Abstract
Targeting of secretory and membrane proteins by the signal recognition particle (SRP) is evolutionarily conserved, and the multidomain protein SRP54 acts as the key player in SRP-mediated protein transport. Binding of a signal peptide to SRP54 at the ribosome is coordinated with GTP binding and subsequent complex formation with the SRP receptor. Because these functions are localized to distinct domains of SRP54, communication between them is essential. We report the crystal structures of SRP54 from the Archaeon Sulfolobus solfataricus with and without its cognate SRP RNA binding site (helix 8) at 4-Å resolution. The two structures show the flexibility of the SRP core and the position of SRP54 relative to the RNA. A long linker helix connects the GTPase (G domain) with the signal peptide binding (M) domain, and a hydrophobic contact between the N and M domains relates the signal peptide binding site to the G domain. Hinge regions are identified in the linker between the G and M domains (292-LGMGD) and in the N-terminal part of the M domain, which allow for structural rearrangements within SRP54 upon signal peptide binding at the ribosome.
Protein transport to or across the plasma membrane in bacteria and the endoplasmic reticulum in eukaryotes is mediated by the signal recognition particle (SRP), a ubiquitous ribonucleoprotein particle (for reviews see refs. 1–3). SRP recognizes amino-terminal signal sequences of newly synthesized polypeptides at the ribosome, and the ribosome nascent chain SRP complex is then targeted to the membrane by an interaction between SRP and its cognate receptor (SR). In the presence of the translocon the signal peptide is released (4–6) and the translating polypeptide is translocated across or inserted into the membrane. The SRP pathway is regulated by the concerted action of GTPases in both the SRP and the SR (7–11). GTP binding is a prerequisite for SRP/SR interaction (7), and GTP hydrolysis in SRP and SR occurs after the signal peptide is released (8) and resolves the SRP/SR complex (12).
Although the SRP pathway is evolutionarily conserved, the composition of SRP and the SR varies widely. The complex mammalian SRP consists of six protein subunits (ranging from 9 to 72 kDa) and a 7SL RNA. In eubacteria SRP consists of only one protein subunit (Ffh, fifty-four-homologue) and a 4.5S RNA. SRP in archaea represents an intermediate between these two as only two polypeptides (homologues of SRP19 and SRP54) have been identified in archaeal genomes, and the SRP RNA of ≈310 nts resembles the mammalian 7SL RNA (for review see ref. 13). The SRP receptor consists of only one protein (FtsY, SRα homologue) in eubacteria and archaea, but two proteins (SRα and SRβ) in eukaryotes.
SRP54 is the only protein subunit that is conserved in all SRPs, and thus it is the key player in protein transport. SRP54 is essential for binding signal peptides (14–16) at the ribosome and for GTP-dependent complex formation with the SR (17, 18). SRP54 is a multidomain protein that consists of an N-terminal N domain (a four-helix bundle), followed by a central G (GTPase) domain and a C-terminal M (methionine-rich) domain. The M domain is responsible for binding signal peptide and the SRP RNA (14–16, 19). Homologues of the N and G domains are also present in the SR proteins (FtsY, SRα). The G domains of SRP54 and SRα define a distinct subfamily within the Ras-like G proteins (20), the SRP GTPases, which are structurally characterized by an insertion (I-box) in the effector region and a close interaction with the N domain (21, 22). The GTPases in both SRP54 and SRα stimulate each other upon complex formation and have been proposed to act as GTPase-activating proteins for each other (8, 9).
SRP function relies on the tightly controlled communication of SRP54 with external regulators (e.g., the ribosome, the SR, and the translocon) and on internal communication between the domains of SRP54. For example, the interaction of SRP with the ribosome increases the affinity of SRP54 for GTP (23). SRP54 interacts with the ribosomal proteins L23a and L35 (24) [and their homologues in Escherichia coli (25, 26)], and the presence of the receptor modulates the interaction between SRP54 and the ribosome (24). A role of SRP RNA in the regulation of the GTPase and the interaction with the SR has been shown (27, 28). Interdomain communication in SRP54 has been demonstrated by mutations in a highly conserved motif in the N domain affecting signal peptide binding in the M domain (29), and the mutation of a universally conserved glycine (Gly-254 in Sulfolobus solfataricus) in the G domain, giving rise to a lethal phenotype and reduced interaction with the SR (30). Both sites are located in the interface between the N and G domains, which has been proposed to be involved in a common intramolecular signaling mechanism (29, 31–33).
A number of structures of the NG domain of Ffh from eubacteria and SRP54 from archaea (22, 31, 32, 34) and the NG domain of FtsY from E. coli (21) have been reported. The structure of a major part of the M domain with and without RNA has also been determined for E. coli Ffh, human SRP54, and Ffh from Thermus aquaticus (35–38). The recent structures of the complex of part of the SRP RNA together with SRP19 and the M domain of human SRP54 (38) and of archaeal SRP19 and part of the SRP RNA (39) have provided insights into the assembly of SRP. However, as the linker region between the G and M domains, including the N-terminal part of the M domain, was not ordered in the only crystal structure of SRP54 available so far (35), there was no information on the 3D domain arrangement of SRP54 and on a SRP54/RNA complex. We have now solved the structure of the SRP54 from the archaeon S. solfataricus alone and in complex with the helix 8 of SRP RNA at 4-Å resolution. These structures reveal the position of SRP54 relative to the RNA, the spatial arrangement of the SRP54 domains, and a contact between the M and N domains. These data give insights into the molecular mechanism of SRP54 function.
Methods
Protein expression, purification, and crystallization were performed by using standard procedures as described (58). SRP54 has been shortened by 15 residues at the C terminus according to previously reported structures (35–37). The RNA includes 43 nts of the conserved RNA helix 8 (nucleotides 181–223 plus two non-native base pairs) (37). Crystals were obtained of both SRP54 alone and in complex with its binding site on SRP RNA. The data were processed with mosf lm/scala (40). The structures of the SRP54/RNA complex and SRP54 alone were solved independently by the method of molecular replacement using the programs beast (41) and amore (42). The NG domain of SRP54 from Acidianus ambivalens (PDB 1J8M, sequence identity 73%) was used as a search model (31). Although in both cases the crystals diffracted only to ≈4.0-Å resolution (Table 1, which is published as supporting information on the PNAS web site), the initial maps contained clear extra density in which the RNA and/or about half of the M domain could be placed by using the structure of RNA helix 8 in complex with the core region of the M domain from E. coli (37). The presence of well defined, previously unaccounted electron density allowed the tracing of the remaining sequence of SRP54, e.g., the linker region between the NG and the M domains as well as the finger loop. Model building was done with the program o (43). The model of the SRP54/RNA complex covers all amino acids in the construct (amino acids 1–432, the His tag is disordered) and complete helix 8 (Fig. 1D). In the structure of SRP54 alone residues 320–326 in the M domain before helix αM1 are not visible in the electron density. The high solvent content (60% for the crystals of SRP54, 80% for the SRP54/RNA complex), application of strict noncrystallographic symmetry and the use of additional RNA restraints allowed for one round of positional refinement by using a simulated annealing protocol in cns (44). Positional refinement has been performed to remove sterical clashes and account for the differences between the search model and our structures.
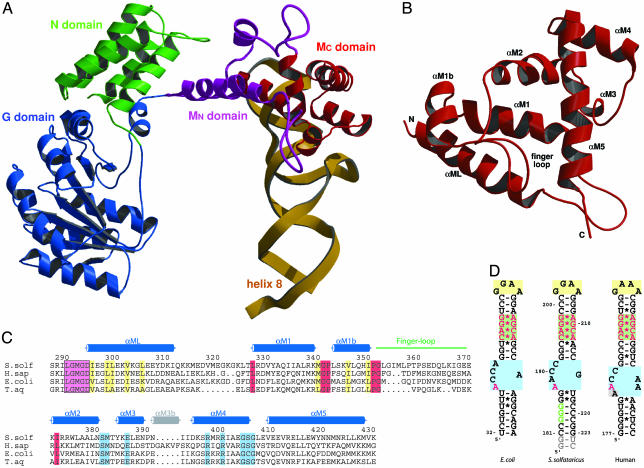
Fig. 1.
Overall structure of the SRP core. (A) Domain arrangement in SRP54 in complex with SRP RNA helix 8 shown in a ribbon representation of the N (green), G (blue), and M (purple/red) domains and the SRP RNA helix 8 (yellow). The novel N-terminal part of the M domain (MN) containing the linker helix (αML) and the closed finger loop is highlighted in purple. (B) Ribbon diagram of the M domain in a top view compared with A. The finger loop on top is folded into the hydrophobic groove, which is lined by helices αM1, αM1b, αM2, and αM5. (C) Structure-based sequence alignment of the M domain from different species (S. solf, S. solfataricus; H. sap, Homo sapiens; T. aq, T. aquaticus). Residue numbering corresponds to S. solfataricus SRP54. The secondary structure elements are indicated above the alignment. The highly conserved LGMGD motif (in a magenta box) and the long amphipathic linker helix (αML) are indicated. Regions involved in adjusting the finger loop for signal peptide binding are marked in red. Highly conserved hydrophobic residues involved in the contact with the N domain are shown in yellow. Residues involved in RNA binding are shown in blue. The short helix αM3b (gray) is found only in higher eukaryotes. (D) Nucleotide sequences of helix 8 of the SRP RNA from S. solfataricus used for this work (Center) compared with E. coli (Left) domain IV and human (Right) helix 8. The conserved features of helix 8 are highlighted: GNRA-type tetraloop (yellow box), symmetric loop (green box), and asymmetric loop (blue box). Nucleotides equivalent to the ones protected by the NG domain in A. fulgidus (179-GGG in A. fulgidus) (52) are in green letters, and nucleotides involved in SRP54M binding are highlighted in red. Non-native nucleotides added from the T7 promotor and for ribozyme cleavage are marked in gray.
Both crystal forms were highly twinned, and detwinning the data as perfectly merohedral twinned was essential to refine the structures (58). Details of the current structures are summarized in Table 1. All ribbon presentations were prepared by using the programs bobscript (45) and raster3d (46). The sequence alignment was done with clustalx (47), and Fig. 1C was done with the program alscript (48).
Results and Discussion
Overall Structure of the SRP Core. The crystal structures of S. solfataricus SRP54 were solved alone and in complex with helix 8 of the SRP RNA (see Methods). The linker region between the NG and the M domain is well ordered, allowing us to describe the thus far unknown arrangement of the domains of SRP54 (Fig. 1 A). SRP54 appears as an L-shaped molecule, with the NG domain represented by the longer arm of the L and the M domain represented by the shorter arm of the L. In the SRP54/RNA structure the RNA helix is approximately parallel to the long axis of the NG domain. This gives the complex the shape of the letter U. The angle between the two arms is ≈90°, giving the protein an overall extended and open conformation.
The structure of the NG domain of S. solfataricus SRP54 is similar to the previously determined structure of the NG domain from the closely related Archaeon A. ambivalens (31), which also belongs to the Crenarchaeota. When compared with the NG domain from T. aquaticus Ffh (22, 32, 34) a number of differences are observed. These are the unique dipeptide GY insertions in Crenarchaeota in the so-called switch II region, a rotation of the N domain relative to the G domain of ≈30° around an axis almost perfectly aligned with helix αN4, the conformation of the two loops connecting the helices αN1–2 and αN3–4 at the distal part of the N domain, and the defined structure of the closing loop (G5 region) in the absence of nucleotide. Recent data suggest that nucleotide binding to SRP54 might not be necessary for complex formation with SR in A. ambivalens (49). Therefore, the structural differences observed between T. aquaticus and the Crenarchaeota A. ambivalens and S. solfataricus might be functionally significant.
The structure of helix 8 (nucleotides 181–223) of the SRP RNA is essentially as reported for the homologous part (domain IV)ofthe E. coli RNA (37) (Fig. 1 A and D). In the S. solfataricus structure the RNA is involved in a crystal contact through stacking of the 5′ ends. Differences are present in the more distal part of the RNA in and below the asymmetric loop, but higher-resolution data would be needed to accurately describe the changes in this region. The interaction between the M domain and the asymmetric and symmetric loops of RNA helix 8 is essentially as described for the homologous part of E. coli SRP (37). The M domain has a unique all-helical fold, containing seven α-helices (Fig. 1 B and C). The C-terminal part of the M domain (MC, helices αM2–5) forms the stable core of this domain, which is involved in RNA binding (37). This region is almost identical to the corresponding part of previously reported M domain structures and the helices αM2–5 superimpose with an rms deviation of ≈1 Å. A comparison of the free and RNA-bound S. solfataricus structures shows that the MC domain binds to the RNA as a rigid body. The complete N-terminal part of the M domain (MN, helices αML-αM1b and the finger loop) is ordered in the SRP54/RNA complex and provides information on three main features of SRP54. (i) A flexible linker region (292-LGMGD) and a long linker helix (αML, residues 297–313) connect the G domain with the M domain, (ii) the finger loop (between αM1b and αM2) folds into the hydrophobic signal peptide binding site and is adjusted by hinge regions, and (iii) a contact exists between the N and M domains. The MN domain superimposes less well with the corresponding parts of other M domain structures, indicating a higher degree of flexibility.
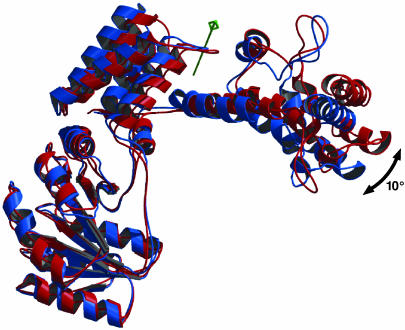
Position of the M Domain. In both S. solfataricus SRP54 structures (with and without RNA) the overall position of the M domain relative to the NG domain and the contact between the N and M domains are conserved despite the different environment in the two crystal forms (Fig. 2). This finding indicates that for S. solfataricus SRP54 the domain arrangement as seen in both structures is not an artifact of crystal packing. However, a superposition of the two S. solfataricus structures based on the G domain shows a movement of the M domain toward the NG domain. A more detailed analysis by the program dyndom (50) identifies a rotation axis close to the linker region, which runs through the interface between the N and M domains (NM interface).
Fig. 2.
Superposition of SRP54 with (red) and without (blue) RNA shown as a ribbon diagram. The RNA is omitted for clarity. A rotation axis (green) has been identified between the N and M domains by the program dyndom (50); the flexibility of SRP54 is indicated by a black arrow.
The observed rotation of ≈10° moves the most distal part of the M domain by ≈10 Å, indicating the flexibility of SRP54. When the RNA is modeled into the free SRP54, the two ends of the U-shaped structure would come in close proximity. A remarkable shape and charge complementarity is observed between the phosphoribose backbone of the minor groove of the RNA and a region in the G domain around residues 121–126 (Fig. 5, which is published as supporting information on the PNAS web site). Positively charged residues are conserved in this region of SRP54 (amino acids 121-KKRGYK), but not in the non-RNA binding SRα homologues (51). The affinity of SPR54 for the RNA decreases significantly upon deletion of the NG domain (52, 53) and a protection of the stem of helix 8 (domain IV) along the proximal strand by the NG domain has been reported for the E. coli (54) and Archaeoglobus fulgidus (52) SRP. The corresponding nucleotides (nucleotides 183–185) of the S. solfataricus RNA would be perfectly shielded by the G domain when the U closes as indicated in Fig. 5. The open structure of the SRP54/RNA complex may be favored by the stacking of the RNA in a crystal contact. Taken together, these findings suggest that the SRP core may be even more dynamic than observed in the S. solfataricus structures.
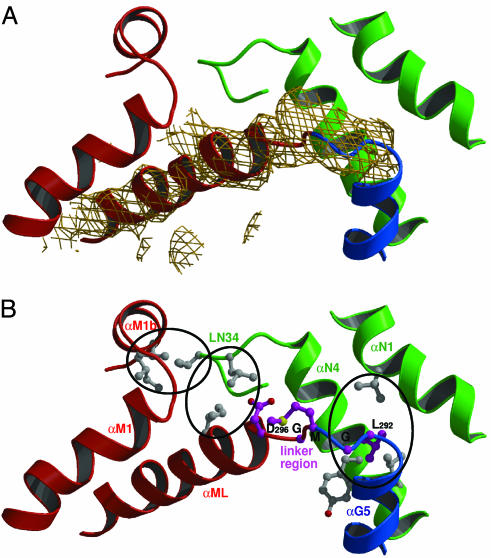
The GM Domain Linker Region. In both S. solfataricus SRP54 structures the linker region between the G and M domain (GM linker) is visible as continuous electron density (Fig. 3A and Fig. 6, which is published as supporting information on the PNAS web site), and the complete fold of the MN domain can now be described. The only direct contact between the G and the M domain is their connection in the protein sequence by the motif 292-LGMGD, which is highly conserved in all SRP54 sequences (51) (Fig. 1C). Based on the observed movement of the M domain relative to the NG domain this motif seems to form a hinge. The linker region packs against helices αN1 and αN4 of the N domain (Fig. 3B) with Leu-292 being involved in a conserved hydrophobic contact. The two glycines may allow for structural rearrangements in SRP54 when it binds to the ribosome nascent chain. This conserved motif is followed by a 17-aa-long α-helix (αML, linker helix), which packs against the second helix of the M domain (αM1) mainly through hydrophobic interactions. The intimate contact between αML and αM1 allows defining the linker helix as part of the M domain. From our data it seems unlikely that αM1 in a complete SRP54 could detach from the M domain and perform a helix swapping with another M domain, as it was reported for the isolated M domain of human SRP54 (36, 38).
Fig. 3.
Features of SRP54. (A) Unbiased 2m Fo – DFc electron density for the linker region and the linker helix (αML) contoured at 1.2 σ. N domain is green; G domain is blue, and M domain is red. The density has been calculated after rigid body refinement of the search model only [A. ambivalens NG domain (PDB ID code 1J8M)] (31). (B) Interactions in the interface of the N and M domains. The highly conserved LGMGD motif is shown in purple, and important hydrophobic residues are in gray and grouped by ovals.
Interaction Between N and M Domains. Only one region of interaction between the N and M domains is observed. The loop connecting the helices αN3 and αN4 (LN34, residues Glu-58 to Arg-66) at the distal end of the N domain is in close contact with the N-terminal region of αML and the C-terminal region of the short α-helix αM1b adjacent to the finger loop (Fig. 3B). In particular, Val-63 packs closely with Val-348 and Ile-352 in αM1b and the conserved Ile-64 interacts with Ile-300 in αML. In the SRP54 structure without RNA the LN34 loop is also involved in a hydrophobic contact with the M domain. The high degree of evolutionary conservation of these residues suggests a functional role of this contact. The helix αML is amphipathic and participates in the interaction with the N domain. It seems to form a “greasy slide” along which the hydrophobic interface may be adjusted to a certain extend, allowing for the observed flexibility of SRP54. In the T. aquaticus Ffh structure the linker between the G and M domains was disordered, and thus, the topological arrangement of M with respect to the NG domain was ambiguous (35). However, these crystals were obtained at high detergent concentrations (above the critical micellar concentration), which most likely break these hydrophobic contacts. None of the three positions described for the M domain corresponds to the one observed in the S. solfataricus structures.
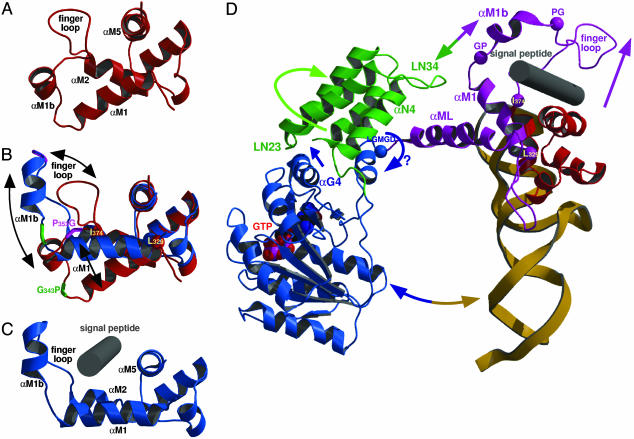
Finger Loop and Proposed Signal Peptide Binding Site. The binding site for signal peptides has been proposed to be contained within the M domain with some contributions from the NG domain (14, 15, 55, 56). A deep hydrophobic groove is formed by the helices αM1, αM2, and the C-terminal helix αM5. The finger loop (between helices αM1b and αM2, residues 353–373) has been implicated in signal peptide binding and could be built in previous structures only when the hydrophobic groove was occupied by parts of an adjacent molecule in the crystal (35–38), and thus was stabilized in an open conformation. In the S. solfataricus SRP54/RNA complex the structure of the proposed signal peptide binding site is different as it is not occupied by a crystallographic neighbor. The finger loop is involved in a crystal contact and folds into the signal peptide binding groove where it forms mainly hydrophobic contacts (Fig. 4A). The closed conformation of the finger loop in S. solfataricus, which differs significantly from previous structures, shields the hydrophobic groove from solvent, and may thus stabilize the protein when the binding site is not occupied. Superposition of the M domain structures of S. solfataricus and T. aquaticus based on the rigid part (MC domain) highlights four regions that may be important for adjusting the MN domain during signal peptide binding (Fig. 4B). Two conserved motifs (343-GP and 353-PG) flank the short helix αM1b. The helix-capping prolines maintain the structural integrity of the helix, whereas the glycines provide flexibility in respect to the adjacent helix αM1 and the finger loop. The MC domain is insulated from the changes in the MN domain at two points. One is at the N terminus of helix αM2 marking the end of the finger loop (Ile-374), and the other is at the beginning of αM1 (Leu-329), which has been implicated in RNA and signal peptide binding (57). However, from the S. solfataricus structure this residue seems to be rather important for anchoring αM1 in the hydrophobic core of the M domain as it superimposes well between the different structures of M domains. The angle of αM1 with respect to the MC domain varies significantly among the different M domain structures and may play a role in adjusting the hydrophobic groove for signal peptide binding. Taken together, superimposing the structures of the two M domains provides snapshots of the binding groove at different stages of signal peptide binding, with S. solfataricus representing a closed (empty) state and T. aquaticus an open state, which would be able to accommodate a hydrophobic signal peptide in a helical conformation as shown in the model presented in Fig. 4C.
Fig. 4.
Structure of the proposed signal peptide binding site. (A) Closed conformation of the hydrophobic groove in the S. solfataricus SRP54/RNA complex. The finger loop is folded into the groove. Helix αML is not shown for clarity. Elements involved in signal peptide binding are named. (B) Superposition of the M domain of S. solfataricus (red) and T. aquaticus (blue) to visualize the different conformations of the signal peptide binding groove including the finger loop and helix αM1b. Movements between structures are indicated by black arrows. The position of the conserved motifs GP (green) and PG (pink) differ significantly, the two “anchor” points (Leu-329 and Ile-374) are marked as spheres. (C) Structure of the M domain of T. aquaticus Ffh with the finger loop in an open conformation. A putative signal peptide (gray cylinder) is modeled into the binding site. (D) Model for the conformational changes in the SRP core. SRP54 is shown in a ribbon diagram; color code is as in Fig. 1 A. Rearrangements in SRP54 upon interaction with a signal peptide at the ribosome (see text) are indicated by arrows, the linker region LGMGD is indicated by a blue sphere, the anchor points Leu-329 and the N terminus of helix αM2 (Ile-374) as well as the GP and PG motifs are shown as pink spheres. The MN domain is adjusted at the four pink spheres to adopt a conformation competent for signal peptide binding as shown in C. The GTP (space-filling model) and the signal peptide (gray cylinder) are placed in their respective binding sites.
Interdomain Communication in SRP54. Because of the high degree of conservation of SRP54 and helix 8 of the SRP RNA between species (51) the structure of S. solfataricus SRP54/RNA complex (Fig. 1 A) can also serve as a model for the SRP core of eubacteria and of the mammalian SRP. The latter one can be derived from a superposition with the ternary complex of human SRP (38) without any sterical clashes (see Fig. 7, which is published as supporting information on the PNAS web site). Besides information on the 3D domain arrangement, the two S. solfataricus SRP54 structures presented here also provide insight into conformational flexibility in SRP54. The two structures suggest the following dynamic changes in SRP54 upon signal peptide binding at the ribosome (Fig. 4D). In the mammalian system SRP54 crosslinks with two ribosomal proteins (L23a and L35) in close proximity to the exit tunnel (24). The ribosome induces a structural rearrangement of SRP54 that leads to an increased GTP binding affinity (23). This may be achieved by resolving the interaction between the N and M domains, which would allow a rotation of the NG domain around the GM linker region. Signal peptide binding into the hydrophobic groove of the M domain of SRP54 opens the finger loop (Fig. 4 B and D). This involves a tilt of αM1 and a movement of helix αM1b, presumably via the GP/PG motifs, which may lead to a structure as observed in the M domain of T. aquaticus (Fig. 4C). The contact between the N and M domains would be directly influenced by this rearrangement because the interaction with the LN34 loop will be changed or lost (Fig. 4D). As a consequence, the N domain would no longer be fixed relative to the M domain and two interdomain rotations seem now to be enabled. One is around the conserved LGMGD linker motif in the loop connecting the G and the M domains. The other one is a rotation around helix αN4. This rotation is observed between the N and G domains of SRP54 from A. ambivalens (31) and T. aquaticus (≈30°), and in T. aquaticus (≈6°) depending on the nucleotide load (33). Helix αG4 in the interface between the N and G domain can sense this rotation when the LN23 loop moves toward the GTP binding site. In E. coli Ffh point mutations in the highly conserved ALLEADVN motif in helix αN2 and in the LN23 loop have indeed an effect on signal peptide binding (29) and the mutation of a conserved glycine in the αG4 helix leads to a loss of complex formation with the receptor (30). Taken together, these findings suggest that signal peptide binding to the M domain may result in a similar structural rearrangement in the NG domain interface as it is needed for, or induced by, GTP binding to the G domain. This would elegantly link signal sequence binding to the M domain with GTP binding to the G domain.
Conclusions
The flexibility of SRP54 described here for S. solfataricus allows for important structural rearrangements during the SRP cycle. The hinge region identified in the GM domain interface may play a crucial role for these rearrangements. The hydrophobic contact observed in SRP54 between the N and M domains relates the signal peptide binding site to the GTPase, which is the basis for signaling between these domains. Regions that have been implicated in the interaction of SRP54 with the ribosome (26) and the SR (31) are still available at the protein surface. The structure analysis of SRP bound to the ribosome is a prerequisite to understanding precisely how the ribosome influences the structure of SRP. In addition, the knowledge of the crystal structure of the SRP/SR complex is essential for understanding the regulation of the GTPases driving the SRP cycle.
Supplementary Material
Acknowledgments
We thank Ralf Moll (University of Lübeck, Lübeck, Germany) for genomic DNA of S. solfataricus; Bernhard Dobberstein (Zentrum fur Molekulare Biologie der Universität Heidelberg), Ivo Tews, and Matthew Groves for stimulating discussions; and the European Synchrotron Radiation Facility staff for providing excellent support at beamlines ID13 and ID29 at the European Synchrotron Radiation Facility in Grenoble, France. K.R.R. gratefully acknowledges support by the Research Council of Norway and the European Molecular Biology Laboratory Ph.D. Program. G.M. was supported by the Peter and Traudl Engelhorn Foundation. The work was supported by European Union-Network Grant QLK-3CT-2000-00082 and Deutsche Forschungsgemeinschaft Grant SFB-352 (to I.S.).
Abbreviations: SRP, signal recognition particle; SR, SRP receptor.
Data deposition: The coordinates and structure factors of the SRP54 and the SRP54/helix 8 complex have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1QZX and 1QZW, respectively).
References
- 1.Lutcke, H. (1995) Eur. J. Biochem. 228, 531–550. [DOI] [PubMed] [Google Scholar]
- 2.Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. (2001) Annu. Rev. Biochem. 70, 755–775. [DOI] [PubMed] [Google Scholar]
- 3.Walter, P. & Johnson, A. E. (1994) Annu. Rev. Cell Biol. 10, 87–119. [DOI] [PubMed] [Google Scholar]
- 4.Nicchitta, C. V. & Blobel, G. (1990) Cell 60, 259–269. [DOI] [PubMed] [Google Scholar]
- 5.Gorlich, D., Prehn, S., Hartmann, E., Kalies, K. U. & Rapoport, T. A. (1992) Cell 71, 489–503. [DOI] [PubMed] [Google Scholar]
- 6.Gorlich, D., Hartmann, E., Prehn, S. & Rapoport, T. A. (1992) Nature 357, 47–52. [DOI] [PubMed] [Google Scholar]
- 7.Connolly, T. & Gilmore, R. (1989) Cell 57, 599–610. [DOI] [PubMed] [Google Scholar]
- 8.Miller, J. D., Wilhelm, H., Gierasch, L., Gilmore, R. & Walter, P. (1993) Nature 366, 351–354. [DOI] [PubMed] [Google Scholar]
- 9.Powers, T. & Walter, P. (1995) Science 269, 1422–1424. [DOI] [PubMed] [Google Scholar]
- 10.Bacher, G., Pool, M. & Dobberstein, B. (1999) J. Cell Biol. 146, 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulga, T. A., Sinning, I., Dobberstein, B. & Pool, M. R. (2001) EMBO J. 20, 2338–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly, T., Rapiejko, P. J. & Gilmore, R. (1991) Science 252, 1171–1173. [DOI] [PubMed] [Google Scholar]
- 13.Zwieb, C. & Eichler, J. (2002) Archaea 1, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romisch, K., Webb, J., Lingelbach, K., Gausepohl, H. & Dobberstein, B. (1990) J. Cell Biol. 111, 1793–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zopf, D., Bernstein, H. D., Johnson, A. E. & Walter, P. (1990) EMBO J. 9, 4511–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutcke, H., High, S., Romisch, K., Ashford, A. J. & Dobberstein, B. (1992) EMBO J. 11, 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, D. I. & Dobberstein, B. (1980) J. Cell Biol. 87, 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilmore, R., Walter, P. & Blobel, G. (1982) J. Cell Biol. 95, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batey, R. T., Sagar, M. B. & Doudna, J. A. (2001) J. Mol. Biol. 307, 229–246. [DOI] [PubMed] [Google Scholar]
- 20.Bourne, H. R., Sanders, D. A. & McCormick, F. (1991) Nature 349, 117–127. [DOI] [PubMed] [Google Scholar]
- 21.Montoya, G., Svensson, C., Luirink, J. & Sinning, I. (1997) Nature 385, 365–368. [DOI] [PubMed] [Google Scholar]
- 22.Freymann, D. M., Keenan, R. J., Stroud, R. M. & Walter, P. (1997) Nature 385, 361–364. [DOI] [PubMed] [Google Scholar]
- 23.Bacher, G., Lutcke, H., Jungnickel, B., Rapoport, T. A. & Dobberstein, B. (1996) Nature 381, 248–251. [DOI] [PubMed] [Google Scholar]
- 24.Pool, M. R., Stumm, J., Fulga, T. A., Sinning, I. & Dobberstein, B. (2002) Science 297, 1345–1348. [DOI] [PubMed] [Google Scholar]
- 25.Ullers, R. S., Houben, E. N., Raine, A., ten Hagen-Jongman, C. M., Ehrenberg, M., Brunner, J., Oudega, B., Harms, N. & Luirink, J. (2003) J. Cell Biol. 161, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, S. Q., Peske, F., Wieden, H. J., Rodnina, M. V. & Wintermeyer, W. (2003) RNA 9, 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peluso, P., Shan, S. O., Nock, S., Herschlag, D. & Walter, P. (2001) Biochemistry 40, 15224–15233. [DOI] [PubMed] [Google Scholar]
- 28.Jagath, J. R., Matassova, N. B., de Leeuw, E., Warnecke, J. M., Lentzen, G., Rodnina, M. V., Luirink, J. & Wintermeyer, W. (2001) RNA 7, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newitt, J. A. & Bernstein, H. D. (1997) Eur. J. Biochem. 245, 720–729. [DOI] [PubMed] [Google Scholar]
- 30.Lu, Y., Qi, H. Y., Hyndman, J. B., Ulbrandt, N. D., Teplyakov, A., Tomasevic, N. & Bernstein, H. D. (2001) EMBO J. 20, 6724–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montoya, G., te Kaat, K., Moll, R., Schafer, G. & Sinning, I. (2000) Struct. Folding Des. 8, 515–525. [DOI] [PubMed] [Google Scholar]
- 32.Freymann, D. M., Keenan, R. J., Stroud, R. M. & Walter, P. (1999) Nat. Struct. Biol. 6, 793–801. [DOI] [PubMed] [Google Scholar]
- 33.Ramirez, U. D., Minasov, G., Focia, P. J., Stroud, R. M., Walter, P., Kuhn, P. & Freymann, D. M. (2002) J. Mol. Biol. 320, 783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Padmanabhan, S. & Freymann, D. M. (2001) Structure (London) 9, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keenan, R. J., Freymann, D. M., Walter, P. & Stroud, R. M. (1998) Cell 94, 181–191. [DOI] [PubMed] [Google Scholar]
- 36.Clemons, W. M., Jr., Gowda, K., Black, S. D., Zwieb, C. & Ramakrishnan, V. (1999) J. Mol. Biol. 292, 697–705. [DOI] [PubMed] [Google Scholar]
- 37.Batey, R. T., Rambo, R. P., Lucast, L., Rha, B. & Doudna, J. A. (2000) Science 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 38.Kuglstatter, A., Oubridge, C. & Nagai, K. (2002) Nat. Struct. Biol. 9, 740–744. [DOI] [PubMed] [Google Scholar]
- 39.Hainzl, T., Huang, S. & Sauer-Eriksson, A. E. (2002) Nature 417, 767–771. [DOI] [PubMed] [Google Scholar]
- 40.Collaborative Computing Project No. 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 41.Read, R. J. (2001) Acta Crystallogr. D 57, 1373–1382. [DOI] [PubMed] [Google Scholar]
- 42.Navaza, J. (1994) Acta Crystallogr. A 50, 157–163. [Google Scholar]
- 43.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 44.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 45.Esnouf, R. M. (1997) J. Mol. Graphics 15, 133–138. [DOI] [PubMed] [Google Scholar]
- 46.Merritt, E. A. & Bacon, D. J. (1997) Macromol. Crystallogr. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barton, G. J. (1993) Protein Eng. 6, 37–40. [DOI] [PubMed] [Google Scholar]
- 49.Moll, R. G. (2003) Biochem. J. 374, 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayward, S., Kitao, A. & Berendsen, H. J. (1997) Proteins 27, 425–437. [DOI] [PubMed] [Google Scholar]
- 51.Rosenblad, M. A., Gorodkin, J., Knudsen, B., Zwieb, C. & Samuelsson, T. (2003) Nucleic Acids Res. 31, 363–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diener, J. L. & Wilson, C. (2000) Biochemistry 39, 12862–12874. [DOI] [PubMed] [Google Scholar]
- 53.Samuelsson, T. & Olsson, M. (1993) Nucleic Acids Res. 21, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lentzen, G., Moine, H., Ehresmann, C., Ehresmann, B. & Wintermeyer, W. (1996) RNA 2, 244–253. [PMC free article] [PubMed] [Google Scholar]
- 55.Cleverley, R. M. & Gierasch, L. M. (2002) J. Biol. Chem. 48, 46763–46768. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, N. & Gierasch, L. M. (1997) Mol. Cell. 1, 79–87. [DOI] [PubMed] [Google Scholar]
- 57.Huang, Q., Abdulrahman, S., Yin, J. & Zwieb, C. (2002) Biochemistry 41, 11362–11371. [DOI] [PubMed] [Google Scholar]
- 58.Rosendal, K. R., Sinning, I. & Wild, K. (2004) Acta Crystallogr. D, in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.