Abstract
Chemotherapy and radiotherapy commonly damage DNA and trigger p53-dependent apoptosis through intrinsic apoptotic pathways. Two unfortunate consequences of this mechanism are resistance due to blockade of p53 or intrinsic apoptosis pathways, and mutagenesis of non-malignant surviving cells which can impair cellular function or provoke second malignancies. Death ligand-based drugs, such as tumor necrosis factor-related apoptosis inducing ligand (TRAIL), stimulate extrinsic apoptotic signaling, and may overcome resistance to treatments that induce intrinsic apoptosis. As death receptor ligation does not damage DNA as a primary mechanism of pro-apoptotic action, we hypothesized that surviving cells would remain genetically unscathed, suggesting that death ligand-based therapies may avoid some of the adverse effects associated with traditional cancer treatments. Surprisingly, however, treatment with sub-lethal concentrations of TRAIL or FasL was mutagenic. Mutations arose in viable cells that contained active caspases, and overexpression of the caspase-8 inhibitor crmA or silencing of caspase-8 abolished TRAIL-mediated mutagenesis. Downregulation of the apoptotic nuclease caspase-activated DNAse (CAD)/DNA fragmentation factor 40 (DFF40) prevented the DNA damage associated with TRAIL treatment. Although death ligands do not need to damage DNA in order to induce apoptosis, surviving cells nevertheless incur DNA damage after treatment with these agents.
Keywords: late effects, second malignancies, Apo-2L, DNA damage, mutagenesis, HPRT
Introduction
Conventional chemotherapy agents interfere with DNA replication and therefore primarily target dividing cells, by either preventing nucleotide biosynthesis, cross-linking DNA, preventing the maintenance of DNA topology or interfering with microtubule dynamics. These processes exert anti-tumor effects by inducing apoptosis, senescence and/or mitotic catastrophe. The pro-apoptotic action of many chemotherapeutic agents involves the generation of DNA damage, whose recognition activates stress kinases that phosphorylate p53 and prevent its proteosomal degradation (Meulmeester and Jochemsen, 2008). p53 induces pro-apoptotic proteins such as PUMA p53-upregulated modulator of apoptosis (PUMA) (Nakano and Vousden, 2001; Yu et al., 2001), which participates in the mitochondrially-controlled ‘intrinsic' apoptosis pathway (Yu and Zhang, 2008). This cell death pathway is regulated by Bcl-2 family proteins (Brunelle and Letai, 2009) and is executed by a molecular machinery including cytochrome-c, Apaf-1 and caspases-9, -3 and -7 (Riedl and Salvesen, 2007). Active downstream caspases cleave a variety of cellular substrates, leading to the demolition of the cell (Timmer and Salvesen, 2007). Inhibitor of caspase-activated DNAse (ICAD) (also known as DNA fragmentation factor 45; DFF45) is one important caspase substrate (Enari et al., 1998, Liu et al., 1998). Its cleavage permits caspase-activated DNAse (CAD)/DNA fragmentation factor 40 (DFF40) to fragment chromosomal DNA (Enari et al., 1998; Liu et al., 1998).
Although chemotherapy drugs can cure the majority of patients diagnosed with particular types of cancer (Evens et al., 2008; Jeha, 2009), many cancers tend to respond poorly, and better treatments are needed for these tumors (Winer et al., 2009). Defects in p53 or its relatives, mutations in components of the intrinsic apoptosis pathway or alterations in their expression, can contribute to chemotherapy insensitivity (Coultas and Strasser, 2003; Muller et al., 2006), so treatments that can bypass p53 and the intrinsic pathway may be effective in treating unresponsive cancers. Another challenge for improving cancer treatment is minimizing adverse effects of anti-neoplastic therapies. In addition to well-documented acute toxic effects of chemotherapy and radiotherapy, cancer survivors sometimes suffer so-called ‘late effects' that can manifest years into decades after successful treatment. These include cardiac, endocrine and central nervous system symptoms, as well as second malignant neoplasms (Dickerman, 2007; Meadows et al., 2009). These new cancers probably result from the genotoxic effects of chemotherapy or radiotherapy on normal cells (Rice et al., 2004).
A major recent focus of development of new cancer therapies has been the extrinsic death receptor apoptosis pathway, particularly drugs based on tumor necrosis factor-related apoptosis inducing ligand (TRAIL, also known as Apo2 L) (Mahalingam et al., 2009). TRAIL and related agents offer potential advantages over conventional chemotherapeutic drugs both in terms of efficacy and minimizing side effects. TRAIL is a type-II membrane protein with an extracellular carboxyl-terminal domain (amino acids 114 to 281) constituting the receptor-binding site (Wiley et al., 1995; Pitt et al., 1996). It induces the extrinsic apoptosis pathway by ligating death receptors (DR4/TRAIL-R1 and DR5/TRAIL-R2) that promote recruitment of FADD, caspase-8 and/or caspase-10 to form the ‘death-inducing signaling complex' (DISC) (Ashkenazi, 2008). Caspases-8 and/or -10 are activated within the DISC. In ‘type-I' cells, these are able to directly cleave and activate executioner caspases-3 and -7; in ‘type-II' cells, mitochondrial amplification of this signal is required for apoptosis to proceed (Knight et al., 2004; Vogler et al., 2008). Because TRAIL induces apoptosis through a distinct pathway from classical anti-cancer treatments, it promises to bypass blocks in intrinsic apoptotic pathways that render some tumor cells resistant to chemotherapy and/or radiotherapy. Pre-clinical studies revealed that ‘soluble' trimeric TRAIL, consisting of the ligand's extracellular domain complexed with zinc, showed a high degree of specificity for tumor cells relative to normal cells (Ashkenazi et al., 1999; Hao et al., 2004; Lawrence et al., 2001), however cross-linked and tagged formulations of TRAIL induced apoptosis in hepatocytes (Jo et al., 2000; Lawrence et al., 2001; Ganten et al., 2006; Ashley et al., 2008) and various other normal cells (Ashley et al., 2008; Leverkus et al., 2000; Nitsch et al., 2000). Soluble TRAIL and agonistic antibodies targeting its pro-apoptotic receptors have shown robust anti-tumor activity in animal models (Chuntharapai et al., 2001; Kelley et al., 2001), and encouraging results are emerging from early phase clinical trials (Ashkenazi, 2008; Holoch and Griffith, 2009; Mahalingam et al., 2009).
This study was designed to investigate the impact of TRAIL exposure on surviving cells. We reasoned that, as TRAIL induces apoptosis through a mechanism that does not require damage to DNA, cells that survive TRAIL treatment would be unlikely to bear mutations that could provoke late effects such as second malignancies in a clinical setting. We, therefore, compared the mutagenic capacity of sub-lethal doses of TRAIL and conventional chemotherapy drugs, using two commonly employed assays for DNA damage. The hypoxanthine-guanine phosphoribosyltransferase (HPRT) enzyme participates in purine salvage pathways. HPRT-proficient cells are killed by the purine analog 6-thioguanine (6-TG), but cells bearing HPRT loss-of-function mutations survive and form colonies. Hence, the number of 6-TG-resistant colonies after treatment with a drug such as TRAIL provides a readout of the mutational capacity of that drug (DeMars, 1971). Generation of double-stranded DNA breaks leads to activation of protein kinases including ataxia-telangiectasia mutated (ATM), whose downstream targets include histone 2AX (H2AX). H2AX phosphorylation (γH2AX) covers large regions of the chromosome surrounding each lesion, hence levels of γH2AX reflects the degree of DNA damage (Kuo and Yang, 2008; Watters et al., 2009). To our surprise, these assays revealed that TRAIL treatment did damage the DNA in surviving cells. This genotoxic effect was caspase-8- and CAD-dependent.
Results
Increased mutation frequency in cells treated with TRAIL and FasL
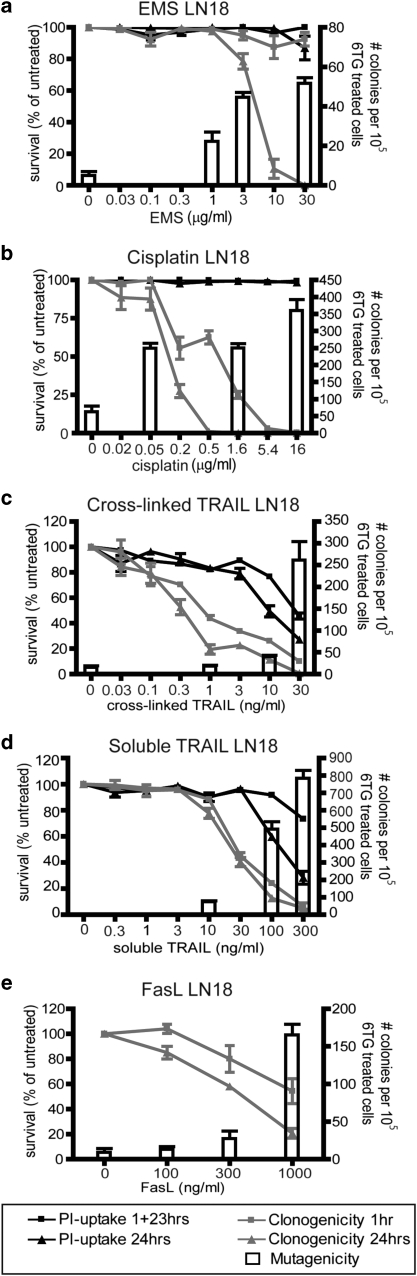
In this study, we compared the potential of TRAIL, FasL and known mutagens to cause acute cell death, impair clonogenic survival and generate mutations in surviving cells. To monitor cell death, propidium iodide-uptake and clonogenicity assays were performed, in which the cells were tested after 1 or 24 h exposure. Resistance to 6-TG was used as a readout of mutations in the HPRT locus. Ethane methyl sulfonate (EMS) is a well-studied mutagen (Gocke et al., 2009) and was used as a positive control drug. Treatment of the LN18 glioblastoma cell line with EMS decreased the cells' clonogenic survival and elicited a corresponding increase in the formation of 6-TG resistant colonies (Figure 1a), implying the presence of mutations in the HPRT locus. Cisplatin is a commonly used chemotherapy drug, which cross-links DNA and causes DNA damage. It showed a similar effect as EMS—an increase in mutation frequency with decreasing cell survival (Figure 1b). TRAIL induces apoptosis through activation of the extrinsic pathway, a mechanism thought not to be involved in DNA damage. Nevertheless, LN18 cells that were treated with a cross-linked TRAIL formulation or a soluble form showed an increase in mutation frequency at the HPRT locus, along with decreased cell survival (Figures 1c and d).
Figure 1.
TRAIL or FasL treatment of LN18 cells provokes generation of 6-TG-resistant colonies. LN18 cells were incubated with various doses of ethane methyl sulfonate (a), cisplatin (b), cross-linked TRAIL (c), soluble TRAIL (d) or Fas ligand (e), either for 24 h or for 1 h followed by 23 h incubation in normal media. Propidium iodide uptake (a–d) or clonogenicity assays (a–e) were then performed, as outlined in the Materials and methods. Cells were exposed to selected doses of the drugs for 1 h for HPRT mutagenicity assays. The left Y-axis and black and gray lines depict survival as measured in acute cell death and clonogenicity assays. The right Y-axis and white columns show the number of 6-TG-resistant colonies in mutational assays. Error bars indicate standard errors of the means from either three (a, b, e), four (c) or five (d) independent experiments.
To determine whether this apparent mutagenic property is specific to TRAIL, or may be a general feature of death ligands, we monitored the impact of FasL exposure on viability and mutation rate in LN18 cells. As for TRAIL, the pro-apoptotic effect of FasL was accompanied by emergence of 6-TG-resistant clones (Figure 1e), implying that it too induces mutations in surviving cells.
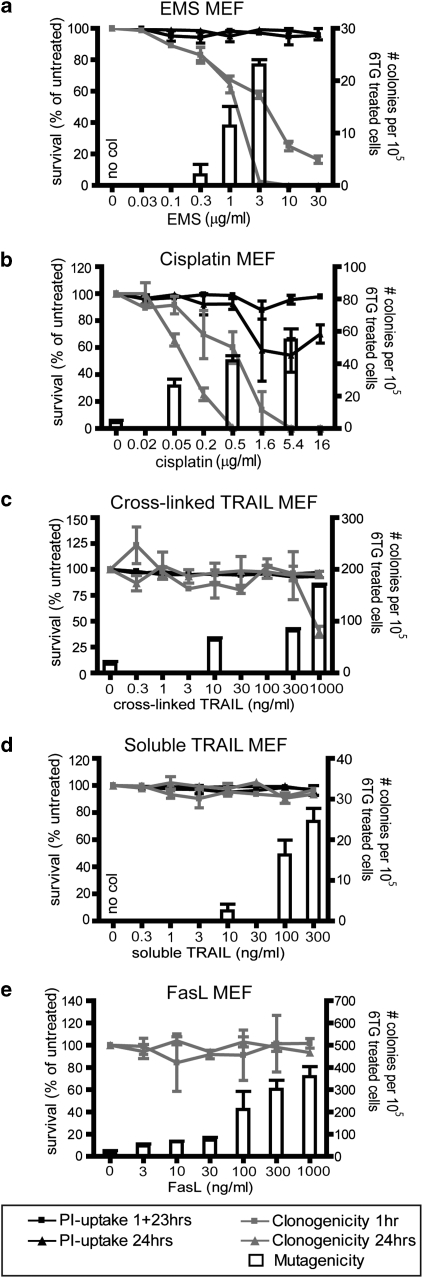
These viability and mutagenesis assays were repeated in a second cell type: mouse embyronic fibroblasts (MEFs). EMS exerted the same effect in MEFs as in LN18 cells: mutation frequency increased with decreasing clonogenic cell survival (Figure 2a). As in LN18 cells, cisplatin provoked a dose-dependent impact on clonogenic survival and increase in mutation frequency in MEFs (Figure 2b).
Figure 2.
TRAIL or FasL treatment of mouse embryo fibroblast (MEF) cells provokes generation of 6-TG-resistant colonies. MEF cells were incubated with various doses of ethane methyl sulphonate (a), cisplatin (b), cross-linked TRAIL (c), soluble TRAIL (d) or Fas ligand (e), for 1hr or 24 h, prior to propidium iodide uptake (a–d) or clonogenicity assays (a–e). Cells were exposed to selected doses of the drugs for 1 h for HPRT mutagenicity assays. The left Y-axis and black and gray lines depict survival as measured in acute cell death and clonogenicity assays. The right Y-axis and white columns show the number of 6-TG-resistant colonies in mutational assays. Doses, which failed to generate any 6-TG-resistant colonies are labeled ‘no col'. Error bars indicate standard errors of the means from either three (a, b, e), four (c) or five (d) independent experiments.
MEF cells were strikingly resistant to the toxic effects of cross-linked TRAIL. After a 1 h exposure, no difference could be observed in clonogenic survival between untreated and TRAIL-treated cells. After 24 h exposure, a decrease in clonogenic potential was observed only at the highest dose (1 μg/ml). 6-TG-resistant colonies emerged after treatment with sub-lethal doses of cross-linked TRAIL, and mutation frequency increased with exposure to increasing concentrations (Figure 2c).
MEF cells were totally resistant to apoptosis induced by soluble TRAIL at the doses used (up to 300 ng/ml), but an increase in mutation frequency could be detected with increasing doses (Figure 2d). An increase in MEF cells bearing HPRT mutations was also detected following exposure to FasL, whereas the cells' clonogenic potential remained unchanged (Figure 2e). These data show that TRAIL and FasL can stimulate mutagenesis in contexts in which these agents do not induce apoptosis. This argues against the possibility that the appearance of 6-TG-resistant clones following death-ligand treatment merely reflects selection for a pre-existing sub-population that is resistant to death induced by both 6-TG and death ligands.
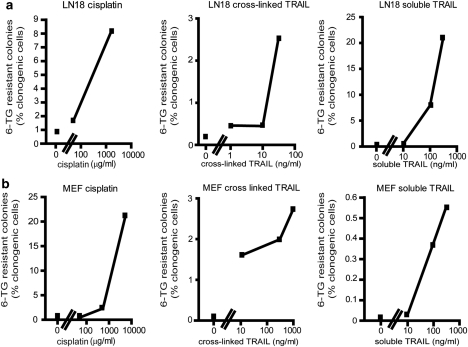
The highest mutation frequency stimulated by TRAIL was experienced by LN18 cells exposed to 300 ng/ml of soluble TRAIL (Figure 3a). This treatment abolished the clonogenic potential of 93% of the cells, and 20.6% of the remaining viable cells formed colonies in the presence of 6-TG, implying that they had sustained loss-of-function mutations in the HPRT locus. In comparison, incubation with 1.6 μg/ml cisplatin prevented clonogenic growth of 75% of LN18 cells, and 8.1% of the cells that retained clonogenic potential acquired HPRT mutations. In MEF cells, cisplatin was more mutagenic than either formulations of TRAIL at the doses analyzed (Figure 3b).
Figure 3.
Mutation frequencies of TRAIL and cisplatin. Mean mutation frequencies of LN18 (a) and mouse embryo fibroblast (b) cells are expressed as the percentage of clonogenically competent cells that generated 6-TG-resistant colonies. The X-axes show the doses analyzed (in log scale). Only doses that enabled the clonogenic survival of more than five cells per experiment were included in this analysis.
Double-stranded DNA damage in cells treated with TRAIL or cisplatin
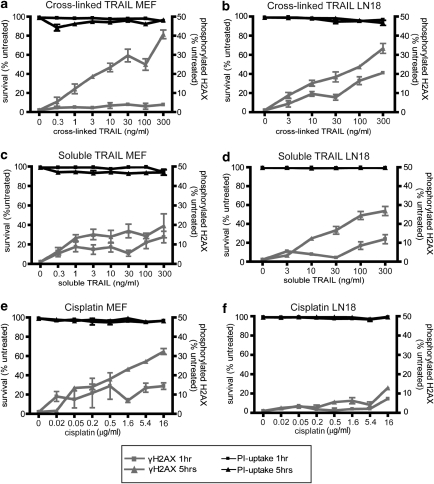
To confirm that the development of 6-TG-resistant clones following TRAIL treatment did reflect mutagenesis (rather than an alteration in nucleotide biosynthesis), we used a second, independent technique to monitor DNA damage. Phosphorylated H2AX proteins surrounding double-stranded breaks promotes the accumulation of DNA repair factors at the site of damage. Flow cytometric quantitation of H2AX phosphorylation provides a robust and sensitive measure of DNA damage (Watters et al., 2009). Exposure to cross-linked TRAIL provoked a dose- and time-dependent increase in the proportion of MEF and LN18 cells containing phosphorylated H2AX (Figures 4a and b). Treatment with the soluble form of TRAIL triggered γH2AX in fewer cells (Figures 4c and d). Incubation with cisplatin, stimulated substantial H2AX phosphorylation in MEF cells (Figure 4e), but much less in LN18 cells (Figure 4f). It has been previously noted that γH2AX foci form in cisplatin-treated cells only following transit through S phase of the cell cycle (Frankenberg-Schwager et al., 2005; Olive and Banath, 2009), so the lower proportion of γH2AX-positive LN18 cells relative to MEF cells may simply reflect differences in proliferation rates.
Figure 4.
TRAIL treatment stimulates phosphorylation of H2AX. The right Y-axes and gray lines show the proportion of MEF cells (a, c, e) or LN18 cells (b, d, f) bearing phosphorylated H2AX after 1 or 5 h exposure to cross-linked TRAIL (a, b), soluble TRAIL (c, d) or cisplatin (e, f). The left Y-axes and black lines represent the percentage of viable cells after same drug exposures. Error bars indicate s.e.m. from three independent experiments.
The mutagenic effect of TRAIL requires caspase-8
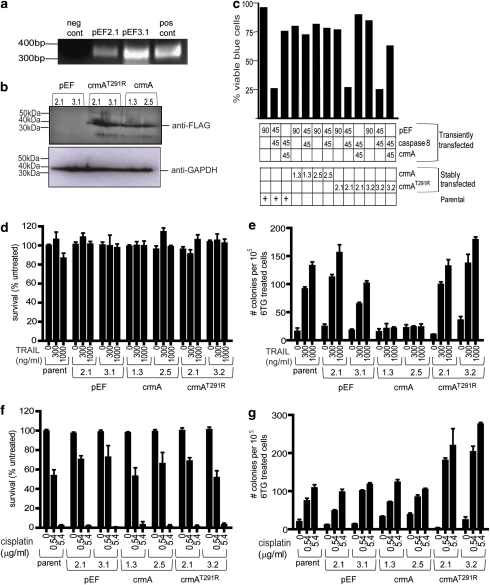
Stable MEF clones expressing wild-type crmA or an inactive crmA mutant (Ekert et al., 1999), or bearing an empty vector (pEF) were generated to investigate whether crmA inhibition of caspase-8 could block TRAIL-induced apoptosis and/or mutagenesis. The presence of plasmid DNA in the empty vector clones was confirmed by PCR (Figure 5a) and expression of wild-type and mutant crmA was visualized by western blot (Figure 5b). Functionality of the stable crmA transfectants was tested by transiently transfecting these and parental MEF cells with a caspase-8 expression plasmid. Overexpression of caspase-8 induced apoptosis in parental MEFs and the crmA mutant MEF clones, but the wild-type crmA MEF clones were resistant to caspase-8 induced death (Figure 5c). As expected, transient co-transfection of a crmA plasmid together with the caspase-8 construct failed to kill parental MEFs and the crmA mutant MEF cell lines (Figure 5c). All stable transfectant lines maintained equivalent clonogenic potential following exposure to cross-linked TRAIL for 1 h (Figure 5d). Parental MEFs, crmA mutant clones and empty vector transfectants formed 6-TG-resistant colonies following TRAIL treatment in a dose-dependent manner, but the crmA-expressing clones only developed background levels of 6-TG-resistant colonies, regardless of TRAIL exposure (Figure 5e). Cisplatin provoked clonogenic cell death in crmA, crmA mutant, pEF clones and in the parental cell line (Figure 5f). The mutagenic effect of cisplatin persisted in the presence of wild-type crmA and was curiously enhanced by expression of mutant crmA (Figure 5g).
Figure 5.
CrmA expression prevents the formation of 6-TG-resistant mouse embryo fibroblast (MEF) colonies following TRAIL treatment. MEF cells were stably transfected with empty vector (pEF), a wild-type FLAG-tagged crmA expression plasmid or a loss-of-function FLAG-crmA expression plasmid. Genomic DNA from pEF clones was amplified using PCR (a). Products of reactions containing either no template or 100 ng of plasmid are denoted ‘pos cont' and ‘neg cont' respectively. CrmA expression was analyzed using anti-FLAG immunoblotting, relative to a GAPDH loading control (b). (c) The phenotypes of crmA wild-type and mutant clones were tested by transiently transfecting the clones with empty vector (pEF), crmA and/or caspase-8 expression plasmids (90% of transfected DNA), along with a β-galactosidase expression plasmid (10%). The transfectants were stained with Xgal and the blue (transfected) cells were scored visually for morphological characteristics of apoptosis. The proportions of each transiently transfected plasmid are indicated under the graph, as are the stable clones into which this DNA was introduced. Clonogenic survival of each clone was monitored following 1 h exposure to cross-linked TRAIL (d) or cisplatin (f). Parental MEF cells and the indicated stable cell lines were treated with cross-linked TRAIL(e) or cisplatin (g) for 1 h then subjected to the HPRT mutational assay, and the 6-TG-resistant clones that emerged were counted.
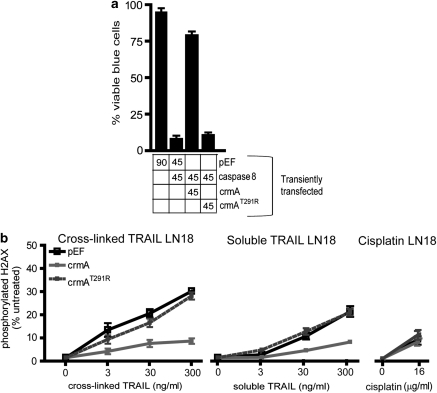
The effect of crmA expression on TRAIL-induced DNA damage was also investigated in LN18 cells. Co-transfection with a green fluorescent protein expression plasmid revealed that transient transfection frequencies of around 80% were consistently achieved in this cell line, obviating the need to generate stable transfectants. As in MEF cells, expression of wild-type crmA protected LN18 cells from apoptosis induced by caspase-8 overexpression (Figure 6a). Significantly fewer LN18 cells expressing wild-type crmA bore phosphorylated H2AX than cells transfected with either the crmA mutant plasmid or empty vector (Figure 6b). The smaller number of γH2AX positive cells in the samples transfected with the crmA plasmid probably represents the sub-population (∼20% of cells) which were not transiently transfected. In contrast, crmA expression had no effect on cisplatin-mediated cell death nor the modest ability of cisplatin to stimulate γH2AX in this cell line (Figure 6b).
Figure 6.
CrmA expression prevents TRAIL-induced DNA damage in LN18 cells. (a) LN18 cells were transiently transfected with the indicated plasmids (90% of transfected DNA), along with a β-galactosidase expression plasmid (10%). The transfectants were stained with Xgal and the blue (transfected) cells were scored visually for apoptosis based on morphological criteria. The percentage of the transiently transfected plasmids are indicated under the graph. (b) LN18 cells were transiently transfected with an empty vector (pEF) or plasmids encoding wild-type or mutant crmA, then incubated with the indicated doses of cross-linked TRAIL, soluble TRAIL or cisplatin for 5 h. The proportions of cells bearing phosphorylated H2AX were quantitated by flow cytometry. Error bars indicate standard errors of the means from three independent experiments.
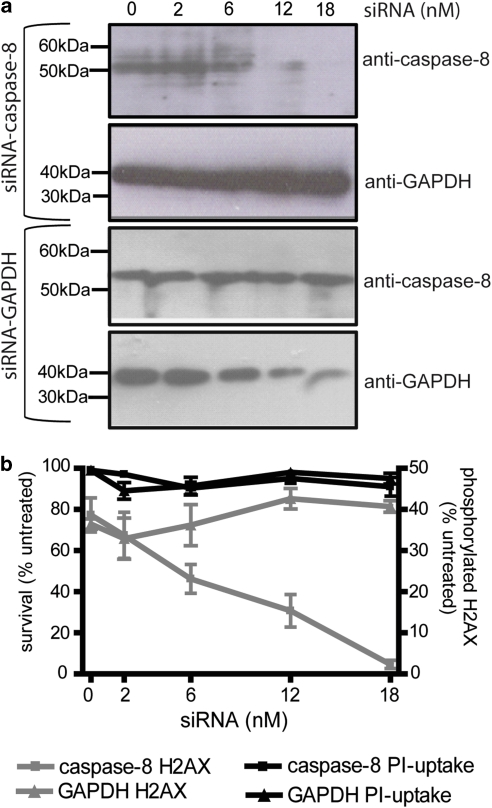
The above results imply that a crmA-sensitive protease is required for TRAIL to stimulate DNA damage. Although caspase-8 is the obvious candidate for this protease, these data could not rule out the possible involvement of other proteases susceptible to crmA inhibition, like caspase-1 (Komiyama et al., 1994). To directly test the requirement for caspase-8 in TRAIL mutagenesis, we downregulated its expression in MEF cells and monitored γH2AX formation after TRAIL treatment. Caspase-8 downregulation (Figure 7a) was mirrored by a corresponding decrease in the proportion of cells bearing phosphorylated H2AX (Figure 7b), whereas silencing of glyceraldehyde 3-phosphate dehydrogenase had no such effect (Figure 7b).
Figure 7.
Downregulation of caspase-8 prevents TRAIL-induced DNA damage. MEF cells were transfected with the indicated concentrations of siRNAs targeting caspase-8 or glyceraldehyde 3-phosphate dehydrogenase then subjected to immunoblotting (a) or treated with 300 ng/ml cross-linked TRAIL and assayed for H2AX phosphorylation (b). The left Y-axis and black lines represent the percentage of viable cells. The right Y-axis and gray lines show the proportion of cells bearing phosphorylated H2AX. Error bars indicate s.e.m. from three independent experiments.
CAD involvement in TRAIL-induced DNA damage
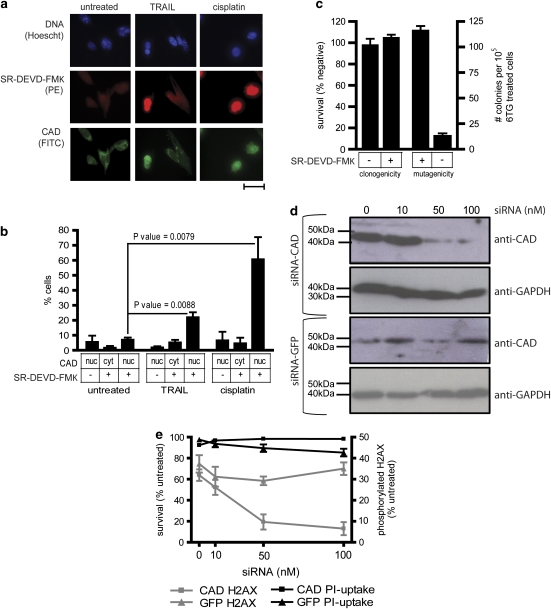
The data above imply that treatment with TRAIL (or FasL) provokes mutagenesis in surviving cells, in a caspase-8 dependent manner. One potential mechanism by which death receptor signaling could stimulate mutagenesis would be caspase-mediated activation of the DNase CAD/DFF40. Activated caspases-3 and -7 can cleave ICAD/DFF45, which keeps CAD inactive and (at least in some cells) sequestered in the cytosol (Enari et al., 1998; Liu et al., 1998). When ICAD is cleaved, CAD becomes activated and can translocate to the nucleus to fragment cellular DNA (Widlak and Garrard, 2005). We examined the effect of TRAIL or cisplatin treatment on caspase activation and CAD translocation (as a measure of activation) in MEF cells. Activation of executioner caspases-3 and -7 was investigated using the fluorescent inhibitor SR-DEVD-FMK. Localization of CAD was measured by fluorescence microscopy. CAD was localized in the nucleus in approximately 24% of TRAIL-treated cells, compared with 13% of untreated cells (Figures 8a and b). Approximately 67% of cisplatin-treated cells bore nuclear CAD (Figures 8a and b). Co-staining with the CAD antibody and fluorescent caspase inhibitor revealed that CAD was nuclearly localized in 78% of cells containing active caspases (Figures 8a and b).
Figure 8.
Caspase-activated CAD is required for TRAIL to provoke mutations in surviving cells. (a) Fluorescent microscopy was used to investigate CAD localization and SR-DEVD-FMK binding, as a measure of caspase-3/7 activation, in untreated MEF cells or cells incubated for 24 h with cisplatin (5.4 μg/ml) or cross-linked TRAIL (300 ng/ml). The scale bar is 100 μm. (b) The proportions of cells bearing nuclear CAD or SR-DEVD-FMK fluorescence after each treatment were counted. At least 250 cells were scored for each treatment per experiment for untreated and TRAIL-treated samples; at least 100 cisplatin-treated cells were scored per experiment. Error bars indicate s.e.m. from four, three or seven replicates of untreated, cisplatin-treated or TRAIL-treated cells respectively. P-values were calculated using a Student's t-test. (c) After exposure to 300 ng/ml cross-linked TRAIL for 24 h, SR-DEVD-FMK positive and negative cells were flow cytometrically sorted and their clonogenicity assayed following treatment with 300 ng/ml cross-linked TRAIL for 1 h. SR-DEVD-FMK positive and negative sorted cells were also subjected to the HPRT mutational assay, and the 6-TG-resistant clones that emerged were counted. Error bars show the s.e.m. from six or nine independent experiments for clonogenicity and mutagenicity experiments respectively. MEF cells were transfected with the indicated concentrations of small interfering RNAs targeting CAD or GFP then (d) subjected to immunoblotting or (e) treated with 300 ng/ml cross-linked TRAIL and assayed for H2AX phosphorylation. The left Y-axis and black lines represent the percentage of viable cells. The right Y-axis and gray lines show the proportion of cells bearing phosphorylated H2AX. Error bars indicate s.e.m. from three independent experiments.
To assess whether CAD and caspase activation correlated with mutagenesis, TRAIL-treated MEF cells were sorted into SR-DEVD-FMK positive and negative populations and cultured. After a week's culturing, clonogenic and HPRT mutagenesis assays were performed with each sub-population. There was no difference in clonogenic potential between SR-DEVD-FMK positive and SR-DEVD-FMK negative cells, but the mutation frequency was 10-fold higher in SR-DEVD-FMK positive cells (Figure 8c).
To directly assess the importance of CAD in TRAIL-induced mutagenesis, small interfering (si)RNA was used to downregulate CAD in MEF cells (Figure 8d). The proportion of MEF cells containing phosphorylated H2AX decreased with increased concentrations of silencer RNA (Figure 8e).
Discussion
TRAIL signaling and its ability to activate extrinsic cell death has been extensively characterized, but the ability of TRAIL to cause mutations has not been explored to date. As TRAIL activates an apoptotic pathway that does not require recognition of DNA damage, unlike many conventional chemotherapeutic agents, we sought to determine whether TRAIL signaling would cause permanent damage to the genomes of surviving cells. It has previously been shown that EMS and cisplatin, drugs that create DNA adducts, can cause DNA damage and mutations (Rabik and Dolan, 2007; Gocke et al., 2009). We have confirmed those results in human and murine cell lines, using a conventional HPRT assay and by detection of phosphorylated H2AX. These techniques and cells were used to evaluate the mutagenic capacity of two formulations of TRAIL: cross-linked (‘superkiller') TRAIL and the extracellular portion (‘soluble' TRAIL). Unlike cross-linked preparations, soluble TRAIL has been previously shown to be non-toxic to normal human cells (Ashkenazi et al., 1999), which would give this formulation an advantage in cancer therapy. To our surprise, both formulations generated mutations in surviving cells. The mutagenic capacity of both preparations of TRAIL was dose-dependent, evident using two independent assays in two cell lines, and manifested in MEF cells at sub-lethal concentrations. Additional research will be needed to explore the susceptibility of additional normal and cancerous cell types to the genotoxic effects of TRAIL.
Fas ligand, another death ligand that activates extrinsic apoptosis signaling, also showed mutagenic potential. This argues against the possibility that TRAIL receptors possess a unique mutagenic signaling mechanism, and implicates molecules downstream of death receptors in transmitting the mutagenic signal.
Expression of wild-type (but not mutant) crmA, or siRNA silencing of caspase-8 abolished the mutagenic effect of TRAIL, indicating that TRAIL-induced mutagenesis is caspase-8 dependent. Apoptotic stimuli provoke caspase-dependent cleavage of ICAD, allowing CAD to fragment DNA during apoptosis (Widlak, 2000). It occurred to us that low levels of CAD activation following sub-lethal TRAIL treatment might underlie TRAIL's genotoxic effect. Consistent with this model, an increase in cells bearing nuclear CAD was observed following exposure to a non-lethal dose of the drug, compared with those untreated cells. Caspases were activated in TRAIL-treated cells bearing nuclear CAD. TRAIL-treated cells containing activated caspases at the time of isolation initially proliferated somewhat slower after sorting, but when reseeded the clonogenicities of the two populations were similar. One explanation for the slow start could be that DNA was damaged in cells bearing active caspases, so they may have arrested and recommenced cycling after their DNA was repaired. Viable TRAIL-treated cells in which caspases had been activated showed a ten-fold increase in mutation frequency compared with caspase-negative cells, which only showed background formation of spontaneous mutations. Although it was originally assumed that cells bearing active caspases would be destined to die, it is increasingly recognized that cells can tolerate a degree of caspase activation without undergoing apoptosis (Abraham and Shaham, 2004), and this study provides further evidence supporting that notion.
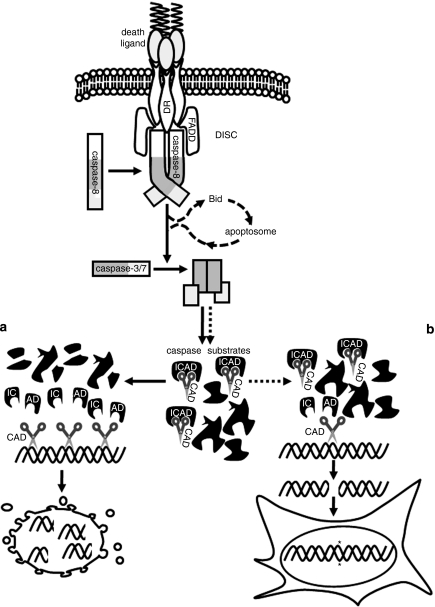
Our study has revealed that cells surviving exposure to TRAIL sustain DNA damage through a caspase-8-dependent mechanism. TRAIL-treated cells with active caspases and nuclearly localized CAD showed markedly higher mutation rates than cells with inactive caspases and cytosolic CAD. This led us to postulate that TRAIL causes mutagenesis in context involving caspase activation that is insufficient to induce apoptosis, but sufficient to cleave ICAD and activate CAD, which damages DNA (Figure 9). Consistent with this model, downregulation of CAD protected TRAIL-treated cells from DNA damage. Our data provide support for a previous speculation that sub-lethal apoptotic signaling may burden surviving cells with mutations caused by misrepair of DNA damage elicited by apoptotic nucleases (Vaughan et al., 2002). CAD cleavage of DNA generates blunt (or nearly blunt) double-stranded breaks (Widlak et al., 2000). These would probably be repaired in surviving cells by previously characterized mechanisms including non-homologous end-joining, which is often non-conservative and can introduce mutations (Hartlerode and Scully, 2009). If the mutagenic capacity of TRAIL we have observed is subsequently confirmed in vivo, the potential for adverse effects resulting from mutations in surviving normal cells should be considered if TRAIL-based drugs become clinically used. Further work will be needed to determine if other anti-cancer agents targeting apoptosis pathways can also provoke mutagenesis in surviving cells.
Figure 9.
TRAIL-induced mutagenesis model. Death receptor ligation promotes formation of the death inducing signalling complex (DISC) and activation of caspase-8, which then activates caspases-3 and/or -7. This can trigger apoptosis (a), through cleavage of numerous cellular substrates including ICAD, whose cleavage provokes activation of CAD which fragments DNA. Data reported herein suggest that in cells bearing only a small number of activated effector caspases, limited caspase-mediated ICAD proteolysis leads to CAD activation and DNA cleavage, but not apoptosis (b). We suggest that inaccurate repair of these lesions creates mutations in surviving cells.
Materials and methods
Cell lines and materials
The glioma cell line LN18 was purchased from ATCC (Manassas, VA, USA). SV-40 transformed MEF were kindly provided by Anissa Jabbour (Jabbour et al., 2009). All cells were cultured in Dulbecco's modified Eagle medium high glucose (Invitrogen, Carlsbad, CA, USA) containing 10% fetal calf serum (Invitrogen). Drugs used in this study were cisplatin (Mayne Pharma, Mulgrave, Victoria, Australia), EMS (Sigma Aldrich, St Louis, MO, USA), 6-TG (Sigma Aldrich), Superkiller (cross-linked) TRAIL (Alexis Biochemicals, Lausen, Switzerland), recombinant human (soluble) TRAIL (Chemicon, Temecula, CA, USA) and SuperFas Ligand soluble (Alexis Biochemicals). The following antibodies were used: anti-FLAG antibody (Sigma Aldrich, clone M2), anti-GAPDH (Chemicon, clone 6C5), anti-CAD (ProSci, Poway, CA, USA; #2007), anti-caspase-8 (ProSci, #3475), anti-H2AX (Ser 139) (Cell Signaling, Danvers, MA, USA; clone 20E3), goat anti-mouse-HRP (Sigma Aldrich), goat anti-rabbit FITC antibody (Chemicon) and donkey anti-rabbit-HRP (Amersham/GE Healthcare, Rydalmere, NSW, Australia).
Plasmids
The pEF, pEF-FLAG-crmA and CMV-LacZ plasmids have been previously described (Hawkins et al., 1996), as was pEF-FLAG-crmA T291R (Ekert et al., 1999). To generate pEF-koz-caspase-8, the coding region of caspase-8 was amplified using primers 1 and 2, and then the product was cut with BamHI and XbaI and inserted into pEF cut with BamHI and XbaI. pEGFP-N1 (Clontech, Mountain View, CA, USA) was used to monitor transfection efficiency.
Transfections
Stable MEF transfectants were made using FuGENE HD transfection reagent (Roche, Basel, Switzerland) and selected in 30 μg/ml puromycin (Sigma Aldrich). Expression was confirmed by immunoblotting as previously published (Ashley et al., 2008). The presence of the empty pEF plasmid was confirmed by PCR amplification using primers 3 and 4 and Taq polymerase (Applied Biosystems, Mulgrave, Victoria, Australia). Genomic DNA was extracted from MEFs using the DNeasy Tissue Kit (Qiagen, Doncaster, Victoria, Australia) according to manufacturer's instructions. Thirty cycles were used to amplify MEF pEF cDNA, according to the following protocol: denaturation (30 s, 94 °C), annealing (30 s, 55 °C) and extension (1 min, 72 °C). LN18 cells were transiently transfected using Lipofectamine Plus (Invitrogen). Transfection efficiency was monitored by performing parallel transfections in which pEGFP-N1 constituted 10% of the transfecting DNA and analyzing the proportion of green cells by flow cytometry using a FACSCanto (BD Biosciences, San Jose, CA, USA). Transient transfection efficiencies for LN18 cells were always greater than 79%. To downregulate gene expression, MEF cells were transfected with X-tremeGENE siRNA transfection reagent (Roche) according to manufacturer's instructions.
Oligonucleotides
1: 5′GCGGGATCCGCCATGGACTTCAGCAGAAATCTTTATG-3′
2:5′-GCTCTAGACTAATCAGAAGGGAAGACAAG-3′
3:5′-GAGACTGAAGTTAGGCCAGCTTGGC-3′
4:5′-CAGCTCTGGCCACCTCTCCTTGTCC-3′
Cell survival assays
Cells were incubated with drugs for 1 and 24 h. Propidium iodide-uptake assays were used to assess acute cell death. Ten thousand cells were pelleted and resuspended in propidium iodide (1 μg/ml in phosphate buffered-saline (PBS)) (Sigma Aldrich) then analyzed using a FACS Canto (BD Biosciences). For all flow cytometry assays, identification of positive populations was achieved using a marker, which designated 1% of untreated cells as positive. For clonogenicity assays, 3000 cells were plated per well of a six-well plate and incubated with drugs for 1 or 24 h. After 6 days, cells were stained with methylene blue (Sigma Aldrich) 1.25 g/l in 50% methanol, incubated for 5 min and washed twice with water and the numbers of colonies counted. CrmA function was tested by transiently co-transfecting MEF or LN18 cells with koz-caspase-8, pEF and/or pEF-FLAG-crmA (90%) and CMV-LacZ (10%). A day after transfection, cells were treated with TRAIL for 24 h then stained with Xgal (Hawkins et al., 1996). Survival of blue (transfected) cells was scored visually based on morphological criteria, as previously described (Hawkins et al., 1996).
HPRT assay
The HPRT assay was based on a previously published method (Op het Veld et al., 1997). Briefly, 106 cells were seeded per 150 mm dish (three dishes per treatment). After culturing for 4 days, 106 cells were re-seeded in fresh 150 mm dishes and grown for an additional 4 days. Cells were seeded at 105 cells per 150 mm dish (three dishes per treatment) in media containing 6-TG (Sigma Aldrich); 20 μg/ml for LN18 cells and 0.6 μg/ml for MEF cells. Colonies were stained with methylene blue and counted after 18 days (LN18) or 13 days (MEF).
DNA damage assay
Staining of γH2AX was performed according to a published protocol (MacPhail et al., 2003), with following changes: cells were exposed to drugs for 1 or 5 h then labeled with anti-H2AX and anti-rabbit FITC antibodies. After washing to remove unbound antibodies, the cells were resuspended in PBS containing propidium iodide (5 μg/ml) to limit the analysis to permeabilized intact cells (excluding cells with sub-G1 DNA content). The γH2AX signals from these cells were analyzed using the FACS Canto (BD Biosciences).
Cell sorting
MEF cells were exposed to 300 ng/ml cross-linked-TRAIL for 24 h. Caspase activity was determined by the red fluorescent caspase activity (SR-DEVD-FMK) assay (Neuromics, Edina, MN, USA) performed according to manufacturer's instructions. Cells were stained with SR-DEVD-FMK and propidium iodide. Cells with disrupted membranes, cellular debris and cell aggregates were eliminated using propidium iodide staining intensity, light scatter and pulse-width parameters. SR-DEVD-FMK positive single viable cells were sorted using a MoFlo (Beckman Coulter, Brea, CA, USA) using a 1–2 drop envelope and high purity sort logic. One thousand cells were plated for clonogenicity assays and 105 cells used in HPRT assays.
Fluorescence microscopy
MEF cells were incubated for 24 h with 300 ng/ml superkiller TRAIL or with 5.4 μg/ml. cisplatin, or normal media. The following day, the cells were washed with PBS and fixed in 4% fresh paraformaldehyde for 15 min. Cells were washed again with PBS three times. Cells were permeabilized by incubating with 0.2% Triton X-100 in PBS for 5 min. Cells were washed for 30 min in PBS 1% bovine serum albumin, then incubated with CAD antibody 1:100 for 1 h. Cells were washed in PBS again three times then incubated in anti-rabbit FITC antibody 1:200 for 2 h in the dark. Cells were washed with PBS containing 1% bovine serum albumin for three times and 1:30 SR-DEVD-FMK reagent (Neuromics) was added and incubated at 37 °C, 5% CO2 for 1 h in the dark. Cells were washed three times with PBS containing 1% bovine serum albumin. Hoechst stain (10 μg/ml) in PBS was added to cells and incubated for 10 min in the dark. Cells were washed with 0.2% Triton X-100 in PBS three times before mounting the coverslip onto a microscope slide for analysis by fluorescent microscopy using an Olympus B202 microscope (Olympus, Mt Waverley, Victoria, Australia).
Small interfering RNA silencing
MEF cells were transfected using the following small interfering RNA constructs: CAD (sc-29872, Santa Cruz Biotechnology, Santa Cruz, CA, USA), caspase-8 (s63395, Applied Biosystems), GFP (sc-45924, Santa Cruz Biotechnology) and glyceraldehyde 3-phosphate dehydrogenase (s234321, Applied Biosystems). Half of the transfected cells were used for immunoblotting and the remaining were exposed to 300 ng/ml cross-linked TRAIL or 5.4 μg/ml cisplatin for 5 h, and γH2AX assays performed.
Acknowledgments
This study was funded by a Cancer Council Victoria Grant-in-Aid (#384400), a La Trobe University Postgraduate scholarship to MML, a National Health and Medical Research Council Career Development Award (#541930) and Australian Research Council Future Fellowship (#FT0991464) to CJH. We are grateful to Anissa Jabbour for the MEF cells and advice, to Rebecca Bilardi for assistance in establishing the γH2AX assay and to Lahiru Gangoda for help with fluorescence microscopy. We thank Tanja Kitevska for proofreading, Delara Pantaki, Sarah Roberts, Sunil Kumar and Livleen Kaur for technical assistance and members of the Hawkins laboratory and Children's Cancer Centre for advice and suggestions.
The authors declare no conflict of interest.
References
- Abraham MC, Shaham S. Death without caspases, caspases without death. Trends Cell Biol. 2004;14:184–193. doi: 10.1016/j.tcb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–331. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley DM, Riffkin CD, Lovric MM, Mikeska T, Dobrovic A, Maxwell JA, et al. In vitro sensitivity testing of minimally passaged and uncultured gliomas with TRAIL and/or chemotherapy drugs. Br J Cancer. 2008;99:294–304. doi: 10.1038/sj.bjc.6604459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuntharapai A, Dodge K, Grimmer K, Schroeder K, Marsters SA, Koeppen H, et al. Isotype-dependent inhibition of tumor growth in vivo by monoclonal antibodies to death receptor 4. J Immunol. 2001;166:4891–4898. doi: 10.4049/jimmunol.166.8.4891. [DOI] [PubMed] [Google Scholar]
- Coultas L, Strasser A. The role of the Bcl-2 protein family in cancer. Semin Cancer Biol. 2003;13:115–123. doi: 10.1016/s1044-579x(02)00129-3. [DOI] [PubMed] [Google Scholar]
- DeMars R. Genetic studies of HG-PRT deficiency and the Lesch-Nyhan syndrome with cultured human cells. Fed Proc. 1971;30:944–955. [PubMed] [Google Scholar]
- Dickerman JD. The late effects of childhood cancer therapy. Pediatrics. 2007;119:554–568. doi: 10.1542/peds.2006-2826. [DOI] [PubMed] [Google Scholar]
- Ekert PG, Silke J, Vaux DL. Inhibition of apoptosis and clonogenic survival of cells expressing CrmA variants: optimal caspase substrates are not necessarily optimal inhibitors. EMBO J. 1999;18:330–338. doi: 10.1093/emboj/18.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Evens AM, Hutchings M, Diehl V. Treatment of Hodgkin lymphoma: the past, present, and future. Nat Clin Pract Oncol. 2008;5:543–556. doi: 10.1038/ncponc1186. [DOI] [PubMed] [Google Scholar]
- Frankenberg-Schwager M, Kirchermeier D, Greif G, Baer K, Becker M, Frankenberg D. Cisplatin-mediated DNA double-strand breaks in replicating but not in quiescent cells of the yeast Saccharomyces cerevisiae. Toxicology. 2005;212:175–184. doi: 10.1016/j.tox.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Ganten TM, Koschny R, Sykora J, Schulze-Bergkamen H, Buchler P, Haas TL, et al. Preclinical differentiation between apparently safe and potentially hepatotoxic applications of TRAIL either alone or in combination with chemotherapeutic drugs. Clin Cancer Res. 2006;12:2640–2646. doi: 10.1158/1078-0432.CCR-05-2635. [DOI] [PubMed] [Google Scholar]
- Gocke E, Burgin H, Muller L, Pfister T. Literature review on the genotoxicity, reproductive toxicity, and carcinogenicity of ethyl methane sulfonate. Toxicol Lett. 2009;190:254–265. doi: 10.1016/j.toxlet.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Hao C, Song JH, Hsi B, Lewis J, Song DK, Petruk KC, et al. TRAIL inhibits tumor growth but is nontoxic to human hepatocytes in chimeric mice. Cancer Res. 2004;64:8502–8506. doi: 10.1158/0008-5472.CAN-04-2599. [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins CJ, Uren AG, Hacker G, Medcalf RL, Vaux DL. Inhibition of interleukin 1-beta-converting enzyme-mediated apoptosis of mammalian cells by baculovirus IAP. Proc Natl Acad Sci USA. 1996;93:13786–13790. doi: 10.1073/pnas.93.24.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch PA, Griffith TS. TNF-related apoptosis-inducing ligand (TRAIL): a new path to anti-cancer therapies. Eur J Pharmacol. 2009;625:63–72. doi: 10.1016/j.ejphar.2009.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour AM, Heraud JE, Daunt CP, Kaufmann T, Sandow J, O'Reilly LA, et al. Puma indirectly activates Bax to cause apoptosis in the absence of Bid or Bim. Cell Death Differ. 2009;16:555–563. doi: 10.1038/cdd.2008.179. [DOI] [PubMed] [Google Scholar]
- Jeha S. New therapeutic strategies in acute lymphoblastic leukemia. Semin Hematol. 2009;46:76–88. doi: 10.1053/j.seminhematol.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Jo M, Kim TH, Seol DW, Esplen JE, Dorko K, Billiar TR, et al. Apoptosis induced in normal human hepatocytes by tumor necrosis factor-related apoptosis-inducing ligand. Nat Med. 2000;6:564–567. doi: 10.1038/75045. [DOI] [PubMed] [Google Scholar]
- Kelley SK, Harris LA, Xie D, Deforge L, Totpal K, Bussiere J, et al. Preclinical studies to predict the disposition of Apo2 L/tumor necrosis factor-related apoptosis-inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J Pharmacol Exp Ther. 2001;299:31–38. [PubMed] [Google Scholar]
- Knight ME, Riffkin CD, Ekert PG, Ashley DM, Hawkins CJ. Caspase-8 levels affect necessity for mitochondrial amplification in death ligand-induced glioma cell apoptosis. Mol Carcinog. 2004;39:173–182. doi: 10.1002/mc.20011. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Ray C, Pickup D, Howard A, Thornberry N, Peterson E, et al. Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J Biol Chem. 1994;269:19331–19337. [PubMed] [Google Scholar]
- Kuo LJ, Yang LX. Gamma-H2AX—a novel biomarker for DNA double-strand breaks. In vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- Lawrence D, Shahrokh Z, Marsters S, Achilles K, Shih D, Mounho B, et al. Differential hepatocyte toxicity of recombinant Apo2L/TRAIL versions. Nat Med. 2001;7:383–385. doi: 10.1038/86397. [DOI] [PubMed] [Google Scholar]
- Leverkus M, Neumann M, Mengling T, Rauch CT, Brocker EB, Krammer PH, et al. Regulation of tumor necrosis factor-related apoptosis-inducing ligand sensitivity in primary and transformed human keratinocytes. Cancer Res. 2000;60:553–559. [PubMed] [Google Scholar]
- Liu X, Li P, Widlak P, Zou H, Luo X, Garrard WT, et al. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc Natl Acad Sci USA. 1998;95:8461–8466. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPhail SH, Banath JP, Yu TY, Chu EH, Lambur H, Olive PL. Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int J Radiat Biol. 2003;79:351–358. doi: 10.1080/0955300032000093128. [DOI] [PubMed] [Google Scholar]
- Mahalingam D, Szegezdi E, Keane M, Jong S, Samali A. TRAIL receptor signalling and modulation: Are we on the right TRAIL. Cancer Treat Rev. 2009;35:280–288. doi: 10.1016/j.ctrv.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Meadows AT, Friedman DL, Neglia JP, Mertens AC, Donaldson SS, Stovall M, et al. Second neoplasms in survivors of childhood cancer: findings from the Childhood Cancer Survivor Study cohort. J Clin Oncol. 2009;27:2356–2362. doi: 10.1200/JCO.2008.21.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Jochemsen AG. p53: a guide to apoptosis. Curr Cancer Drug Targets. 2008;8:87–97. doi: 10.2174/156800908783769337. [DOI] [PubMed] [Google Scholar]
- Muller M, Schleithoff ES, Stremmel W, Melino G, Krammer PH, Schilling T. One, two, three–p53, p63, p73 and chemosensitivity. Drug Resist Updat. 2006;9:288–306. doi: 10.1016/j.drup.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- Nitsch R, Bechmann I, Deisz RA, Haas D, Lehmann TN, Wendling U, et al. Human brain-cell death induced by tumour-necrosis-factor-related apoptosis-inducing ligand (TRAIL) Lancet. 2000;356:827–828. doi: 10.1016/S0140-6736(00)02659-3. [DOI] [PubMed] [Google Scholar]
- Olive PL, Banath JP. Kinetics of H2AX phosphorylation after exposure to cisplatin. Cytometry B Clin Cytom. 2009;76:79–90. doi: 10.1002/cyto.b.20450. [DOI] [PubMed] [Google Scholar]
- Op het Veld CW, van Hees-Stuivenberg S, van Zeeland AA, Jansen JG. Effect of nucleotide excision repair on HPRT gene mutations in rodent cells exposed to DNA ethylating agents. Mutagenesis. 1997;12:417–424. doi: 10.1093/mutage/12.6.417. [DOI] [PubMed] [Google Scholar]
- Pitt RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice SC, Vacek P, Homans AH, Messier T, Rivers J, Kendall H, et al. Genotoxicity of therapeutic intervention in children with acute lymphocytic leukemia. Cancer Res. 2004;64:4464–4471. doi: 10.1158/0008-5472.CAN-03-3940. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- Vaughan AT, Betti CJ, Villalobos MJ. Surviving apoptosis. Apoptosis. 2002;7:173–177. doi: 10.1023/a:1014374717773. [DOI] [PubMed] [Google Scholar]
- Vogler M, Walczak H, Stadel D, Haas TL, Genze F, Jovanovic M, et al. Targeting XIAP bypasses Bcl-2-mediated resistance to TRAIL and cooperates with TRAIL to suppress pancreatic cancer growth in vitro and in vivo. Cancer Res. 2008;68:7956–7965. doi: 10.1158/0008-5472.CAN-08-1296. [DOI] [PubMed] [Google Scholar]
- Watters GP, Smart DJ, Harvey JS, Austin CA. H2AX phosphorylation as a genotoxicity endpoint. Mutat Res. 2009;679:50–58. doi: 10.1016/j.mrgentox.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Widlak P. The DFF40/CAD endonuclease and its role in apoptosis. Acta Biochim Pol. 2000;47:1037–1044. [PubMed] [Google Scholar]
- Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem. 2005;94:1078–1087. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- Widlak P, Li P, Wang X, Garrard WT. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J Biol Chem. 2000;275:8226–8232. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- Winer E, Gralow J, Diller L, Karlan B, Loehrer P, Pierce L, et al. Clinical cancer advances 2008: major research advances in cancer treatment, prevention, and screening--a report from the American Society of Clinical Oncology. J Clin Oncol. 2009;27:812–826. doi: 10.1200/JCO.2008.21.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27 (Suppl 1:S71–S83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]