Summary
An Immersed Boundary method is developed in which the fluid's motion is calculated using the lattice Boltzmann method. The method is applied to explore the experimentally-observed lateral redistribution of platelets and platelet-sized particles in concentrated suspensions of red blood cells undergoing channel flow. Simulations capture red-blood-cell-induced lateral platelet motion and the consequent development of a platelet concentration profile that includes an enhanced concentration within a few microns of the channel walls. In the simulations, the near-wall enhanced concentration develops within approximately 400 msec starting from a random distribution of red blood cells and a uniform distribution of platelet-sized particles.
Keywords: lattice Boltzmann methods, immersed boundary method, platelet near wall excess, red blood cell membrane mechanics
1. Introduction
Under arterial and arteriolar flow conditions, platelets have an enhanced concentration near the blood vessel walls. The enhanced near-wall concentration has been seen in vivo using intravital fluorescent microscopy [1, 2], and in vitro using platelet-sized latex beads in flowing red blood cell suspensions [3, 4]. This non-uniform cell distribution depends on the fluid dynamics of blood as a heterogeneous medium. Blood is a suspension composed of plasma, red blood cells, platelets, and white blood cells. Plasma is a water-based solution of proteins and electrolytes. A red blood cell is a highly deformable, nucleus-free, fluid-filled capsule whose membrane consists of a lipid bilayer connected to a hexagonal spectrin protein mesh underneath it. Platelets are small, rigid cells that play an important role in blood clotting. They survey the status of the vascular wall lining and respond to diseased or damaged tissue by adhering to it and releasing into the plasma chemicals which can cause other platelets to become activated and capable of cohering to the already wall-adherent platelets. In order to adhere, platelets must first come into contact with the injured vessel wall. Platelets constitute only 0.3% of the blood's volume and are too large to experience significant Brownian motion. In fact, individual platelets in a simple fluid (e.g., saline solution) move essentially along flow streamlines and seldom contact the flow chamber walls. In contrast, platelets in suspension of transparent red blood cell ‘ghosts’ make substantial excursions transverse to the bulk flow [5]. Using the results of experiments of platelet adhesion to the flow chamber wall, it was inferred that, in flowing whole blood, platelets experience a complex motion that was roughly characterized as ‘enhanced diffusion’ [6]. More recently, an enhanced concentration of platelets or platelet-sized particles within a few microns of the vascular wall or flow chamber wall, respectively, were observed in whole blood flowing under arterial or arteriolar flow conditions [1, 3].
To better determine which factors affect the cross-sectional distribution of platelets in flowing blood, Eckstein et al. [3] performed a series of experiments in a 50μm × 1000μm rectangular channel. Suspensions of red cell ghosts and fluorescent platelet-size latex beads (diameter 2.38 μm) were perfused through the channel at a constant flow rate controlled by a syringe pump. The shear rates studied were in the range of 50 to 3180 sec−1 with hematocrits (volume fraction of red blood cells) ranging from 7% to 45%. Eckstein examined the concentration profiles of the platelet-size beads after the flow was fully developed and saw that at higher hematocrits, the near-wall excess of beads was greater. A near-wall excess of beads was seen for shear rates exceeding 430 sec−1. The highest concentration of beads occurred at about 3 μm from the channel wall, and the enhanced concentration region was about 5-8 μm wide. A schematic of these results is displayed in Figure 1. Eckstein fit his experimental results with a phenomenological ‘drift-diffusion’ model involving net drift of beads toward the flow chamber walls that balances the enhanced diffusion of beads to produce the observed concentration profiles [7, 8, 9].
Figure 1.
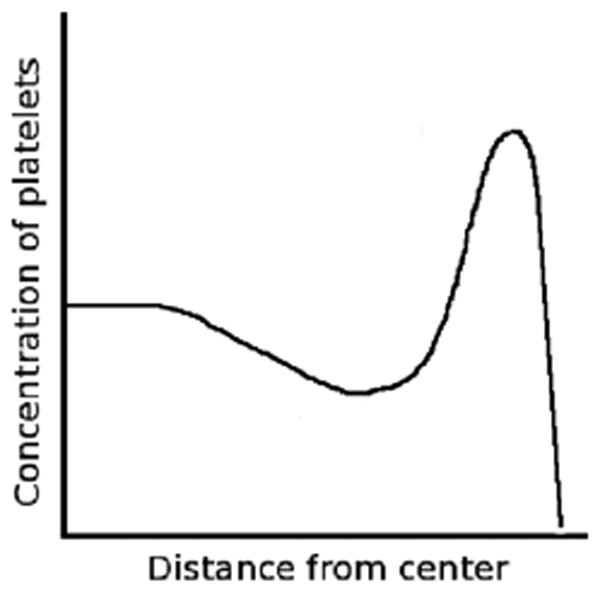
In flowing blood, platelets are highly concentrated in a narrow region near the channel wall.
Although lateral platelet motion has been well documented, the fluid dynamics involved are poorly understood. The deformability of red blood cells, the hematocrit, and the wall shear rate all affect this motion [10, 4, 3], but how they do so is unclear. A physically-based mechanism that quantitatively accounts for the observed platelet motion and cross-lumen distribution in rapidly flowing red blood cell suspensions is not available. It is not possible at present to accurately predict the extent of platelet concentration enhancement near vessel walls under a given set of conditions. A computational model that tracks individual red blood cells and platelets and that realistically accounts for the mechanics of the red blood cells and platelets and their interactions with the suspending fluid should be a useful tool in trying to understand the physics behind the platelet lateral motion and in making quantitative predictions about it under specified conditions. Such a tool would also be useful in predicting how the growth of a wall-bound platelet aggregate in an injured vessel would perturb the distribution of platelets, which, in turn, might significantly influence the further growth of the aggregate into the vessel lumen. These insights could lead to significant improvements in cell-based platelet deposition models [11, 12, 13].
In this paper, we present a computational model that allows simulation of the flow of concentrated suspensions of red blood cells in which platelets are initially uniformly dispersed. The model is described and has been implemented in two spatial dimensions, but can be extended to three dimensions straightforwardly except for the issue of computational cost. Our model is formulated in terms of the Immersed Boundary (IB) method which is a modeling and computational framework based on the coupled equations of motion of a viscous fluid and one or more deformable elastic objects suspended in or in contact with the fluid. The IB method views the elastic material and the fluid as a composite; it uses an Eulerian description of the fluid, while using a Lagrangian description of the elastic objects to enable calculation of forces generated by deformation of those objects. The elastic forces are determined by the instantaneous positions of Lagrangian points in the elastic material and by the configuration of elastic connections among these points. The integral of the fluid force density over any region of space is identified with the sum of the elastic forces for the Lagrangian points within that region. Through its presence in the fluid dynamics equations, the fluid force density helps determine the fluid velocity, and the Lagrangian points then move in the resulting velocity field. In traditional IB-based models, the fluid's dynamics are described by the Navier-Stokes equations.
For actual calculations, the fluid dynamics equations are discretized on a fixed and uniform Cartesian grid over the entire domain. Each elastic object is represented by a set of discrete Lagrangian IB points which are not constrained to lie on the fluid grid. To advance the system by one time step (1) the elastic force is calculated at each IB point, (2) each of these forces is transmitted from the corresponding IB point to nearby grid points by means of a discrete approximate delta-function, (3) the fluid dynamics equations are solved on the grid, and (4) the discrete delta-function is used to interpolate the fluid velocity from the grid to the IB points in order to update their positions. The IB method greatly facilitates solving the equations which govern the interactions of a viscous fluid with multiple immersed elastic objects, even when those objects are of complex shape, move substantially, or grow. In all cases, the routines which solve the fluid dynamics equations ‘see’ only a force density function. Therefore, simple and fast fluid solvers can be used. In our context this means that there is no need to re-grid even as the red blood cells move and deform substantially. Another feature of the IB is that there is no requirement for an inter-particle collision detection mechanism. Particle interactions are mediated completely through the fluid and the method guarantees that fluid does not cross the boundaries of the immersed objects and that immersed objects cannot overlap. These statements hold for a sufficiently small time step and a spacing between IB points of less than half the lattice spacing.
In the computational model we present, the description of the fluid dynamics uses the lattice Boltzmann method rather than the discretized Navier Stokes equations. The lattice Boltzmann method relies entirely on localized interactions, while the Navier Stokes equations involve the solution of linear systems that couple all points of the grid. The structure of the lattice Boltzmann method therefore facilitates parallelizing the calculations. We will present a parallelized lattice Boltzmann IB method for treating concentrated suspensions of cells in a subsequent paper. We note that our blending of the lattice Boltzmann method with the IB method is not unique; several such efforts have been described in recent papers [14, 15, 16].
The structure of the remainder of the paper is as follows: In Section 2, we describe the traditional lattice Boltzmann method and IB method as well as the combined lattice Boltzmann-IB method. In Section 3, we describe the models of red blood cell and platelet mechanics that we use in our simulations. In Section 4, we present two types of numerical simulations. First, we demonstrate that our model produces results in agreement with other simulations and experiments for the behavior of single red blood cells in shear flow and suspensions of red blood cells in Poiseuille flow. Then, we present simulations that show that our model is able to capture the development of a near-wall enhancement of platelet concentration for an arteriolar sized vessel, at a particular arteriolar shear rate, and two different hematocrits. These preliminary results set the stage for a systematic computational exploration of the near-wall enhancement that will be reported in a later paper.
2. Numerical Method
2.1. Lattice Boltzmann Method
The incompressible Navier Stokes equations are conventionally used to define the motion of a fluid in terms of its velocity and pressure. These equations conserve mass and describe the change in momentum due to inertial, viscous and external forces. However, since the incompressible Navier Stokes equations enforce incompressibility using a Poisson equation, the velocity and pressure of the entire domain must be solved for simultaneously. For this reason, the Navier Stokes equations do not lend themselves well to parallel implementation and using them to determine the flow dynamics for complex fluid problems can be time consuming. An alternative approach to computational fluid dynamics is the lattice Boltzmann method, which describes the evolution of fictitious microscopic particles living on a discrete lattice and whose dynamics depend only on interactions between particle populations on neighboring lattice points. Hence all lattice Boltzmann calculations are local ones. At the macroscopic fluid scale, viscous flow dynamics emerge from these particle dynamics as is described below.
The lattice Boltzmann method (LBM) was originally developed during the late 1980's [17]. It removed the stochastic fluctuations from earlier particle-based lattice gas automata methods [18]; instead of tracking discrete particles, it tracks particle distribution functions at points on a lattice. The LBM can also be thought of as a finite-difference approximation to a particular version (the BGK version) of the continuous Boltzmann equation from statistical mechanics [19, 20]. The lattice Boltzmann method is a deterministic method that gives the correct macroscopic behavior of a Newtonian fluid in an appropriate limit. Letting the Mach number approach zero and the hydrodynamic length scale (the ratio of the macroscopic fluid length scale to the lattice spacing) approach infinity, the incompressible Navier Stokes equations can be derived from the lattice Boltzmann equations using a multi-scale expansion known as the Chapman Enksog procedure [21].
There are many variants of the LBM. Here we describe the D2Q9 (2-D 9-velocity) version that we use in our two-dimensional simulations. Consider a lattice placed over the rectangular two-dimensional domain and consisting of square cells of size h. The variable of interest in the LBM is f(x, ei, t) which is the particle density function for the lattice point x, discrete velocity vector ei, and time t. Each particle described by the density function is assumed to carry unit mass. The governing equations for the LBM, in the absence of external forces, are
| (1) |
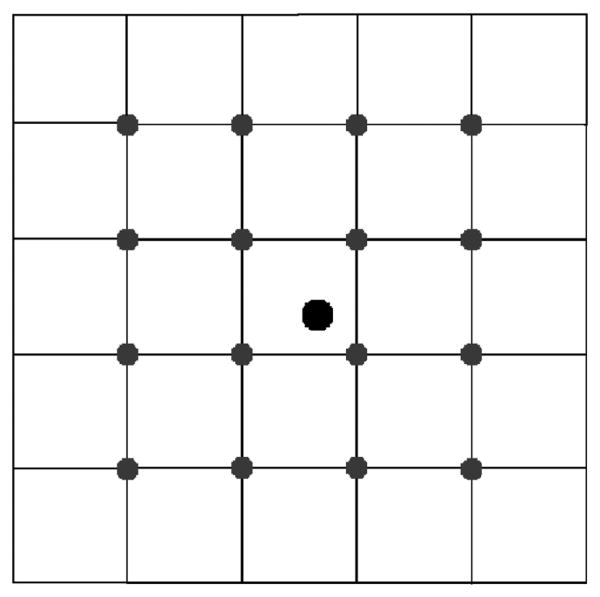
where we use the short-hand notation fi(x, t) = f(x, ei, t) to denote only the density of particles moving with particle velocity ei. The time step of the method δt, the relaxation time τ, the equilibrium distribution , and the macroscopic variables ρ and u are described below. For the D2Q9 method, nine discrete velocities are considered, and i ranges over the integers 0, …, 8. The nine velocity vectors are
and are shown in Figure 2. In these formulas c = h/δt is the particle speed for particles moving horizontally or vertically. Particles moving obliquely do so at speed . Equation 1 describes the two-step update procedure of the LBM. First, in the collision step, each particle distribution function relaxes toward a local equilibrium distribution, at rate 1/τ. Next, in the streaming step, the new distribution fi(x, t) is advected from x at the corresponding velocity ei to the lattice point located at x + δtei. The equilibrium distributions are defined by
Figure 2.
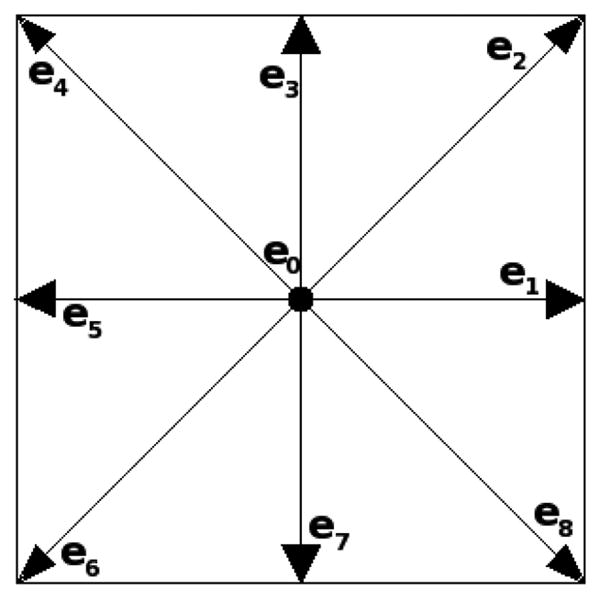
The discrete velocity vectors for the 2D9Q model.
| (2) |
where the weights, wi, are
The equilibrium state at lattice point x depends on the macroscopic quantities ρ(x, t) and u(x, t) which are moments of the nine particle distribution functions at x. These quantities turn out to be approximations to the macroscopic fluid density and velocity in the Navier Stokes equations. The equilibrium function defined above is a discrete approximation to the Maxwellian distribution function used in the BGK Boltzmann equation [22]. The form of the equilibrium function is chosen to ensure that the LBM conserves mass and momentum.
The equilibrium distributions given in Equation 2 have the properties that and . These properties guarantee that the collision step of the LBM update conserves mass and momentum. Since the advection step does also, the overall LBM is conservative. In terms of the particle density functions fi(x, t), we define the macroscopic density ρ(x, t), velocity u(x, t), and inviscid momentum tensor [18, 19] by the formulas
| (3) |
| (4) |
| (5) |
respectively. Consistent with these definitions, the pressure is defined by the adiabatic equation of state [18]
| (6) |
Hence can be interpreted as the speed of sound propagation in the fluid. Finally, the macroscopic fluid viscosity is given in terms of the relaxation time τ and other parameters by the formula
We note that the LBM does not treat the fluid as incompressible. However, provided we ensure that the ratio of the macroscopic velocity u(x, t) to the speed of sound cs is sufficiently small, the fluid behaves as nearly incompressible because small variations in density induce large gradients in the pressure defined above. The LBM is based on a very simplified view of microscopic mechanics, but still captures the macroscopic hydrodynamic behavior of the nearly-incompressible Newtonian fluid we wish to simulate.
2.2. Immersed Boundary Method
To simulate the behavior of platelets and red blood cells in blood, we utilize a numerical scheme known as the Immersed Boundary method. This method was originally created by Peskin in 1972 to analyze the fluid dynamics of heart valves [23]. Since then it has been applied to many other fluid-structure interaction problems, including platelet aggregation [24], deformation of red blood cells [14, 15], motion of cilia and swimming organisms [25].
The Immersed Boundary (IB) method is a specific way of coupling fluid dynamics to the mechanics of one or more objects located within the fluid. In our case the elastic object is the membrane of a red blood cell or platelet. We treat the membrane of each cell as a thin, massless, elastic material and assume that the interior cytoplasm and surrounding plasma behave like incompressible Newtonian fluids.
In the traditional IB method, the incompressible Navier Stokes equations
are assumed to govern the behavior of the fluid. The variables u(x, t) and p(x, t) represent the fluid velocity and pressure, respectively. The fluid is assumed to have constant density ρ and viscosity μ. Deformation of the cells results in elastic cellular forces that are transmitted to the adjacent fluid and contribute to the force density term Ff(x, t) in the momentum equation. Note that the Navier Stokes equations provide an Eulerian description of how the flow evolves at each spatial point x.
Each elastic IB object is represented by a Lagrangian variable X(q, t) where the parameter q labels a material point on the object. We let q be arclength in a reference configuration. At any fixed time t, the set of points X(q, t) defines the current configuration of that immersed boundary. From this configuration and specified elastic properties of the object, a distribution of forces along the immersed boundary is determined. For example, if it were specified that the membrane only resisted forces due to stretching or compression, then the elastic force (per unit q) at point X(q, t) is
| (7) |
where S is the membrane stiffness, T is the tension, and is the unit tangent vector along the membrane. Note that this force ‘lives’ at the Lagrangian IB points. Because the membrane is treated as massless, this is also the force that is applied to the fluid by the membrane at location X(q, t). The mathematical expression of the transmission of force from the elastic membrane to the fluid is given by the equation
| (8) |
where δ(x) is the Dirac delta function. The function Ff(x, t) is the force density applied to the fluid by the elastic membrane, and is concentrated spatially along the membrane. When there is more than one elastic object, the pth object contributes a term of the form to the fluid force density. Here, Xp(q, t) and are the coordinates and IB force density for the pth object. The total fluid force density is obtained by summing their contributions. It is an important consequence of the above definition of Ff(x, t) that for any region of the domain, the integral of Ff(x, t) over that domain equals the integral of FIB(q, t) over the parts of the elastic membranes that are contained in that region.
The final equation in the IB method comes from the no-slip condition applied at each point of an elastic membrane. The requirement that each such point moves at the same velocity as the immediately adjacent fluid can be expressed as
| (9) |
To summarize, the traditional IB method is based on four sets of equations: the Navier Stokes equations for the fluid motion, a constitutive law that defines the elastic forces on each IB object as a function of its current configuration, and the two equations that ‘communicate’ information, forces and velocities, between the Eulerian fluid description and the Lagrangian descriptions of the IB objects.
For actual computations, the IB equations are discretized. The fluid equations are solved on a Cartesian spatial grid and each IB object is represented by a discrete grid of Lagrangian IB points. Discrete versions of the constitutive laws are specified to determine the elastic IB forces. The IB points are not constrained to be at the grid points of the fluid mesh, and communication between the IB points and the fluid mesh is carried out with discrete versions of Equations 8 and 9. In these equations, the integrals are replaced by sums and the Dirac delta function is replaced by a smooth approximate delta function δh(x) that has finite support. Criteria for defining a good approximate delta function for the IB problem are discussed in Peskin [26]. A common choice for the discrete delta function, and the one we use, is
| (10) |
Note that with this choice, each IB point transmits force to and interpolates velocity from sixteen nearby Eulerian grid points (see Figure 4).
Figure 4.
An immersed boundary point spreads forces to and interpolates velocities from sixteen nearby Eulerian grid points.
The cost of an IB calculation depends on the size of the Eulerian fluid grid, the number of IB objects, and the number of discrete IB points used the represent each of them. An attraction of replacing the traditional Navier Stokes solver with a lattice Boltzmann fluid solver is that, because the latter involves only local interactions, it facilitates parallel implementation and potentially substantially reduces the time required to do IB simulations with large numbers of cells.
2.3. Lattice Boltzmann-Immersed Boundary Method
A lattice Boltzmann-Immersed Boundary method is created by replacing the discretized Navier Stokes equations with the lattice Boltzmann equations to determine the fluid motion. This change results in purely localized evolution equations for the macroscopic fluid variables, which in turn facilitates parallelization. The governing equations for the elastic membrane forces and the communications between the fluid grid and the IB objects are the same in the lattice Boltzmann-IB method as in the traditional IB method.
The lattice Boltzmann equations (given in Equation 1) are for the situation in which no external force is applied to the fluid. In the IB context, a spatially and temporally varying force, calculated from the IB objects, acts on the fluid and Equation 1 must be modified to account for this. Using a Chapman Enskog expansion, Guo et al. [27] showed that the following lattice Boltzmann equations give a second-order-accurate approximation to v, the Navier Stokes velocity in the presence of a spatially varying, time dependent force:
| (11) |
Here, v is defined as and the weights wi are defined in Equation 2. The momentum at equilibrium ρv now includes an additional term due to the external force applied to the fluid, and is an approximation to the momentum of the fluid at time t + δt/2. Equation 11 also includes two additive terms. The addition of the first term (proportional to Ff · ei) results in the presence of an external force term (Ff) in the Navier Stokes approximation. The second, higher order term removes higher order spurious terms in the Navier Stokes approximation that result from the presence of the first additive term in the lattice Boltzmann equations. Because v is the approximation to the Navier Stokes velocity, v (not u) is used to update the IB point location in the discrete version of Equation 9.
2.4. Boundary Conditions
In our numerical simulations, we impose no-slip boundary conditions on the tube walls in our lattice Boltzmann-IB simulations using the modified bounce-back scheme. In the modified bounce-back scheme, the wall is located at a distance h/2 from lattice nodes adjacent to it. For a particle density function that would be carried from such a node into the wall during the advection step, the distribution instead remains at that node but reverses direction as if the particles had “bounced back” from the wall. He et al. [28] showed that the modified bounce-scheme is second-order accurate for both Poiseuille and Couette flow. Including solid walls into our numerical scheme raises additional issues.
The discrete delta-function used in the IB method is nonzero in a 4h × 4h region around each IB point. This region overlaps the physical boundary of the fluid domain when an IB point is within distance 2h of the solid wall. To handle this situation, we extend the fluid lattice with two rows of ‘ghost’ lattice points outside each solid boundary. For an IB point within distance 2h of the boundary the forces are spread in the usual way to this extended lattice, and only the forces within the physical domain actually influence the dynamics. Before determining the velocities of the IB points, the lattice velocity is extended to the ghost points from the first two rows of lattice points inside the corresponding wall. A velocity is defined at each ghost point by using the velocity at its image point on the other side of the wall, but with change of sign. This ensures that the velocity as interpolated to an IB point tends to the solid wall's velocity if that IB point approaches the wall. Most IB applications which use the discrete delta-function embed the physical domain in a larger periodic fluid box and construct ‘physical’ boundaries inside that box using arrays of IB points tethered to fixed spatial points along the desired boundary location. Hence, the issue of overlap of the delta function and a solid wall does not arise. Research by one of the authors and his coworkers into the best way to deal with this issue is under way. Preliminary results suggest that the method used in this paper is reasonable in that it gives near-wall IB particle motions very much like those obtained when solid walls are simulated by arrays of tethered IB points.
3. Membrane Mechanics
3.1. Red Blood Cells
The red blood cell membrane is only a few molecules thick, and hence it is typically treated as a two-dimensional sheet with some inherent, constant thickness. The elastic properties of a cellular membrane can be described in terms of three independent elastic moduli: shear modulus G, area expansion modulus K, and bending modulus EB. Typical values of these moduli for red blood cell membranes are given in Table I.
Table I. Red blood cell parameters.
When exposed to blood flow, the shape of a red blood cell may change dramatically, although both its volume and surface area remain fairly constant due to its lipid-bilayer membrane structure [31, 32]. The Skalak tension law [32, 33] was developed specifically to describe the stress-strain relationship of red blood cell membranes, and the parameters in Table I are based on this model. The Skalak tension law for a two-dimensional membrane is
where the tension vector lies in the plane tangent to the membrane and represents the principle stretch ratio for i = 1, 2. The term ds represents an infintesimal length element after some deformation and dS is the length element prior to the deformation. For the two-dimensional computational model developed in this paper, a reduction is made in the dimension of the red blood cell membrane. Only in-plane membrane tension need be considered because we assume that the membrane tension is isotropic (T1 = T2 or λ1 = λ2). The corresponding one-dimensional Skalak tension is
| (12) |
In the absence of external forces, a red blood cell has a biconcave discoidal shape. This shape is due to the hexagonal spectrin protein mesh that lies directly beneath the bilipid membrane. To incorporate a preferred red blood cell resting shape into our model, we define a bending energy . The force associated with minimizing this energy acts in the normal direction to restore the membrane to its preferred shape. This bending force is bn, where b, the transverse shear tension [34], is
| (13) |
Here EB is the bending modulus, κ is the instantaneous curvature, and κ0 is the curvature of minimum bending energy. In our simulations, the curvature of minimum bending energy is set to that of a biconcave disk with resting shape parameterized by the curve [15],
| (14) |
| (15) |
where the scaling parameter a is set so that the red blood cell has a diameter of 8 μm. Putting together the Skalak tension law and this bending force expression, we see that for a red blood cell
| (16) |
where t is tangent, n is normal to the membrane, and q is arclength in the reference configuration.
3.2. Platelets
Unactivated platelets in vivo are ellipsoids that measure 1.5-3.0 μm in diameter and it is straightforward with the IB method to construct approximately rigid elliptical model platelets. For the current studies, we compare our simulation results to Eckstein's spherical latex bead experiments, and so we model each ‘platelet’ as an approximately rigid circular object. We choose a one-dimensional uniaxial reduction of the two-dimensional linear Hookean law for platelet membrane tension
and set the membrane stiffness S such that a platelet's membrane length remains fairly constant over the range shear rates it will experience in our numerical simulations (100-1200 sec−1). We also include bending resistance in the platelet membrane force equation so that the rest shape is a circle with constant curvature, and choose a bending modulus, EB that is large enough to prevent more than 1% deformation away from a circle during our simulations. The equation governing the force on a platelet membrane is identical to Equation 16, but with Tsk replaced by TH and parameters given in Table II.
Table II. Platelet parameters.
| S = 1 × 10−1 dynes/cm |
| EB = 2 × 10−11 dynes-cm |
4. Numerical Simulations
4.1. Model Validation
A red blood cell rotates and tumbles like a rigid particle under low shear conditions but when exposed to high levels of shear stress, it orients itself at some constant angle between 5° and 25° relative to the direction of flow [31]. In addition, under high shear, the membrane rotates around the fluid inside the cell, a process known as tank treading [35]. Sui et al. [14] simulated a single red blood cell in a linear shear flow using a two-dimensional lattice Boltzmann method with a refined mesh near the moving cell boundary. The boundary was coupled to the fluid by the immersed boundary method. The tension in the red blood cell membrane was assumed to be governed by the Skalak formula given in Equation 12, but no resistance to membrane bending was included. The red blood cell shape was initialized to be a biconcave disk. This study found that red blood cells equilibrate to an elongated ellipse with a major to minor axis ratio of about 5:1 and an inclination angle of 15° in simple shear flow. These results are quite similar to those from a simulation (shown in Figure 5) with our method at a constant shear rate of 1200 sec−1.
Figure 5.
Simple shear experiment. A simulated red blood cell exhibits tank treading and obtains an equilibrium shape with a major to minor axis ratio of 5.3:1 and a inclination angle of 17° The x and y axes shown are in microns. The lattice spacing is such that in its undeformed state, the red blood cell is approximately 40 lattice points along its major axis.
Bagchi [36] used a traditional two-dimensional IB method that also included a non-uniform viscosity to account for the correct viscosity ratio (≈ 5) between red blood cell cytoplasm and plasma. The viscosity was updated using a front-tracking method. He assumed that red blood cell mechanics were governed by a neo-Hookean law for stretch and by Equation 13 above for bending. Bagchi's simulations showed the presence of a tank-treading-to-tumbling bifurcation of a single red blood cell in Poiseuille flow as the deformability of the cell was decreased. In a later paper that included bending tension, Sui et al. saw a transition between tumbling and tank treading at a bending modulus of EB = 3 × 10−11dyne-cm [15] in simple shear flow. We also observe this qualitative change in behavior by varying either the bending stiffness EB or the shear rate in our simple shear numerical experiments.
In single cell experiments performed in a cylindrical tube with constant flow rate (at a hematocrit < 2%), red blood cells drift towards the center of the tube [31]. This behavior has also been observed numerically. Bagchi [36] demonstrated lateral motion of red blood cells in dilute suspensions and observed that red blood cells drift towards the center of a two-dimensional channel. Even at high concentrations, red blood cells move toward the center of a blood vessel, an effect attributed to their deformability [5]. Additional experimental studies by Aarts et al. show that red blood cells in whole blood migrate towards the center axis of a vessel and leave a peripheral plasma layer near the walls. The size of this cell free layer depends on hematocrit and flow rate, but in general varies in thickness on the order of about 10 μm for a tube with a diameter of 300 μm [10]. In cylindrical tubes with diameters between 40 and 83 μm, the cell free layer can be estimated to be roughly 100/Hct [37]. This means that at 20% hematocrit the cell free layer should be about 5 μm wide (and 2.5 μm for the 40% hematocrit case). In Figure 6 we simulate a 50 μm wide channel with hematocrits of 20% (a) and 40% (b). The pressure gradient is set so that the wall shear rate in both simulations is 1100 sec−1 once the flow is developed. We see the appearance of a cell free layer of thickness 5-6 μm for a hematocrit of 20% and 2-3 μm for a hematocrit of 40%, in agreement with experiments. Bagchi [36] reports a 4 μm cell free layer under comparable conditions for a 80 μm wide channel. He seems to be using a definition based on the location of the red blood cells' centers of mass because his simulation plot (Bagchi [36], Figure 5a) does not show an obvious cell free layer as in our Figure 6.
Figure 6.
High concentrations of red blood cells in channel flow after 500 milliseconds of simulated time. Note the cell free layer (2-6 μm) near channel walls. Lengths are in microns.
4.2. Whole Blood Simulations
Numerical experiments presented in this section were performed on a rectangular domain representing a 50 micron wide channel. The left and right boundary conditions were periodic with no-slip bounce-back boundary conditions at the top and bottom to represent blood vessel walls. The flow was driven by a constant pressure gradient in the x direction, which was included in the external force along with the immersed boundary force. Lattice Boltzmann particle density functions were initially set to their equilibrium values for a given initial parabolic fluid velocity whose magnitude was determined from the pressure gradient. Membrane parameters for each red blood cell and platelet are given in Table I and Table II, respectively.
The grid resolution of the following simulations is 350 × 700, comparable to the resolution chosen by Bagchi for his simulations [36]. Parallelization with OpenMP was used to improve the performance of our method since the time step is restricted to be 2 × 10−8 seconds. To represent a nearly-incompressible fluid, the ratio u/cs must be small enough that the density ρ does not vary more than approximately 0.1% in space and time. Since and we have already chosen a spatial discretization μm, to increase the speed of sound cs sufficiently, we must choose a small time step δt.
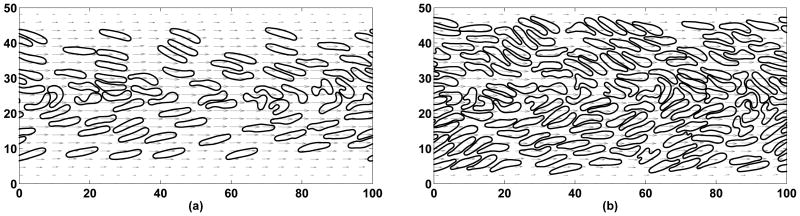
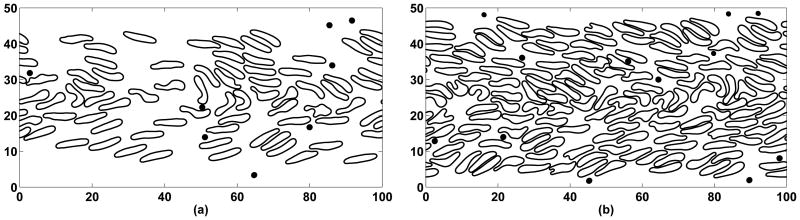
We include details of numerical experiments performed at hematocrits of 20% and 40%. In both cases, red blood cells were initially placed in a structured way with some amount of randomness in both position and rotation such that no overlapping occurs. Platelets were then distributed in the domain so that their initial lateral distribution was roughly uniform and there was no intersection with red blood cells (see Figure 7). The pressure gradient was set so that once the flow has developed, the wall shear rate would be approximately 1100 sec−1 for both cases. Shear rate in our simulations was found by computing a time average of the gradient of the computed velocity at the wall.
Figure 7.
Initial positions of red blood cells and platelets at 20% hematocrit (a) and at 40% hematocrit (b). Lengths are in microns.
Within the first 400 milliseconds of simulated time, the red blood cells drift away from the walls and the cell free layer develops in both numerical simulations. In Figure 8 we also see what could be a near-wall excess of platelets. However, in order to show that we do indeed see this phenomenon we need to consider a much larger data set. By sampling from Eckstein's empirical bead distribution function [7] and forming a histogram of the sample results, we infer that 50-60 samples are needed to see clear evidence of a near-wall enhanced concentration, and that additional samples are required to adequately characterize the height and width of the region of enhanced concentration.
Figure 8.
After 400 milliseconds cells have laterally redistributed: 20% hematocrit (a) and 40% hematocrit (b). A cell free layer has developed in both cases and platelets have become more concentrated near the channel walls. Lengths are in microns.
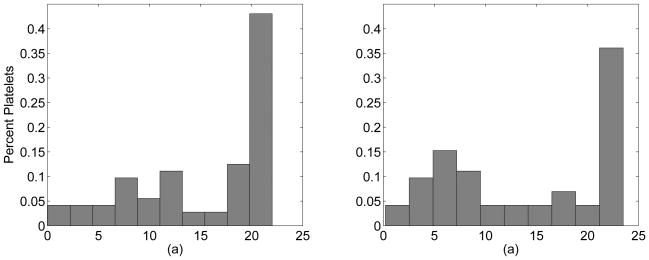
Figure 9 shows histograms of 72 platelets for the 20% case and 72 platelets for the 40% case after 400 msec of simulated time. This data was obtained from a series of similar experiments with different random initial placements of the cells. Although it is not clear that the profile is in equilibrium yet, Figure 9 illustrates that a non-uniform distribution of platelets has developed in a short amount of time. The data here is still quite noisy and more data is necessary to make firm conclusions about the actual shape of this non-uniform distribution. Even so, this near-wall excess is very similar to what has been seen experimentally [3]. The heights of peaks in both cases are about three to four times larger than the platelet concentration at the center of the channel. It is also apparent that peaks occur at different distances from the wall depending on hematocrit; this is most likely due to the size of the cell free layer.
Figure 9.
Histograms of data from 72 platelets at 20% hematocrit (a) and 72 platelets at 40% hematocrit (b) after 400 msec of simulated time. The x-axis is distance from the channel center in microns. The wall lies 25 microns away from the center.
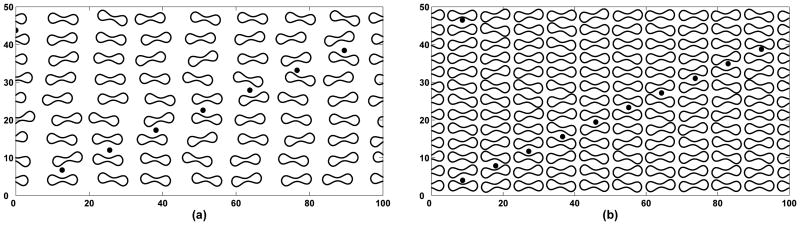
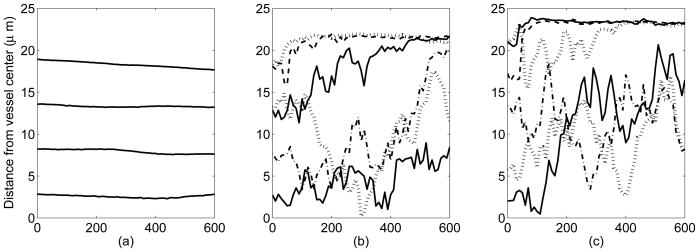
Figure 10 shows lateral positions of selected platelets over the course of 600 milliseconds. In the case where there are no red blood cells in the flow affecting the motion of platelets (a) there is only a negligible amount of lateral motion. In fact, we see platelets move slightly away from the walls when they are a few microns from the wall, a region in which they experience high levels of shearing. Unless platelets interact with each other, they have only a small affect on the fluid, and appear to simply advect with the flow. Contrast this to the 20% and 40% hematocrit cases shown in (Figure 10(b-c)); platelets are either pushed toward the walls into the cell free layer or move much more erratically in the center of the vessel due to interactions with the deformable red blood cells. The platelets' tranverse excursions seem to be larger for higher volume fraction (40%) suspensions than for lower ones (20%), but this conclusion is preliminary and requires more extensive study.
Figure 10.
Lateral platelet motion in: (a) the absence of red blood cells, (b) presence of red blood cells at 20% hematocrit, and (c) presence of red blood cells at 40% hematocrit. Time (horizontal axis) is measured in milliseconds.
5. Discussion
In this paper, we describe a hybrid lattice Boltzmann-Immersed Boundary computational model for studying the behavior of concentrated suspensions of red blood cells and other particles. The Immersed Boundary method is well suited to this problem which involves interactions, through the suspending fluid, among highly deformable red blood cells that move and change shape substantially during a simulation. In the IB method, the fluid equations are solved on a simple Cartesian grid, while each IB object, such as a red blood cell, is represented by a grid of Lagrangian points. Elastic forces generated within the cell because of its deformation can be calculated solely in terms of the current configuration of the Lagrangian points that make up the cell. The elastic forces are then communicated from the Lagrangian IB points to the Cartesian fluid grid, using a discrete approximation to the Dirac delta function, to define the fluid force density, and it is only through the influence of this force density (rather than an explicit change in geometry of the fluid domain) that the fluid ‘feels’ the presence of the suspended cells. The same discrete delta function is used to define the velocity at each IB point so that the position of that point can be updated consistent with there being no-slip between points of the cell boundary and the adjacent viscous fluid.
In the computational model we describe, the motion of the fluid is determined using the lattice Boltzmann method. This method involves only local interactions among lattice points and therefore offers the potential through parallel computation of substantial reduction in the time required to carry out extensive simulations. The IB method facilitates the use of realistic models of cellular mechanics, such as the Skalak model for red blood cell membrane tension. It also has the advantage, compared to many other particle tracking methods, of not requiring special procedures for detecting and handling particle-particle collisions and for preventing particles from overlapping with one another. In the IB formulation, particles cannot interpenetrate provided the spacing of the Lagrangian IB points is sufficiently fine (less than about h/2, where h is the fluid lattice spacing) and the time step is sufficiently small.
In simulations of single red blood cells in a shear flow we see the expected behaviors: tumbling at low shear rates or if the red cell membrane bending rigidity is high, orientation at a fixed angle to the flow accompanied by membrane tank treading for high shear rates or lower bending rigidity. For suspensions of red blood cells flowing through a tube, we see the development of a near-wall cell free layer. The thickness of the layer depends on the volume fraction of red blood cells in the suspension in a way that accords with experimental observations reported in Blackshear et al. [37]. In these simulations, the red blood cells that are in the higher shear regions near the walls orient at an angle to the bulk flow and their membranes undergo tank treading.
To explore the influence of red blood cells on the motion and distribution of platelet-sized beads, we added a number of small circular particles to the suspensions of red blood cells. For each of the hematocrits 20% and 40%, we performed a series of computational experiments, that differed only in the initial random placement of the red blood cells, in order to get sufficient data from which to begin to draw statistically meaningful conclusions. Following the motion of a total of 48 beads (6 simulations with 8 beads/simulation) at 20% hematocrit, and 72 beads (6 simulations with 12 beads/simulation) at 40% hematocrit, we see clearly the development of a near-wall enhancement of bead concentration at both hematocrits within the first 400 msec of suspension flow at a shear rate of about 1100 sec−1. The general features of the bead concentration profile are similar to those seen by Eckstein et al. [3] and we have the new information that this profile can develop quickly. We do not yet know whether the profiles we see would develop further in longer simulations. It remains to do more and longer simulations for a range of shear rates, hematocrits, and bead sizes to see how well the simulation results match experimental observations.
Our study shares features with the work of Sui et al. [14, 15], Bagchi [36], and AlMomani et al. [38]. Sui and co-workers used a lattice Boltzmann Immersed Boundary method much like that we present, but with local refinement of the lattice around the cell, to look at the behavior of single red blood cells in different flow situations. For the dense suspension of cells we consider, a uniformly fine grid is required. Bagchi used a traditional Navier-Stokes-based Immersed Boundary method to look at the rheological properties of concentrated suspensions of red blood cells, but did not explore the influence of red blood cell motion on the transluminal distribution of platelets. AlMomani et al. used a different fluid-particle interaction method to look at the effect of red blood cells on platelet motion and on the forces experienced by the platelets. Their approach requires detection of particle proximity to neighboring particles and the use of repulsive forces obtained from a ‘soft’ potential. Their model of red blood cells constrains the cell's shape to be elliptical. Their level-set representation of cell boundaries seems to preclude tracking tangential motions of cell membranes making it impossible to simulate tank-treading. They look at several low hematocrit cases and present evidence of platelet displacement toward the walls. The paper gives no information about the number of platelets considered and so allows no assessment of the statistical power of the results.
Our Immersed Boundary-based whole blood simulation model allows us to simulate the complex fluid-structure interactions that take place in flowing blood. By allowing observation and tracking of each red blood cell and platelet (or platelet-sized bead) during their interactions, our simulations offer access to a wealth of information unavailable from experiments. The ability to track individual cells and to perturb physical properties on a cell-by-cell basis, if desired, makes our computational model a unique and powerful tool for elucidating the physical mechanisms that underly the lateral redistribution of platelets. That said, these simulations are computationally intensive and currently take a long time even in two spatial dimensions. To overcome this limitation, we are developing an MPI-based distributed-memory parallel algorithm for carrying out the simulations. Because the lattice Boltzmann method uses only local information, that portion of the method is straightforward to parallelize. We have also devised and are testing parallelization strategies for the Immersed Boundary parts of the calculations: the IB force calculation, the transmission of the IB forces to the Eulerian fluid grid, and the interpolation of fluid velocities to the IB points in order to update their positions. When completed, we expect that the new parallel version of our method will give us the ability to carry out a much more extensive and systematic exploration of the conditions under which platelets undergo the substantial lateral motion that leads to the development of a near-wall enhanced platelet concentration. Once we have confidence that the simulations produce results in agreement with experiment, we will use the unique capabilities of our method to begin to dissect the physical mechanisms that underly these phenomena.
Figure 3.
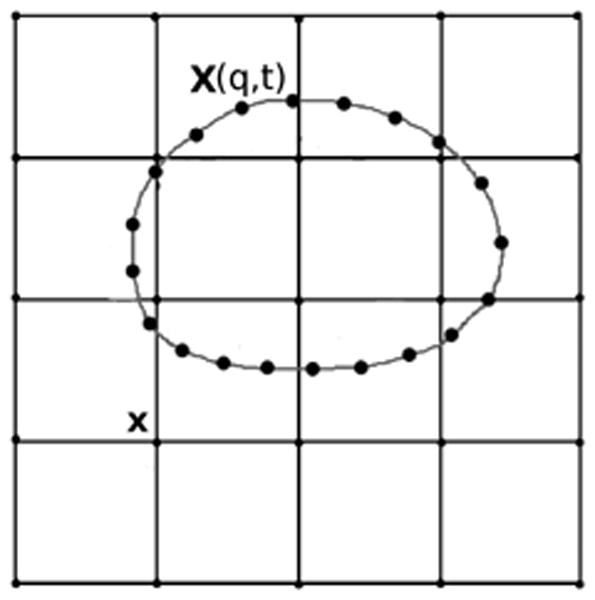
An immersed boundary object: The Lagrangian points X(q, t) located on the immersed boundary are allowed to move anywhere in the fluid domain; they are not constrained to points of the Eulerian grid.
Acknowledgments
This work was supported in part by NSF grants DMS-0354259 and DMS-0540779. The authors are grateful to J.P. Keener, R.M. Kirby, R.D. Guy, K.M. Leiderman, E.C. Eckstein, and P. Bagchi for helpful discussions.
References
- 1.Tangelder GJ, Teirlinck HC, Slaaf DW, Reneman RS. Distribution of blood platelets flowing in arterioles. American Journal of Physiology - Heart and Circulatory Physiology. 1985;248(3):H318–H323. doi: 10.1152/ajpheart.1985.248.3.H318. [DOI] [PubMed] [Google Scholar]
- 2.Woldhuis B, Tangelder GJ, Slaaf DW, Reneman RS. Concentration profile of blood platelets differs in arterioles and venules. American Journal of Physiology - Heart and Circulatory Physiology. 1992;262(4):H1217–H1223. doi: 10.1152/ajpheart.1992.262.4.H1217. [DOI] [PubMed] [Google Scholar]
- 3.Tilles AW, Eckstein EC. The near-wall excess of platelet-Sized particles in blood flow: Its dependence on hematocrit and wall shear rate. Microvascular Research. 1987;33:211–223. doi: 10.1016/0026-2862(87)90018-5. [DOI] [PubMed] [Google Scholar]
- 4.Eckstein EC, Tilles AW, Millero FJ., III Conditions for the occurrence of large near-wall excesses of small particles during blood flow. Microvascular Research. 1988;36:31–39. doi: 10.1016/0026-2862(88)90036-2. [DOI] [PubMed] [Google Scholar]
- 5.Goldsmith H. Red cell motions and wall interactions in tube flow. Federation Proceedings. 1971;30(5):1578–1588. [PubMed] [Google Scholar]
- 6.Turitto V, Benis A, Leonard E. Platelet diffusion in flowing blood. Industrial and Engineering Chemistry Fundamentals. 1972;11(2):216–223. [Google Scholar]
- 7.Eckstein EC, Belgacem F. Model of platelet transport in flowing blood with drift and diffusion terms. Biophysical Journal. 1991;60:53–69. doi: 10.1016/S0006-3495(91)82030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh C, Calvez AC, Eckstein EC. An estimated shape function for drift in a platelet-transport model. Biophysical Journal. 1994;67:1706–1716. doi: 10.1016/S0006-3495(94)80595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh C, Eckstein EC. Transient lateral transport of platelet-sized particles in flowing blood suspensions. Biophysical Journal. 1994;66:1706–1716. doi: 10.1016/S0006-3495(94)80962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aarts PA, van den Broek SA, Prins GW, Kuiken GD, Sixma JJ, Heethaar RM. Blood platelets are concentrated new the wall and red blood cells, in the center in flowing blood. Arteriosclerosis, Thrombosis, and Vascular Biology, Journal of the American Heart Association. 1988;8(6):819–824. doi: 10.1161/01.atv.8.6.819. [DOI] [PubMed] [Google Scholar]
- 11.Fogelson AL. A mathematical model and numerical method for studying platelet adhesion and aggregation during blood clotting. Journal of Computational Physics. 1984;56:111–134. [Google Scholar]
- 12.Fogelson AL, Kuharsky A, Yu Haoyu. Computational modeling of blood clotting: Coagulation and three-dimensional platelet aggregation. In: Alt W, Chaplain M, Griebel M, Lenz J, editors. Polymer and Cell Dynamics: Multicsale Modeling and Numerical Simulations. Birkhaeuser-Verlag; Basel: 2003. pp. 145–154. [Google Scholar]
- 13.Fogelson AL, Guy RD. Immersed-boundary-type models of intravascular platelet aggregation. Computer Methods in Applied Mechanics and Engineering. 2008;197:2087–2104. [Google Scholar]
- 14.Sui Y, Chew YT, Low HT. A lattice Boltzmann study on the large deformation of red blood cells in shear flow. International Journal of Modern Physics C. 2007;18(6):993–1011. [Google Scholar]
- 15.Sui Y, Chew YT, Roy P, Chen XB, Low HT. Transient deformation of elastic capsules in shear flow: Effect of membrane bending stiffness. Physical Review E. 2007;75:1–10. doi: 10.1103/PhysRevE.75.066301. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Johnson P, Popel A. Red blood cell aggregation and dissociation in shear flows simulated by lattice Boltzmann method. Journal of Biomechanics. 2007;41:47–55. doi: 10.1016/j.jbiomech.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNamara GR, Zanetti G. Use of the Boltzmann equation to Ssmulate lattice-gas automata. Physical Review Letters. 1988;61(20):2332–2335. doi: 10.1103/PhysRevLett.61.2332. [DOI] [PubMed] [Google Scholar]
- 18.Ladd A, Verberg R. Lattice-Boltzmann simulations of particle-fluid suspensions. Journal of Statistical Physics. 2001;104(5/6):1191–1251. [Google Scholar]
- 19.He X, Luo L. A priori derivation of the lattice Boltzmann equation. Physical Review E. 1997;55(6):R6333–R6336. [Google Scholar]
- 20.He X, Luo L. Lattice Boltzmann model for the incompressible Navier-Stokes equation. Journal of Statistical Physics. 1997;88(3/4):927–944. [Google Scholar]
- 21.Frisch U, Hasslacher B, Pomeau Y. Lattice-Gas automata for the Navier-Stokes equation. Physical Review Letters. 1986;56(14):1505–1508. doi: 10.1103/PhysRevLett.56.1505. [DOI] [PubMed] [Google Scholar]
- 22.He X, Luo L. Theory of the lattice Boltzmann method: From the Boltzmann equation to the lattice Boltzmann equation. Physical Review E. 1997;56(6):6811–6817. [Google Scholar]
- 23.Peskin C. Flow patterns around heart valves: A numerical method. Journal of Computational Physics. 1972;10:252–271. [Google Scholar]
- 24.Fogelson AL, Guy R. Platelet-wall interactions in continuum models of platelet aggregation: Formulation and numerical solution. Mathematical Medicine and Biology. 2004;21(4):293–334. doi: 10.1093/imammb21.4.293. [DOI] [PubMed] [Google Scholar]
- 25.Cortez R, Cowen N, Dillon R, Fauci L. Simulation of swimming organisms: Coupling internal mechanics with external fluid dynamics. Computing in Science and Engineering. 2004;6(3):38–45. [Google Scholar]
- 26.Peskin C. The immersed boundary method. Acta Numerica. 2002;11:1–39. [Google Scholar]
- 27.Guo Z, Zheng C, Shi B. Discrete lattice effects on the forcing term in the lattice Boltzmann method. Physical Review E. 2002;65:1–6. doi: 10.1103/PhysRevE.65.046308. [DOI] [PubMed] [Google Scholar]
- 28.He X, Zou Q, Luo L, Dembo M. Analytic solutions of simple flows and analysis of nonslip boundary conditions for the lattice Boltzmann BGK model. Journal of Statistical Physics. 1997;87(1/2):115–136. [Google Scholar]
- 29.Pozrikidis C. Modeling and Simulations of Capsules and Biological Cells. Chapman and Hall/CRC; Boca Raton: 2003. [Google Scholar]
- 30.Hochmuth R, Waugh R. Erythrocyte membrane elasticity and viscosity. Annual Review of Physiology. 1987;49:209–219. doi: 10.1146/annurev.ph.49.030187.001233. [DOI] [PubMed] [Google Scholar]
- 31.Goldsmith H, Marlow J. Flow behavior of erythrocytes. I. Rotation and deformation in dilute suspensions. Proceedings of the Royal Society of London Series B, Biological Sciences. 1972;182(1068):351–384. [Google Scholar]
- 32.Skalak R, Tozeren A, Zarda R, Chein S. Strain energy function of red blood cell membranes. Biophysical Journal. 1973;13(3):245–264. doi: 10.1016/S0006-3495(73)85983-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barthes-Biesel D, Diaz A, Dhenin E. Effect of constitutive laws for two-dimensional membranes on flow-induced capsule deformation. Journal of Fluid Mechanics. 2002;460:211–222. [Google Scholar]
- 34.Pozrikidis C. Effect of membrane bending stiffness on the deformation of capsules in simple shear flow. Journal of Fluid Mechanics. 2001;440:269–291. [Google Scholar]
- 35.Fischer TM. Tank-tread frequency of the red Cell membrane: Dependence of the viscosity of the suspending medium. Biophysical Journal. 2007;93(14):2553–2561. doi: 10.1529/biophysj.107.104505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagchi P. Mesoscale simulation of blood flow in small vessels. Biophysical Journal. 2007;92:1858–1877. doi: 10.1529/biophysj.106.095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blackshear P, Jr, Forstrom R, Dorman F, Voss G. Effect of flow on cells near walls. Federal Proceedings. 1971;30(5):1600–1609. [PubMed] [Google Scholar]
- 38.AlMomani T, Udaykumar HS, Marshall JS, Chandran KB. Micro-scale dynamic simulation of erythrocyte-platelet interaction in blood flow. Annals of Biomedical Engineering. 2008;36:905–920. doi: 10.1007/s10439-008-9478-z. [DOI] [PubMed] [Google Scholar]