenografts. Values are means (±standard deviation).
Modulation of HMGA1 expression had no impact on cellular proliferation in monolayer culture (data not shown). We previously demonstrated that modulation of HMGA1 expression did not affect the cellular proliferation in standard monolayer culture.12
DISCUSSION
At the time of diagnosis, most patients with pancreatic adenocarcinoma have metastatic or locally advanced disease that precludes surgical resection. Even among the few patients who are able to undergo successful resection, most are destined to die from recurrent cancer. Therefore, the identification of novel prognostic markers and molecular targets for this disease is of high priority. In this study, we focused on 1 such molecule: HMGA1. First, we examined the clinical relevance of HMGA1 expression in pancreatic adenocarcinomas. HMGA1 expression was observed in >90% of pancreatic adenocarcinoma specimens. Although HMGA1 expression was not associated significantly with any of the clinicopathologic variables that were studied, negative HMGA1 status predicted improved survival in patients with pancreatic cancer, and this correlation persisted even after adjusting for other confounding variables. More important, our study demonstrated that HMGA1 expression may help to identify subsets of patients with distinct clinical outcomes despite similar pathologic characteristics. The median survival was significantly longer (up to 3-fold) in HMGA1-negative patients than in HMGA1-positive patients. Although it has been demonstrated that HMGA1 is indicative of a poor prognosis in patients with other cancers, our current study is novel because, to our knowledge, it is the first study to demonstrate that HMGA1 is an independent prognostic indicator in patients with pancreatic adenocarcinoma. Clearly, the identification of HMGA1 as a prognostic indicator potentially may be useful, because it allows the identification of patients who would benefit from more aggressive treatment of their disease. Although our current study represents 1 of the largest immunohistochemical studies of pancreatic adenocarcinoma, our relatively modest sample size limits interpretation beyond the results presented. Our preliminary data on the value of HMGA1 as a prognostic marker is promising and warrants future study with a larger series of pancreatic cancer patients.
Having established the clinical relevance of HMGA1 in patients with pancreatic cancer, we embarked on studies to elucidate the roles of HMGA1 in the aggressive phenotype of pancreatic cancer cells by using in vitro and in vivo experiments. In this study, our findings indicate that HMGA1 promotes anchorage-independent proliferation by pancreatic cancer cells in vitro and tumorigenesis in vivo. In contrast, our previous observation suggested that modulating HMGA1 expression in pancreatic cancer cells had no impact on their proliferation under standard monolayer culture conditions.12 These results suggest that HMGA1 does not act simply as a mitogenic stimulus. The findings are not surprising, because the kinetics of 3-dimensional colony formation in vitro more closely approximate those of in vivo tumor growth than cells in monolayer culture.16 The normal cellular response to deprivation from appropriate contact with substratum is to undergo apoptosis, which, in this context, is termed anoikis.17 A defining feature of transformed cells is resistance to anoikis and the ability to proliferate under anchorage-independent conditions (eg, in soft agar).18–20 Anchorage-independent growth probably is a result of the ability to proliferate in absence of substratum and to resist apoptosis because of the loss of substratum. To address the question of which aspect is modulated by HMGA1, our group previously investigated the specific roles of HMGA1 in anoikis by investigating the apoptotic status of pancreatic adenocarcinoma cells grown in polyhydroxyethylmethacrylate-coated plates.12 We observed that HMGA1 overexpression promoted anoikis resistance in pancreatic adenocarcinoma cells. Taken this together, the findings indicate that HMGA1 mediates its function in tumorigenesis through 2 aspects: 1) by enhancing resistance to anoikis, as demonstrated in our previous study,12 and 2) by allowing continued 3-dimensional proliferation, as demonstrated in the current study. Our in vivo data provide corroborating findings: HMGA1 silencing was associated with reductions in cellular proliferation (Ki-67 index) and increases in apoptosis (TUNEL staining), corresponding to overall reductions in tumor growth.
Our study further builds on this observation by defining a novel mechanism through which HMGA1 mediates anchorage-independent cellular proliferation: PI-3K/Akt signaling. Previously, we demonstrated that HMGA1 silencing is associated with reductions in Akt phosphorylation at Ser473 (a marker of Akt activation), whereas forced HMGA1 overexpression is associated with increases in Akt kinase activity and in Akt phosphorylation. Neither HMGA1 silencing nor HMGA1 overexpression had any impact on levels of total Akt expression.11 Our current data clearly demonstrate that intact PI-3K/Akt signaling is necessary for HMGA1 overexpression to promote colony formation in soft agar. The findings also demonstrate that constitutively active PI-3K/Akt signaling is sufficient to maintain the capacity for colony formation in soft agar in the context of HMGA1 silencing. In our previous study, we confirmed the effects of modulating HMGA1 expression on the functional status of Akt-dependent pathways by assessing its effects on mammalian target rapamycin (mTOR) phosphorylation, a well known downstream target of Akt.11 We observed that modulation of HMGA1 expression has a direct effect on mTOR phosphorylation, indicating that HMGA1 does have a functional effect on the PI3-K/Akt/mTOR pathway. Although the mechanisms by which HMGA1 modulates the activity of PI3-K/Akt pathway remain unknown, clues can be obtained from a previous study that identified which genes are regulated by HMGA1 using cDNA microarray analysis.21 Among the list of genes, multiple fibroblast growth factor (FGF) pathway components (eg, FGF receptor 1 [FGFR1], FGF2b, FGF6, FGF7, and FGF9) appear to be regulated positively by HMGA1. It is plausible that induction of the FGF pathway, by binding of FGF to its receptors, could result in downstream stimulation of survival signaling pathways like the PI3-K/Akt pathway, as demonstrated in this study.22
Our group11 and others23 previously described a role for HMGA1-dependent MEK/ERK signaling. Thus, in the current study, we tested the effects of inhibiting this pathway. We observed that the inhibition of this pathway with the small-molecule inhibitor PD98059 had no impact on the proliferation in soft agar of cells with ectopic HMGA1 overexpression. MEK/ERK inhibitor did not even have a basal effect (as demonstrated by control cells after inhibition of MEK/ERK) in cells that overexpressed HMGA1. These findings suggest that HMGA1 overexpression reduced the sensitivity of the cells to MEK/ERK inhibition. First, this implies that HMGA1-induced colony formation is not dependent on the ERK pathway and highlights the relative unimportance of this pathway in the context of HMGA1 over-expression. Second, HMGA1 overexpression likely has effects on several pro-oncogenic pathways (1 of which is described in this study: the PI3-K/Akt pathway). The effects on these other pathways may be more crucial in promoting soft agar growth and, hence, rendering the inhibition of a single pathway like the MEK/ERK pathway ineffective in reducing colony formation. In our previous study,11 we demonstrated that HMGA1 silencing had no impact on ERK phosphorylation, although HMGA1 silencing clearly resulted in reductions in cellular proliferation in soft agar. Taken together, these findings suggest that HMGA1-induced cellular proliferation in soft agar growth is independent of MEK/ERK signaling.
It is clear now that HMGA1 exerts its effects not only by altering the conformational structure of DNA. More important, accumulating evidence suggests that HMGA1 exerts its functions by other mechanisms. Recent study has suggested that HMGA1 is capable of inhibiting the functions of p53, a well known oncosuppressor gene, by cytoplasmic relocalization of its proapoptotic activator HIPK2.24,25 Nuclear HMGA1 also has been described to directly influence mitochondrial functions.26 Further study is likely to reveal increasing complexity in the mechanisms that mediate the biologic actions of HMGA1.
In summary, HMGA1 represents a novel prognostic marker in pancreatic cancer. Functionally, HMGA1 mediates tumor progression by promoting anchorage-independent proliferation by pancreatic cancer cells through a PI-3K/Akt-dependent mechanism. Given the minimal or absent expression of HMGA1 in normal adult tissues, HMGA1 warrants further investigation as a tumor cell-specific therapeutic target.
Acknowledgments
S.-S. Liau is in receipt of the International Hepato-Pancreato-Biliary Association (IHPBA) Kenneth W. Warren Fellowship, the Pancreatic Society of Great Britain and Ireland Traveling Fellowship, an Aid for Cancer Research Grant, and Cancer Research UK Core Skills Bursary.
Supported by grants from the National Institutes of Health (RO1 CA114103), the American Cancer Society (RSG-04221-01-CCE), and the National Pancreas Foundation.
We gratefully acknowledge the secretarial assistance of Jan D. Rounds.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Friedmann M, Holth LT, Zoghbi HY, Reeves R. Organization, inducible-expression and chromosome localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res. 1993;21:4259–4267. doi: 10.1093/nar/21.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 4.John S, Reeves RB, Lin JX, et al. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-kappa B family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 6.Sarhadi VK, Wikman H, Salmenkivi K, et al. Increased expression of high mobility group A proteins in lung cancer. J Pathol. 2006;209:206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- 7.Chang ZG, Yang LY, Wang W, et al. Determination of high mobility group A1 (HMGA1) expression in hepatocellular carcinoma: a potential prognostic marker. Dig Dis Sci. 2005;50:1764–1770. doi: 10.1007/s10620-005-2934-9. [DOI] [PubMed] [Google Scholar]
- 8.Chiappetta G, Botti G, Monaco M, et al. HMGA1 protein overexpression in human breast carcinomas: correlation with ErbB2 expression. Clin Cancer Res. 2004;10:7637–7644. doi: 10.1158/1078-0432.CCR-04-0291. [DOI] [PubMed] [Google Scholar]
- 9.Czyz W, Balcerczak E, Jakubiak M, Pasieka Z, Kuzdak K, Mirowski M. HMGI(Y) gene expression as a potential marker of thyroid follicular carcinoma. Langenbecks Arch Surg. 2004;389:193–197. doi: 10.1007/s00423-004-0479-6. [DOI] [PubMed] [Google Scholar]
- 10.Abe N, Watanabe T, Masaki T, et al. Pancreatic duct cell carcinomas express high levels of high mobility group I(Y) proteins. Cancer Res. 2000;60:3117–3122. [PubMed] [Google Scholar]
- 11.Liau SS, Jazag A, Whang EE. HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 2006;66:11613–11622. doi: 10.1158/0008-5472.CAN-06-1460. [DOI] [PubMed] [Google Scholar]
- 12.Liau SS, Jazag A, Ito K, Whang EE. Overexpression of HMGA1 promotes anoikis resistance and constitutive Akt activation in pancreatic adenocarcinoma cells. Br J Cancer. 2007;26(96):993–1000. doi: 10.1038/sj.bjc.6603654. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Liau SS, Ashley SW, Whang EE. Lentivirus-mediated RNA interference of HMGA1 promotes chemosensitivity to gemcitabine in pancreatic adenocarcinoma. J Gastrointest Surg. 2006;10:1254–1262. doi: 10.1016/j.gassur.2006.06.011. discussion 1263. [DOI] [PubMed] [Google Scholar]
- 14.Yao Z, Okabayashi Y, Yutsudo Y, Kitamura T, Ogawa W, Kasuga M. Role of Akt in growth and survival of PANC-1 pancreatic cancer cells. Pancreas. 2002;24:42–46. doi: 10.1097/00006676-200201000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Tong WG, Ding XZ, Talamonti MS, Bell RH, Adrian TE. LTB4 stimulates growth of human pancreatic cancer cells via MAPK and PI-3 kinase pathways. Biochem Biophys Res Commun. 2005;335:949–956. doi: 10.1016/j.bbrc.2005.07.166. [DOI] [PubMed] [Google Scholar]
- 16.Demicheli R, Foroni R, Ingrosso A, Pratesi G, Soranzo C, Tortoreto M. An exponential-Gompertzian description of LoVo cell tumor growth from in vivo and in vitro data. Cancer Res. 1989;49:6543–6546. [PubMed] [Google Scholar]
- 17.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430:1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 19.Yawata A, Adachi M, Okuda H, et al. Prolonged cell survival enhances peritoneal dissemination of gastric cancer cells. Oncogene. 1998;16:2681–2686. doi: 10.1038/sj.onc.1201792. [DOI] [PubMed] [Google Scholar]
- 20.Takaoka A, Adachi M, Okuda H, et al. Anti-cell death activity promotes pulmonary metastasis of melanoma cells. Oncogene. 1997;14:2971–2977. doi: 10.1038/sj.onc.1201147. [DOI] [PubMed] [Google Scholar]
- 21.Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol. 2001;21:575–594. doi: 10.1128/MCB.21.2.575-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wente W, Efanov AM, Brenner M, et al. Fibroblast growth factor-21 improves pancreatic beta-cell function and survival by activation of extracellular signal-regulated kinase 1/2 and Akt signaling pathways. Diabetes. 2006;55:2470–2478. doi: 10.2337/db05-1435. [DOI] [PubMed] [Google Scholar]
- 23.Treff NR, Pouchnik D, Dement GA, Britt RL, Reeves R. High-mobility group A1a protein regulates Ras/ERK signaling in MCF-7 human breast cancer cells. Oncogene. 2004;23:777–785. doi: 10.1038/sj.onc.1207167. [DOI] [PubMed] [Google Scholar]
- 24.Pierantoni GM, Rinaldo C, Mottolese M, et al. High-mobility group A1 inhibits p53 by cytoplasmic relocalization of its proapoptotic activator HIPK2. J Clin Invest F. 2007;117:693–702. doi: 10.1172/JCI29852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Frasca F, Rustighi A, Malaguarnera R, et al. HMGA1 inhibits the function of p53 family members in thyroid cancer cells. Cancer Res. 2006;66:2980–2989. doi: 10.1158/0008-5472.CAN-05-2637. [DOI] [PubMed] [Google Scholar]
- 26.Dement GA, Maloney SC, Reeves R. Nuclear HMGA1 non-histone chromatin proteins directly influence mitochondrial transcription, maintenance, and function. Exp Cell Res. 2007;313:77–87. doi: 10.1016/j.yexcr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]