Abstract
Oligoarginine and guanidinium-rich molecular transporters have been shown to facilitate the intracellular delivery of a diverse range of biologically relevant cargos. Several such transporters have been suggested to interact with cell surface heparan sulfate proteoglycans as part of their cell entry pathway. Unlike other guanidinium-rich transporters, the cellular uptake of guanidinoglycosides at nanomolar concentrations is exclusively heparan sulfate dependent. As distinct cells differ in their expression levels and/or composition of cell-surface heparan sulfate proteoglycans, one may be able to exploit such differences to selectively target certain cell types. To systematically investigate the nature of their cell surface interactions, monomeric and dimeric guanidinoglycosides were synthesized using neomycin, paromomycin, and tobramycin as scaffolds. These transporters differ in the number and 3-dimensional arrangement of guanidinium groups. Their cellular uptake was measured by flow cytometry in wild type and mutant Chinese hamster ovary cells after generating the corresponding fluorescent streptavidin-phycoerythrinCy5 conjugates. All derivatives showed negligible uptake in mutant cells lacking heparan sulfate. Decreasing the number of guanidinium groups diminished uptake, but the three dimensional arrangement of these groups was less important for cellular delivery. Whereas conjugates prepared with the monomeric carriers showed significantly reduced uptake in mutant cells expressing heparan sulfate chains with altered patterns of sulfation, conjugates prepared with the dimeric guanidinoglycosides could overcome this deficiency and maintain high levels of uptake in such deficient cells. This finding suggests that cellular uptake depends on the valency of the transporter and both the content and arrangement of the sulfate groups on the cell surface receptors. Competition studies with chemically desulfated or carboxy-reduced heparin derivatives corroborated these observations. Taken together, these findings show that increasing the valency of the transporters retains heparan sulfate specificity and provides reagents that could distinguish different cell types based on the specific composition of their cell surface heparan sulfate proteoglycans.
Keywords: Heparan sulfate, Cellular uptake, Molecular transporters, Molecular recognition, Cooperative effects
Introduction
High molecular weight biomolecules, such as certain proteins and nucleic acids, display therapeutic potential.[1–4] Their limited cellular uptake, however, hampers their utility and has prompted the development of diverse delivery technologies including viromes,[5] liposomes,[6] lipoplexes,[7] and molecular transporters.[8] The latter largely relied on arginine-containing peptides[9] and the installation of guanidinium groups on diverse polyfunctional scaffolds derived from peptides,[10] peptoids,[11] carbohydrates,[12] and dendrimers.[13] These guanidinium-rich transporters have been demonstrated to effectively deliver otherwise non-permeable cargos.[2, 14–16] Their mechanism of cell entry is, however, not fully understood. Multiple uptake pathways are likely to operate, including clathrin-mediated endocytosis and macropinocytosis.[17–27]
Association of guanidinium-rich transporters with heparan sulfate has been observed, suggesting a significant role for this abundant cell surface glycosaminoglycan in either recognition or internalization of these carriers.[3, 28–31] The prominent arginine-rich oligomers (e.g., Arg9) display cellular uptake, albeit less effectively, in heparan sulfate-deficient cell lines, indicating the contribution of heparan sulfate-independent entry pathways.[25, 32] In this respect, guanidinylated aminoglycosides or guanidinoglycosides, a family of synthetic derivatives where all the ammonium groups of aminoglycoside antibiotics were converted into guanidinium groups, stand out.[33–34] At low carrier concentrations, their uptake is exclusively heparan sulfate-dependent, suggesting unique cell-surface interactions with these multivalent cell surface receptors.[32]
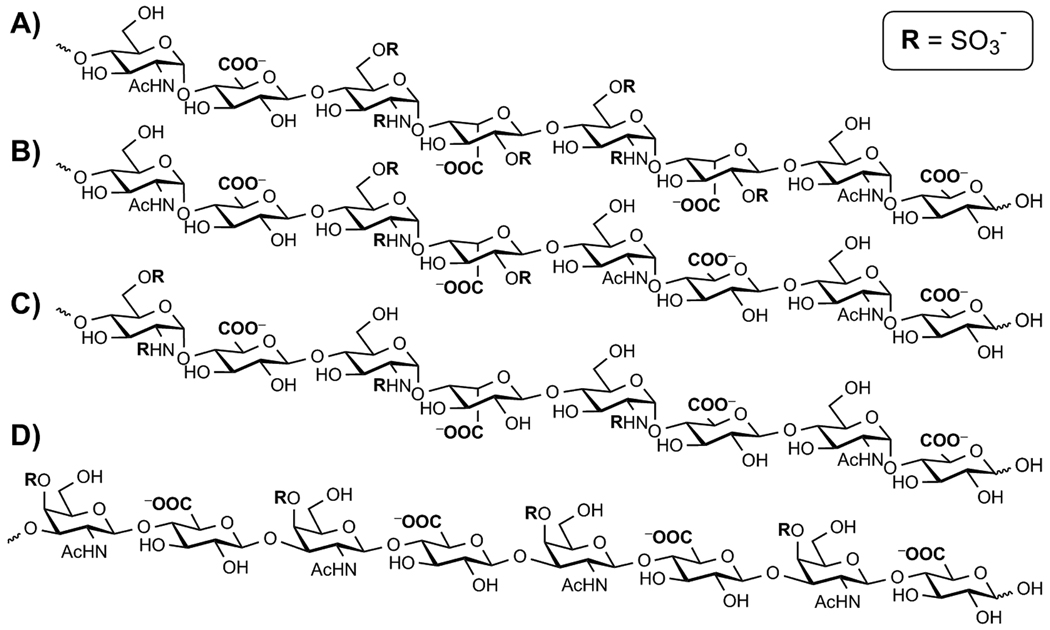
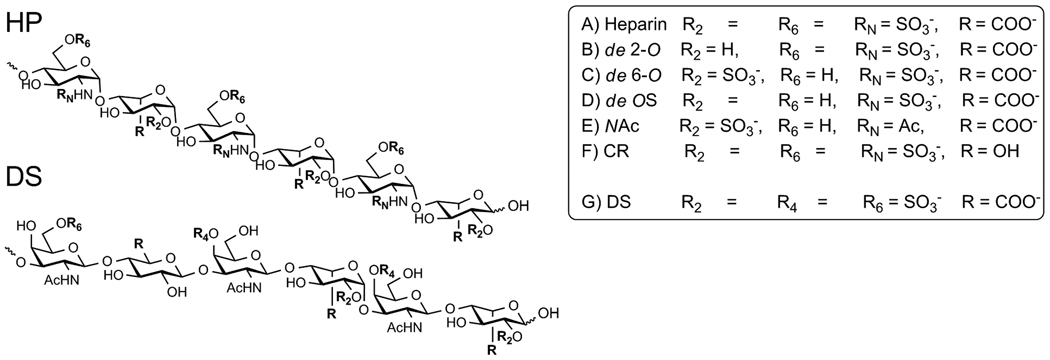
Heparan sulfate proteoglycans are expressed by virtually all multicellular organisms and play essential roles in human physiology.[35] Extending from a core protein, heparan sulfate glycosaminoglycans protrude into the extracellular matrix, coating the cell. Heparan sulfate itself is a linear polymer comprised of repeating disaccharide units of glucosamine and uronic acid, which are heterogeneously N-and O-sulfated (Scheme 1). While the overarching structure is conserved, individual heparan sulfate chains maintain a high level of diversity, differing in chain length, extent of sulfation, and degree of epimerization.[36] Furthermore, distinct cells differ in their expression levels and/or composition of cell-surface heparan sulfate proteoglycans.[35] This may allow one to exploit differences in this proteoglycan landscape for selective targeting by guanidinoglycosides, provided that their cell-surface interactions are better understood.
Scheme 1.
Representative hexasaccaride segments of heparan sulfate oligosaccharides as they are expressed in the cell lines utilized: A) wild type heparan sulfate from CHO K1 cells, B) pgsE, C) pgsF whereas D) pgsD, are represented by chondroitin sulfate as these mutants do not express heparan sulfate. All negatively charged moieties are highlighted in bold. Note that although only a 6-saccharide segment is schematically depicted, full length heparan sulfate is heterogeneous and typically 40–60 saccharide units.[36]
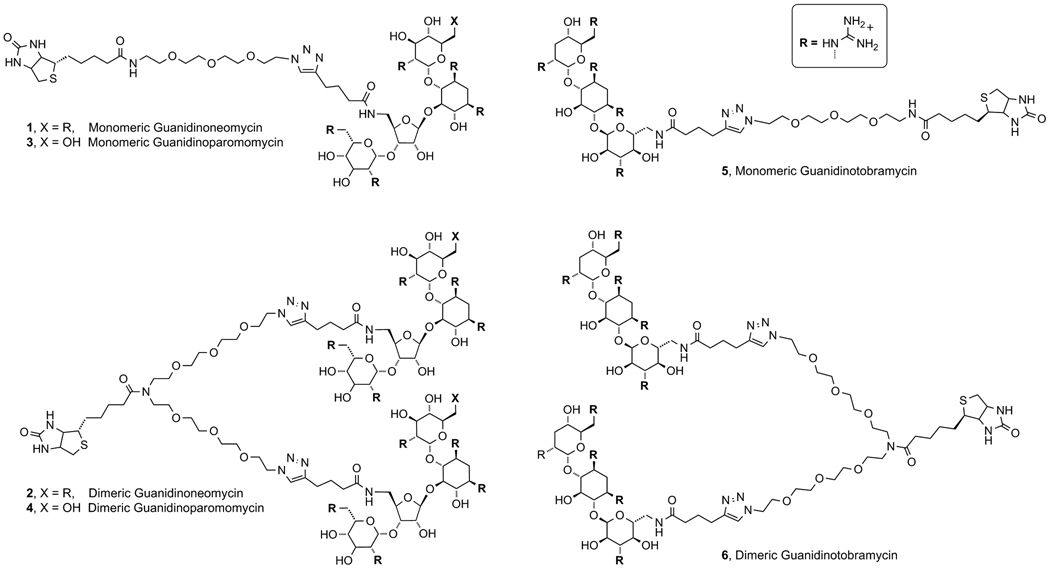
In this report we explore the impact and significance of three key parameters on cellular uptake: a) The number and spatial distribution of guanidinium groups on guanidinoglycosides, b) the degree and pattern of sulfate groups on cell-surface glycosaminoglycans, and c) potential cooperativity governed by the guanidinoglycoside scaffold, being either monomeric or dimeric. We have synthesized both monomeric and dimeric guanidinoglycoside carriers, derived from three different aminoglycosides (Scheme 2). Their uptake was evaluated in five unique Chinese hamster ovary cell lines, each differing in its expression of heparan sulfate, with the aim of deciphering the nature of heparan sulfate-guanidinoglycoside interactions and the possibility of exploiting their specificity to increase the efficacy and versatility of the guanidinoglycoside transporter. Our findings illustrate the significance of the number of guanidinium groups on uptake, although their spatial distribution plays a relatively minor role. Importantly, the dimeric guanidinoglycoside derivatives maintain considerably higher levels of cellular uptake than the monomeric carriers in cells expressing poorly sulfated heparan sulfate chains.
Scheme 2.
The guanidinoglycoside scaffolds utilized in both monomeric and dimeric forms, compounds 1–6.
Results and Discussion
To explore the subtle features of the heparan sulfate-selective uptake of guanidinoglycosides, we evaluated the impact of the number and arrangement of guanidinium groups in both monomeric and dimeric guanidinoglycoside constructs on their cellular uptake. The use of cell lines expressing biosynthetically altered heparan sulfate,[36] in addition to cell lines that do not express heparan sulfate, facilitated the systematic investigation of the fundamental interactions between this important cell surface charged biopolymer and these unique carriers.
Carrier Design
To explore differences between monomeric and dimeric guanidinoglycosides, structurally related, flexible, and water soluble monomeric and dimeric linkers were prepared (Scheme 3A). Bifunctional poly-ethylene glycol (PEG) chains incorporating an azide and an amine were designed to facilitate the “clicking” of a guanidinoglycoside and the coupling of biotin, respectively. Conjugation of the biotinylated-guanidinoglycoside to a fluorescently labeled streptavidin enabled comparative flow cytometry measurements.[32] Note that under these conditions the monomeric guanidinoglycoside constructs become tetra-valent and the dimeric constructs yield octa-valent conjugates, as four molecules of biotin bind to a single streptavidin.
Scheme 3.
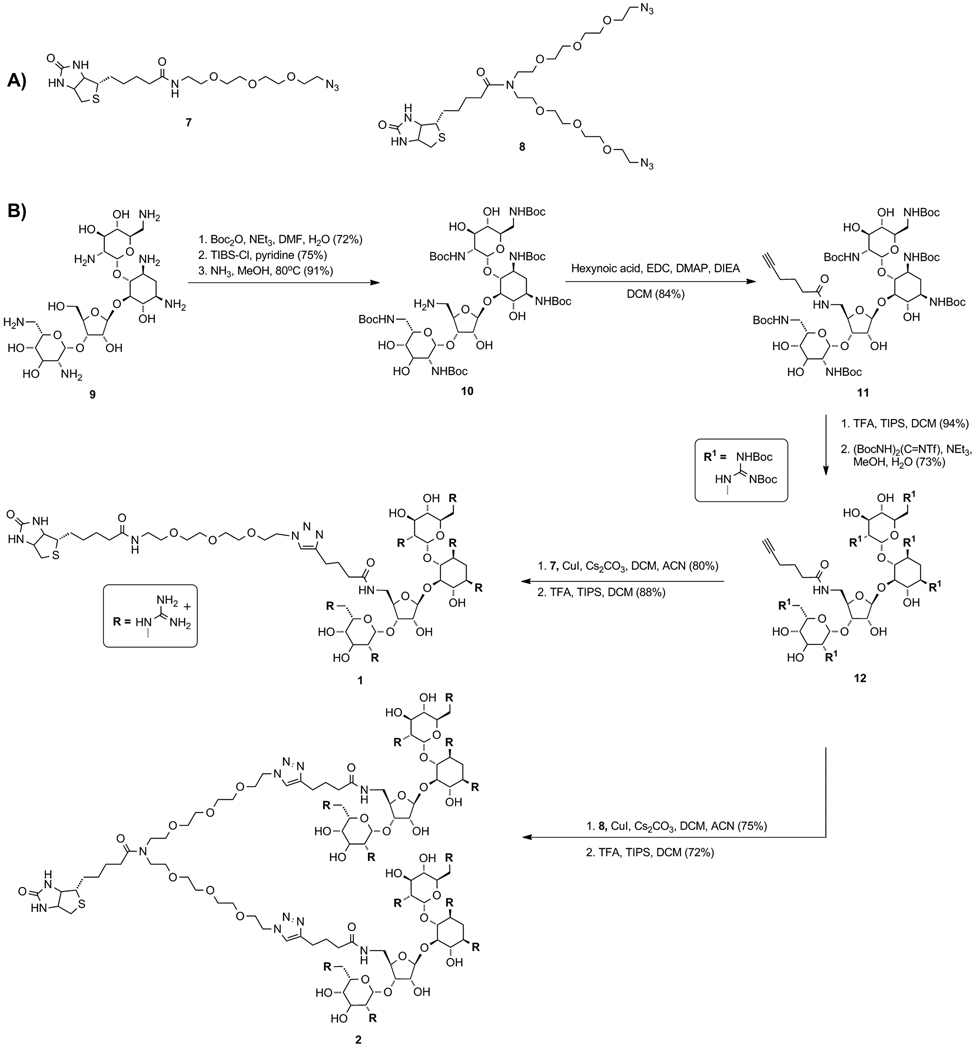
Synthesis of biotinylated monomeric and dimeric guanidinoneomycin carriers (1 and 2, respectively).
Synthesis of guanidinoglycoside conjugates
Using previously reported procedures, neomycin B (9) was Boc-protected and treated with 2,4,6-triisopropylbenzenesulfonyl chloride (TIBS-Cl) to selectively activate the primary hydroxymethyl group (Scheme 3B).[32, 37] Treatment with methanolic ammonia provided the aminomethyl derivative (10),[38–39] which was coupled to hexynoic acid using standard peptide coupling conditions to give 11. The Boc groups were removed using trifluoroacetic acid and the resulting modified aminoglycoside was treated with N,N’-di-tertbutoxycarbonyl-N’’-triflylguanidine to yield the protected guanidinoglycoside (12).[33–34] The alkyne-linked 12 was conjugated to linkers 7 or 8 using Cu(I)-mediated “click chemistry”, and subsequently treated with trifluoroacetic acid to give the corresponding monomeric (1) or dimeric construct (2) as a guanidinium-TFA salt. The guanidinoparomomycin (3, 4) and guanidinotobramycin derivatives (5, 6) were prepared in a similar manner (Figures S4 and S5).[40–42]
Selection of cell lines with varied heparan sulfate expression
Chinese hamster ovary cells (CHO-K1), expressing natural levels of heparan sulfate, as well as specific mutants, were utilized to investigate the effect of heparan sulfate-sulfation patterns on cellular uptake (Scheme 1A). Several well-behaved CHO mutants, including pgsA, pgsD, pgsE, and pgsF, were utilized to systemically evaluate the role of sulfation levels in the recognition/uptake of guanidinoglycosides. Two undersulfated mutants were employed, pgsE and pgsF, each deficient in specific enzymes that catalyze sulfation of the heparan sulfate oligosaccharide during its biosynthesis.[36] The pgsE mutants lack GlcNAc N-deacetylase/N-sulfotransferase resulting in poor N-sulfation and a significantly decreased level of overall sulfation (Scheme 1B).[43] Mutant pgsF cells are deficient in uronyl-2-O-sulfotransferase, which results in under expression of 2-O sulfation and a slightly increased level of 6-O sulfation (Scheme 1C).[44–46] Comparatively, the pgsE mutants are less sulfated than then the pgsF mutants. The two remaining cell lines serve as important controls, as neither pgsA nor pgsD express heparan sulfate.[47–49] The pgsD mutant cells, however, do express elevated levels of chondroitin sulfate (Scheme 1D), a related highly charged proteoglycan, which differs from heparan sulfate primarily in its uniform β-linked oligosaccharide chains.[36, 50]
Quantifying cellular uptake
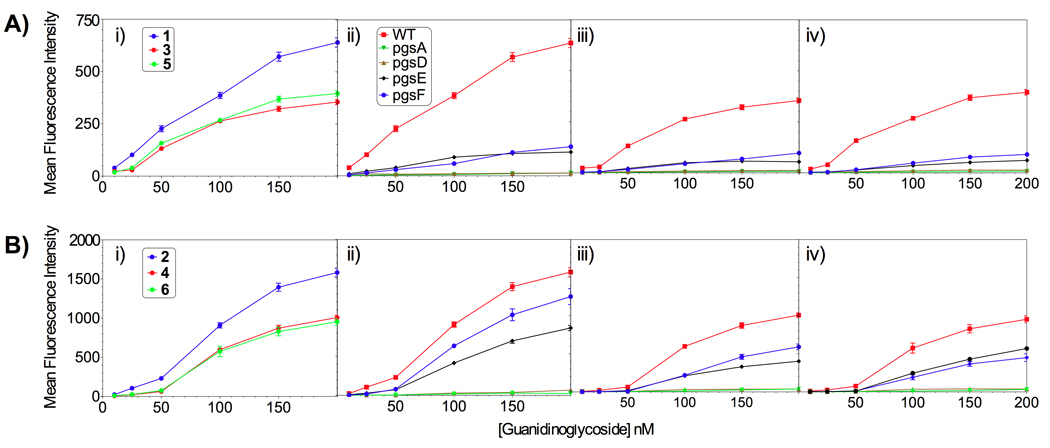
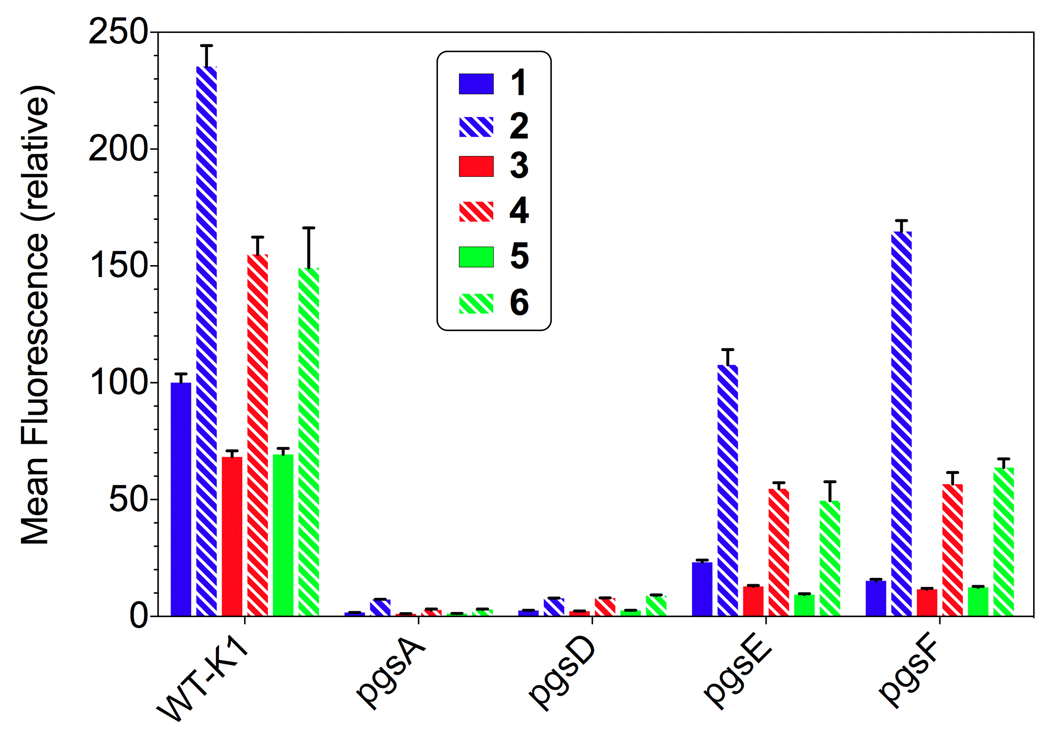
The biotinylated guanidinoglycosides were conjugated to streptavidin PE-Cy5 and diluted into F-12 media. Cells were incubated in a solution of the corresponding monomeric or dimeric transporter for 2 hours at 37°C and were then washed, detached using trypsin/EDTA, and analyzed using flow cytometry. Under these conditions, cell surface bound material is entirely cleaved and the FACS signal represents internalization of the conjugates.[51] For any given cell line, the mean fluorescence intensity was plotted against the concentration of the transporter. Figure 1 highlights the uptake data for the monomeric and dimeric constructs, while Figure 2 summarizes the data at 100 nM carrier concentration.
Figure 1.
Comparing the cellular uptake of (A) monomeric and (B) dimeric guanidinoglycoside constructs: i) CHO K1 (WT) cells, (ii) guanidinoneomycin, iii) guanidinoparomomycin, and iv) guanidinotobramycin against the cell lines tested.[52–53] Key: i) guanidinoneomycin (blue), guanidinoparomomycin (red), guanidinotobramycin (green), ii–iv) wild type CHO-K1 cells (red), pgsA (green), pgsD (brown), pgsE (blue), pgsF (black).
Figure 2.
Normalized cellular uptake at 100 nM comparing the monomeric (solid) and dimeric (dashed) constructs with respect to the a) scaffold and b) various cell lines investigated.[52] Key: guanidinoneomycin (blue), guanidinoparomomycin (red), and guanidinotobramycin (green).
Effect of number and distribution of guanidinium groups
Cellular internalization was dependent on the number of guanidinium groups on the guanidinoglycoside scaffolds. For both the monomeric and dimeric constructs, the uptake of guanidinoneomycin (1/2) was 30% higher than that of guanidinoparomomycin (3/4) and guanidinotobramycin (5/6) (Figure 1 Ai and Bi). This has been observed with other guanidinium containing transporters.[54–55] Additionally, the guanidinoparomomycin and guanidinotobramycin conjugates, for both the monomeric and dimeric constructs (compounds 3/5 and 4/6, respectively), demonstrated similar uptake behavior in all cell lines (Figure 1). As both guanidinoglycoside scaffolds contain five guanidinium groups, this observation suggests that the 3-dimensional architecture of the guanidinium groups plays a limited role in cellular uptake. The inability of the cells, and more specifically cell surface glycans, to distinguish between the different arrangements suggests that the binding/recognition inherent to the uptake process could be quite plastic. We note, however, that guanidinoglycosides, in contrast to linear guanidinium-rich carriers such as HIV TAT and poly-Arg, possess a high density of charged groups, and that the subtle differences in the architecture of these transporters might have been overshadowed by the inherent tetrameric nature of the streptavidin core.
Significance of glycan sulfation levels
Heparan sulfate deficient pgsA and pgsD mutant cells showed poor uptake, less than 5%, compared to uptake in wild type CHO cells (Figure 1). This observation confirms the heparan sulfate-dependent nature of guanidinoglycoside cellular uptake observed before using different constructs, demonstrating that this is an inherent trait of the guanidinoglycoside scaffold and is not linker or construct dependent. For the monomeric constructs (Figure 1A), uptake in the undersulfated pgsE and pgsF cells was reduced to less than 20% of that observed in the wild type cells. This behavior was not observed for the dimeric constructs (Figure 1B), as high uptake levels, between 50–75% compared to the uptake seen in wild type cells, were observed in both pgsE and pgsF mutant cells. These trends were observed at all concentrations tested, and suggest that there is a significant relationship between glycan sulfation and either the level of guanidinylation or the carrier valency.
Dimeric constructs illicit cooperative response
Minimal internalization was observed at concentrations lower than 50 nM (particularly for the guanidinoparomomycin and guanidinotobramycin derivatives) suggesting a switch-like or cooperative mechanism (Figure 1). Possibly, the enhanced uptake observed as concentration increases is due to aggregation of proteoglycan receptors on the cell surface and/or the activation of endocytosis.[56–57] This suggests that a critical concentration of the transporter might be necessary to induce effective uptake. The multivalent and heterogeneous nature of heparan sulfate makes it difficult to determine the underlying mechanism. Interestingly, this cooperative-like effect was accentuated in the dimeric constructs, consistent with the idea that the guanidinoglycosidic ligand induces clustering of heparan sulfate proteoglycans.
In wild type cells, all the dimeric constructs showed 2.5-fold enhanced uptake over the corresponding monomeric carriers, while a 5-fold increase was observed in the undersulfated cell lines pgsE and pgsF at 100 nM concentrations (Figure 2). For the guanidinoneomycin constructs (1 and 2), a 10-fold increase in uptake of the dimeric construct over monomeric was observed in pgsF cells at the same concentration (Figure 2). This trend demonstrates that the dimeric constructs are increasingly able to overcome undersulfation and maintain high levels of cellular uptake. Likely, this behavior is due to the apparent increase in avidity of the dimeric constructs, enabling cooperative cell-surface binding.[58] The modest increase in uptake of the dimers in wild type cells compared to their monomeric counterparts speaks to the efficiency of the guanidinoglycoside-mediated uptake mechanism. A guanidinoglycoside monomer appears to enter the cell with high efficiency, negating the contribution from the second arm of the dimeric constructs when heparan sulfate is abundant. This is reinforced by the increased response in the undersulfated cells, where the poor sulfation is insufficient to facilitate adequate uptake of the monomeric carriers. In this case, the dimeric constructs demonstrate a chelate-like effect enabling increased interactions between the carrier and cell surface glycans, thus facilitating effective uptake.
Competition experiments using model oligomeric glycans
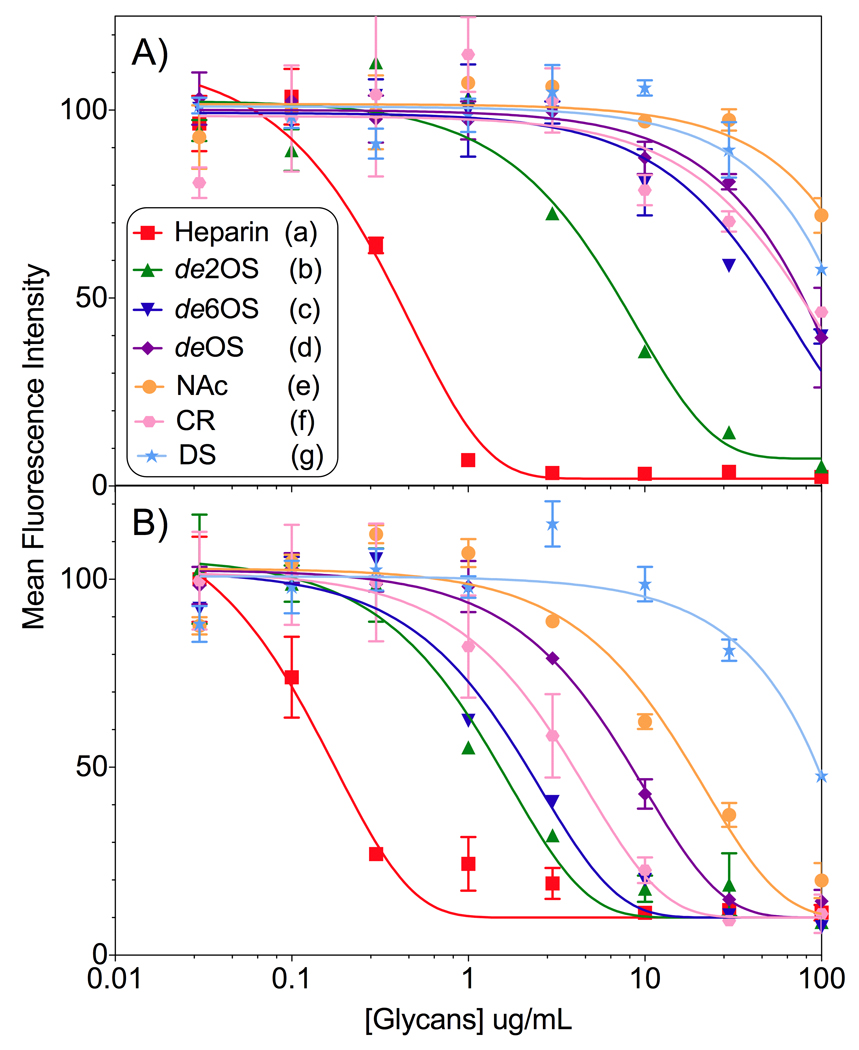
To further assess the significance of glycan sulfation levels and support the observations described above, the cellular uptake of the guanidinoneomycin constructs, compounds 1 and 2, was evaluated in the presence of competing model glycans derived from heparin. In this fashion, coloring of the results due to subtle variations between mutant cell lines, in addition to the specific glycosaminoglycan modification as described above, can be eliminated. Specifically, heparin, de 2-O sulfated heparin, de 6-O sulfated heparin, de O-sulfated heparin, N-desulfo-N-acetylated heparin, and heparin containing reduced uronic acids were tested (Scheme 4). Dermatan sulfate, an analog of chondroitin sulfate which is over expressed in the pgsD mutant cells, was also explored as a competitor.[36, 48] Wild type cells were treated with increasing concentrations of the competing glycans, along with a fixed concentration of the guanidinoneomycin conjugates, 1 and 2 respectively, and the cellular uptake was quantified using flow cytometry as outlined above. The mean fluorescence intensity was plotted against the concentration of the glycans utilized (Figure 3), and IC50 values were extracted (Table 1).
Scheme 4.
Competition studies were performed with glycans derived from heparin (HP) or dermatan sulfate (DS), each differing in its sulfation pattern as well as other key features. A–F) Modified heparin derivatives, and g) dermatan sulfate. Note that full length glycans were utilized (typically 12 kDa, ca. 40–60 saccharide units), although only a 6-saccharide segment is schematically depicted.[36]
Figure 3.
Normalized inhibition curves for A) monomeric guanidinoneomycin 1 and B) dimeric guanidinoneomycin 2. See Scheme 4 for structures of glycans A–G.[52] Key: heparin (red), de 2-O sulfated heparin (green), de 6-O sulfated heparin (navy), de O sulfated heparin (purple), N-Acetylated heparin (orange), carboxy reduced heparin (pink), and dermatan sulfate (teal).
Table 1.
IC50values for soluble competitors inhibiting the uptake of monomeric and dimeric guanidinoneomycin 1 and 2, respectively (in µg/mL). See Scheme 4 for structures of glycans A–G.
| Glycan | 1 | 2 |
|---|---|---|
| Heparin (A) | 0.33 (± 0.1) | 0.11 (± 0.07) |
| de 2-O sulfate (B) | 6.3 (± 3.5) | 0.94 (± 0.8) |
| de 6-O sulfate (C) | > 100 | 1.7 (± 0.8) |
| de O sulfate (D) | > 100 | 6.5 (± 2) |
| N-Acetyl (E) | > 100 | 11 (± 10.5) |
| Carboxy reduced (F) | > 100 | 3.2 (± 3) |
| Dermatan sulfate, DS (G) | > 100 | > 100 |
Reaffirming observed cooperativity
Inhibition of guanidinoglycoside uptake with native heparin elicited the most pronounced inhibitory response (Figure 3), highlighting a 3-fold increase of sensitivity of the dimeric over the monomeric construct. This appears intuitive as heparin contains the highest sulfation levels of the competing glycans, and mirrors the modest cooperative affect observed for cellular uptake in wild type cells. For monomeric guanidinoneomycin 1, only heparin and de 2-O sulfate heparin showed inhibition at the tested concentrations.[57] For dimeric guanidinoneomycin 2, inhibition was observed for all the heparin derivatives. Dermatan sulfate failed to inhibit cellular uptake of either construct. The inhibition response from undersulfated heparin glycans demonstrated a dramatic 10-fold difference in efficiency between constructs. These observations parallel the uptake data, which showed that the dimeric scaffold increases the ability of the guanidinoglycoside to interact with undersulfated cell surface glycans. Notably, the observed 3–10-fold cooperative effect between the monomeric and dimeric guanidinoneomycin constructs was similar for both the uptake and the competition experiments. Additionally, the competition experiment enabled quantification of the binding interactions between the conjugates and the model glycans, as well as between the monomeric and dimeric constructs.
Not all charge is created equal
The IC50 values obtained from the competition study indicate that all charged functional groups of the glycosaminoglycans are important for binding/recognition toward guanidinoglycosides. However, as indicated by the high level of inhibition maintained for de 2-O sulfate heparin (Figure 3), some positions and functional groups may be more vital than others. This is likely the result of the overall structure and presentation of these charged groups. Additionally, dermatan sulfate displays no significant inhibition, further supporting that there is little to no interaction between this glycan and guanidinoglycosides. This corroborates and complements the lack of uptake observed in the pgsD mutant cells, which overexpress an analogous glycan. As the sulfation levels of heparan sulfate and dermatan/chondroitin sulfate are similar, our observations suggest that the interactions governing this highly selective recognition process are more complicated than strict electrostatic interactions. Moreover, these glycans differ significantly in their incorporations of α versus β glycosidic linkages, suggesting that the folded 3-dimensional structures of the glycans, and not the transporter scaffold, are of prominence in governing the uptake process of guanidinoglycosides.[3] This hypothesis could explain the perceived disparity in the field of guanidinium rich transports, as different carriers have shown distinct degrees of involvement by these glycans during the cellular uptake process.[59–60]
Implications and Summary
Taken together, the data demonstrate that cellular uptake of the monomeric guanidinoglycoside constructs is limited in cells expressing undersulfated heparan sulfate, whereas the additional cell surface binding ability of the dimeric guanidinoglycoside transporters enables them to overcome these deficiencies. This trend suggests that by manipulating the valency of the transporter, one can discern the “quality” of cell surface heparan sulfate chains. Additionally, the concept of glycan sulfation affecting overall uptake may have universal significance, as it suggests an additional variable when evaluating the behavior of molecular transporters. This implies that the cell lines utilized in uptake experiments can have a profound effect on the outcome, as different cell lines express varying amounts of heparan sulfate proteoglycans.[35] These finding highlight the versatility and promise of a guanidinoglycoside-based molecular transporter, as well as the increased applicability involved in targeting cell surface heparan sulfate as a gateway into the cell.
Methods
Materials
Unless otherwise specified, materials obtained from commercial suppliers were used without further purification. Neomycin, paromomycin, and tobramycin were purchased as their sulfate salts from Sigma-Aldrich and were converted into the corresponding neutral form by passing through DOWEX MONOSPHERE 550 Å (OH) anion exchange resin. Deuterated NMR solvents were purchased from Cambridge Isotope Laboratories (Andover, MA). The heparin derivatives utilized in the competition study were purchased from Neoparin. PBS (Dulbecco’s phosphate buffered saline), HBSS (Hanks’ balanced salt solution), and F-12K Media were purchased from Invitrogen (Gibco). FACS buffer (Isotonic solution 0.85% w/v, phosphate buffered pH 7.1–7.3) and streptavidin-PE-Cy5 were purchased from BD Biosciences. Trypsin/EDTA was purchased from VWR (Mediatech). The 24-well plates used are Costar 3524 (Corning).
Instrumentation
NMR spectra were recorded on either a Varian Mercury 400 MHz or 500 MHz spectrometer. Mass spectra were recorded at the UCSD Chemistry and Biochemistry Mass Spectrometry Facility, utilizing either an LCQDECA (Finnigan) ESI with a quadrapole ion trap or an MAT900XL (ThermoFinnigan) FAB double focusing mass spectrometer. Reversed phase HPLC (Vydac C18 column) purification and analysis were carried out using an Agilent 1200 series instrument. Flow cytometry studies were performed on a BD FACSCalibur, with excitation at 635 nm and emission monitored at 670 nm.
Synthesis
Detailed synthetic procedures and characterization data are described in the Supporting Information.
Quantifying cellular uptake
The guanidinoglycoside derivatives were dissolved into HBSS and treated with fluorescently labeled streptavidin (ST-PE-Cy5) in a 10:1 molar ratio. After 15 minutes, the unbound biotinylated-guanidinoglycoside conjugates were removed using a desalting spin column (Amicon Ultra-4 Centrifugal Filter with a 10 KDa threshold from Millipore), leaving only the guanidinoglycoside-streptavidin conjugate in the column. The purified conjugates were diluted into media to form 10, 25, 50, 100, 150, and 200 nM solutions. The media used in these experiments was treated with 10% fetal bovine serum (FBS).
150,000 cells were counted using a hemocytometer for each of the cell lines examined and transferred to 24-well plates, and incubated in 300 µL of Media (with a 1% solution of penicillin/streptomycin and 10% FBS) overnight at 37°C. The cells were then washed with PBS and treated with a 150 µL solution of the corresponding conjugate (in F-12 Media containing 10% FBS) and incubated for 2 hours. Following this, the cells were washed twice with PBS to remove any remaining extracellular conjugates. The cells were then detached with 50 µL trypsin/EDTA, diluted with 50 µL media and 200 µL FACS buffer, and analyzed using flow cytometry. Cellular uptake was quantified by the mean fluorescence intensity; the crude data was interpreted using FlowJo v8.8.6 wherein the median value was determined and later plotted and further analyzed using GraphPad Prism v5.0a.
Competition study
50,000 WT CHO-K1 cells were counted using a hemocytometer, seeded onto 48-well plates, and incubated in 300 µL of media (with a 1% solution of penicillin/streptomycin and 10% FBS) overnight at 37°C. The cells were then washed with PBS and treated with a 75 uL solution (in media with 10% FBS) of the various glycans. To each well, 75 uL of a 1.5 nM solution (in media with 10% FBS) of the guanidinoglycoside conjugates was added (note, the conjugates were prepared and purified in the same way as for the quantification study). The cells were then incubated for two hours, at which point they were washed twice with PBS and detached using 30 uL trypsin/EDTA. The lifted cells were diluted with 30 µL media and 200 µL of FACS buffer and analyzed by flow cytometry.
Supplementary Material
Acknowledgment
We thank the National Institutes of Health for the generous support (GM 077471), the National Science Foundation (instrumentation grant CHE-0741968), the Fulbright program for a fellowship (Lucile Fischer), and Yun Xie for her assistance with preparation of the manuscript.
Contributor Information
Jeffrey D. Esko, Email: jesko@ucsd.edu.
Yitzhak Tor, Email: ytor@ucsd.edu.
References
- 1.Dietz GPH. Curr. Pharm. Biotechnol. 2010;11:167–174. doi: 10.2174/138920110790909731. [DOI] [PubMed] [Google Scholar]
- 2.Gump JM, Dowdy SF. Trends in Mol. Med. 2004;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Nakase I, Takeuchi T, Tanaka G, Futaki S. Adv. Drug Delivery Rev. 2008;60:598–607. doi: 10.1016/j.addr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Wu W, Hsiao SC, Carrico ZM, Francis MB. Angew. Chem., -Int. Ed. 2009;48:9493–9497. doi: 10.1002/anie.200902426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinmetz NF, Mertens ME, Taurog RE, Johnson JE, Commandeur U, Fischer R, Manchester M. Nano Letters. 2010;10:305–312. doi: 10.1021/nl9035753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lentacker I, Geers B, Demeester J, De Smedt SC, Sanders NN. Mol. Ther. 2010;18:101–108. doi: 10.1038/mt.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pack DW, Hoffman AS, Pun S, Stayton PS. Nat. Rev. Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 8.Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. Adv. Drug Delivery Rev. 2008;60:452–472. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubikovskaya EA, Thorne SH, Pillow TH, Contag CH, Wender PA. Proc. Natl Acad. Sci. U.S.A. 2008;105:12128–12133. doi: 10.1073/pnas.0805374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potocky TB, Silvius J, Menon AK, Gellman SH. Chembiochem. 2007;8:917–926. doi: 10.1002/cbic.200600563. [DOI] [PubMed] [Google Scholar]
- 11.Cooley CB, Trantow BM, Nederberg F, Kiesewetter MK, Hedrick JL, Waymouth RM, Wender PA. J. Am. Chem. Soc. 2009;131:16401–16403. doi: 10.1021/ja907363k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiti KK, Lee WS, Takeuchi T, Watkins C, Fretz M, Kim DC, Futaki S, Jones A, Kim KT, Chung SK. Angew. Chem., -Int. Ed. 2007;46:5880–5884. doi: 10.1002/anie.200701346. [DOI] [PubMed] [Google Scholar]
- 13.Chung HH, Harms G, Seong CM, Choi BH, Min CH, Taulane JP, Goodman M. Biopolymers. 2004;76:83–96. doi: 10.1002/bip.10597. [DOI] [PubMed] [Google Scholar]
- 14.Hassane FS, Ivanova GD, Bolewska-Pedyczak E, Abes R, Arzumanov AA, Gait MJ, Lebleu B, Gariepy J. Bioconjugate Chem. 2009;20:1523–1530. doi: 10.1021/bc900075p. [DOI] [PubMed] [Google Scholar]
- 15.Maiti KK, Jeon OY, Lee WS, Chung SK. Chem. -- Eur. J. 2007;13:762–775. doi: 10.1002/chem.200600898. [DOI] [PubMed] [Google Scholar]
- 16.Wang YH, Chen CP, Chan MH, Chang M, Hou YW, Chen HH, Hsu HR, Liu K, Lee HJ. Biochem. Biophys. Res. Comm. 2006;346:758–767. doi: 10.1016/j.bbrc.2006.05.205. [DOI] [PubMed] [Google Scholar]
- 17.Brooks H, Lebleu B, Vives E. Adv. Drug Delivery Rev. 2005;57:559–577. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari A, Pellegrini V, Arcangeli C, Fittipaldi A, Giacca M, Beltram F. Mol. Ther. 2003;8:284–294. doi: 10.1016/s1525-0016(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 19.Futaki S. Adv. Drug Delivery Rev. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Prochiantz A. Adv. Drug Delivery Rev. 2005;57:491–493. doi: 10.1016/j.addr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Rothbard JB, Jessop TC, Wender PA. Adv. Drug Delivery Rev. 2005;57:495–504. doi: 10.1016/j.addr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Tyagi M, Rusnati M, Presta M, Giacca M. Biol. Chem. 2001;276:3254–3261. doi: 10.1074/jbc.M006701200. [DOI] [PubMed] [Google Scholar]
- 23.Vives E. J. Mol. Recognit. 2003;16:265–271. doi: 10.1002/jmr.636. [DOI] [PubMed] [Google Scholar]
- 24.Chung SK, Maiti KK, Lee WS. Int. J. Pharm. 2008;354:16–22. doi: 10.1016/j.ijpharm.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Gump JM, June RK, Dowdy SF. Biol. Chem. 2010;285:1500–1507. doi: 10.1074/jbc.M109.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Futaki S, Nakase I, Taclokoro A, Takeuchi T, Jones AT. Biochem. Soc. Trans. 2007;35:784–787. doi: 10.1042/BST0350784. [DOI] [PubMed] [Google Scholar]
- 27.Melikov K, Chernomordik L. Cell. Mol. Life Sci. 2005;62:2739–2749. doi: 10.1007/s00018-005-5293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Biol. Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 29.Ziegler A, Seelig J. Biophys. J. 2004;86:254–263. doi: 10.1016/S0006-3495(04)74101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Console S, Marty C, Garcia-Echeverria C, Schwendener R, Ballmer-Hofer K. Biol. Chem. 2003;278:35109–35114. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Futaki S, Niwa M, Tanaka S, Ueda K, Sugiura Y. Biol. Chem. 2002;277:2437–2443. doi: 10.1074/jbc.M110017200. [DOI] [PubMed] [Google Scholar]
- 32.Elson-Schwab L, Garner OB, Schuksz M, Crawford BE, Esko JD, Tor Y. Biol. Chem. 2007;282:13585–13591. doi: 10.1074/jbc.M700463200. [DOI] [PubMed] [Google Scholar]
- 33.Luedtke NW, Baker TJ, Goodman M, Tor Y. J. Am. Chem. Soc. 2000;122:12035–12036. [Google Scholar]
- 34.Baker TJ, Luedtke NW, Tor Y, Goodman M. J. Org. Chem. 2000;65:9054–9058. doi: 10.1021/jo001142e. [DOI] [PubMed] [Google Scholar]
- 35.Bishop JR, Schuksz M, Esko JD. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 36.Varki A, Cumming DR, Esko J, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. 2 ed. CSH Press; 2009. [PubMed] [Google Scholar]
- 37.Luedtke NW, Carmichael P, Tor Y. J. Am. Chem. Soc. 2003;125:12374–12375. doi: 10.1021/ja0360135. [DOI] [PubMed] [Google Scholar]
- 38.Wang NX, Yu AG, Wang GX, Zhang XH, Li QS, Li Z. Synthesis. 2007:1154–1158. [Google Scholar]
- 39.Ugarkar BG, Castellino AJ, DaRe JS, Ramirez-Weinhouse M, Kopcho JJ, Rosengren S, Erion MD. J. Med. Chem. 2003;46:4750–4760. doi: 10.1021/jm030230z. [DOI] [PubMed] [Google Scholar]
- 40.See Supporting Information for additional synthetic schemes and characterization data.
- 41.Michael K, Wang H, Tor Y. Bioorg. Med. Chem. 1999;7:1361–1371. doi: 10.1016/s0968-0896(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 42.Blount KF, Tor Y. Chembiochem. 2006;7:1612–1621. doi: 10.1002/cbic.200600109. [DOI] [PubMed] [Google Scholar]
- 43.Bame KJ, Zhang LJ, David G, Esko JD. Biochemical J. 1994;303:81–87. doi: 10.1042/bj3030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence R, Olson SK, Steele RE, Wang LC, Warrior R, Cummings RD, Esko JD. Biol. Chem. 2008;283:33674–33684. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai XM, Bame KJ, Habuchi H, Kimata K, Esko JD. Biol. Chem. 1997;272:23172–23179. doi: 10.1074/jbc.272.37.23172. [DOI] [PubMed] [Google Scholar]
- 46.Bai XM, Esko JD. Biol. Chem. 1996;271:17711–17717. doi: 10.1074/jbc.271.30.17711. [DOI] [PubMed] [Google Scholar]
- 47.Miao HQ, Fritz TA, Esko JD, Zimmermann J, Yayon A, Vlodavsky I. J. Cell. Biochem. 1995;57:173–184. doi: 10.1002/jcb.240570202. [DOI] [PubMed] [Google Scholar]
- 48.Lidholt K, Weinke JL, Kiser CS, Lugemwa FN, Bame KJ, Cheifetz S, Massague J, Lindahl U, Esko JD. Proc. Nat. Acad. Sci. U.S.A. 1992;89:2267–2271. doi: 10.1073/pnas.89.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The pgsA mutants lack the xylosyltransferase that initiates the two major sulfated polysaccharides in CHO cells, heparan sulfate and chondroitin/dermatan sulfate. Thus, both classes of glycosaminoglycans are missing. The cells also make sialylated oligosaccharides found on glycoproteins (Asn-linked and Ser-linked) and glycolipids, and a small amount of sulfated glycolipid, but the charge density of these relatively small glycans is quite different than the heavily sulfated, uronic acid containing glycosaminoglycans. Apparently, these other glycans do not bind GNeo based on flow cytometry data shown in the paper.
- 50.The pgsD mutants lack both N-acetylglucosaminyltransferase and glucuronosyltransferase, enzymes required for the polymerization of heparan sulfate chains.
- 51.Sarrazin S, Wilson B, Sly WS, Tor Y, Esko JD. Mol. Ther. 2010;18:1268–1274. doi: 10.1038/mt.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.See Supporting Information for additional graphics.
- 53.Note that in some cases the symbol size is larger than the error for a given data point.
- 54.Sakai N, Takeuchi T, Futaki S, Matile S. Chembiochem. 2005;6:114–122. doi: 10.1002/cbic.200400256. [DOI] [PubMed] [Google Scholar]
- 55.Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. J. Am. Chem. Sci. 2004;126:9506–9507. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- 56.Lambert S, Bouttier M, Vassy R, Seigneuret M, Petrow-Sadowski C, Janvier S, Heveker N, Ruscetti FW, Perret G, Jones KS, Pique C. Blood. 2009;113:5176–5185. doi: 10.1182/blood-2008-04-150342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorensen HP, Vives RR, Manetopoulos C, Albrechtsen R, Lydolph MC, Jacobsen J, Couchman JR, Wewer UM. Biol. Chem. 2008;283:31920–31932. doi: 10.1074/jbc.M804113200. [DOI] [PubMed] [Google Scholar]
- 58.Kawamura KS, Sung M, Bolewska-Pedyczak E, Gariepy J. Biochemistry. 2006;45:1116–1127. doi: 10.1021/bi051338e. [DOI] [PubMed] [Google Scholar]
- 59.Jiao CY, Delaroche D, Burlina F, Alves ID, Chassaing G, Sagan S. Biol. Chem. 2009;284:33957–33965. doi: 10.1074/jbc.M109.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakase I, Tadokoro A, Kawabata N, Takeuchi T, Katoh H, Hiramoto K, Negishi M, Nomizu M, Sugiura Y, Futaki S. Biochemistry. 2007:492–501. doi: 10.1021/bi0612824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.