Abstract
Genomic imprinting results in preferential gene expression from paternally versus maternally inherited chromosomes. We used a genome-wide approach to uncover sex-specific parent-of-origin allelic effects in the adult mouse brain. Our study identified preferential selection of the maternally inherited X chromosome in glutamatergic neurons of the female cortex. Moreover, analysis of the cortex and hypothalamus identified 347 autosomal genes with sex-specific imprinting features. In the hypothalamus, sex-specific imprinted genes were mostly found in females, suggesting parental influence over the hypothalamic function of daughters. We show that Interleukin 18, a gene linked to diseases with sex-specific prevalence, is subject to complex, regional, and sex-specific parental effects in the brain. Parent of origin effects thus provide new avenues for investigation of sexual dimorphism in brain function and disease.
Genomic imprinting is an epigenetic mode of gene regulation involving preferential expression of the paternally or maternally inherited allele (1). Sexual dimorphism is a central characteristic of mammalian brain function and behavior that influences major neurological diseases in humans (2). Here we address the potential existence of differential genomic imprinting in the brain according to the sex of individuals. Imprinting refers to gene expression differences between maternal and paternal chromosomes (3), and is also used more strictly to define complete allele-specific silencing (4). Our analysis encompasses sex differences in parent-of-origin allelic effects involving all-or-none allele-specific expression and parental biases in gene expression.
Three processes may underlie sexually dimorphic genomic imprinting (fig. S1A–C). Non-random X inactivation, such as the imprinted X-inactivation observed in marsupials and the mouse extra-embryonic lineages, could result in the preferential silencing of one of the parentally inherited X chromosomes in females (fig. S1A) (5). In addition, imprinting of individual X-linked loci in females results in gene expression from the active paternally-inherited X that differs from the active maternally-inherited X (fig. S1B). Studies of Turner Syndrome suggested imprinting of X chromosome loci with relevance to brain function (6), and X-linked imprinted genes have indeed been identified in the brain (7, 8). Finally, autosomal genes might be imprinted in one sex, but not the other (fig. S1C). A recent study of quantitative trait loci influencing growth and body composition in mice suggests that such mechanisms may exist (9).
We have used Illumina transcriptome sequencing of F1 hybrid mice generated from initial (F1i) and reciprocal (F1r) crosses of CASTEiJ (CAST) and C57BL6J (C57) mice to investigate genomic imprinting in the brain with high resolution (10). Here we compare parental effects in the transcriptome of the adult male versus female preoptic area (POA) of the hypothalamus and medial prefrontal cortex (mPFC). Detailed methods are described in the supplementary material and in our companion paper (10).
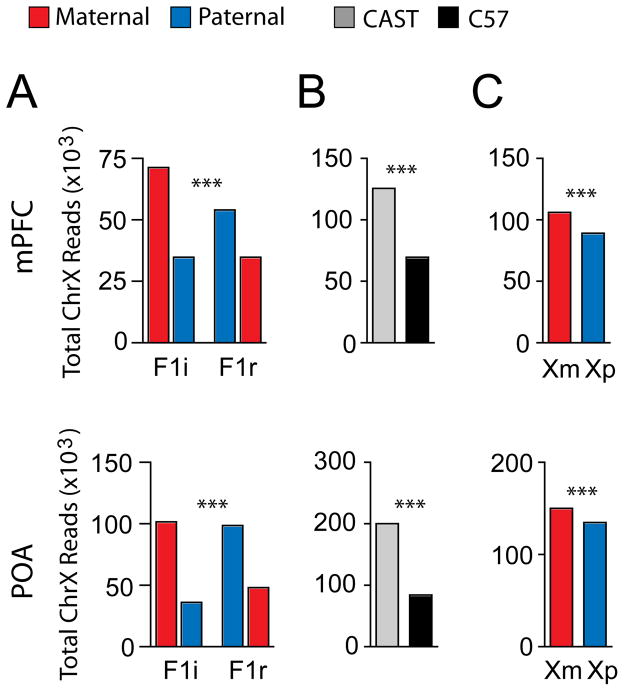
We first assessed global levels of X-linked gene expression from the maternal X chromosome (Xm) versus the paternal X chromosome (Xp) in the adult female POA and mPFC. A significant strain-effect favoring expression from the CAST X was observed in F1 females (Fig. 1A and B). This difference is likely due to preferential selection of the CAST X-chromosome in the hybrids (11). In addition, we identified a parent-of-origin effect (Fig. 1A and C), such that total levels of expression from the Xm were increased by 19% and 11% relative to the Xp in the mPFC and POA, respectively. The Xm bias was significantly greater in the mPFC, than the POA (P<0.0001, two-tailed Fisher’s Exact Test).
Fig. 1.
Sex specific imprinting and preferential expression of the Xm in the female brain. (A) Total maternal and paternal X-linked reads for the adult female mPFC and POA in F1i and F1r crosses reveals a highly significant association between strain and cross (mPFC P<0.0001; POA: P<0.0001, two-tailed Fisher’s Exact Test). (B) Identification of a significant strain effect favoring CAST X-chromosome expression (χ2 analysis). (C) Preferential expression of the Xm in the mPFC and POA (χ2 analysis). ***P <0.001; *P<0.05.
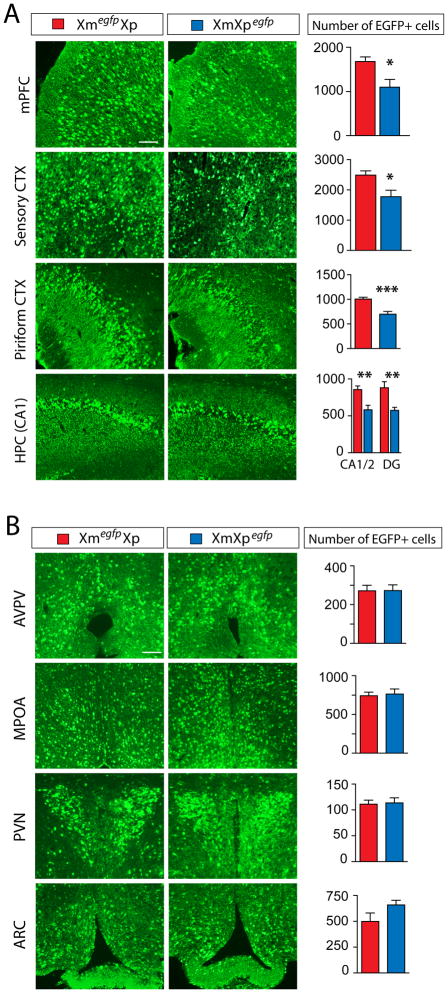
This elevated expression from the Xm versus the Xp (Fig. 1A and C) may indicate a bias in X-inactivation in the brain, a hypothesis further investigated with a transgenic mouse line expressing X-linked egfp under the control of the CMV promoter as a reporter of the active X chromosome (12). Control studies confirmed that the egfp transgene reports X-inactivation (fig. S2) and egfp expression was found restricted to a subpopulation of vGLUT2+ glutamatergic neurons (~72%) (fig. S3). We compared the number of Xm- versus Xp-expressing glutamatergic neurons in adult Xmegfp/Xp and Xm/Xpegfp females (Fig. 2). In cortical regions, 40–50% more neurons expressed the Xm than the Xp in the mPFC, the sensory CTX, and the piriform CTX (Fig. 2A). We also observed a significant Xm bias in the CA1/2 and DG regions of the HPC (Fig. 2A). In contrast, no difference in the number of Xm versus Xp expressing cells was detected in the hypothalamus (Fig. 2B). We then asked whether the bias observed in cortical versus hypothalamic glutamatergic neurons could be generalized to all or few neuronal populations in these brain regions. We summed the Xm and Xp reads for 7 well-characterized neuron-specific X-linked genes and found a significant Xm expression bias in both the mPFC (21% Xm bias; P<0.0001) and POA (15% Xm bias; P<0.0001, two-tailed Fisher’s Exact Test) (fig. S4). Therefore, whereas Xegfp+ glutamatergic neurons in POA do not preferentially select the Xm, some other neuronal populations of the hypothalamus likely do (fig. S4).
Fig. 2.
Preferential expression of Xm in female cortical regions indicated by XmegfpXp and XmXpegfp transgenic mice. (A, B) The number of EGFP+ cells in different cortical (A) and hypothalamic (B) brain regions of XmegfpXp (red) versus XmXpegfp (blue) 5 week old females. Cortical regions: medial prefrontal CTX mPFC, sensory CTX, piriform CTX, and CA1/2 regions and dentate gyrus (DG) of the hippocampus (HPC CA1). Hypothalamic regions: anteroperiventricular nucleus (AVPV), medial preoptic area (MPOA), periventricular nucleus (PVN), or arcuate nucleus (ARC). Two-tailed unpaired t-test; n=7; ***P< 0.001; **P< 0.01; *P< 0.05. Red bars, XmegfpXp; Blue bars, XmXpegfp. Scale bar is 50 μm.
We then assessed X-linked imprinting at the level of individual genes using a χ2 test in which the expected value was adjusted for strain and maternal X selection biases. Using the stringent cutoff of P<0.05 in the F1i and F1r cross used in our companion study (10) to assess imprinting, we failed to identify X-linked imprinted loci. Using a less stringent cutoff (P<0.1), the previously known MEG, Xrl3b (7, 8) was correctly identified and this approach further identified 9 candidate imprinted genes in the POA and 3 in the mPFC (Table S1), such as yipf6, which was identified in the POA (maternal bias) and mPFC (paternal bias).
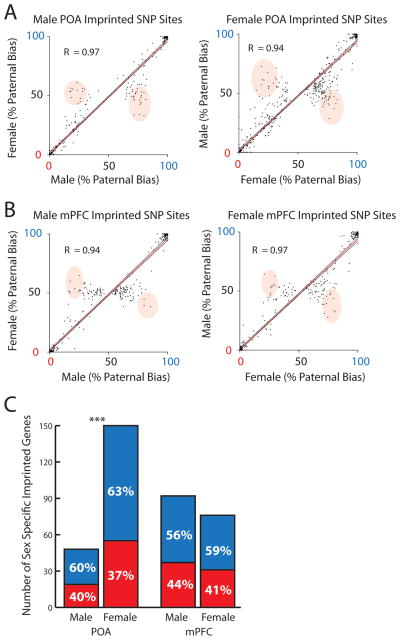
Finally, we searched for sex-specific parental allelic effects on the autosomes. As reported in our companion paper, parental expression biases in the male and female datasets were highly correlated (Fig. 3A and B). However, SNP sites that exhibited a strong parental bias in one sex, but not the other, were also apparent in the data (Fig. 3A and B). A χ2 test was applied in the F1i and F1r cross to identify SNP sites (cutoff P<0.05) significantly imprinted in one sex, but not the other (P>0.05).
Fig. 3.
Sex-specific imprinted autosomal genes were uncovered in the adult male and female POA and mPFC. (A and B) Scatterplots and Pearson’s correlation analysis (R) of the paternal expression bias exhibited by imprinted SNP sites identified in male versus female data. Note some SNP sites exhibit parental effects that are not reproduced (shaded regions). (C) Analysis of the proportion of sex specific PEGs and MEGs in the female versus male POA (χ2 Analysis). ***P<0.001. Red, maternal expression; Blue, paternal expression.
This study identified 347 candidate genes associated with sex specific parental allelic effects in the adult brain, as defined by the presence of one or more SNP sites statistically imprinted in one sex, but not the other (Table S2 and S3). The average parental expression bias exhibited by sex specific imprinted SNP sites was 73% (POA) and 68% (mPFC), whereas the average bias for the same sites in the opposite sex was 52% (POA) and 51% (mPFC). Females had 3 times the total number of genes with sex specific imprinted features (Fig. 3C; 150 genes (1.3% of 11241 genes assessed)) as males in the POA (48 genes (0.5% of 9235 genes assessed)), but no difference was observed in the mPFC. This correlates well with the fact that the POA is a highly sexually dimorphic region of the brain involved in the control of maternal and mating behaviors, and that imprinting is known to influence maternal behavior (13). We noted a paternal bias in the number of sex specific genes identified in all samples (Fig. 3C).
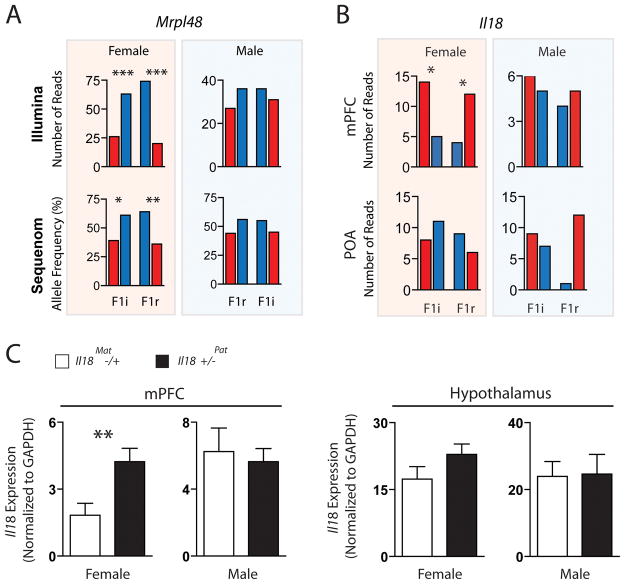
We carried out an in depth analysis of two candidate genes subject to sex-specific parental effects: Mitochondrial ribosomal protein 48 (Mrpl48) and Interleukin-18 (Il18). Mitochondria are strictly maternally inherited and mitochondrial ribosomal proteins regulate translation in mitochondria, but are encoded in the nuclear DNA (14). Mrpl48 is one of 4 Mrpl genes found in our companion studies, indicating parental control over the bioenergetics of neural cells. In the current study, Mrpl48 was identified as paternally expressed in the female POA, but not the male POA (Fig. 4A and fig. S5A). In the female POA, 8 of 9 Mrpl48 SNP sites exhibited a paternal expression bias in the F1i and F1r cross, 4 of which achieved statistical significance (P<0.05) (Fig. 4A). In contrast, none of the 9 SNP sites exhibited a paternal expression bias in the male POA. The female-specific paternal expression bias was confirmed in the POA by Sequenom (Fig. 4A).
Fig. 4.
Sex specific imprinted expression of Mrpl48 and Il18 in the female brain. (A) Illumina read data for an imprinted SNP in the 3′ UTR of Mrpl48 (highlighted in blue in (A); SNP_ID: uc009inh.1_801) indicates preferential expression of the paternal allele in the female, but not male POA (χ2 Analysis). Sequenom analysis confirmed the result (average allele frequency from 3 biological and 3 technical replicates). (B) Illumina read data for the imprinted SNP in ll18 (SNP_ID: uc009inh.1_801) indicates preferential expression of the maternal allele in the female mPFC, but not the male mPFC or the POA (χ2 Analysis). (C) QPCR analysis of Il18 expression in maternal versus paternal deletion Il18 heterozygous mice on C57 background reveals reduced expression in the mPFC of female maternal deletion mice relative to paternal deletion mice (n= 10, two-tailed, unpaired t-test, P = 0.0086). No difference was observed in the male mPFC (n=5) or the hypothalamus (females: n=5, males: n = 6). ***P<0.001; **P<0.01; *P<0.05. Red, maternal expression; Blue, paternal expression.
Il18 encodes a cytokine expressed by neurons, astrocytes and microglia, which modulates neuroinflammation, as well as homeostatic processes and behavior (15). Il18 has been linked to Multiple Sclerosis, a highly sexually dimorphic disease that predominates in women and is associated with parent-of-origin effects through the maternal lineage (16). We found Il18 to be preferentially expressed from the maternal allele in the female, but not male mPFC or the POA. We identified 2 SNP sites (3 bases apart) in one exon of Il18 in the female mPFC that indicate 74% of transcription from this region of the locus arises from the maternal allele (Fig. 4B and fig. S5B).
Il18 signaling has anorectic effects and heterozygous Il18 female, but not male mice, exhibit hyperphagia (17). We used qPCR to assess Il18 levels in the mPFC and the hypothalamus of ll18 heterozygous mice on a C57 background (Fig. 4C and fig. S5B). Loss of the maternal allele in the mPFC of Il18−/+ females, but not males, resulted in a 2.3-fold reduction in the level of Il18 expression relative to animals in which the paternal allele was deleted (Fig. 4C). No significant parent-of-origin effects were observed in the hypothalamus in males or females (Fig. 4C). These results are consistent with the preferential expression of the maternal allele in the female mPFC uncovered by the Illumina analysis.
Il18 is adjacent to SDHD (succinate dehydrogenase complex, subunit D) and Bcdo2 (Beta, beta-carotene 9′, 10′-dioxygenase variant 2) in mouse and human. Mutations in SDHD lead to head and neck paragangliomas in humans only when paternally inherited, yet previous studies have failed to detect imprinting at this locus (18). We found evidence for sex specific parent-of-origin effects in the mPFC, but not the POA, for both SDHD (male maternal bias) and Bcdo2 (female paternal bias) (fig. S6), suggesting a putative gene cluster with highly complex, region-specific and sex-specific parent-of-origin-effects. Future studies will be required to determine the existence an imprinting control region or other defining features of bona fide imprinted gene clusters.
Our data present evidence for epigenetic mechanisms by which parents may differentially influence gene expression in the brain of daughters versus sons, providing insights into sexually dimorphic epigenetic pathways recently uncovered in the brain (19). Some of the genes identified have known relevance to behavior and disease, although the mechanisms and functions of these parental effects are unclear. Previous analysis of X inactivation in the brain focused on very early stages of neural development and failed to observe any parental bias (20). The Xm bias may emerge during development through differential cell proliferation or survival, although, a few studies have suggested that X-inactivation in female somatic lineages favors selection of the Xm (21–23). The Xm enrichment contrasts with the paternal bias found among autosomal genes subject to sex specific imprinting and the 70% paternal bias of autosomal genes identified in our companion study (10). The X chromosome is enriched for genes involved in brain function (24, 25) and theoretical work has postulated that the maternally biased inheritance of the X selects for maternal interests (26, 27). Investigating the potential relationships between maternal and paternal gene expression programs may shed light on brain function, evolution, and disease.
Supplementary Material
Acknowledgments
We thank L. Luo, T. Maniatis, R. Losick, S. Hippenmeyer, and members of the Maniatis and Dulac labs for critical comments on the manuscript. We thank R. Hellmiss for help with figures and R. Jaenisch for Xegfp mice. This work was supported by the Klarman Foundation for Eating Disorders and the Howard Hughes Medical Institute (HHMI). CG is supported by a Human Frontiers long-term fellowship and Alberta Heritage Foundation for Medical Research Incentive Award. CD is an HHMI investigator. All data can be found in GEO: GSE22131.
References and Notes
- 1.Reik W, Walter J. Nat Rev Genet. 2001;2:21. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.BJB, BKJ, GN, HE, HJP, YE . Sex Differences in the Brain: From Genes to Behavior. Oxford University Press, Inc; New York, New York: 2008. [Google Scholar]
- 3.Hall JG. Am J Hum Genet. 1990;46:857. [PMC free article] [PubMed] [Google Scholar]
- 4.Bartolomei MS, Tilghman SM. Annu Rev Genet. 1997;31:493. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 5.Payer B, Lee JT. Annu Rev Genet. 2008;42:733. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 6.Skuse DH, et al. Nature. 1997;387:705. doi: 10.1038/42706. [DOI] [PubMed] [Google Scholar]
- 7.Raefski AS, O’Neill MJ. Nat Genet. 2005;37:620. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- 8.Davies W, et al. Nat Genet. 2005;37:625. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- 9.Hager R, Cheverud JM, Leamy LJ, Wolf JB. BMC Evol Biol. 2008;8:303. doi: 10.1186/1471-2148-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregg C, et al. Science companion study. 2010 [Google Scholar]
- 11.Heard E, Clerc P, Avner P. Annu Rev Genet. 1997;31:571. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, et al. Nat Genet. 2001;28:371. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- 13.Keverne EB. Adv Genet. 2007;59:217. doi: 10.1016/S0065-2660(07)59008-5. [DOI] [PubMed] [Google Scholar]
- 14.Koc EC, et al. J Biol Chem. 2001;276:43958. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 15.Alboni S, Cervia D, Sugama, Conti J Neuroinflammation. 2010;7:9. doi: 10.1186/1742-2094-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrera BM, et al. Neurology. 2008;71:799. doi: 10.1212/01.wnl.0000312377.50395.00. [DOI] [PubMed] [Google Scholar]
- 17.Zorrilla, et al. Proc Natl Acad Sci USA. 2007;104:11097. [Google Scholar]
- 18.Baysal BE. American journal of medical genetics Part C, Seminars in medical genetics. 2004;129C:85. doi: 10.1002/ajmg.c.30018. [DOI] [PubMed] [Google Scholar]
- 19.McCarthy MM, et al. J Neurosci. 2009;29:12815. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan SS, Williams EA, Tam P. Nat Genet. 1993;3:170. doi: 10.1038/ng0293-170. [DOI] [PubMed] [Google Scholar]
- 21.Chadwick LH, Willard HF. Mamm Genome. 2005;16:691. doi: 10.1007/s00335-005-0059-2. [DOI] [PubMed] [Google Scholar]
- 22.Fowlis DJ, Ansell JD, Micklem HS. Genet Res. 1991;58:63. doi: 10.1017/s001667230002961x. [DOI] [PubMed] [Google Scholar]
- 23.Falconer DS, Isaacson JH, Gauld IK. Genet Res. 1982;39:237. doi: 10.1017/s0016672300020930. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen DK, Disteche CM. Nat Genet. 2006;38:47. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 25.Zechner U, et al. Trends Genet. 2001;17:697. doi: 10.1016/s0168-9525(01)02446-5. [DOI] [PubMed] [Google Scholar]
- 26.Haig D. Evolution. 2006;60:440. [PubMed] [Google Scholar]
- 27.Haig D. Cytogenet Genome Res. 2006;113:68. doi: 10.1159/000090816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.