Abstract
Rationale
Establishment of a functional vasculature requires the interconnection and remodeling of nascent blood vessels. Precise regulation of factors that influence endothelial cell (EC) migration and function is essential for these stereotypical vascular patterning events. The secreted Slit ligands and their Robo receptors constitute a critical signaling pathway controlling the directed migration of both neurons and vascular ECs during embryonic development, but the mechanisms of their regulation are incompletely understood.
Objective
To identify microRNAs regulating aspects of the Slit-Robo pathway and vascular patterning.
Methods and Results
Here we provide evidence that microRNA (miR) -218, which is encoded by an intron of the Slit genes, inhibits the expression of Robo1 and Robo2 and multiple components of the heparan sulfate (HS) biosynthetic pathway. Using in vitro and in vivo approaches, we demonstrate that miR-218 directly represses the expression of Robo1, Robo2, and glucuronyl C5-epimerase (GLCE), and that an intact miR-218–Slit–Robo regulatory network is essential for normal vascularization of the retina. Knockdown of miR-218 results in aberrant regulation of this signaling axis, abnormal EC migration and reduced complexity of the retinal vasculature.
Conclusions
Our findings link Slit gene expression to the post-transcriptional regulation of Robo receptors and heparan sulfate biosynthetic enzymes, allowing for precise control over vascular guidance cues influencing the organization of blood vessels during development.
Keywords: Angiogenesis, gene regulation, developmental biology
Introduction
Blood vessels and nerves are organized in nearly identical patterns throughout the body1, however the mechanisms that direct these disparate cell types to follow similar migratory tracts during development have not been fully defined. Recent studies have revealed that vessels and nerves possess a common set of transmembrane receptors that respond to molecular cues governing cell migration and pathfinding. These “axon guidance molecules” influence the behavior of both nerves and endothelial cells (ECs), resulting in similar patterning of each cell type 1, 2.
Roundabout (Robo) receptors and their Slit ligands function as crucial regulators of axon and vascular guidance. Slit-Robo signaling provides a short-range directional cue for the tangential migration of neurons, ejecting axons from the midline and preventing re-crossing 3–5. Slit-Robo signaling also controls EC function and the process of vessel sprouting, called angiogenesis, and contributes to the stability of the vascular network 6–11. The Robo receptor may impart a positive or negative influence on cell migration and directionality depending on the isoform composition and cellular context 9,12. Although it is apparent that the expression and activity of the various Slit and Robo genes exerts a major influence on the ultimate form of the vessels and nerves, the rules governing Slit-Robo dependent cellular behavior are not clear.
The biological activity of many angiogenic growth factors, including Slit, vascular endothelial growth factor (VEGF)-A, fibroblast growth factor (FGF)-2, and platelet derived growth factor (PDGF)-BB, is modulated by the sulfation state of heparan sulfate proteoglycans (HSPGs) 13, 14. HSPGs are transmembrane or secreted proteins that are covalently linked to heparan sulfate (HS) chains. HSPGs are essential for Slit-Robo signaling, affecting the diffusion radius of Slit or acting as co-receptors that strengthen the interaction between Slit and Robo 15–17. Modification of HSPGs by epimerization and sulfation potentiates Slit binding in vitro, while enzymatic digestion of cell surface HS can abolish Slit-Robo activity 17–19. Slit-Robo signaling in vivo is blunted by genetic ablation of enzymes involved in the HS biosynthetic pathway 20, 21.
Recently, microRNAs (miRNAs) have been documented as key contributors to the process of angiogenesis 22–25. miRNAs are small non-coding RNAs that inhibit translation or promote mRNA degradation by binding to the 3′ untranslated region (UTR) of target mRNAs, resulting in the “fine-tuning” of gene expression 26, 27. It is becoming evident that a common regulatory mechanism for miRNA action involves the modest repression of many mRNAs with similar function or within a common molecular pathway by an individual miRNA 28. This creates an additive effect and reduces the dependence on a single molecular target to elicit a functional response. It also appears that miRNAs encoded by introns of protein-coding genes commonly regulate processes related to the host gene function 29. In this way, transcriptional regulation of the host gene results in the post-transcriptional regulation of a set of target genes that enhance or diminish the host gene activity.
In the present study, we describe an additional level of control over the Slit-Robo signaling pathway, whereby the expression and activity of the Robo receptors is dependent upon a miRNA encoded within an intron of the Slit genes. The Slit2 and Slit3 genes encode miR-218, which directly represses the expression of Robo1, Robo2, and multiple components of the HSPG biosynthetic pathway. Antisense mediated knockdown of miR-218 relieves repression of Robo1, Robo2, and the heparan-sulfate modifying enzyme glucuronyl C5-epimerase (GLCE or HSEPI), resulting in alterations in Slit-Robo signaling and, consequently, EC migration. We also show that miR-218 plays a role in retinal angiogenesis, controlling the density of the capillary plexus, at least partially via promotion of secondary vascular interconnections. Our results reveal an intricate regulatory network between Slit genes, Robo receptors and HSPG biosynthetic proteins allowing for the precise control over the expression level and activity of Slit-Robo signaling molecules during vascular patterning.
Methods
RNA isolation and analyses
Total RNA was isolated using Trizol (Invitrogen) and manufacturer’s protocol. microRNA and mRNA levels were quantified using Northern blot or with TaqMan microRNA real time probes against miR-218, Robo1, Robo2, and GLCE, Hs3st3B1, Hs6st3, KDR, Flt1, and Hrt1 (Applied Biosystems).
Cell culture and transfection
Antisense oligonucleotides possessing 2-O’Me modifications on the outermost 6 nucleotides (IDT, final concentration of 10nM) or LNA modified antisense oligonucleotides (final concentration of 50nM) were transfected using lipofectamine 2000 (Invitrogen). 48 hrs later, total RNA or protein was isolated. 2-O’Me modified oligonucleotide sequences: anti miR-218, 5′-AACCACATGGTTAGATCAAGCACAA-3′; control, 5′-CCTCTTACCTCAGTTACAATTTATA-3′. LNA modified oligonucleotide sequences: anti miR-218, 5′-GTTAGATCAAGCACA-3′; control, 5′-CCTAGAAAGAGTAGA-3′.
3′UTR reporter construction
The 3′UTRs of the mouse Robo1, Robo2 and GLCE genes were amplified and cloned into the SacI/HindIII sites of the pMIR-Report luciferase vector. The seed region of the miR-218 target sites in the Robo1 and GLCE 3′UTRs were mutated using the Quickchange II site directed mutagenesis kit (Stratagene). Primer sequences are in the supplemental methods.
Western blot
Antibodies directed against Robo1 (University of Iowa Hybridoma Bank), Robo2 (Abcam) and GLCE (Abcam) were used to determine protein level by Western blot. GAPDH (Calbiochem) was detected as loading control. Band intensity was quantified using NIH ImageJ software.
Analysis of postnatal retinal angiogenesis
In vivo injection and siRNA knockdown in the mouse retina was performed primarily as previously described 30 and detailed in the supplemental methods. Briefly, 1 μl of a 5 mg/ml solution of LNA modified anti-miR-218 or universal control oligonucleotides was unilaterally injected in the subretinal space of postnatal day (P) 2 mice in the ICR background. Mice were allowed to develop for 3 days prior to RNA and protein isolation at P5, and 5 days prior to isolation of retinas at P7 for histological analysis. Proliferating cells were visualized by staining for phospho-histone H3 (Cell Signaling). Visualization of the vasculature was performed by isolectin (Molecular Probes) staining of retinal flatmounts. Quantification of vessel density was performed using NIH ImageJ software. The thickness of the vascular plexus was calculated from confocal z-stacks taken at the vascular migration front. The radial length of the vascular network was calculated by measuring the distance form the optic disc to the periphery of the vascular plexus. Student’s t-tests were used to determine statistical significance between groups.
In situ hybridization
Wholemount microRNA in situ hybridization was performed essentially as previously described 31. 5′ and 3′ DIG labeled antisense LNA probes directed against miR-218 and miR-133 (Exiqon) were hybridized overnight at 49°C and 57°C, respectively.
Institutional compliance and animal care
All experiments utilizing animals were previously approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center.
Results
Expression of miR-218 from within the Slit2 and Slit3 genes
We and others have recently described important roles for intronic miRNAs in the control of cardiovascular development and disease 24, 25, 29, 32. Using a bioinformatics approach, we therefore searched for additional miRNAs within introns of protein-coding genes involved in cardiovascular development and function. We identified miR-218–1 and miR-218–2, within intron 14 of the mouse Slit2 and Slit3 genes, respectively (Figure 1A). The miR-218 stem-loop shares a high level of sequence conservation across species, displaying 100% identity across the mature miRNA from human and mouse to zebrafish and Xenopus (Online Figure I), suggesting this miRNA has an evolutionarily beneficial function.
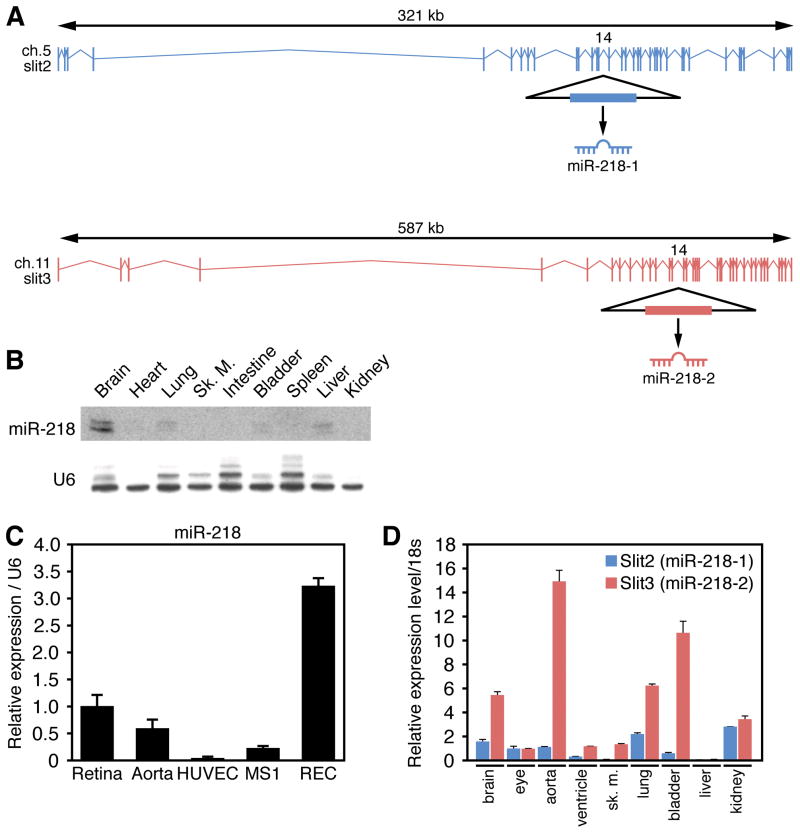
Figure 1. Co-expression of miR-218 and Slit genes.
A. Slit2 and Slit3 genomic structure and location of the miR-218-1 and miR-218-2 genes within intron 14. B. Multiple tissue Northern Blot to detect miR-218 in the adult mouse. U6 was detected as loading control. C. Real Time RT-PCR comparing the expression of miR-218 in the retina, aorta, HUVECs, retinal ECs (REC) and MS1 ECs. D. Real Time RT-PCR reveals the expression of Slit2 and Slit3 in multiple adult mouse tissues.
Northern blot analysis of adult mouse tissues revealed relatively high levels of miR-218 expression in the brain, the predominant domain of Slit gene expression. Lower levels of miR-218 were also detected in various endothelial cell (EC)-rich organs and tissues such as the heart and lungs (Figure 1B). miR-218 was detected in the retina and various EC lines (Figure 1C), as previously reported for members of the Slit family 33. We also found that the expression profile of miR-218 was similar to that of the host genes Slit2 and Slit3, as determined by Real Time RT-PCR, although the relative levels of expression sometimes differed (Figure 1D).
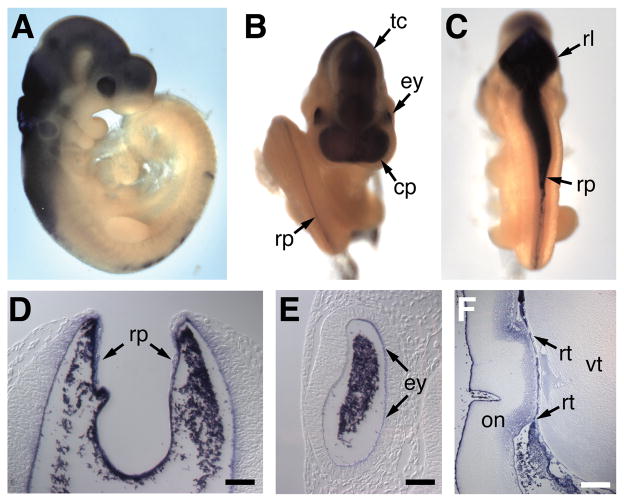
Whole-mount in situ hybridization was performed on mouse embryos using digoxygenin-labeled antisense oligonucleotides designed to detect miR-218 (Figure 2), or miR-133 as a control for tissue specificity (Online Figure II). This analysis confirmed miR-218 expression in the brain, neural tube and the eyes (Figure 2A-E). At embryonic day (E) 9.5, miR-218 displayed robust expression in the dorsal aspects of the midbrain and diencephalon (Figure 2C), and particularly strong expression in the roofplate of the neural tube (Figure 2C, D). Histological sections of stained embryos further demonstrated specific staining in the roofplate (Figure 2D). Expression of miR-218 was also observed in the embryonic eye (Figure 2B and E), persisting in the retinal layer of the eye into the neonatal period (Fig. 2F). These expression domains are similar to those of Slit2 and Slit 3, which are highly expressed in neural midline structures and the retina 33, 34. The similar expression and genomic localization of miR-218 and the Slit2 and Slit3 genes strongly suggest that miR-218 is generated by processing of the Slit2/3 pre-mRNAs. The expression levels of miR-218 and the Slit2/3 mRNAs are not identical, however, possibly reflecting variation in processing of the miRNA in different tissues.
Figure 2. Embryonic expression of miR-218.
In situ hybridization of E9.5 mouse embryos reveals the expression of miR-218. A. Lateral view reveals miR-218 expression in the eye, midbrain and hindbrain. B. Rostral view reveals miR-218 expression in the eyes, roof plate of the neural tube, tectum and commissural plate. C. Ventral view reveals miR-218 expression in the rhombic lip and roof plate of the neural tube. Histological sections of miR-218 stained embryos demonstrating expression in the roof plate (D), eye (E). Arrows point to staining, probe trapping is observed in lumen. F. Histological section of the adult eye demonstrates miR-218 expression in the retina. commissural plate (cp), eye (ey), optic nerve (on), retina (rt), rhombic lip (rl), roof plate (rp), tectum (tc), vitreous (vt).
Predicted targets of miR-218
To begin to elucidate the potential functions of miR-218, we examined its predicted targets using a bioinformatics approach consisting of algorithms designed to group targets based on functional classification (DIANA-miRPath) 35. Interestingly, the biological pathways that displayed the most significant enrichment in miR-218 targeted mRNAs included proteins involved in axon guidance (p < 3 × 10−6) and heparan sulfate (HS) biosynthesis (p < 4 × 10−4) (Figure 3A). The gene categories predicted to be targets of miR-218 include components of the Slit-Robo pathway, Robo1, Robo2, and SRGAP2 and the HS biosynthetic molecules Hs3st3b1, Hs6st3, and GLCE. We examined the efficiency of interaction with and evolutionary conservation of predicted targets using Targetscan5.0 36. Most of the predicted targets from within the top two most enriched groups contained at least one highly conserved 8-mer binding site for miR-218 within the 3′ UTR (Figure 3B). Like miR-218, the top predicted targets are conserved from humans to Xenopus (Online Figure III). These results suggested the possibility that miR-218 might link the expression of the Slit genes to post-transcriptional regulation of components of the Slit-Robo signaling axis.
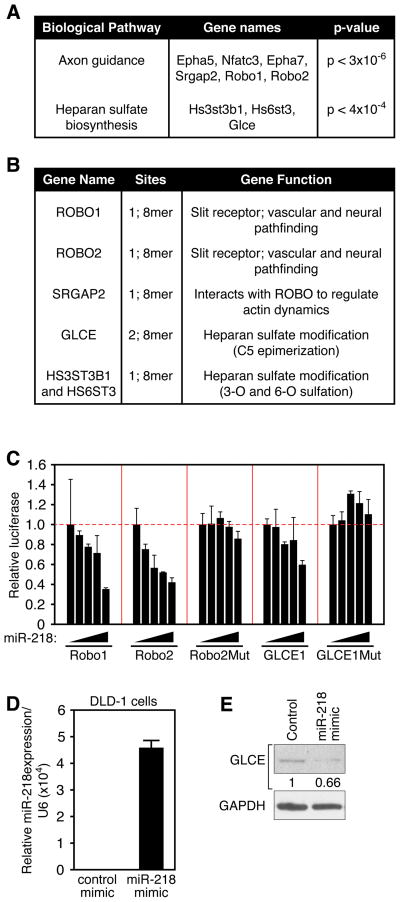
Figure 3. Identification of predicted miR-218 targets.
A. Top two most enriched biological pathways containing an over-representation of genes predicted to be targeted by miR-218 using DIANAlab analysis. B. Components of the axon guidance and HS biosynthesis pathways were analysed by TargetScan 5.0. Number and quality of sites within the various predicted targets and target function are highlighted. C. COS cells were transfected with a luciferase reporter linked to designated 3′UTRs, increasing amounts of miR-218 expression vector, and 20ng of lacZ expression vector. Total plasmid amount was kept constant with empty pcDNA3.1 vector and results were normalized to β-galactosidase activity. Mut designates 3′UTR with a mutation in the seed region of the miR-218 target site. D. Real Time RT-PCR reveals overexpression of miR-218 in DLD1 cells upon transfection with 2-O’Me modified miR-218 mimic oligonucleotides. miR-218 expression is normalized to U6. E. Western blot to detect GLCE in DLD1 cells transfected with 2-O’Me modified miR-218 mimic or control oligonucleotide. GAPDH is detected as loading control. Numbers refer to GLCE band intensity, normalized to GAPDH and relative to control.
Repression of Robo and HS biosynthetic genes by miR-218
We next conducted reporter assays to determine whether miR-218 can directly inhibit predicted target genes. Plasmids consisting of the 3′UTR of predicted targets linked to a luciferase reporter driven by a constitutive promoter were transfected into COS cells along with increasing amounts of a miR-218 expression plasmid. The activity of the luciferase reporter linked to the Robo1, Robo2, and GLCE 3′UTRs, normalized to β-galactosidase activity, displayed dose-dependent repression by miR-218 (Figure 3C). This effect was dependent upon the miR-218 target site, as mutation of this site attenuated repression by miR-218 (Figure 3C).
Repression of endogenous target proteins by administration of 2-O’Me-modified oligonucleotides that mimic miR-218 activity was next examined in cultured cells. For these experiments, we utilized the DLD1 colon cancer cell line, which expresses GLCE but not miR-218. Transfection of miR-218 mimic oligonucleotides resulted in significantly reduced levels of endogenous GLCE protein, as demonstrated by Western blot (Figure 3D and E). These results indicate that miR-218 links the Slit genes to the regulation of components of the Slit-Robo signaling pathway. We hypothesized that miR-218 may impinge upon Slit-Robo regulated processes such as EC migration and angiogenesis.
miR-218 affects endothelial cell migration
miR-218 expression was detected to varying degrees in HUVEC, MS1 and retinal ECs (Figure 1C and 4A). In addition to Robo1, which has previously been documented in ECs 11, we also detected endogenous expression of GLCE in ECs (data not shown). These results confirm reports of a role for Slit-Robo signaling in EC function 10, 11 and suggested miR-218 might impinge upon this process. To begin to decipher the potential influence of miR-218 on EC function, we used locked nucleic acid (LNA)-modified oligonucleotides to knockdown the expression of miR-218 and 2-O’Me modified mimic oligonucleotides to overexpress miR-218 in HUVECs and MS1 cells (Figure 4A and data not shown).
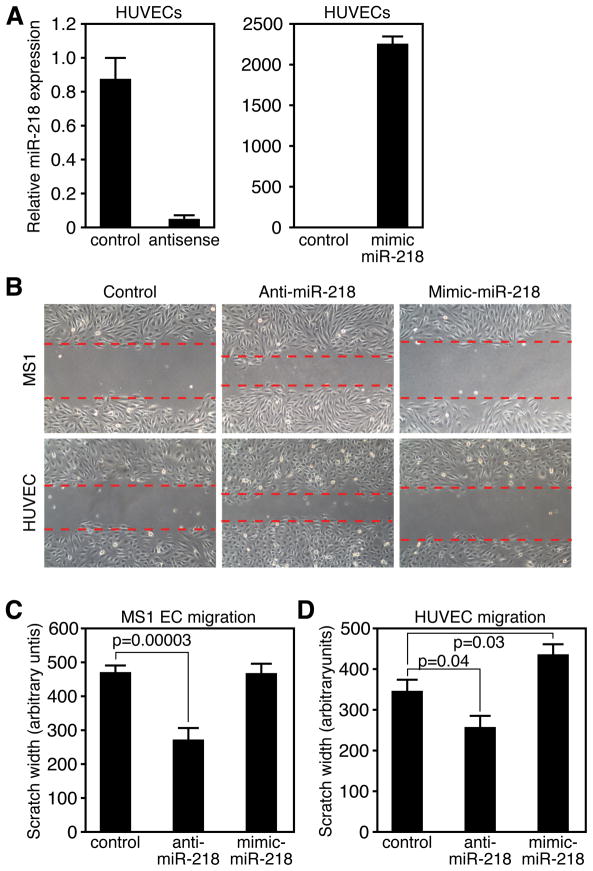
Figure 4. miR-218 contributes to EC migration.
A. Real Time RT-PCR reveals knockdown or overexpression of miR-218 in HUVECs transfected with 2-O’Me modified antisense or mimic oligonucleotides. B. Images of scratch wound following ~12 hours of incubation. MS1 cells (top row) or HUVECs (bottom row) were transfected with control, anti miR-218 or miR-218 mimic for 48 hours prior to scratch assay. Dotted lines denote front of EC migration. C, E. Quantification of migration in MS1 ECs (D) or HUVECs (E). Y-axis is the width of the “wound” 12 hours after scratching. Measurements are taken at 4 locations per well in duplicate wells in 3 independent experiments. Error bars represent SEM.
Since the Slit-Robo pathway modulates EC migration across a scratch “wound”, we examined the effect of altering miR-218 levels on this process. Upon miR-218 knockdown, we observed an increase in the distance of EC migration in a monolayer scratch wounding assay (Figure 4B). Quantification of the distance of migration revealed a significant increase upon miR-218 knockdown in both HUVECs and MS1 cells (Figure 4C and D). Conversely, overexpression of miR-218 resulted in a significant reduction of HUVEC migration (Figure 4D). Initial filipodia extension and actin stress fiber formation was not noticeable altered by miR-218 knockdown (Online FigureIV). The alterations in migration by modulation of miR-218 levels are likely to be direct, rather than secondary to alterations in proliferation, since the doubling time of HUVECs and MS1 cells is greater or equal to 24 hours, while the migration assay was performed following an overnight incubation. These observations support our hypothesis that modulation of Slit-Robo signaling by miR-218 affects EC migration. Furthermore, our results suggest that Robo1/2 and GLCE positively influence the migration rate of EC cells in culture and that miR-218 counteracts this process.
miR-218 contributes to retinal angiogenesis
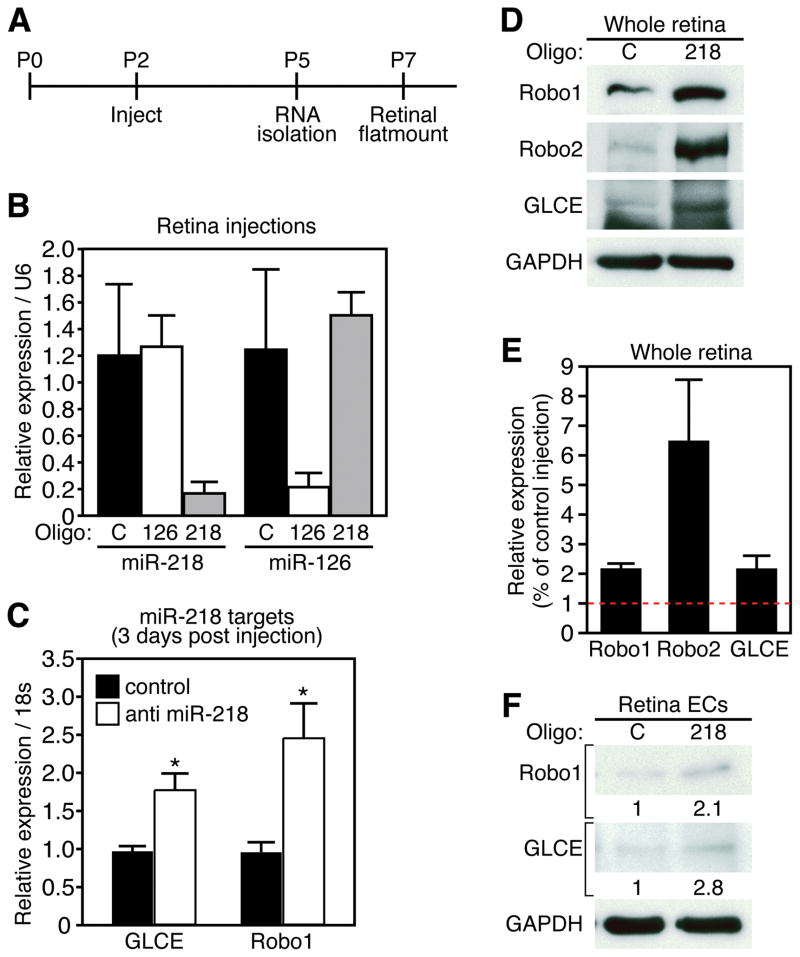
The previous results suggested that miR-218 might influence EC migration during vascular patterning. To further address this potential role, we perturbed miR-218 expression in the retina of neonatal mice. Retinal vascular development is a well-characterized model of angiogenesis that is highly amenable to siRNA mediated knockdown of gene expression 30. Importantly, miR-218 is highly expressed in the retina, as demonstrated by Real Time RT-PCR and in situ hybridization (Figure 1C and 2F). We therefore used LNA-modified antisense oligonucleotides to silence the expression of miR-218 during early postnatal development by injection into the subretinal space. Injection was performed 2 days after birth (P2) and retinas were isolated for RNA and protein analyses 3 days later (P5). An additional subset of animals was allowed to develop until P7 and retinas were isolated for immunohistochemistry to examine the developing vascular network (Figure 5A).
Figure 5. miR-218 modulates expression of Slit-Robo axis genes in the retina.
A. Experimental setup of retinal injections. Postnatal (P) age is denoted above line. Timepoints of injection and sample isolation are denoted below line. B. Real Time RT-PCR demonstrates knockdown of miR-218 or miR-126 in retinal samples 3 days after injection with LNA modified antisense miR-218 or miR-126 oligonucleotides. Expression is relative to control oligonucleotide and normalized to U6. C. Real time RT-PCR reveals expression levels of GLCE and Robo1 from retinal samples 3-days after injection. Error bars are SEM (n = 3, * denotes p < 0.05). D. Western for Robo1, Robo2, and GLCE from a pool of 5 retinas, 3 days after injection with control or anti miR-218 oligonucleotides. GAPDH is detected as loading control. C = control, 218 = anti miR-218. E. Quantification of protein levels from retinal explant pools. F. Western blot detecting Robo1 and GLCE from retinal ECs 48 hrs after transfection with anti miR-218 oligonucleotides. GAPDH is detected as loading control. Numbers refer to Western band intensity, normalized to GAPDH and relative to control.
Retinal injection of LNA-anti-miR-218 resulted in ~ 90% reduction in miR-218 levels compared with either uninjected or control LNA oligonucleotide injected retinas (Figure 5B). LNA miR-218 injection did not inhibit the unrelated miRNA, miR-126, nor did injection of an LNA modified oligonucleotide directed against miR-126 inhibit miR-218 expression in the retina (Figure 5B). These results demonstrate a high level of efficacy and specificity of LNA oligonucleotides, and imply limited potential for off-target effects.
Importantly, the mRNA and protein levels of Robo1, Robo2, and GLCE were enhanced by miR-218 knockdown in the retina (Figure 5C and D), although Robo2 mRNA levels were not significantly affected by miR-218 knockdown (data not shown). Quantification of Western band intensity confirmed the significant enrichment of Robo1, Robo2 and GLCE protein levels upon silencing of miR-218 (Figure 5F). The predicted miR-218 targets, Hs3st3b1 and Hs6st3, did not change at the mRNA level (Online Figure V), although we did not have an adequate antibody to examine the protein levels. The alterations in Slit-Robo pathway genes was not accompanied by changes in the Notch target Hrt1, or the VEGF receptors, KDR and Flt1 (Online Figure VI)
In order to dissect the cell type responsible for Slit-Robo modulation by miR-218, we utilized a retinal EC line. Transfection of retinal ECs with antisense LNA modified miR-218 oligonucleotides resulted in elevated levels of Robo1 and GLCE protein levels (Figure 5E and F), implying miR-218 expression in ECs contributes to the effect on Slit-Robo pathway gene expression.
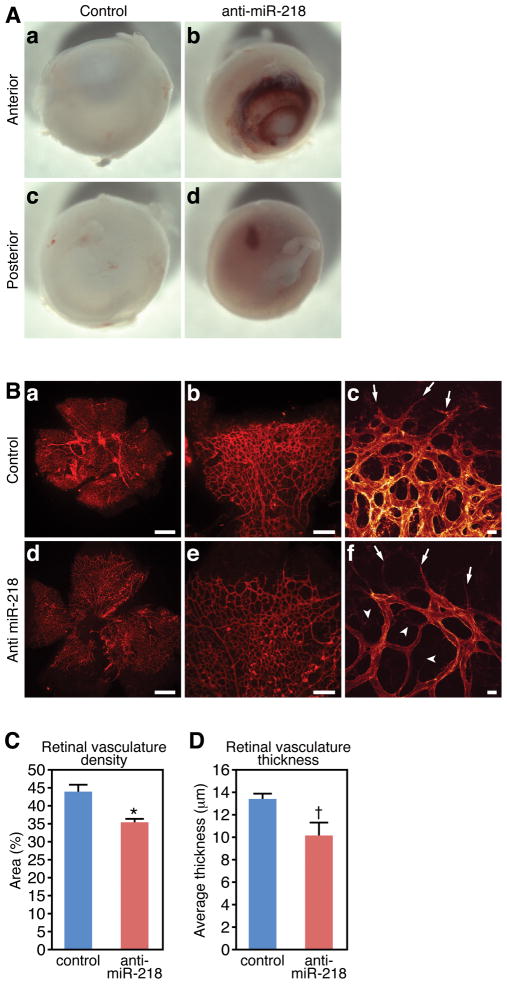
The modulation of the Slit-Robo axis by miR-218 in the mammalian retina suggests this regulatory network may constitute a conserved mechanism controlling blood vessel patterning. Indeed, gross examination revealed vascular leakage in LNA-anti miR-218 injected retinas, while control injected retinas rarely exhibited hemorrhage (Figure 6A). We therefore harvested LNA-anti miR-218 and control injected retinas at 5 days post-injection (P7) and examined the vascular network by staining of retinal flatmounts with isolectin (Figure 6B). This experiment revealed that antisense mediated knockdown of miR-218 levels in the retina during the neonatal period resulted in aberrant patterning and reduced complexity of the retinal vascular plexus (compare Figure 6B(a-c to d-f)). Quantification of vascular density and the thickness of the vascular bed revealed significant attenuation of both in anti-miR-218 injected retinas (Figure 6C and D).
Figure 6. miR-218 contributes to retinal angiogenesis.
A. Hemmorhage in miR-218 knockdown retinas. Control eyes (a, b) display hemmorhage at a frequency of 14%, while hemmorhage is observed in 67% of LNA anti miR-218 injected retinas (c, d) at P7. N = 10 anti-miR-218 and 7 controls (p<0.05). B. Vasculature of the retina at P7, as visualized by isolectin staining in flatmount preparation. Top row (a-c) is representative control oligonucleotide injected retina and lower row (d-f) is LNA anti-miR-218 injected. High magnification confocal images of migration front of control injected (c) and LNA miR-218 injected (f) retinas reveals altered EC plexus morphology. Arrows mark tip cells and arrowheads mark EC projections failing to make interconnections. (Scale bars = 500 μm in A and D, 200 μm in b and e, and 20 μm in c and f). C. Quantification of retinal vascular density. Error bars represent SEM (measurements were taken in four quadrants from 7 control injected retinas and 5 anti miR-218 injected retinas, * denotes p < 0.01). D. Quantification of retinal vascular thickness from confocal Z-stacks. Error bars represent SEM (n = 6 control and 6 anti miR-218 injected retinas, t denotes p < 0.05).
High resolution confocal imaging of the endothelial tip cells at the migratory front revealed relatively normal filopodia formation, however the number and diameter of capillaries within vascular plexus appeared to be dramatically diminished following miR-218 knockdown (compare Figure 6B(c to f)). EC protrusions emanated from the stalk cells within the migrating front of the vascular plexus (arrowheads in Figure 6B(f)), but there was a reduction in interconnections between adjacent capillaries, which may explain the lower density and reduced vascular thickness observed in miR-218 deficient retinas. Taken together, our study suggests miR-218 represents a regulatory link between the Slit genes and other components of the Slit-Robo signaling pathway during EC pathfinding.
Discussion
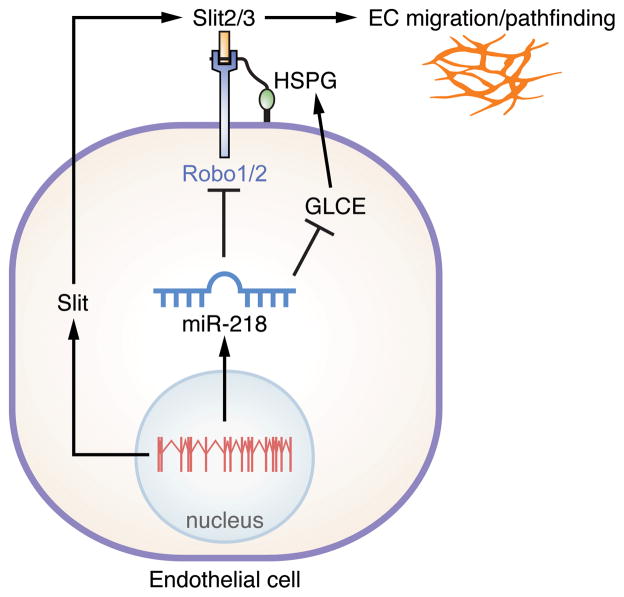
Here we describe an evolutionarily conserved miRNA located within an intron of the Slit genes, miR-218, which controls the expression of various Slit-Robo pathway components during development. Dysregulation of Slit-Robo signaling by altered miR-218 levels affects EC function in vitro. We also provide evidence that deficiency in miR-218 dependent Slit-Robo regulation results in aberrant vascular patterning in the mouse retina. Taken together, this study identifies miR-218 as an important regulatory hub, linking transcriptional regulation of Slit genes to post-transcriptional regulation of multiple components of the Slit-Robo pathway, which ultimately influence angiogenesis (Figure 7).
Figure 7. Proposed mechanism of miR-218 activity during blood vessel patterning.
The Slit2/3 genomic locus gives rise to both the Slit2/3 mRNAs and miR-218. Slit proteins are secreted and interact with the Robo receptor. miR-218 targets the Robo1/2 receptors and GLCE for inhibition. Reduced levels of GLCE alter HSPG biosynthesis, ultimately impinging upon the interaction between Slit and Robo. miR-218 links transcriptional activation of the Slit locus to the efficiency of Slit Robo signaling, ultimately controlling vascular patterning.
These data are consistent with previous reports of intronic miRNA function, in which the miRNA regulates the same biological process as the protein encoded by the host gene29. miR-218 may contribute to “fine-tuning” of Slit-Robo pathway genes or generate negative feedback in response to Slit gene activation. It is interesting to speculate that miR-218 may serve to repress the expression of the Robo1/2 receptors in the Slit ligand expressing cells, thereby spatially separating ligand from receptor. Since Robo4 is not a target of miR-218 regulation, it also is possible that miR-218 affects the ratio of Robo1/2 and Robo4 proteins, thereby influencing vascular patterning.
It is currently debated whether the Robo1 and 2 receptors provide a positive or negative influence on EC migration, although Robo4 is generally thought of as a repulsive or stabilizing cue during neural and vascular pathfinding 9, 37. In our hands, it appears that repression of Robo1/2 and HSPG biosynthetic molecules by miR-218 negatively affects EC migration. In line with these findings, miR-218 was recently reported to also inhibit tumor cell migration and metastasis via repression of Robo138. The disorganization and reduction of retinal vascular density observed upon miR-218 knockdown appears to be a consequence of migration or cellular adhesion since we did not detect aberrant proliferation (Online Figure VII).
The second most enriched biological process targeted by miR-218 includes components of the HS biosynthetic pathway. Here, we show that miR-218 directly targets the C5 epimerase, GLCE, for degradation during vascular development. miR-218 is also predicted to repress 2-O and 6-O sulfotransferases, both of which are critical enzymes, along with GLCE, in regulating the biological activity of HSPGs 13. HSPGs interact with and regulate the function of many angiogenic growth factors. For example, HSPGs contribute to transport of signaling molecules through the extracellular matrix, formation of growth factor gradients, and stability of ligand-receptor interactions, including Wnt, VEGF, FGF, and Slit-Robo pathways 19, 21, 39–41. It is conceivable that miR-218 dependent modulation of HSPGs may influence Slit-Robo signaling via multiple mechanisms, including alteration of Slit-Robo affinity or modulating the diffusion radius of Slit from the secreting cell.
Another level of HSPG-dependent regulation involves the localization of HS in the cellular microenvironment. Recent evidence has revealed an important distinction between HSPG presentation to a transmembrane receptor in cis or in trans. For example, HS in trans potentiates VEGFR-mediated angiogenesis; it is hypothesized that HS presented by an adjacent cell delays VEGFR internalization and increases the efficiency and duration of activity of the bound receptor 14. It is interesting to speculate that miR-218 might influence the HSPG microenvironment, thereby affecting the activity of various growth factor receptors on neighboring cells.
The Slit-Robo signaling pathway was initially identified as a guidance cue controlling the crossing of the midline by axons 42. Multiple studies report the interdependence of vascular and neural patterning during development 1. Although our study reveals a direct role of miR-218 in EC migration in vitro, it remains possible that the effect of miR-218 on retinal angiogenesis is at least partially due to an influence on axonal pathfinding. It is also possible that miR-218 contributes to the dynamic regulation of the Robo receptors at the neural midline, directly influencing axonal pathfinding. While miR-218 is robustly expressed in the brain and eye, it is interesting to note that miR-218 is expressed at much lower levels than the Slit1 and 2 genes in bladder, kidney and lungs. This suggests the possibility that miR-218 is inefficiently processed in these tissues, which may result in altered miR-218-dependent Slit-Robo activity.
Slit1 and Robo1 were recently identified as part of the transcriptional network activated during the switch to tumor angiogenesis 43. This process appears to be especially sensitive to miRNA control. In addition to the present study, miRNA-126, –378, –296, and the 17–92 cluster have been shown to regulate various aspects of vessel formation, remodeling and tumor angiogenesis 44. Other physiological processes that might respond to alterations in miR-218 levels and Slit-Robo signaling include oxygen-induced retinopathy, light-induced choroidal angiogenesis, VEGF-induced vascular permeability and tumor angiogenesis. Further analysis of miR-218 during vascular pathologies and tumor angiogenesis may define miR-218 as an intriguing candidate for therapeutic manipulation in the treatment of vascular diseases.
Novelty and Significance.
What is known?
microRNAs are small non-coding RNAs that inhibit the expression of target mRNAs.
Individual microRNAs are often predicted to repress large numbers of target mRNAs.
Control of gene expression by microRNAs influences many physiological processes.
What new information does this article contribute?
miR-218 is expressed within an intron of the Slit genes.
miR-218 regulates the expression of multiple genes that contribute to the Slit-Robo signaling pathway.
miR-218 dependent regulation of Slit-Robo signaling controls vascular patterning in the retina.
Summary.
microRNAs are short non-coding RNAs that often inhibit the expression of a large number of mRNA targets via interaction with complimentary sequences in the 3′ untranslated region. It is hypothesized that a common mode of microRNA action involves targeting multiple mRNAs that encode proteins contributing to the same biological process. Here we describe the repression of multiple components of the Slit-Robo signaling pathway by miR-218, which is encoded by an intron of the Slit genes. Furthermore, we demonstrate that the miR-218-Slit-Robo signaling pathway is necessary for normal patterning of the retinal vasculature in mice. Taken together our study describes the coordinated function of a microRNA and its host gene, and advances the understanding of how microRNAs control biological processes by modest inhibition of an organized array of target mRNAs.
Supplementary Material
Acknowledgments
We thank Ondine Cleaver and Stryder Meadows for helpful discussions and critical reading of the manuscript, Eva van Rooij for the LNA modified oligonuceotides, John Shelton for technical assistance, Jose Cabrera for graphics and Jennifer Brown for editorial assistance.
Sources of Funding
Work in the lab of E.N.O. was supported by grants from the NIH, the Donald W. Reynolds Center for Clinical Cardiovascular Research, The Robert A. Welch Foundation (grant number I-0025), the Fondation Leducq’s Transatlantic Network of Excellence in Cardiovascular Research Program, the American Heart Association and the Jon Holden DeHaan Foundation. E.M.S. was supported by a Scientist Development Grant from the American Heart Association.
Non-standard Abbreviations and Acronyms
- EC
endothelial cell
- GLCE
glucuronyl C5-epimerase
- HS
heparan sulfate
- HSPG
heparan sulfate proteoglycan
- HUVEC
human umbilical vein endothelial cells
- LNA
Locked Nucleic Acid
- miR
microRNA
- PDGF
platelet derived growth factor
- Robo
Roundabout
- UTR
untranslated region
- VEGF
vascular endothelial growth factor
Footnotes
Disclosures
E.N.O holds equity in miRagen Therapeutics, which is developing miRNA-based therapies for muscle disease.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 2.Cleaver O, Krieg PA. Vascular Development. In: Rosenthal N, Harvey RP, editors. Heart Development and Regeneratoin. Vol. 1. New York: Elsevier; 2010. pp. 487–528. [Google Scholar]
- 3.Nguyen Ba-Charvet KT, Brose K, Marillat V, Kidd T, Goodman CS, Tessier-Lavigne M, Sotelo C, Chedotal A. Slit2-Mediated chemorepulsion and collapse of developing forebrain axons. Neuron. 1999;22:463–473. doi: 10.1016/s0896-6273(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 4.Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 5.Plump AS, Erskine L, Sabatier C, Brose K, Epstein CJ, Goodman CS, Mason CA, Tessier-Lavigne M. Slit1 and Slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron. 2002;33:219–232. doi: 10.1016/s0896-6273(01)00586-4. [DOI] [PubMed] [Google Scholar]
- 6.Jones CA, Nishiya N, London NR, Zhu W, Sorensen LK, Chan AC, Lim CJ, Chen H, Zhang Q, Schultz PG, Hayallah AM, Thomas KR, Famulok M, Zhang K, Ginsberg MH, Li DY. Slit2-Robo4 signalling promotes vascular stability by blocking Arf6 activity. Nat Cell Biol. 2009;11:1325–1331. doi: 10.1038/ncb1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ypsilanti AR, Zagar Y, Chedotal A. Moving away from the midline: new developments for Slit and Robo. Development. 2010;137:193–1952. doi: 10.1242/dev.044511. [DOI] [PubMed] [Google Scholar]
- 8.Park KW, Morrison CM, Sorensen LK, Jones CA, Rao Y, Chien CB, Wu JY, Urness LD, Li DY. Robo4 is a vascular-specific receptor that inhibits endothelial migration. Dev Biol. 2003;261:25–267. doi: 10.1016/s0012-1606(03)00258-6. [DOI] [PubMed] [Google Scholar]
- 9.Legg JA, Herbert JM, Clissold P, Bicknell R. Slits and Roundabouts in cancer, tumour angiogenesis and endothelial cell migration. Angiogenesis. 2008;11:1–21. doi: 10.1007/s10456-008-9100-x. [DOI] [PubMed] [Google Scholar]
- 10.Sheldon H, Andre M, Legg JA, Heal P, Herbert JM, Sainson R, Sharma AS, Kitajewski JK, Heath VL, Bicknell R. Active involvement of Robo1 and Robo4 in filopodia formation and endothelial cell motility mediated via WASP and other actin nucleation-promoting factors. FASEB J. 2009;23:513–522. doi: 10.1096/fj.07-098269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B, Dietrich UM, Geng JG, Bicknell R, Esko JD, Wang L. Repulsive axon guidance molecule Slit3 is a novel angiogenic factor. Blood. 2009;114:430–4309. doi: 10.1182/blood-2008-12-193326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Xiao Y, Ding BB, Zhang N, Yuan X, Gui L, Qian KX, Duan S, Chen Z, Rao Y, Geng JG. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003;4:19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 13.Jia J, Maccarana M, Zhang X, Bespalov M, Lindahl U, Li JP. Lack of L-iduronic acid in heparan sulfate affects interaction with growth factors and cell signaling. J Biol Chem. 2009;284:15942–15950. doi: 10.1074/jbc.M809577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakobsson L, Kreuger J, Holmborn K, Lundin L, Eriksson I, Kjellen L, Claesson-Welsh L. Heparan sulfate in trans potentiates VEGFR-mediated angiogenesis. Dev Cell. 2006;10:625–634. doi: 10.1016/j.devcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Hussain SA, Piper M, Fukuhara N, Strochlic L, Cho G, Howitt JA, Ahmed Y, Powell AK, Turnbull JE, Holt CE, Hohenester E. A molecular mechanism for the heparan sulfate dependence of slit-robo signaling. J Biol Chem. 2006;281:39693–39698. doi: 10.1074/jbc.M609384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuhara N, Howitt JA, Hussain SA, Hohenester E. Structural and functional analysis of slit and heparin binding to immunoglobulin-like domains 1 and 2 of Drosophila Robo. J Biol Chem. 2008;283:16226–16234. doi: 10.1074/jbc.M800688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu H. Cell-surface heparan sulfate is involved in the repulsive guidance activities of Slit2 protein. Nat Neurosci. 2001;4:695–701. doi: 10.1038/89482. [DOI] [PubMed] [Google Scholar]
- 18.Ronca F, Andersen JS, Paech V, Margolis RU. Characterization of Slit protein interactions with glypican-1. J Biol Chem. 2001;276:29141–29147. doi: 10.1074/jbc.M100240200. [DOI] [PubMed] [Google Scholar]
- 19.Johnson KG, Ghose A, Epstein E, Lincecum J, O’Connor MB, Van Vactor D. Axonal heparan sulfate proteoglycans regulate the distribution and efficiency of the repellent slit during midline axon guidance. Curr Biol. 2004;14:499–504. doi: 10.1016/j.cub.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Lee JS, von der Hardt S, Rusch MA, Stringer SE, Stickney HL, Talbot WS, Geisler R, Nusslein-Volhard C, Selleck SB, Chien CB, Roehl H. Axon sorting in the optic tract requires HSPG synthesis by ext2 (dackel) and extl3 (boxer) Neuron. 2004;44:947–960. doi: 10.1016/j.neuron.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 21.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 22.Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 24.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 28.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweetman D, Rathjen T, Jefferson M, Wheeler G, Smith TG, Wheeler GN, Munsterberg A, Dalmay T. FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev Dyn. 2006;235:2185–2191. doi: 10.1002/dvdy.20881. [DOI] [PubMed] [Google Scholar]
- 32.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan W, Zhou L, Chen JH, Wu JY, Rao Y, Ornitz DM. The mouse SLIT family: secreted ligands for ROBO expressed in patterns that suggest a role in morphogenesis and axon guidance. Dev Biol. 1999;212:290–306. doi: 10.1006/dbio.1999.9371. [DOI] [PubMed] [Google Scholar]
- 34.Holmes GP, Negus K, Burridge L, Raman S, Algar E, Yamada T, Little MH. Distinct but overlapping expression patterns of two vertebrate slit homologs implies functional roles in CNS development and organogenesis. Mech Dev. 1998;79:57–72. doi: 10.1016/s0925-4773(98)00174-9. [DOI] [PubMed] [Google Scholar]
- 35.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 36.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 37.Jones CA, London NR, Chen H, Park KW, Sauvaget D, Stockton RA, Wythe JD, Suh W, Larrieu-Lahargue F, Mukouyama YS, Lindblom P, Seth P, Frias A, Nishiya N, Ginsberg MH, Gerhardt H, Zhang K, Li DY. Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nat Med. 2008;14:448–453. doi: 10.1038/nm1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, Guo X, Wang B, Gang Y, Zhang Y, Li Q, Qiao T, Zhao Q, Nie Y, Fan D. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6:e1000879. doi: 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gitay-Goren H, Soker S, Vlodavsky I, Neufeld G. The binding of vascular endothelial growth factor to its receptors is dependent on cell surface-associated heparin-like molecules. J Biol Chem. 1992;267:6093–6098. [PubMed] [Google Scholar]
- 40.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 41.Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP., Jr Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 42.Kidd T, Brose K, Mitchell KJ, Fetter RD, Tessier-Lavigne M, Goodman CS, Tear G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- 43.Abdollahi A, Schwager C, Kleeff J, Esposito I, Domhan S, Peschke P, Hauser K, Hahnfeldt P, Hlatky L, Debus J, Peters JM, Friess H, Folkman J, Huber PE. Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proc Natl Acad Sci U S A. 2007;104:12890–12895. doi: 10.1073/pnas.0705505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Olson EN. AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.