Abstract
Upon encountering microbial pathogens, CD4+ T cells become activated and differentiate to various subsets with distinct patterns of gene expression and profiles of cytokine production. These cells are critical for host defense, but are also major drivers of immune-mediated disease. Although classically viewed as distinct lineages, recent work calls into question whether helper CD4+ T cell subsets are more appropriately view as terminally differentiated cells or “works in progress”. Herein, we review recent advances that pertain to this topic and the mechanisms that contribute to helper CD4+ T cell commitment and plasticity. The therapeutic implications of these new findings are also considered.
Faced with innumerable microbial pathogens, antigen-inexperienced, “naïve” CD4+ T cells orchestrate immune responses by differentiating into T helper (Th) cell populations that secrete distinct sets of cytokines. In this manner, they tailor their responses to the character of the threat encountered, providing help to B lymphocytes and CD8+ cytotoxic T cells, and activating the cells of the innate immune system. The importance of CD4+ T cells is vividly illustrated in the range of infections that afflict HIV-infected individuals when their blood CD4+ T cell numbers decline. CD4+ T cells also play critical roles in the pathogenesis of autoimmunity, asthma, allergy and likely cancer. The concept of distinct Th cell lineages has been a useful paradigm for the conceptualization of CD4+ T cell differentiation but immunologists are now rethinking how Th cell subsets should be viewed. Are distinct Th subsets really different lineages and if so, how plastic are they in altering or modifying their committed state? Or, do Th cells lack features of “end stage” commitment and readily alter their profile of secreted products? Certainly, there is increasing evidence in vitro that differentiated CD4+ T cell populations can alter the range of cytokines they produce although whether this occurs as readily in vivo remains unclear. Understanding the molecular basis of CD4+ Th cell differentiation and of the process through which cells alter their cytokine-producing potential, will likely provide interesting insights into subset specification and gene regulation. Equally, these insights may allow the development of strategies to alter Th cell function in circumstances of autoimmunity or allergy or alternatively, when a response is mounted against a pathogenic microbe or a tumor cell that is appropriate in specificity but inappropriate in type.
Why should we think of subsets of cytokine-producing CD4+ T cells as differentiated lineages?
Nearly a quarter of a century ago, it was recognized that cytokine production by Th cells was not stochastic(1). Rather, T cell clones could be divided into two subsets, Th1 and Th2, which respectively produced the signature cytokines interferon (IFN)-γ or interleukin (IL)-4 and IL-13. Importantly, selective cytokine production was a stable feature of multiply-passaged Th1 and Th2 cell lines. Th1 and Th2 cells also preferentially expressed particular cytokine and chemokine receptors. Both subsets were thought to provide help to B cells, although they “instructed” the class switching mechanism toward different immunoglobulin (Ig) classes (see discussion of T follicular helper (Tfh) cells in the next section). Their selective cytokine production was important for the proper elimination of microbial pathogens, intracellular microbes for Th1 cells and helminths for Th2 cells (2). These subsets also expressed lineage-specifying transcription factors (T-bet and GATA3), and overexpression of these “master regulators” induced the production of the stereotypic cytokine (3). The concept of stable, terminally differentiated Th cells made sense – after exposure to a particular pathogen, memory T cells would “remember” to make responses not only of the correct specificity but also of the appropriate type.
How does increasing subset complexity fit with existing models?
Although this paradigm was a useful construct for understanding immunoregulation and to define the molecular mechanisms that shaped CD4+ T cell differentiation, the dualistic view of Th cell lineages was complicated by the recognition of new subsets of Th cells. CD4+ CD25+ regulatory T cells (Tregs) were identified and found to be critical for the preservation of immune tolerance (4-6). Besides thymic-derived Tregs (natural or nTregs), it was shown that naïve T cells in the periphery could acquire immunosuppressive properties and become induced Tregs (iTregs). Both Treg subsets express the forkhead transcriptional repressor, Foxp3 (5, 6). Absence of Foxp3 results in widespread autoimmunity and absence of Tregs, whereas its over-expression confers immunosuppressive functions (5, 6). Consequently, Foxp3 was designated as the Treg master regulator. Although the biologic significance of Tregs is well established, the relative importance of nTregs and iTregs is still unresolved. No decisive tests to differentiate their function in physiologic settings and particularly in humans have yet been devised.
The next “lineage” recognized was cells that selectively produced IL-17 (Th17 cells), which also seemed to have their own master transcription factor, the orphan retinoid receptor, Rorγt (7, 8). The recognition of Tregs and Th17 cells improved our understanding of immunoregulation and provided insights into the pathogenesis of autoimmunity, but whether Tregs and Th17 cells behaved like polarized Th1 and Th2 cells was unclear. For one, Tregs and Th17 cells are both induced by transforming growth factor (TGF)β (9). Tfh cells are yet another CD4+ T cell population with a particular function, helping B cells make antibody responses to T cell-dependent antigens (10-12). They aid in the development of germinal centers (specialized regions within secondary lymphoid tissues that promote B cell immumity), promote immunoglobulin class switch recombination and affinity maturation. Tfh cells express Bcl6, their master regulator treanscription factor (13, 14), but unlike T-bet and GATA3, Bcl6 is a transcriptional repressor. Whether Tfh cells are truly a population (lineage) parallel to Th1, Th2, Th17 or iTreg cells or a particular state of some or all of these cells is unresolved (10 -12). Tfh cells have distinct properties and phenotypes (e.g. expression of CXCR5, ICOS, and Bcl6); however, individual Tfh cells may produce Th1 or Th2 signature cytokines, depending on the conditions of their initial activation (10-12) Thus, are IL-4- or IFNγ- producing Tfh cells members of a Tfh lineage that then acquires a distinct cytokine-producing potential or is “Tfh-ness” a property of Th1, Th2, and possibly Th17 cells acquired much in the same way that these cells become central memory cells, effector memory cells or tissue-seeking effector cells? Of course, if the latter is the case, than a discussion of plasticity for Tfh cells is the same as discussing plasticity among the conventional effector lineages. Although these new discoveries increase the complexity of the Th response and raise questions of how decision-making in the differentiation process occurs, they could still be accommodated within the end stage differentiation paradigm. T cell fates need not be restricted to two or even four Th cell lineages; on the contrary, given the many microbial pathogens, one could imagine the need for multiple specialized subsets. But how the system determines the character of the threat and makes appropriate decisions as to what the dominant response will be grows increasingly complicated with each additional “lineage”.
To what extent is cytokine production plastic?
More challenging than the identification of new Th cell subsets was the emergence of evidence that cytokine expression is not as stable as initially thought; indeed, there are now many examples of flexible cytokine production (9,10,15). Once thought to be a Th2 cytokine, it is now recognized that IL-10 is produced by multiple cell subsets– Th1, Th2, Tregs and Th17 cells. Similarly, Th2 cells can acquire IL-9-producing capacity in the presence of TGFβ, (16), but IL-9 can also be produced by Th17 cells(17). Acquisition of IFNγ-producing potential by Th17 cells, particularly the simultaneous production of IL-17 and IFN-γ, is a common occurrence – especially in vivo (18, 19). Th17 cells can even extinguish production of their cytokine signature, becoming selective IFN-γ producers (20-22). Although Th1 cells do not become IL-17 producers, under the right circumstances they can make IL-13 (23). Th17 cells produce IL-22, but cells that make IL-22 and not IL-17 have recently been identified (24, 25). Simultaneous production of IL-22 and IFN-γ also occurs and indeed, IL-22 was originally viewed as a Th1 cytokine. More concerning for the “firmly fixed” notion of cytokine production, however, is the finding that in vitro differentiated, IL-4-producing Th2 cells specific for lymphocytic choriomeningitis virus (LCMV), produce IFN-γ when transferred into mice subsequently infected with LCMV (26). Whether cells primed in vivo under robust Th2-inducing conditions show an equal propensity to acquire IFNγ-producing capacity has not been established.
Collectively, these findings argue for much flexibility in cytokine production, but the true frequency with which Th cells alter their cytokine-producing potential in vivo is still uncertain. More precisely, to what extent do cells really change? Not just by expressing a cytokine that they formerly did not, but rather by altering the major properties that define Th cells. To the extent that such plasticity is a major feature of immune responses, the stakes are raised in the effort to understand how the system reads pathogen threats and “orders up” the right response.
Is expression of master regulators stable?
What is clearly problematic for a strict lineage commitment model of Th cell differentiation is that the expression of master regulators that drive differentiation is not fixed. For instance, it now appears that Foxp3+ Treg cells are heterogeneous and transient expression of Foxp3 is a common event (27). Many studies show that Foxp3 expression can be extinguished, and that former Tregs can acquire the ability to produce pro-inflammatory cytokines (27-30). This phenomenon is enhanced in the setting of autoimmunity. More disturbing is that Th cells can also express more than one “master regulator”. Cells expressing Foxp3 and T-bet are present at sites of inflammation and limit inflammation (31). Tregs can simultaneously express Foxp3, RORγt and the transcriptional repressor Runx3 (32,33). Alternatively, Foxp3+ cells can differentiate into Tfh cells in Peyer's patches in the gut, implying that they acquire Bcl6 expression (34).
So what does all this mean? Recent studies indicate that flexibility in expression of master regulators and cytokines is relatively common. Whether this will be true for Th cell populations primed in vivo will require much additional analysis and whether this potentiality translates into change in phenotype under physiologic conditions also remains to be established. Such plasticity may have evolutionarily selective value: to the extent that aging humans and mice rely more and more on memory CD4+ T cells to respond to newly encountered pathogens, freezing the phenotype of memory cells could lead to inappropriate responses to new threats. So, perhaps our earlier “evolutionary” argument for stability is not so compelling after all.
What mechanisms underlie commitment and plasticity?
Flexibility in cytokine production by CD4+ T cells does not easily square with conventional models of helper cell lineage commitment and raises the question as whether these notions are still useful. If flexibility and/or plasticity is the rule, what governs such changes in production of individual cytokines or wholesale changes in the properties of cells? In this context, it is useful to revisit the concept of lineage commitment and consider recent advances that shed new light on terminal differentiation.
Defining factors that regulate lineage commitment and plasticity is not new, nor is it unique to T cells. Arguments regarding determinism and flexibility in developmental biology arise from the earliest experiments in model organisms. These lessons show that morphogens drive expression of lineage-specifying transcription factors, a classic example being the TGFβ-related cytokine, activin, that promotes mesoderm differentiation by inducing the T-box protein Brachyury. Other transcription factors of the homeobox, STAT, NF-κB, Notch and Forkhead families have similar, critical roles in development. A key aspect of commitment is that not only are genes turned on by transcription factors, but also that genes conferring alternative fates are repressed. Preferential gene expression is heritable and diverse epigenetic modifications ensure that this is the case. As a result, cell identity remains stable without continued external signals. Cell stability is also controlled by microRNA expression (35). These short single stranded non-coding RNAs repress target mRNAs by inhibiting translation and enhancing mRNA decay. From this perspective, Th cell differentiation resembles classical lineage commitment. Cytokines provide morphogen-like signals, inducing expression of lineage-specifying transcription factors that belong to the same families that are critical for development in general. The stability of cytokine production is also preserved by epigenetic modifiation. Specifically, lineage-defining cytokine genes (Ifng, Il4 and Il17) have the predicted permissive and repressive epigenetic modifications when the cells are differentiated in vitro(28, 36). Both the Il4 and Ifng genes also show striking CpG demethylation, an epigenetic modification that is associated with gene expression, in their promoters and in enhancer regions during the process of differentiation to Th2 and Th1 cells, respectively. The region upstream of exon-1 of the Foxp3 gene is also strongly methylated in non-Tregs, but is largely demethylated in nTregs (37). Thus, the ability of different Th cells to selectively produce cytokines looks like commitment; however, even classical lineage commitment does not exclude flexibility. Although commitment indicates that the developmental fate of a cell and its progeny is restricted, it is divided into two phases: specification and determination: The former implies reversibility whereas the latter connotes irreversible commitment. It might be reasonable to posit then, that many CD4+ T cells subsets are specified but rarely become fully determined. This makes sense because unlike sessile cells in organs like the brain or heart, T cells migrate; irreversible commitment might be disadvantageous under changing conditions. To completely discard the notion of determination, however, would be premature. Cells grown in tissue culture over long periods of time can act like determined cells, but to what extent does this mimic physiology? Nonetheless, the demonstration that terminally differentiated cells can be become inducible pluripotent stem cells and generate a new organism just by expressing four transcription factors, puts all determination “at risk” (38). Thus, plasticity is relative, not absolute. Importantly, there are also other examples of reprogramming in which de-differentiation back to stem cells is not required (39). Such examples represent extraordinary examples of plasticity, but they are artificial. Regardless, these insights establish the principle that transcription factor expression dramatically alters cell fate – even in highly differentiated cells. One implication is that rather than assuming that expression of a master regulator implies a discrete function, we should be thinking more about gradients of transcription factors such as Rorγt, Foxp3 and T-bet and repressors like Bcl6 and Blimp1. As we better understand the extrinsic and intrinsic signals that influence expression of transcription factors and transcriptional repressors, we might be surprised by the plasticity that emerges.
As indicated above, epigenetic modifications help ensure that phenotype does not alter, even in the absence of extrinsic signals; however, epigenetic marks are not immutable. Intriguingly, the epigenetic modifications of the genes encoding T-bet and GATA3 are more complex than one might expect a priori. Unlike the signature cytokine genes, the genes encoding the master regulators for Th1 and Th2 cells have both repressive and permissive marks in opposing lineages (28). This pattern has been denoted as bivalency and is typical of genes in stem cells that are poised for induction (40). Furthermore, epigenetic modifications are dynamic and H3K27 demethylases (Jumonji d3 and UTX) can remove repressive marks (41). Of note, when isolated Foxp3+ cells were transferred into autoimmune recipients and Foxp3 expression was downregulated, the Foxp3 locus became re-methylated (27).
miRNAs are another factor that influences the stability of Foxp3 gene expression. Deleting the genes encoding the miRNA processing enzymes Dicer and Drosha in Tregs results in loss of Foxp3 expression and the resultant T cells have the capacity to become effectors (42).
Similarly, deletion of the transcriptional repressor Bcl-6 results in the expression of a large number of miRNAs (13). Thus, regulation of miRNAs has the potential of being an important mechanism in preserving and altering helper cell phenotype but we need to understand when and how expression of key miRNAs is altered physiologically and how they influence epigenetic regulation.
Lineages versus subsets - who cares?
For those not directly involved, the question of stability versus plasticity of subpopulations might be viewed as a tempest in a teapot – who cares whether a Th cell represents a determined or potentially plastic lineage or a subset? From a cell biologist's perspective, the distinction might seem arbitrary or even pretentious; however, this is not just a pedantic distinction and there are very pragmatic reasons for carefully considering this question. In fact, the prospect of lineage commitment versus flexible programs for T cells has direct implications for disease pathogenesis and the success of therapeutic interventions. A variety of autoimmune and allergic inflammatory disorders are associated with the presence of Th cells of particular types and these cells have a major influence on, even controlling the pathophysiology of these disorders. If T cell responses are plastic, one should be able to “reset the clock” therapeutically. The allergen-specific Th2 cells of asthmatics and others might be altered in vivo and thus interrupt the disorder. In multiple sclerosis and early type I diabetes, the destructive Th1 and/or Th17 cells might be altered to more “benign” Th2 cells. A better understanding of the molecular mechanisms that stabilize committed cytokine production may provide new therapeutic opportunities or revise our approaches for treating such diseases. Of course, there is also a downside. Administering Tregs in some mice with autoimmunity ameliorates disease. Administration of human Tregs is being considered as a treatment for various severe human autoimmune diseases. It could be disastrous if these cells were to become inflammatory, Th17 cells.
The issue is important for another reason - the question of commitment versus flexibility is a fundamentally important and interesting cell biological problem. Th cells represent outstanding models for understanding how extrinsic factors in the microenvironment influence intrinsic factors, to ultimately control gene expression. T cell biologists have generated a wealth of tools (e.g. numerous knockout mice) and with any luck, studies from T cell differentiation might provide valuable lessons for other biologists.
What are the challenges for future research?
It has been unquestionably useful in advancing our understanding of immune cell function to recognize that B cells, T cells and different T cell subsets (CD4+, CD8+, α/β, γ/δ and NK T cells) constitute distinct lineages that do not interconvert. Although new subsets of Th cells continue to be elevated to the status of “lineages”, however, there are really no accepted criteria for what qualifies a cell for this august designation. The selective production of cytokines is the sine qua non for a subset, but this definition is tautologic and consequently leads to considerable confusion. Th1 cells make IFN-γ but are all IFN-γ-producing CD4+ T cells Th1 cells? IL-22 producing cells make IFN-γ, but are they then Th1s or Th22s? Can a cell be a Th1/Th22 or should this be a new “lineage”? In the simplest view, Th cell lineages express lineage-defining transcription factors, but, as discussed, we know that transcription factor expression is dynamic; a particular subset can express more than one master regulator and expression of these factors can be lost or induced. It is impractical to suggest that it is time to call a moratorium on Th lineages, and it is equally unlikely that all immunologists will agree on the criteria for lineage vs. subset - there will always be lumpers and splitters. Minimally though, concepts such as fate determination and lineage commitment should not be used lightly; they do have biological implications albeit imprecise. Immunologists need to recognize that in most cases, this is just shorthand and we should not become entangled by our own semantic distinctions. We should certainly not infer stable function based on the presence of a transcription factor or the production of cytokine; in fact, it might be safer to view cytokine- producing subsets in probabilistic terms. Certain factors increase the likelihood of stably producing a cytokine, but many factors ranging from transcription factor expression to epigenetic modifications and miRNA expression need to be quantified to predict with more accuracy that a helper cell will behave more like a differentiated cell.
It is also timely to gain a more sophisticated understanding of T cell-expressed transcription factors and repressors. Reductionist approaches are of course essential in trying to make sense of complex biological processes; however, the risk is that the concepts that emerge may be overly simplistic. Although it has been a convenient to refer to master regulators, the limitations of such an approach are becoming increasingly evident and a more nuanced view of transcription factor function is appropriate. For instance, GATA3 is important in the acquisition of IL-4-producing capacity, but once Th2 status has been achieved, GATA3 can be deleted without abolishing the capacity of a Th2 cell to produce IL-4. Even more interestingly, although GATA3 appears essential to achieve Th2 status, this does not necessarily mean that GATA3 expression has to rise for a cell to become a Th2 cell. In vitro, introducing constitutively active STAT5a by retroviral transduction into cells being differentiated under Th1 conditions results in repression of T-bet and acquisition of IL-4-producing capacity (43). GATA3 levels in these STAT5a overexpressors are as low or lower than in Th1 cells but that modest amount of GATA3 is essential. Constitutively active STAT5a fails to cause Th2 differentiation in conditional GATA3 deleted mice. Thus, it may well be that the concentration of the required transcription factor may vary depending on the concentration of other transcription factors and once a differentiated state is induced, that factor may be dispensable. Similarly, we have long known that although T-bet and STAT4 are important for Th1 differentiation, absence of either does not abrogate IFN-γ production. We also know that IFN-γ acting through STAT1 promotes T-bet expression (44,45). Given our present level of sophistication and technological capabilities, we should be more able to appreciate that transcription factors and repressors work in complex regulatory circuits; embracing their complexity will be the key to really understanding how they work. Rather than thinking about presence or absence of a transcription, it will be more fruitful to consider levels and ratios as they change during the course of immune stimulation. In fact, like other aspects of hematopoietic cell differentiation, one needs to think of an array of transcription factors participating in concert with one another (46, 47). Here's the good news: Improved computational approaches and more sophisticated assessment of dynamic interactions between transcription factors are in sight (48). High throughput, comprehensive approaches, such as chromatin immunoprecipitation with massive parallel sequencing will facilitate more profound and global views of transcription factor function. This technique also allows genome-wide analysis of epigenetic modifications; thus, the interplay between transcription factors and epigenetic changes can be assessed. High throughput sequencing can also be used to measure RNA expression including mRNA, miRNAs and long noncoding mRNAs. Thus, we can comprehensively measure transcription factor binding, epigenetic modifications, and expression of miRNA and mRNA; however, we will have to deal with all these data. Modern technology allows a comprehensive set of data to be acquired but to put the massive number of “pixels” into a true picture of the status of the cell is a daunting task. The sheer immensity of the challenge makes this a logical area for an integrated systems approach involving a concerted and organized effort among several labs.
Adding yet another dimension of difficulty is the cell heterogeneity even among cells that have been subjected to identical differentiation regimes. Ideally, it would be desirable to obtain information about gene expression and epigenetic modifications on individual cells although this is not technically possible at present. The classical studies on helper T cell differentiation have relied heavily on in vitro manipulation; indeed, this has been a criterion for a lineage – stability of cytokine production after extended in vitro passage with resistance to cytokines that might alter fate decision–but this is both a strength and weakness. Although the new Th lineages have not been subjected to this standard, stability based on in vitro passage is surely not the gold standard that we would like to apply; in vivo clearly trumps in plastico. Moreover, there is little standardization regarding in vitro differentiation – each lab does things a little differently. The critical issue will be to define whether Th cells generated in vivo during the course of infection or autoimmune disease behave as lineages or not. This will need to be done over substantial time, not just a few days or weeks. Fate mapping studies using genetically engineered mice in which deletion of “STOP” is determined by cytokine expression and thus cells acquire a permanent marker based on an initial differentiation decision can be of particular value for the assessment of the in vivo differentiation. Coupling this technology with intravital imaging will certainly improve our chances of understanding physiologic helper cell fates, as anatomical considerations undoubtedly contribute to T cell differentiation. Advances in basic immunology have led to many new therapies including biologics and small molecules that influence the actions of cytokines. These new therapies provide an exciting opportunity to probe whether such precise therapies can influence Th differentiation and plasticity. Similarly, as new vaccines are developed, it will be useful to assess their impact with more sophisticated measures of helper cell function and differentiation. Hopefully, these advances can be useful in the prevention or treatment of human disease.
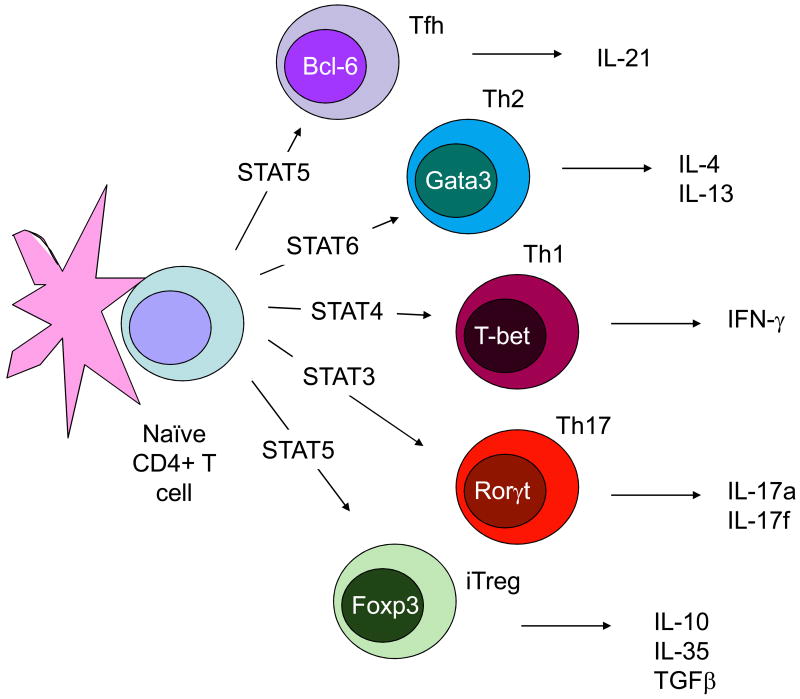
Fig. 1.
The Classical Monolithic View of Helper T Cell Differentiation: Lineages and Master Regulators. Initial studies arising from in vitro cultured Thelper1 (Th1) and Th2 cells led to the idea that these subsets behaved like lineages, meaning their phenotype (i.e. selective cytokine production) was inflexible. Accordingly, these subsets expressed lineage-defining transcription factors that were sufficient to impart this selective cytokine production. As newer subsets of cytokine producing cells were identified, they too were viewed as stable lineages.
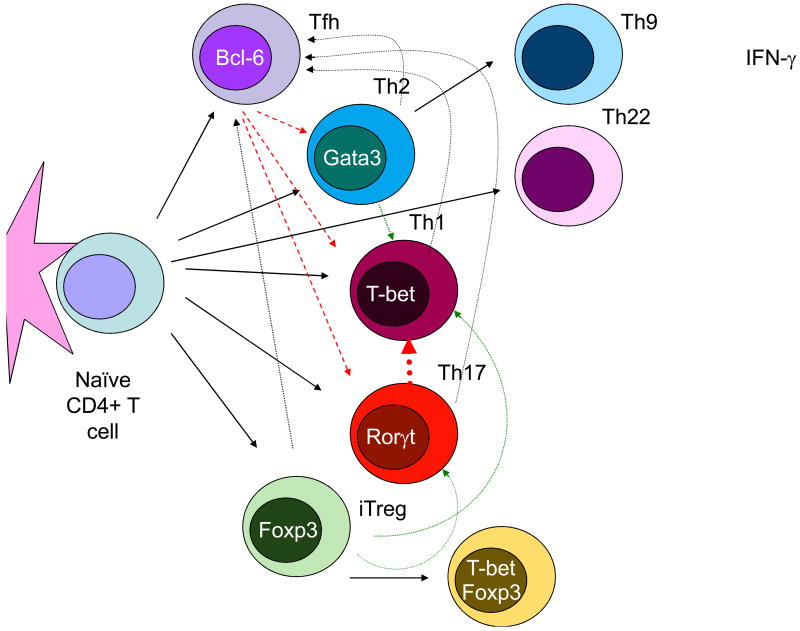
Fig. 2.
Flexibility and Plasticity of Helper T cells. Recent studies of Th cells have revealed more flexibility in cytokine production than predicted by earlier work and there are now many examples of plasticity of Th cell phenotype. CD4+ T cells can change their profile of cytokine production (dotted lines) and there are now circumstances in which the expression of master regulators is transient or instances where cells express more than one master regulator. New subsets are also recognized such as Th9 and Th22 cells, which selectively produce IL-9 and IL-22 respectively.
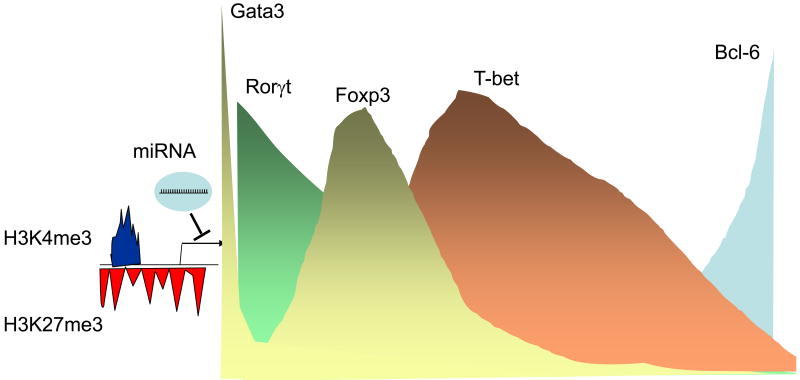
Fig. 3.
A More Nuanced View of Th Differentiation and Regulated Expression of Transcription Factors. Recent insights suggest that it is probably more appropriate to view the process of Th cell differentiation in the context of varying ratios of transcription factors, whose expression is regulated by an array extrinsic and intrinsic factors. That is, some factors like GATA3 and Rorγt have roles in thymic T cell development, but in some subsets, the expression of these factors is further induced (Th2 and Th17 respectively), whereas in other subsets (e.g. Th1 cells), expression declines. Foxp3 is highly expressed in T reg cells, but it now appears that it can be expressed more widely in T cells in which its expression is transient. Rorγt and Foxp3 can be expressed in the same cells and associate with one another. Other Foxp3+ Tregs express T-bet, which seems to be important for the proper trafficking to sites of inflammation. Th17 cells, which express Rorγt, can also express T-bet and acquire the ability to make IFN-γ. Follicular helper T cells express the transcriptional repressor Bcl6. Tfh can arise independently from other subsets, can also be generated from other T helpers or can even develop to preferentially express cytokines typically expressed by helper cell subsets. Cytokines that drive helper cell differentiation induce expression of these different transcription factors, but in addition epigenetic factors also influence expression. However, the epigenetic modifications of the genes encoding T-bet and GATA3 suggest that they are “poised” for expression. In addition, microRNAs and another factor that can tune gene expression in helper cell subsets. Thus, the bottomline is that transcription factor expression is more dynamic and fluid than originally recognized. Not surprisingly, the cytokines regulated by these factors also seem to have more plasticity in their regulation. Although, a few transcription factors are shown, this is undoubtedly a vast oversimplification and likely a panoply of the factors contribute to the specialized function of T cells.
References
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. J Immunol. 1986 Apr 1;136:2348. [PubMed] [Google Scholar]
- 2.Abbas AK, Murphy KM, Sher A. Nature. 1996 Oct 31;383:787. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 3.Dong C, Flavell RA. Sci STKE. 2000 Sep 12;2000:PE1. doi: 10.1126/stke.2000.49.pe1. [DOI] [PubMed] [Google Scholar]
- 4.Shevach EM. Nat Rev Immunol. 2002 Jun;2:389. doi: 10.1038/nri821. [DOI] [PubMed] [Google Scholar]
- 5.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Cell. 2008 May 30;133:775. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Josefowicz SZ, Rudensky A. Immunity. 2009 May;30:616. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stockinger B, Veldhoen M, Martin B. Semin Immunol. 2007 Dec;19:353. doi: 10.1016/j.smim.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Miossec P, Korn T, Kuchroo VK. N Engl J Med. 2009 Aug 27;361:888. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 9.Lee YK, Mukasa R, Hatton RD, Weaver CT. Curr Opin Immunol. 2009 Jun;21:274. doi: 10.1016/j.coi.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Zhou L, Chong MM, Littman DR. Immunity. 2009 May;30:646. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Fazileau N, Mark L, McHeyzer-Williams LJ, McHeyzer Williams MG. Immunity. 2009 Mar 20;30:324. doi: 10.1016/j.immuni.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King C, Tangye SG, Mackay CR. Annu Rev Immunol. 2008;26:741. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 13.Awasthi A, Kuchroo VK. Science. 2009 Aug 21;325:953. doi: 10.1126/science.1178752. [DOI] [PubMed] [Google Scholar]
- 13.Johnston RJ, et al. Science. 2009 Aug 21;325:1006. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nurieva RI, et al. Science. 2009 Aug 21;325:1001. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Bailey-Bucktrout S, Jeker LT, Bluestone JA. Curr Opin Immunol. 2009 Jun;21:281. doi: 10.1016/j.coi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veldhoen M, et al. Nat Immunol. 2008 Dec;9:1341. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 17.Elyaman W, et al. Proc Natl Acad Sci U S A. 2009 Aug 4;106:12885. doi: 10.1073/pnas.0812530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson NJ, et al. Nat Immunol. 2007 Sep;8:950. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Tato CM, Muul L, Laurence A, O'Shea JJ. Arthritis Rheum. 2007 Sep;56:2936. doi: 10.1002/art.22866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YK, et al. Immunity. 2009 Jan 16;30:92. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bending D, et al. J Clin Invest. 2009 Feb 2; [Google Scholar]
- 22.Shi G, et al. J Immunol. 2008 Nov 15;181:7205. doi: 10.4049/jimmunol.181.10.7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi N, et al. Proc Natl Acad Sci U S A. 2007 Sep 11;104:14765. doi: 10.1073/pnas.0706378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Nat Immunol. 2009 Jul 5; doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 25.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Nat Immunol. 2009 Jul 5; [Google Scholar]
- 26.Lohning M, et al. J Exp Med. 2008 Jan 21;205:53. doi: 10.1084/jem.20071855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, et al. Nat Immunol. 2009 Sep;10:1000. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L, Kitani A, Fuss I, Strober W. J Immunol. 2007 Jun 1;178:6725. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 29.Wei G, et al. Immunity. 2009 Jan 16;30:155. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komatsu N, et al. Proc Natl Acad Sci U S A. 2009 Feb 10;106:1903. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch MA, et al. Nat Immunol. 2009 Jun;10:595. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L, et al. Nature. 2008 May 8;453:236. [Google Scholar]
- 33.Zhang F, Meng G, Strober W. Nat Immunol. 2008 Nov;9:1297. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuji M, et al. Science. 2009 Mar 13;323:1488. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 35.Cordes KR, et al. Nature. 2009 Aug 6;460:705. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson CB, Rowell E, Sekimata M. Nat Rev Immunol. 2009 Feb;9:91. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 37.Floess S, et al. PLoS Biol. 2007 Feb;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamanaka S. Philos Trans R Soc Lond B Biol Sci. 2008 Jun 27;363:2079. doi: 10.1098/rstb.2008.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q, Melton DA. Cell Stem Cell. 2008 Oct 9;3:382. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein BE, et al. Cell. 2006 Apr 21;125:315. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, et al. J Exp Med. 2008 Sep 1;205:1983. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnston RJ, et al. Science. 2009 Aug 21;325:1006. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J, et al. Nat Immunol. 2004 Nov;5:1157. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 44.Lighvani AA, et al. Proc Natl Acad Sci U S A. 2001 Dec 18;98:15137. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thieu VT, et al. Immunity. 2008 Nov 14;29:679. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Georgescu C, et al. Proc Natl Acad Sci, USA. 2008 Dec 23;105:20100. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothenberg EV, Moore JE, Yui MA. Nat Rev Immunol. 2008 Jan;8:9. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barski A, et al. Cell. 2007 May 18;129:823. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]