Abstract
Sphingobium chlorophenolicum completely mineralizes PCP (pentachlorophenol). Two GSTs (glutathione transferases), PcpC and PcpF, are involved in the degradation. PcpC uses GSH to reduce TeCH (tetrachloro-p-hydroquinone) to TriCH (trichloro-p-hydroquinone) and then to DiCH (dichloro-p-hydroquinone) during PCP degradation. However, oxidatively damaged PcpC produces GS-TriCH (S-glutathionyl-TriCH) and GS-DiCH (S-glutathionyl-TriCH) conjugates. PcpF converts the conjugates into TriCH and DiCH, re-entering the degradation pathway. PcpF was further characterized in the present study. It catalysed GSH-dependent reduction of GS-TriCH via a Ping Pong mechanism. First, PcpF reacted with GS-TriCH to release TriCH and formed disulfide bond between its Cys53 residue and the GS moiety. Then, a GSH came in to regenerate PcpF and release GS–SG. A TBLASTN search revealed that PcpF homologues were widely distributed in bacteria, halobacteria (archaea), fungi and plants, and they belonged to ECM4 (extracellular mutant 4) group COG0435 in the conserved domain database. Phylogenetic analysis grouped PcpF and homologues into a distinct group, separated from Omega class GSTs. The two groups shared conserved amino acid residues, for GSH binding, but had different residues for the binding of the second substrate. Several recombinant PcpF homologues and two human Omega class GSTs were produced in Escherichia coli and purified. They had zero or low activities for transferring GSH to standard substrates, but all had reasonable activities for GSH-dependent reduction of disulfide bond (thiol transfer), dehydroascorbate and dimethylarsinate. All the tested PcpF homologues reduced GS-TriCH, but the two Omega class GSTs did not. Thus PcpF homologues were tentatively named S-glutathionyl-(chloro)hydroquinone reductases for catalysing the GSH-dependent reduction of GS-TriCH.
Keywords: glutathione conjugate, glutathione transferase (GST), glutathione-dependent reductase, pentachlorophenol, S-glutathionyl-hydroquinone
INTRODUCTION
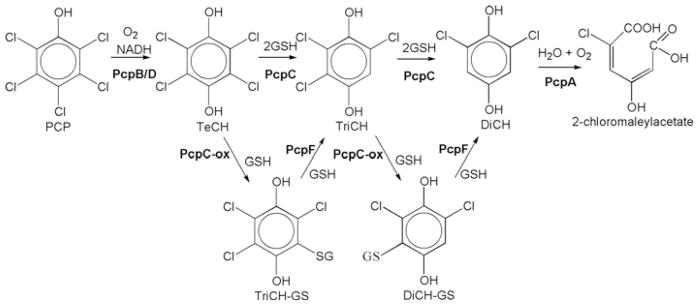
PCP (pentachlorophenol) was first synthesized in 1872 [1], but its massive release into the environment started in the late 1930s, associated with its use as a wood preservative [2]. Since then there has been no evidence of natural production of PCP, although some bacteria have evolved the ability to degrade it within the past 100 years. Sphingobium chlorophenolicum ATCC39723 was isolated in 1985 [3]. The genes and enzymes involved in the degradation pathway of PCP have been identified and characterized from the bacterium (Figure 1) [4]. PcpB, a flavin-containing mono-oxygenase, oxidizes PCP to tetrachloro-p-benzoquinone, which can be chemically or enzymatically reduced by PcpD to TeCH (tetrachloro-p-hydroquinone) [5,6]. PcpC, a GST (glutathione transferase), uses GSH to reductively dechlorinate TeCH to TriCH (trichloro-p-hydroquinone) and then to DiCH (2,6-dichloro-p-hydroquinone) [7,8]. PcpA, a dioxygenase, cleaves the aromatic ring of DiCH to produce 2-chloromalaylacetate, a relatively common metabolic intermediate in the degradation of chlorinated aromatic compounds [9]. A cysteine residue at the active site of PcpC can be oxidatively damaged, and the damaged PcpC produces GS (S-glutathionyl)-TriCH and GS-DiCH conjugates [10]. PcpC cannot transform the conjugates, but PcpF converts the GS-TriCH and GS-DiCH back into TriCH and DiCH respectively [11]. When the pcpF gene is disrupted, S. chlorophenolicum degrades PCP more slowly and becomes more sensitive to PCP. Consequently, the accumulation of the conjugates is detrimental to the bacterium. Thus PcpF plays two roles: channelling the conjugates back to the metabolic pathway and preventing the accumulation of the harmful conjugates.
Figure 1. Pentachlorophenol degradation pathway of S. chlorophenolicum ATCC 39723.
PcpB, PCP 4-monooxygenase; PcpD, tetrachloro-p-benzoquinone reductase; PcpC, TeCH reductive dehalogenase; PcpC-ox, oxidatively damaged PcpC; PcpA, DiCH 1,2-dioxygenase; PcpE, chloromaleylacetate reductase; PcpF, GS-(chloro)hydroquinone reductase.
PcpC and PcpF fall into different classes of GSTs. The two proteins share 17.7 % sequence identity with a global alignment score of −48. A BLASTP search shows the N-terminus of PcpC is most similar to the Zeta class of GSTs and the C-terminus is highly homologous with the Beta class of GSTs. PcpC has been assigned to the Zeta class, but it does not have high sequence identity with any GSTs. The closest relative is a hypothetical GST (GenBank® accession number AAZ25344) of Colwellia psychrerythraea 34H (27.3 % sequence identity) as determined by a TBLASTPN search of bacterial genomes. In contrast, PcpF is highly conserved in bacteria and in Saccharomyces cerevisiae [12]. S. cerevisiae has three PcpF homologues, and ECM4 (extracellular mutant 4), a protein involved in cell-surface biosynthesis and architecture, is the most similar to PcpF. Although ECM4 has been characterized as an Omega class GST (GTO2) for its ability to use some substrates of the Omega class GSTs [12], it shares less than 20 % sequence identity with other Omega class GSTs. The conserved domain database has an ECM4 as group COG0435, which is not assigned to the Omega class of GSTs [13] (COGs are assigned for clusters of orthologous groups of proteins, and orthologues occupy the same functional niche in different species).
In the present study, we determined that PcpF, ECM4 and two bacterial PcpF homologues used GS-TriCH as a substrate, whereas the two human Omega class GSTs did not. The substrate difference and sequence analysis suggest that PcpF and homologues, including ECM4, belong to a new class of GSTs: S-glutathionyl-(chloro)hydroquinone reductases. The reaction mechanism for GS-TriCH reduction was also investigated.
EXPERIMENTAL
Chemicals and enzymes
All chemicals were obtained from Sigma–Aldrich or Fisher Scientific. S. cerevisiae glutathione reductase was obtained from Sigma–Aldrich. Restriction enzymes were obtained from New England Biolabs. PCR was performed with Taq DNA polymerase and primers from Invitrogen.
Bacterial strains, plasmids and culture conditions
Escherichia coli strains BL21(DE3) and M15, containing the pRep4 (Qiagen) antibiotic resistance plasmid, were grown in LB (Luria–Bertani) medium or on LB agar plates at 37 °C or as specified. Kanamycin (30 μg/ml) was added to the LB medium when required. The pET30-LIC plasmid carrying the pcpF-C53A mutant gene has been described previously [11]. The genes coding for PcpF, YqjG (GenBank® accession number NP_417573) of E. coli, YqjG (GenBank® accession number YP_295669) of Ralstonia eutropha (also known as Cupriavidus necator), JMP134 and ECM4 (GenBank® accession number NP_013002) of S. cerevisiae were cloned into pET30-LIC with a C-terminal His6-tag. The cloning was essentially the same as described previously [11]. Briefly, genomic DNA was isolated with PureGene DNA Isolation Kit (Gentra). The target gene was amplified from the genomic DNA with designed primers (available upon request from L.X.). The PCR product was digested by NdeI and HindIII (or another restriction enzyme). The digested PCR products were ligated into pET30-LIC vector (Novagen). The ligation products were electroporated into E. coli BL21(DE3). Clones were confirmed by colony PCR and sequencing. The correct clones were directly used for protein production. N-terminally His6-tagged hGSTO (human glutathione transferase Omega class) 1 and hGSTO2-C4 {a stabilized variant that contains five cysteine residue substitutions (C80S, C121S, C136S, C140S, C170S and C214S) and a deletion of last four amino acid residues at the C-terminus (Phe240, Gly241, Leu242 and Cys243); the modified enzyme retains 70 % of the dehydroascorbate reductase activity of the wild-type enzyme [14]} were cloned in to the pQE30 vector with E. coli M15 (carrying pRep4) as host as described previously.
Protein purification
E. coli stains carrying the cloned gene on an expression vector were grown in 1 litre of LB medium at 37 °C to D = 0.6 at 600 nm, induced with 0.2 mM isopropyl-β-D-thiogalactopyranoside, and then incubated at room temperature (23 °C) for 5 h. PcpF-C14S was purified as described previously [11]. All other proteins were purified with Ni-NTA (Ni2+-nitrilotriacetate)–agarose resin, according to the manufacture’s instructions (Qiagen). Targeted proteins were concentrated and the buffer was changed to potassium phosphate buffer [40 mM potassium phosphate, pH 7, containing 2 mM DTT (dithiothreitol) with Centriprep Ultracel YM-10 (Millipore)]. Glycerol was added to 10 % (v/v) before storage at −80 °C.
Enzyme assays
GS-TriCH reduction was analysed by HPLC as previously described [11]. For kinetic analysis, a spectrometric assay was developed, using 1 ml of 70 mM potassium phosphate buffer, pH 6.5, containing 50 μM GS-TriCH, 2 mM ascorbic acid, 5 mM GSH, 300 μM NADPH and 1 μl of glutathione reductase (G3664–500UN; Sigma). The reaction was monitored at 340 nm. Then, 1–5 μl of PcpF, or other GSTs, was added and the decrease at 340 nm for NADPH consumption was continuously monitored. The change in slope after PcpF addition was used to calculate the enzymatic rate. For kinetic analysis, GSH was varied from 0.2 to 10 mM and GS-TriCH from 5 to 200 μM. GS-TriCH was prepared by using ultracentrifuged E. coli cell extracts containing overproduced PcpC-C14S in essentially the same manner as described previously [11] with some modifications. Specifically, reactions were carried out in 1 ml of 70 mM potassium phosphate buffer, pH 6.5, containing 2 mM ascorbic acid, 0.5 mM GSH, 200 μM TeCH and 20 μl of PcpC-C14S (ultracentrifuged cell extracts of overexpressing E. coli, 27.6 mg/ml). The sample was incubated at 30 °C for 10 min and then heated at 65 °C for 10 min. The quantitative conversion of TeCH into GS-TriCH was confirmed by HPLC analysis as described previously [11]. The denatured proteins were removed by centrifugation (20 000 g for 2 min), and the supernatant was stored at 4 °C, which was stable for at least 1 week.
Similar spectrometry assays were used to analyse the kinetic parameters with cysteine, 2-mercaptoethanol and DTT as co-substrate rather than GSH. Again, the reactions were started and NADPH consumption was monitored for 1 min. Then, PcpF was added. The slope difference was used to calculate PcpF activity.
The GST activity was measured with 0.2 mM CDNB (1-chloro-2,4-dinitrobenzene), or ethacrynic acid, as the substrate in 100 mM potassium phosphate buffer, pH 6.5, containing 0.2 % Triton X-100, and the continuous increase in absorbance at 340 nm (or 270 nm) was monitored [15]. Thiol transferase activity was measured with HED (2-hydroxyethyl disulfide) as a substrate [16]. DHA (dehydroascorbate) reductase assay and DMAV (dimethylarsinate) reductase assay were preformed using established protocols [17]. The S-(2′,4′-dichlorophenacyl)glutathione reductase activity was assayed by following the decrease in absorption at 276 nm as described previously [18], except that the GSH concentration was increased to 5 mM for all the assays except for with ethacrynic acid, which had a very high non-enzymatic reactivity at 5 mM GSH. The rates before and after enzyme addition were recorded. The difference was used to calculate the enzyme activities.
Static light-scattering
The average molecular mass of individual GSTs was measured by combined size-exclusion chromatography and multi-angle laser light scattering. Briefly, 100 μg of GST in a freshly prepared buffer (1 × PBS) was loaded on to a BioSep-SEC-S2000 column (Phenomenex) and eluted isocratically with a flow rate of 0.5 ml · min−1. The elute was passed, in tandem, through a UV detector and a Dawn EOS laser light-scattering detector (Wyatt Technology). Scattering results were analysed using the Zimm-fitting method with software (ASTRA) provided by the instrument manufacturer.
Protein sequence analysis
TBLASTN (searching a translated nucleotide database using a protein query) was used to identify PcpF homologues in sequenced genomes at the website of PubMed. Protein sequences were aligned by using ClustalW [19] and a phylogenetic tree was generated by using the neighbour-joining method of MEGA version 4.0.2 [20]. The evolutionary distances were computed using Poisson correction and were in units of the number of amino acid substitutions per site. Optimal global alignment of two protein sequences was performed with the ALIGN program of Biology Workbench (http://workbench.sdsc.edu). All of the analyses were performed with default parameters.
RESULTS
BLAST search of PcpF homologues
BLAST searches revealed that PcpF had homologues in selected species in all domains of life. A TBLASTN search of archaeal genomes revealed the presence of PcpF homologues in every genome (nine in total) in the order of Halobacteriales. However, no significant similarities were found in any other orders within the archaea. In bacteria, PcpF had many homologues in the phyla of actinobacteria, cyanobacteria, firmicutes (containing orders of Bacillales, Clostridia and Lactobacillales) and proteobacteria. The top 100 hits against bacterial genomes showed sequence identity from 55 % to 65 %. In eukaryotes, PcpF homologues were well conserved in fungi and plants, but not in animals. The top 100 hits against fungal genomes showed sequence identity from 36 % to 50 %. Some species had more than one homologue, e.g. S. cerevisiae had three homologues, GTO1, ECM4 and GTO3 [12]. PcpF homologues were present in every sequenced plant genome, and some plants had several PcpF homologues. For example, Arabidopsis thaliana had four homologues (GenBank® accession numbers, NP_199315, NP_001031671, NP_199312 and NP_568632). These conserved homologues had at least 30 % sequence identity with PcpF by global alignment. PcpF homologues were not found by TBLASTN search of the sequenced genomes in most archaea, some bacteria and all animal species.
Sequence comparison of PcpF with selected proteins by global alignment
The proteins identified by TBLASTN were further compared by global alignment. The R. eutropha JMP134 homologue [Re-YqjG (R. eutropha YqjG) hypothetic protein] was the fourth most homologous with PcpF (64 % identity by TBLASTN) and the E. coli homologue [Ec-YqjG (E. coli YqjG) hypothetic protein] was not on the top-100 list of TBLASTN search of bacterial genomes. Ec-YqjG was selected for further analysis because it was from E. coli, a model organism. PcpF and Re-YqjG shared 60.7 % sequence identity with a global alignment score of 1355. The two YqjGs had 63 % identity and a global alignment score of 1455. PcpF and Ec-YqjG had 51.7 % sequence identity with a global alignment score of 1107. A. thaliana had four PcpF homologues listed as ECM4-like proteins with sequence identity ranging from 31.2 to 41.8 % by global alignment. ECM4a (GenBank® accession number NP_199315) of A. thaliana was the most similar to PcpF, sharing 41.8 % sequence identity with a global alignment score of 845. S. cerevisiae had three PcpF homologues, and ECM4 was the most homologous to PcpF with 35.4 % sequence identity and a global alignment score of 625. As the three yeast homologues are characterized as Omega class GSTs [12], we compared the sequence of PcpF with that of hGSTO1-1, the most characterized Omega class GST [21]. The two proteins had a sequence identity of 20.1 % with a negative global alignment score of −15. PcpF and hGSTO2-2 shared only 18.6 % sequence identity with a global alignment score of −24. Furthermore, ECM4 and hGSTO1-1 had 15.7 % sequence identity with a global alignment score of −128.
Phylogenetic analysis of GSTs with a catalytic cysteine residue
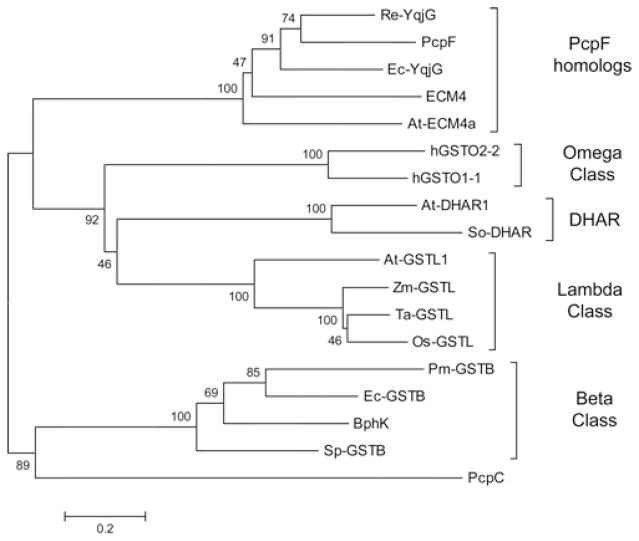
Sequence analysis is the primary basis for the classification of GSTs. Well characterized classes include: Alpha, Mu, Pi and Kappa of mammalian GSTs [22]; Phi, Tau, Lambda, and DHAR (dehydroascorbate reductase) of plant GSTs [23]; Delta from insects [24]; and Beta of bacterial origin. In addition, Theta, Zeta and Omega classes of GSTs are widely present in a variety of life forms [21,24]. The GSTs can also be grouped on the basis of active-site residues [22]. The Alpha, Mu and Pi classes of GSTs have an N-terminal tyrosine residue that interacts with GSH to stabilize the thiolate anion; a serine residue fulfils the function in the Theta, Zeta, Kappa, Phi and Tau classes [25]; and an N-terminal cysteine residue plays the catalytic role in Beta, Omega, Lambda and DHAR classes [23]. PcpF contains a catalytic cysteine residue at its N-terminus [11]. Therefore, PcpF and representative GSTs from each class containing the active cysteine residue were analysed by multi-sequence alignment, and the relationship was presented in a phylogenetic tree (Figure 2). The PcpF homologues selected for alignment were Re-YqjG and Ec-YqjG of bacteria, At-ECM4a (A. thaliana ECM4a) of plant and ECM4 of yeast. PcpF and homologues formed a distinct group, more related to the Omega, Lambda and DHAR classes than to the Beta class. PcpC was also included in the analysis because it catalyses GSH-dependent dechlorination of TeCH [7] and an N-terminal cysteine residue is involved in the catalysis [10]. PcpC has been characterized as a Zeta class GST [26].
Figure 2. Dendrogram of inferred phylogenetic relationships among GSTs with an N-terminal cysteine residue known or assumed to interact with the thiol group of bound GSH.
The tree was constructed using the neighbour-joining method. The numbers on the branches are bootstrap values (percentage of 1000 runs), indicating the frequency of grouping in the cluster. The distance correlates to the number of amino acids substituted per site. The GSTs and corresponding GenBank® protein accession numbers are as follows: Ec-YqjG, NP_417573; Re-YqjG, YP_295669; ECM4, NP_013002; PcpF, AAM96671; At-ECM4a, NP_199315; Zm-GSTL, CAA41447; Os-GSTL, AAF70831; At-GSTL1, AL162973; Ta-GSTL, CAA76758; hGSTO1-1, NP_004823; hGSTO2-2; NP_899062; So-DHAR, AAG24945; At-DHAR1, AAF98403; BphK, YP_556402; Sp-GSTB, 1F2ED; Pm-GSTB, 2PMTA; Ec-GSTB, 1A0F_A; PcpC, AAM96673 Os, Oryza sativa; Pm, Proteus mirabilis; So, Spinacia oleracea; Ta, Triticum aestivum; Zm, Zea mays.
Multiple sequence alignment and amino acid residues in substrate binding
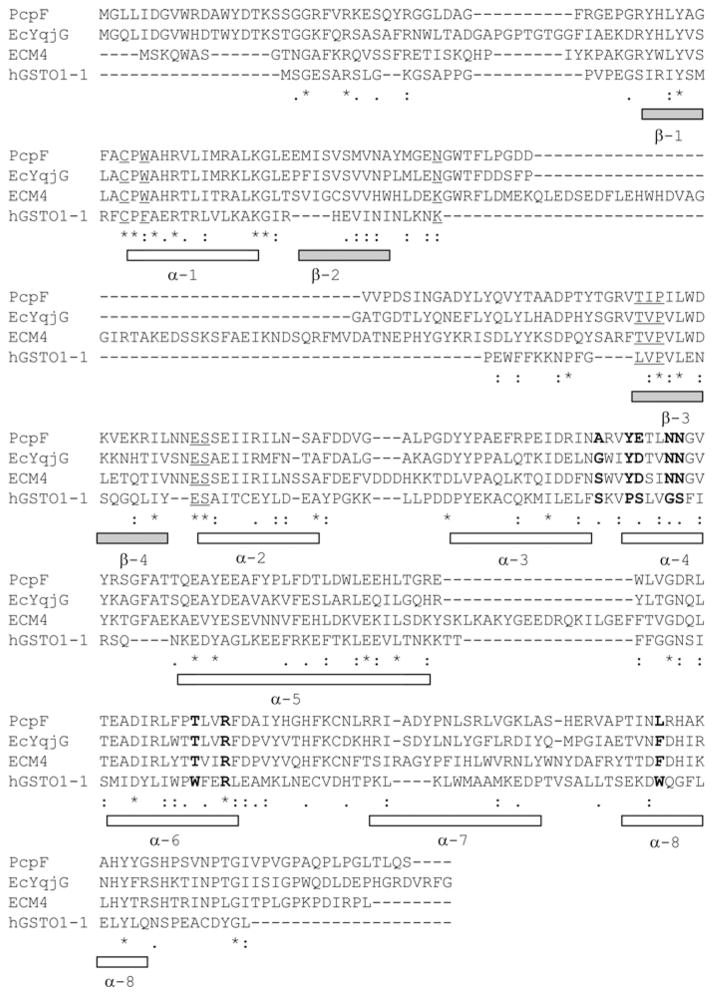
Among the GSTs in the Omega, Lambda, DHAR and PcpF homologues, only the structure of hGSTO1-1 is available. Three PcpF homologues and hGSTO1-1 were aligned by ClustalW. The secondary structure of hGSTO1-1 is presented for reference (Figure 3). Despite the fact that hGSTO1-1 was much smaller than PcpF homologues, the amino acid residues (Cys32, Phe34, Lys59, Leu71, Val72, Pro73, Glu85 and Ser86; Figure 3, underlined residues) involved in the binding of GSH in hGSTO1-1 were well conserved in the other proteins. The amino acid residues (Ser121, Pro124, Ser125, Gly128, Ser129, Trp180, Arg183 and Trp222; Figure 3, bold residues), positioned for the binding of the second substrate, were not conserved between hGSTO1-1 and PcpF homologues, except for Arg183, but the corresponding amino acid residues were highly conserved among the three PcpF homologues (Figure 3, bold residues). The variation in key residues for substrate binding implies that PcpF homologues may have a different substrate range from that of hGSTO1-1.
Figure 3. Alignment of peptide sequences of PcpF homologues with hGSTO1-1.
GenBank® accession numbers for the indicated protein sequences are given Figure 2. Underlined amino acid residues are involved in GSH-binding, and bold residues are involved in the binding of the second substrate. The secondary structures (α-helixes and β-sheets) of hGSTTO1-1 are shown for reference.
Recombinant enzyme production and purification
The pcpF gene was re-cloned as a C-terminally His6-tagged protein that was over-produced as a major soluble protein in E. coli. The over-produced protein was purified by Ni-NTA–agarose resin. The His6 tag did not affect the specific activity of PcpF for GS-TriCH reduction [11]. Thus the His6-tagged PcpF was used to further characterize its activities. ECM4 has been characterized as an Omega class GST [12], but phylogenetic analysis grouped ECM4 with PcpF homologues rather than the Omega class GSTs (Figure 2). In order to compare PcpF homologues with the Omega class of GSTs, Re-YqjG, Ec-YqjG and ECM4 were cloned in expression vector pET30-LIC, as C-terminally His6-tagged proteins. The GSTs and hGSTO1-1 and hGSTO2-2 were produced in E. coli as His6-tagged proteins and purified by Ni-NTA–agarose resin.
Substrate specificities of PcpF and related GSTs
PcpF reduces GS-TriCH and GS-DiCH to TriCH and DiCH with GSH as a co-substrate; however, little is known about PcpF as a GST [11]. The most highly related GST that has been characterized is ECM4, assigned to the Omega class [12]. Thus the substrates of Omega class GSTs were tested with PcpF. For comparison, ECM4, GSTO1-1, GSTO2-2 and two bacterial PcpF homologues (Ec-YqjG and Re-YqjG) were tested. Among the substrates tested (Table 1), the human GSTs did not use GS-TriCH as a substrate, whereas the other enzymes did. In addition, only the human GSTs used CDNB as a substrate, but the activity was very weak. In comparison, Proteus mirabilis GSTB1–1 (Pm-GSTB) has a specific activity of 1.1 μmol · min−1 · mg−1 of protein [27], which is 50 times higher than that of GSTO1-1 (Table 1). All the enzymes reduced the disulfide bond (thiol transfer), DHA and DMAV with good activities. Most of them had highest activity as a thiol transferase, using HED as the substrate, except for GSTO2-2, which had the highest activity for DHA reduction. Furthermore, all the enzymes had very low activity for transferring GSH to ethacrynic acid. S-(Phenacyl)glutathiones are specific substrates of GSTO1-1; hGST2-2 does not use them [18]. Among the PcpF homologues, only PcpF used S-(2′,4′-dichlorophenacyl)glutathione. Thus S-(phenacyl)glutathiones are not specific substrates for either the Omega class or PcpF homologues. A mutant PcpF-C53A, cannot reduce GS-TriCH [11]. We further tested PcpF-C53A with the substrates listed in Table 1. The mutant protein showed a similar activity compared with PcpF for transferring GSH to ethacrynic acid, but lost the activities towards all the other tested substrates.
Table 1.
Activities of recombinant GSTs from various species
| Enzyme substrate | PcpF | PcpF C53S | Re-YqjG | Ec-YqjG | ECM4 | hGSTO1-1 | hGSTO2-2 |
|---|---|---|---|---|---|---|---|
| CDNB | ND | ND | ND | ND | ND | 0.02 ± 0.01 | 0.01 ± 0.01 |
| Ethacrynic acid | 0.09 ± 0.01 | 0.11 ± 0.01 | 0.26 ± 0.05 | 0.07 ± 0.01 | 0.13 ± 0.01 | 0.02 ± 0.01 | 0.37 ± 0.06 |
| GS-TriCH | 2.66 ± 0.12 | ND | 1.67 ± 0.13 | 1.76 ± 0.07 | 3.18 ± 0.06 | ND | ND |
| HED | 23.1 ± 0.8 | ND | 37.9 ± 0.8 | 9.54 ± 0.23 | 22.9 ± 0.3 | 3.91 ± 0.09 | 9.73 ± 0.08 |
| DMAV | 2.49 ± 0.31 | ND | 2.28 ± 0.13 | 1.30 ± 0.24 | 1.42 ± 0.17 | 0.074 ± 0.008 | 0.46 ± 0.02 |
| DHA | 1.91 ± 0.14 | ND | 1.71 ± 0.16 | 0.27 ± 0.03 | 0.88 ± 0.02 | 0.57 ± 0.01 | 26.5 ± 0.53 |
| DiCPGS | 0.22 ± 0.01 | ND | ND | ND | ND | 1.56 ± 0.01 | ND |
Specific activities are reported as substrate consumed (μmol · min−1 · mg−1) of protein at 23 ± 1 °C and are means ± S.D. Assays were performed with 5 mM GSH except for the ethacrynic acid assay which used 1 mM GSH. ND, no apparent activity was detected; DiCPGS, S-(2′4′-dichlorophenacyl)glutathione.
Kinetic analysis of PcpF
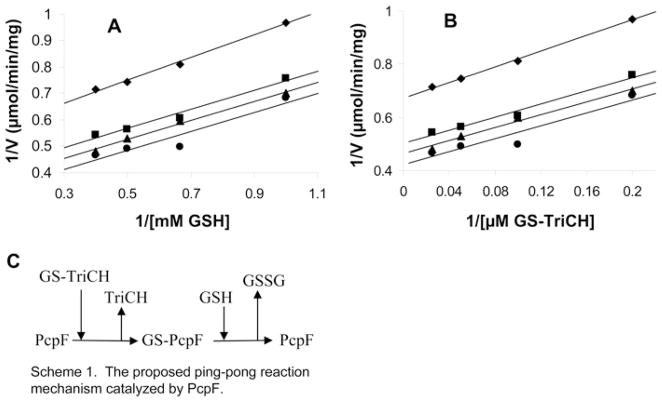
The GS-TriCH reduction mechanism was also analysed with PcpF. The Km values were 4.4 ± 0.6 μM for GS-TriCH and 1.3 ± 0.4 mM for GSH with a Vmax of 3.4 ± 0.4 μmol · min−1 · mg−1 of protein. Given the high Km values for GSH, we increased GSH from 1 mM to 5 mM in most assays when the specific activities were determined (Table 1). However, for the ethacrynic acid assay, 1 mM GSH was used because of the spontaneous reaction between GSH and ethacrynic acid being too high at 5 mM GSH. Kinetic analysis with various concentrations of GSH and GS-TriCH produced parallel lines in double reciprocal plots (Figures 4A and 4B), suggesting that PcpF catalyses GS-TriCH reduction via a Ping Pong mechanism. We further tested whether PcpF could use other small thiols for GS-TriCH reduction. Besides GSH, PcpF used L-cysteine, 2-mercaptoethanol and DTT to reduce GS-TriCH. The formation of TriCH was confirmed by HPLC analysis. The end-products were expected to be GS–S-DTT, GS–SG, GS–S-CH3-CH2OH, or GS–S-Cys, depending on the small thiol used. As glutathione reductase is known to effectively reduce asymmetric substrate analogues, including GS–S-nitrobenzoate and GS–S-nitropyridine [28] as well as GS–S-CH3 [29], we applied glutathione reductase to reduce the mixed disulfide bond. The consumption of NADPH was observed and used to determine the kinetic parameters (Table 2). As judged by kcat/Km values, PcpF used the substrates in descending order: DTT, GSH, 2-mercaptoethanol and L-cysteine; however, GSH supported the fastest catalytic turnover.
Figure 4. Double reciprocal plots of initial velocities of PcpF against substrate concentrations.
A GS-TriCH enzymatic assay was performed [a coupled assay of PcpF (23 μg · ml−1 at 2.66 μmol · min−1 · mg−1) and glutathione reductase (2.7 μg · ml−1 at 168 μmol · min−1 · mg−1)]. Results are means of two measurements. (A) Enzyme activity was plotted against various concentrations of GSH with four sets of constant concentrations of GS-TriCH. (B) Enzyme activity was plotted against various concentrations of GS-TriCH with four sets of constant concentrations of GSH. (C) Scheme of deduced Ping Pong mechanism of PcpF from kinetic analysis. The GSH in the scheme can be replaced by DTT, L-cysteine and 2-mercaptoethanol.
Table 2.
Kinetic parameters with different thiols
| Substrate | kcat | Km (mM) | kcat/Km (s−1 · M−1) |
|---|---|---|---|
| DTT | 0.89 ± 0.02 | 0.26 ± 0.03 | 3430 |
| GSH | 2.08 ± 0.25 | 1.3 ± 0.4 | 1600 |
| 2-MerC | 1.09 ± 0.06 | 0.93 ± 0.15 | 1172 |
| L-Cysteine | 0.37 ± 0.09 | 1.97 ± 0.15 | 187 |
GS-TriCH reduction was measured with a spectrometric assay using different small thiols. 2-MerC, 2-mercaptoethanol.
Native forms of GSTs
Most GSTs are dimers, including Omega class GSTs; DHAR and Lambda classes are both monomers [23]. We used both size-exclusion chromatography and a light-scattering detector to determine the oligomeric nature of the native forms of PcpF and ECM4. The retention times of the size-exclusion chromatography were 17.84 min for PcpF and 17.49 min for ECM4, correlating to estimated native molecular masses of 65 and 70 kDa. Static light-scattering data estimated the corresponding molecular masses as 79.7 and 101.2 kDa for PcpF and ECM4 respectively. The calculated dimers of PcpF and ECM4 were 73.8 kDa and 89.6 kDa. The results from both methods suggested that the enzymes are dimers.
DISCUSSION
PcpF and homologues belong to a new class of GSTs
Phylogenetic analysis grouped PcpF homologues into a distinct class that is closely related to the Omega, Lambda and DHAR classes (Figure 2); they are more related to the Omega class because both have the typical dimeric forms of GSTs, and the Lambda and DHAR classes are monomers [23]. The four groups of GSTs all have poor or no activities for transferring GSH to standard GST substrates, e.g. CDNB (Table 1), but are able to catalyse GSH-dependent reductions. The Lambda class GSTs can only catalyse thiol transfer (disulfide bond reduction) and the DHAR class is known to perform both thiol transfer and DHA reduction [23]. The limited substrate range may be attributable to the monomeric nature of DHAR and Lambda class GSTs because the dimer interface of GSTs is involved in substrate binding [22]. The Omega class GSTs catalyse additional reactions, including DMAV reduction [21]. The PcpF homologues can further reduce GS-TriCH (Table 1). Thus both phylogenetic and biochemical evidence separates PcpF homologues from the Omega class GSTs. ECM4 is grouped with PcpF homologues, although it only has 15.7 % sequence identity with hGSTO1-1. It is generally accepted that GSTs with less than 30 % sequence identity are assigned to separate classes [22].
ECM4 is a well-known protein among PcpF homologues
Among the PcpF-related proteins, yeast ECM4 is the best characterized. ECM4 is a protein involved in cell-surface biosynthesis and architecture. ECM4 was identified as a transposon mutant hypersensitive to Calcofluor White (a negatively charged fluorescent dye with a molecular mass of 960.9 kDa) that binds to nascent chains of chitin, a component of yeast cell walls. S. cerevisiae ecm4 mutants show increased amounts of chitin, as determined by the N-acetylglucosamine contents [30]. The expression of the ECM4 gene is up-regulated when yeast is exposed to genotoxic agents, such as methyl methanesulfonate, cisplatin and bleomycin [31]. Methyl methanesulfonate alkylates guanine to form 7-methylguanine adducts, cisplatin is a DNA cross-linking agent and bleomycin induces oxidative damage to DNA. It is unclear how ECM4 is involved in cell-wall synthesis and in protection against genotoxic agents. ECM4 has recently been characterized as an Omega class GST (GTO2) for its ability to use the same substrates of the Omega class GSTs [12]. In the present study we regroup ECM4 with PcpF homologues, into a new class of GSTs, on the basis of phylogenetic analysis (Figure 2) and their abilities to reduce GS-TriCH (Table 1). The establishment of a new GST class is in agreement with the conserved domain database that has an ECM4 group, COG0435, which is not assigned to the Omega class GSTs [13]. The four PcpF homologues in A. thaliana have all been identified as ECM4-like proteins. The listed representatives of COG0435 (ECM4 group) contain both ECM4 and Ec-YqjG.
Reaction mechanism
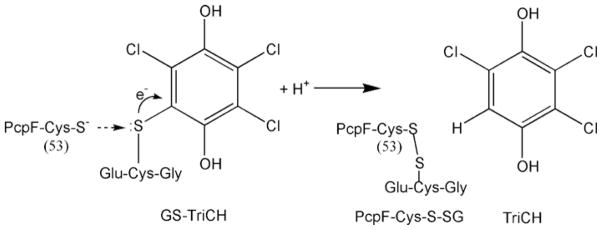
Combining the kinetic data (Figure 4) and the catalytic role of Cys53 of PcpF [11], a Ping Pong reaction mechanism for GS-TriCH reduction is proposed (Figure 4C). First, PcpF reacts with GS-TriCH to release TriCH and to form a disulfide bond of PcpF-Cys53-S–SG. Then, a GSH molecule comes in to regenerate PcpF-Cys53-SH with the concomitant formation of GS–SG. DTT, 2-mercaptoethanol (HS-CH3-CH2OH) and L-cysteine also react with PcpF-Cys53-S–SG to regenerate PcpF-Cys53-SH, with the commitment production of GS–S-DTT, GS-S-CH3-CH2OH and GS-S-Cys respectively. As DTT and 2-mercaptoethanol are not present in the cytoplasm, GSH is the natural substrate for the reaction with a catalytic efficiency (kcat/Km) approx. 10-fold that when using L-cysteine. hGSTO1-1 catalysed a similar reaction by using GSH to reduce S-(phenacyl)glutathiones, and 2-mercaptoethanol can be used to replace GSH for the reaction [18].
Proposed role of Cys53 and Pro54 in catalysis
The phylogenetic tree (Figure 2) is in agreement with the active-site motifs, which also grouped PcpF and homologues separately from the Beta class. The conserved N-terminal active-site motif of the Beta class is Cys-Ser, the cysteine residue of which is involved in interacting with the thiol group of the bound GSH [32]. PcpF homologues, and the Omega, Lambda and DHAR class GSTs, all have an N-terminal active-site motif of Cys-Pro. The two residues, Cys32 and Pro33 of hGSTO1-1, are located at the start of α-helix 1 (Figure 3), and the proximity of Cys32 to the positive end of the α-helix dipole promotes the formation of Cys32 thiolate [21]. The Cys-Pro residues are Cys53 and Pro54 in PcpF (Figure 3). The Cys53 thiolate should be a strong nucleophile, and the sulfur atom of GS-TriCH should be slightly electron-deficient due to the strong electron-withdrawing effect of the aromatic ring of TriCH (Figure 5). Thus a typical bimolecular nucleophilic substitution (SN2 reaction) takes place at the active site of PcpF: the thiolate attacks the sulfur atom, forming PcpF-Cys53-S–SG and expelling TriCH. The apparent difference in key amino acid residues for substrate binding (Figure 3) may prevent the two human Omega class GSTs from binding GS-TriCH.
Figure 5. Proposed SN2 reaction between PcpF-Cys53 thiolate and GS-TriCH.
The Cys53 thiolate, created by its location next to α-helix-1, attacks the sulfur atom of GS-TriCH, which should be slightly electron-deficient due to the strong electron-withdrawing effect of the aromatic TriCH. The result is the formation of PcpF-Cys53-S–SG, releasing TriCH. The mixed disulfide bond (PcpF-Cys53-S–SG) is reduced by small thiols, including GSH, DTT, 2-mercaptoethanol and L-cysteine.
Physiological roles of GS-TriCH reductases
A potential role as thiol transferase for PcpF and homologues is uncertain as the activities with HED, although significant (Table 1), are much lower than the activity of glutaredoxin 1 of the alga Chlamydomonas (Vmax = 710 μmol · min−1 · mg−1) [33]. PcpF homologues all have good DHAR activities (Table 1), but are much lower than that (at 936 μmol · min−1 · mg−1) of At-DHAR1 [23]. Among the activities catalysed by PcpF homologues, only GS-TriCH reduction is unique to the group (Table 1). Thus PcpF homologues are tentatively called S-glutathionyl-(chloro)hydroquinone reductases. PcpF was previously referred as a glutathionyl-chlorohydroquinone lyase for its ability for reducing GS-TriCH and GS-DiCH conjugates [11]. As PcpF and homologues catalyse GSH-dependent reduction of disulfide bond (thiol transfer), DHA, DMAV and GS-TriCH, naming them as reductases, instead of lyases, is more appropriate. The proximity of pcpC and pcpF genes on chromosome of S. chlorophenolicum ATCC 29723 has led to the identification of the role of PcpF in PCP degradation. However, the activity for GS-TriCH reduction is not limited to PcpF. All of the tested homologues reduced GS-TriCH at similar rates (Table 1). The wide presence of PcpF homologues in bacteria, archaea, fungi and plants suggests a physiological role beyond the degradation of PCP, a chemical only introduced into the environment recently [2]. As S-glutathionyl-(chloro)hydroquinone reductases are common, but GS-TriCH is not, it may not be far-fetched to hypothesize that S-glutathionyl-(chloro)hydroquinone reductases can use other substituted GS-hydroquinones as their substrates. Both o- and p-substituted benzoquinones can react with GSH to form substituted GS-hydroquinones, and the reaction can be spontaneous or catalysed by GSTs [34,35]. The physiological advantage of reducing some GS-hydroquinones needs further investigation; however, the maintenance role of PcpF in PCP degradation is clearly beneficial to S. chlorophenolicum [11].
Evolution
GSTs share strong structure similarity despite the low sequence identity (for a review, see [22]). The GST fold consists of an N-terminal glutaredoxin-fold domain, a C-terminal α-helical domain and a cleft between the two domains that houses the active site. The N-terminal domain contains most of the residues for GSH binding, whereas the C-terminal domain is usually responsible for the binding of the second substrate. The glutaredoxin-fold belongs to the thioredoxin-fold superfamily. It has been suggested that conjugating GSTs are evolved from oxidoreductases with GSH as the reductant [36]. Apparently, S-glutathionyl-(chloro)hydroquinone reductases, DHAR, and Omega class and Lambda class GSTs have weak or no conjugation abilities, and all have the thiol transferase activity of glutaredoxin [16]. They may represent a branch of GSTs that catalyse mainly GSH-dependent oxidoreduction (Table 1), rather than conjugation.
Acknowledgments
FUNDING
The research was supported by the U.S.A. National Science Foundation [grant number MCB-0323167]; and by the Australian National Health and Medical Research Council. S.M.B. was a fellowship recipient of an National Institutes of Health Biotechnology Training Grant.
Abbreviations used
- At-
Arabidopsis thaliana
- CDNB
1-chloro-2,4-dinitrobenzene
- DHA
dehydroascorbate
- DHAR
DHA reductase
- DiCH
dichloro-p-hydroquinone
- DMAV
dimethylarsinate
- DTT
dithiothreitol
- Ec-
Escherichia coli
- ECM4
extracellular mutant 4
- GS-DiCH
S-glutathionyl-DiCH
- GST
glutathione transferase
- GSTO
glutathione transferase Omega class
- GS-TriCH
S-glutathionyl-TriCH
- h
human
- HED
2-hydroxyethyl disulfide
- LB
Luria–Bertani
- Ni-NTA
Ni2+-nitrilotriacetate
- PCP
pentachlorophenol
- Re-
Ralstonia eutropha
- TeCH
tetrachloro-p-hydroquinone
- TriCH
trichloro-p-hydroquinone
Footnotes
AUTHOR CONTRIBUTION
Luying Xun conceived and co-ordinated the study. He also conducted the majority of the experiments together with Sara Belchik, Randy Xun and Yan Huang. Huina Zhou and Philip Board generated and purified hGSTO2-2 protein and synthesized S-(2′,4′-dichlorophenacyl)glutathione. Emiliano Sanchez and ChulHee Kang determined the native molecular masses of PcpF and ECM4. All authors contributed to the writing and pre-acceptance editing of the manuscript.
References
- 1.Merz V, Weith W. On the characteristics of pentachlorophenol. Ber Dtsch Chem Ges. 1872;5:458–463. [Google Scholar]
- 2.Crosby DG. Environmental chemistry of pentachlorophenol. Pure Appl Chem. 1981;53:1052–1080. [Google Scholar]
- 3.Saber DL, Crawford RL. Isolation and characterization of Flavobacterium strains that degrade pentachlorophenol. Appl Environ Microbiol. 1985;50:1512–1518. doi: 10.1128/aem.50.6.1512-1518.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai M, Xun L. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J Bacteriol. 2002;184:4672–4680. doi: 10.1128/JB.184.17.4672-4680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai M, Rogers JB, Warner JR, Copley SD. A previously unrecognized step in pentachlorophenol degradation in Sphingobium chlorophenolicum is catalyzed by tetrachlorobenzoquinone reductase (PcpD) J Bacteriol. 2003;185:302–310. doi: 10.1128/JB.185.1.302-310.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xun L, Orser CS. Purification and properties of pentachlorophenol hydroxylase, a flavoprotein from Flavobacterium sp. strain ATCC 39723. J Bacteriol. 1991;173:4447–4453. doi: 10.1128/jb.173.14.4447-4453.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orser CS, Dutton J, Lange C, Jablonski P, Xun L, Hargis M. Characterization of a Flavobacterium glutathione S-transferase gene involved in reductive dechlorination. J Bacteriol. 1993;175:2640–2644. doi: 10.1128/jb.175.9.2640-2644.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xun L, Topp E, Orser CS. Purification and characterization of a tetrachloro-p-hydroquinone reductive dehalogenase from a Flavobacterium sp. J Bacteriol. 1992;174:8003–8007. doi: 10.1128/jb.174.24.8003-8007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xun L, Bohuslavek J, Cai M. Characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) of Sphingomonas chlorophenolica ATCC 39723. Biochem Biophys Res Comm. 1999;266:322–325. doi: 10.1006/bbrc.1999.1805. [DOI] [PubMed] [Google Scholar]
- 10.McCarthy DL, Navarrete S, Willett WS, Babbitt PC, Copley SD. Exploration of the relationship between tetrachlorohydroquinone dehalogenase and the glutathione S-transferase superfamily. Biochemistry. 1996;35:14634–14642. doi: 10.1021/bi961730f. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Xun R, Chen G, Xun L. Maintenance role of a glutathionyl-hydroquinone lyase (PcpF) in pentachlorophenol degradation by Sphingobium chlorophenolicum ATCC 39723. J Bacteriol. 2008;190:7595–7600. doi: 10.1128/JB.00489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcera A, Barreto L, Piedrafita L, Tamarit J, Herrero E. Saccharomyces cerevisiae cells have three Omega class glutathione S-transferases acting as 1-Cys thiol transferases. Biochem J. 2006;398:187–196. doi: 10.1042/BJ20060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmuck EM, Board PG, Whitbread AK, Tetlow N, Cavanaugh JA, Blackburn AC, Masoumi A. Characterization of the monomethylarsonate reductase and dehydroascorbate reductase activities of Omega class glutathione transferase variants: implications for arsenic metabolism and the age-at-onset of Alzheimer’s and Parkinson’s diseases. Pharmacogenet Genomics. 2005;15:493–501. doi: 10.1097/01.fpc.0000165725.81559.e3. [DOI] [PubMed] [Google Scholar]
- 15.Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases: the first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 16.Holmgren A, Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 17.Whitbread AK, Masoumi A, Tetlow N, Schmuck E, Coggan M, Board PG. Characterization of the omega class of glutathione transferases. Methods Enzymol. 2005;401:78–99. doi: 10.1016/S0076-6879(05)01005-0. [DOI] [PubMed] [Google Scholar]
- 18.Board PG, Anders MW. Glutathione transferase omega 1 catalyzes the reduction of S-(phenacyl)glutathiones to acetophenones. Chem Res Toxicol. 2007;20:149–154. doi: 10.1021/tx600305y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 21.Board PG, Coggan M, Chelvanayagam G, Easteal S, Jermiin LS, Schulte GK, Danley DE, Hoth LR, Griffor MC, Kamath AV, et al. Identification, characterization, and crystal structure of the Omega class glutathione transferases. J Biol Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 22.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon DP, Davis BG, Edwards R. Functional divergence in the glutathione transferase superfamily in plants: identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J Biol Chem. 2002;277:30859–30869. doi: 10.1074/jbc.M202919200. [DOI] [PubMed] [Google Scholar]
- 24.Board PG, Baker RT, Chelvanayagam G, Jermiin LS. Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem J. 1997;328:929–935. doi: 10.1042/bj3280929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Xia Z, Ding J. Thioredoxin-like domain of human Kappa class glutathione transferase reveals sequence homology and structure similarity to the Theta class enzyme. Protein Sci. 2005;14:2361–2369. doi: 10.1110/ps.051463905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anandarajah K, Kiefer PMJ, Donohoe BS, Copley SD. Recruitment of a double bond isomerase to serve as a reductive dehalogenase during biodegradation of pentachlorophenol. Biochemistry. 2000;39:5303–5311. doi: 10.1021/bi9923813. [DOI] [PubMed] [Google Scholar]
- 27.Casalone E, Allocati N, Ceccarelli I, Masulli M, Rossjohn J, Parker MW, Di Ilio C. Site-directed mutagenesis of the Proteus mirabilis glutathione transferase B1-1 G-site. FEBS Lett. 1998;423:122–124. doi: 10.1016/s0014-5793(98)00080-5. [DOI] [PubMed] [Google Scholar]
- 28.Wong KK, Vanoni MA, Blanchard JS. Glutathione reductase: solvent equilibrium and kinetic isotope effects. Biochemistry. 1988;27:7091–7096. doi: 10.1021/bi00418a063. [DOI] [PubMed] [Google Scholar]
- 29.Miller H, Claiborne A. Peroxide modification of monoalkylated glutathione reductase. Stabilization of an active-site cysteine-sulfenic acid. J Biol Chem. 1991;266:19342–19350. [PubMed] [Google Scholar]
- 30.Lussier M, White AM, Sheraton J, di Paolo T, Treadwell J, Southard SB, Horenstein CI, Chen-Weiner J, Ram AF, Kapteyn JC, et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caba E, Dickinson DA, Warnes GR, Aubrecht J. Differentiating mechanisms of toxicity using global gene expression analysis in Saccharomyces cerevisiae. Mutat Res. 2005;575:34–46. doi: 10.1016/j.mrfmmm.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Nishida M, Harada S, Noguchi S, Satow Y, Inoue H, Takahashi K. Three-dimensional structure of Escherichia coli glutathione S-transferase complexed with glutathione sulfonate: catalytic roles of Cys10 and His106. J Mol Biol. 1998;281:135–147. doi: 10.1006/jmbi.1998.1927. [DOI] [PubMed] [Google Scholar]
- 33.Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem. 2008;283:8868–8876. doi: 10.1074/jbc.M709567200. [DOI] [PubMed] [Google Scholar]
- 34.Dagnino-Subiabre A, Cassels BK, Baez S, Johansson AS, Mannervik B, Segura-Aguilar J. Glutathione transferase M2-2 catalyzes conjugation of dopamine and dopa o-quinones. Biochem Biophys Res Commun. 2000;274:32–36. doi: 10.1006/bbrc.2000.3087. [DOI] [PubMed] [Google Scholar]
- 35.Ross D, Thor H, Orrenius S, Moldeus P. Interaction of menadione (2-methyl-1,4-naphthoquinone) with glutathione. Chem Biol Interact. 1985;55:177–184. doi: 10.1016/s0009-2797(85)80126-5. [DOI] [PubMed] [Google Scholar]
- 36.Caccuri AM, Antonini G, Allocati N, Di Ilio C, De Maria F, Innocenti F, Parker MW, Masulli M, Lo Bello M, Turella P, et al. GSTB1-1 from Proteus mirabilis: a snapshot of an enzyme in the evolutionary pathway from a redox enzyme to a conjugating enzyme. J Biol Chem. 2002;277:18777–18784. doi: 10.1074/jbc.M201137200. [DOI] [PubMed] [Google Scholar]