Abstract
Negative transcriptional feedback loops are a core feature of eukaryotic circadian clocks and are based on rhythmic interactions between clock-specific repressors and transcription factors. In Drosophila, the repression of dCLOCK (dCLK)-CYCLE (CYC) transcriptional activity by dPERIOD (dPER) is critical for driving circadian gene expression. Although growing lines of evidence indicate that circadian repressors such as dPER function, at least partly, as molecular bridges that facilitate timely interactions between other regulatory factors and core clock transcription factors, how dPER interacts with dCLK-CYC to promote repression is not known. Here, we identified a small conserved region on dPER required for binding to dCLK, termed CBD (for dCLK binding domain). In the absence of the CBD, dPER is unable to stably associate with dCLK and inhibit the transcriptional activity of dCLK-CYC in a simplified cell culture system. CBD is situated in close proximity to a region that interacts with other regulatory factors such as the DOUBLETIME kinase, suggesting that complex architectural constraints need to be met to assemble repressor complexes. Surprisingly, when dPER missing the CBD (dPER(ΔCBD)) was evaluated in flies the clock mechanism was operational, albeit with longer periods. Intriguingly, the interaction between dPER(ΔCBD) and dCLK is TIM-dependent and modulated by light, revealing a novel and unanticipated in vivo role for TIM in circadian transcription. Finally, dPER(ΔCBD) does not provoke the daily hyperphosphorylation of dCLK, indicating that direct interactions between dPER and dCLK are necessary for the dCLK phosphorylation program but are not required for other aspects of dCLK regulation.
Introduction
Circadian (≅24 h) rhythms are driven by cell autonomous clocks that are generally composed of interconnected transcriptional and translational feedback loops (Dunlap, 1999). Studies using Drosophila have made seminal contributions to our understanding of clock mechanisms in general and those of animals in particular (Edery, 2000; Allada et al., 2001). In Drosophila, the major transcriptional negative feedback loop is comprised of dCLOCK (dCLK) and CYCLE (CYC), that heterodimerize to activate the daily transcription of target genes, including the core clock genes period (dper) and timeless (tim), whose protein products participate in the repression of dCLK-CYC-mediated gene expression (Hardin, 2006). dPER and TIM interact in the cytoplasm, an event that promotes their nuclear entry (Vosshall et al., 1994; Myers et al., 1996; Saez and Young, 1996; Meyer et al., 2006), where it is thought that the binding of dPER to dCLK is a crucial step in blocking the transcriptional activity of the dCLK-CYC complex (Darlington et al., 1998; Lee et al., 1998, 1999; Bae et al., 2000). More recent evidence indicates that dPER does not directly inhibit dCLK-CYC-mediated transcription but likely functions as a scaffold to promote the interaction between largely uncharacterized inhibitory factors and dCLK-CYC (Kim and Edery, 2006; Yu and Hardin, 2006; Yu et al., 2006, 2009; Kim et al., 2007; Chen et al., 2009).
How TIM contributes to repressing dCLK-CYC activity is more enigmatic but is mainly thought to be primarily due to its effects on the subcellular localization and stability of dPER. In addition to stimulating the nuclear entry of dPER, TIM also acts to stabilize dPER in the cytoplasm and nucleus by attenuating the ability of the DOUBLETIME (DBT; Drosophila homolog of CK1ε/δ) kinase to evoke the rapid degradation of dPER (Price et al., 1995, 1998; Kloss et al., 1998, 2001; Ko et al., 2002). The observation that TIM is present in a complex with dCLK-CYC during the night while peak activity of transcriptional inhibition occurs (Lee et al., 1998) raised the possibility that TIM might have a more direct role as a repressor. However, dPER has been shown to inhibit dCLK-CYC-mediated transcription independent of TIM in vitro and in vivo (Rothenfluh et al., 2000; Ashmore et al., 2003; Chang and Reppert, 2003), casting doubt on a direct physiological role for TIM in transcriptional repression.
To better understand how dPER inhibits the transactivation potential of dCLK-CYC, we identified a small conserved region of dPER required for its binding to dCLK, termed CBD (for dCLK binding domain). dPER missing the CBD (dPER(ΔCBD)) is unable to inhibit the transcriptional activity of dCLK-CYC in a simplified cell culture system. Surprisingly, when dPER(ΔCBD) was evaluated in flies the clock mechanism was very robust, although it did have a longer period. Intriguingly, despite the inability of dPER(ΔCBD) to directly bind dCLK, it was still associated with dCLK. We show that this association is TIM-dependent and modulated by light, revealing a novel and unanticipated role for TIM in regulating circadian transcription.
Materials and Methods
Plasmids and methods for S2 cell-based assays.
The pAct-dper, pAct-tim, pAct-dper-V5, pMT-dClk-V5, pMT-dbt-V5, and pAct-tim-3HA plasmids were described previously (Ceriani et al., 1999; Ko et al., 2002; Kim and Edery, 2006; Kim et al., 2007). pAct-dperΔC3-V5, pAct-dperΔNC3-V5, pAct-dperΔC4-V5, and pAct-dperΔNC4-V5 (Fig. 1A) were generated by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). All final constructs were verified by DNA sequencing.
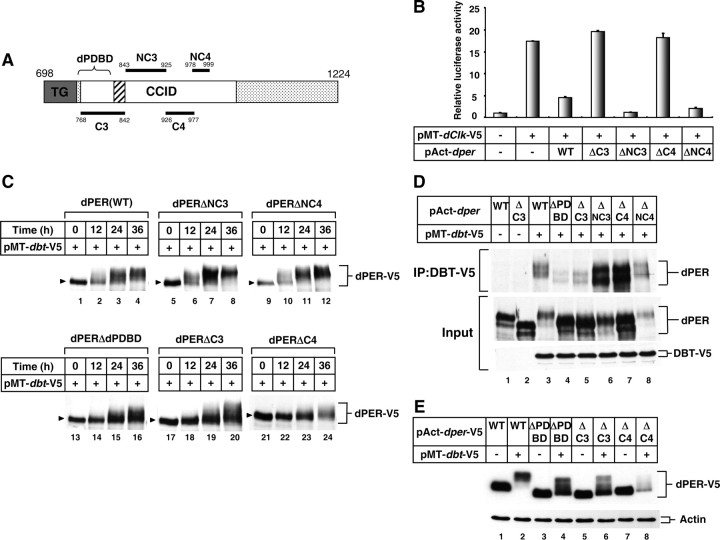
Figure 1.
dPER(ΔC4) is defective in transcriptional repression and DBT-dependent progressive phosphorylation in S2 cells. A, Schematic diagram of functional domains in the C-terminal half of dPER protein (accession no. P07633). Horizontal lines denote regions internally deleted from full-length dPER to generate mutants analyzed in this study. Domains are as follows: Thr-Gly repeats (dark gray box with label TG); dCLK:CYC inhibitory domain (CCID; amino acids 764-1034; white box) (Chang and Reppert, 2003); dPER DBT binding domain (dPDBD; amino acids 755–809; bracket) (Kim et al., 2007); putative nuclear localization sequence (amino acids 813-840, hatched box) (Chang and Reppert, 2003). B, S2 cells were transiently transfected in the presence (+) or absence of (−) pMT-dClk-V5. In addition, some cells were cotransfected with different versions of pAct-dper, as indicated. Shown are the average values from three independent experiments for relative luciferase activity. Luc activity in the absence of pMT-dClk-V5 was set to 1, and all other values were normalized. C–E, S2 cells were transiently transfected with different versions of V5-tagged pAct-dper (C, E) or nontagged pAct-dper (D), as indicated (top of panels). In addition, some cells were cotransfected with pMT-dbt-V5, as indicated (+). Exogenous DBT was induced 36 h after transfection by adding 500 μm CuSO4 to the medium. Cells were harvested at the indicated times (C), 24 h (D), or 36 h (E) after induction, and protein extracts were either directly analyzed by immunoblotting (C–E) or first subjected to immunoprecipitation in the presence of anti-V5 antibodies (D, top). Arrowheads indicate hypophosphorylated isoforms in each variants of dPER. D, “Input” indicates protein lysates used for immunoprecipitation. dPER was visualized with anti-dPER antibodies, whereas recombinant DBT with anti-V5 antibodies.
S2 cells were obtained from Invitrogen and transfected using effectene following the manufacturer's protocol (Qiagen). dCLK-dependent transactivation using a luciferase (luc) reporter assay was performed as described previously (Darlington et al., 1998), with slight modifications (Chang and Reppert, 2003; Kim and Edery, 2006). Briefly, S2 cells were placed in 24-well plates and cotransfected with 10–20 ng of control pAct-dper and/or different modified versions of pAct-dper plasmids, along with 10 ng of perEluc, 30 ng of pAct-β-gal-V5/His, and 2 ng of pMT-dClk-V5. In some experiments, 20–100 ng of pAct-tim plasmids were cotransfected, as indicated. One day after transfection, dClk expression was induced with 500 μm CuSO4 (final in the media), and after another day cells were washed in PBS, followed by lysis in 300 μl of Reporter Lysis Buffer (Promega). Aliquots of cell extracts were assayed for β-galactosidase and luciferase activities using the Luciferase Assay System and protocols supplied by the manufacturer (Promega).
Fly strains and behavioral assays.
To generate transgenic flies that produce the dPERΔCBD protein we used a previously described CaSpeR-4-based transformation vector containing a 13.2 kb genomic dper insert that was modified with sequences encoding for the HA epitope tag and a stretch of histidine residues just upstream of the dper translation stop signal, termed 13.2(per+-HA10His) (Lee et al., 1998). Deletion of sequences encoding amino acids 926–977 from dPER was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) with an appropriate dper genomic subfragment, confirmed by DNA sequencing and reconstructed into the above mentioned transformation vector to yield 13.2(perΔCBD-HA10His) (herein referred to as dper(ΔCBD)). Transgenic flies were generated by BestGene Inc. using standard P element-mediated transformation techniques and w1118 embryos as hosts. Two independent germ-line transformants bearing the dper(ΔCBD) transgene in a per+ background were obtained and then crossed into a wper01 genetic background to yield wper01;p{dper(ΔCBD)}-HAHis (herein referred to as wper01;p{dper(ΔCBD)}). Generation of transgenic flies expressing a wild-type version of the recombinant dPER protein was described in a previous report (Kim et al., 2007), and one of the lines (M16) in a wper01 genetic background (herein referred to as wper01;p{dper(WT)}) was used as control transgenic flies.
The locomotor activities of individual flies were measured as previously described using the monitoring system from Trikinetics (Hamblen-Coyle et al., 1992). Briefly, young adult flies were used for the analysis and kept in incubators at 25°C, exposed to at least 4 d of 12 h light followed by 12 h dark [12:12LD; where zeitgeber time 0 (ZT0) is defined as the time when the light phase begins] and subsequently kept in constant dark conditions for 5–8 d. The locomotor activity data for each individual fly was analyzed using the FaasX software (Fly Activity Analysis Suite for MacOSX), which was generously provided by F. Rouyer (Centre National de la Recherche Scientifique, Paris, France). Periods were calculated for each individual fly using χ2 periodogram analysis and pooled to obtain a group average for each independent transgenic line or genotype. Power is a measure of the relative strength of the rhythm during DD. Individual flies with a power ≥10 and a “width” value of 2 or more (denotes number of peaks in 30 min increments above the periodogram 95% confidence line) were considered rhythmic.
Immunoblotting and immunoprecipitation.
Protein extracts from S2 cells were prepared as previously described (Kim et al., 2007). Briefly, the cells were lysed using modified-RIPA (radioimmunoprecipitation assay) buffer (50 mm Tris-HCl, pH7.5, 150 mm NaCl, 1% NP-40, 0.25% sodium deoxycholate) with the addition of Protease Inhibitor Cocktail (Roche) and PhosSTOP (Roche). For detection of recombinant dCLK, extracts were prepared in harsher conditions using RIPA buffer (25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.5% sodium deoxycholate, 0.5% NP-40, 0.1% SDS) and were sonicated briefly as previously described (Kim and Edery, 2006). In the case of fly material, flies were collected by freezing at the indicated times in LD or DD and total head extracts prepared using either modified-RIPA or RIPA buffer with sonication depending on which proteins we sought to detect; i.e., modified-RIPA was used for dPER and TIM, whereas RIPA with sonication was used for dCLK. We also raised novel anti-dPER antiserum in guinea pigs using the services of Cocalico Biologicals. The immunogen was the same as previously described (Sidote et al., 1998) and in this study we used the anti-dPER antibody called GP339, which showed the highest dPER staining intensity with lowest background (data not shown). Primary antibodies were used at the following dilutions; anti-V5 (Invitrogen), 1:10,000; anti-PER (GP339), 1:3000; anti-HA (3F10; Roche), 1:2000; anti-TIM (TR3), 1:3000 (Sidote et al., 1998); anti-dCLK (GP208), 1:3000 (Kim et al., 2007). Gels (6%) were used to resolve dCLK, dPER and TIM, and in the case of dCLK 3–8% Tris-acetate Criterion gels (Bio-Rad) were also used.
For immunoprecipitation, protein extracts generated from either S2 cells or fly heads were prepared using modified-RIPA buffer with the addition of Protease Inhibitor Cocktail (Roche). To the extracts, 2 μl of anti-HA (12CA5), anti-V5, anti-PER (GP73 or GP339), or anti-dCLK (GP208) antibody was added, as indicated, and incubated with gentle rotation for 3–5 h at 4°C followed by the addition of 25 μl of Gammabind G-Sepharose (GE Healthcare) with a further incubation of 1–2 h. Beads were collected by light centrifugation and immune complexes were mixed with 30 μl of 1× SDS-PAGE sample buffer, incubated for 5 min at 95°C and the resulting supernatants resolved by immunoblotting as described above.
Quantitative real-time PCR.
The relative levels of dper and tim mRNA were measured by quantitative real-time PCR. Total RNA was isolated from frozen heads using TRI Reagent (Molecular Research Center). Five hundred nanograms of total RNA were reverse transcribed with oligo-dT primer using amfiRivert reverse transcriptase (GenDEPOT) and real-time PCR was performed using a Corbett Rotor Gene 6000 (Corbett Life Science) in the presence of Quantitect SYBR Green PCR kit (Qiagen). Primer sequences used here for quantitation of dper and tim RNAs were as described by Yoshii et al. (2007) and are as follows; dper forward: 5′-GACCGAATCCCTGCTCAATA-3′; dper reverse: 5′-GTGTCATTGGCGGACTTCTT-3′; tim forward: 5′-CCCTTATACCCGAGGTGGAT-3′; tim reverse: 5′-TGATCGAGTTGCAGTGCTTC-3′. We also included primers for the noncycling mRNA coding for CBP20 as previously described (Majercak et al., 2004), and sequences are as follows; cbp20 forward: 5′-GTCTGATTCGTGTGGACTGG-3′; cbp20 reverse: 5′-CAACAGTTTGCCATAACCCC-3′. Results were analyzed with software associated with the Rotor Gene 6000 machine, and relative mRNA levels quantitated using the 2−ΔΔCt method.
Immunohistochemistry.
Confocal imaging of adult brains was performed as described previously (Ko et al., 2007). Briefly, adult flies were dissected in ice-cold PBS, heads were cut open, and fixed for 1 h in 4% paraformaldehyde on ice. Subsequently heads were thoroughly washed with PBS containing 1% Triton X-100 and brains were dissected out. Brains were moved to a blocking solution comprised of PBT solution (PBS containing 0.5% Triton X-100) containing 10% horse serum and incubated for 30 min to a few hours. Primary antibodies were directly added to the blocking solution and incubated overnight at 4°C. The following antibodies and final dilutions were used; anti-HA antibody (3F10, Roche), 1:100; anti-PDF antibody (C7), 1:200 (Cyran et al., 2005). Subsequently, the brains were washed with PBT, followed by blocking solution containing secondary antibodies and incubated overnight at 4°C. The secondary antibodies used were Alexa Fluor 488-conjugated anti-rat IgG (Invitrogen) or Alexa Fluor 555-conjugated anti-mouse IgG (Invitrogen), both at a final dilution of 1:200. After several washes with PBT, brains were transferred onto slides and mounted with Vectashield (Vector Laboratories). Confocal images were obtained with a LSM700 Confocal Microscope (Zeiss) and processed with ZEN LE software (Zeiss).
Results
A small conserved region on dPER (amino acids 926-977) is necessary for its transcriptional repressor function in cultured Drosophila cells
Prior work using a simplified Drosophila Schneider 2 (S2) cell culture assay identified a region of dPER that is required for strong inhibition of dCLK-CYC-mediated transcription, termed the dCLK-CYC inhibition domain (CCID) (Chang and Reppert, 2003). The CCID encompasses amino acids 764-1034 of dPER, which includes previously identified conserved (C3 and C4) and non-conserved (NC3 and NC4) regions (Colot et al., 1988) (Fig. 1A). To explore the possible function(s) of these regions, we generated a series of dPER variants wherein each region was deleted. The four variants were named dPER(ΔC3) (conserved region 3; aa768-842), dPER(ΔNC3) (non-conserved region 3; aa843-925), dPER(ΔC4) (conserved region 4; aa926-977) and dPER(ΔNC4) (non-conserved region 4; aa978-999).
We first evaluated the ability of each dPER variant to inhibit dCLK-CYC-mediated transactivation using the standard luciferase (luc) reporter-based assay in S2 cells (Darlington et al., 1998; Chang and Reppert, 2003; Kim and Edery, 2006). While dPER(ΔNC3) and dPER(ΔNC4) manifested repressor activity comparable to that observed for wild-type dPER, removal of either conserved region resulted in severely impaired repressor activity (Fig. 1B). Variations in the protein levels of the different dPER variants cannot explain the differential capabilities in transcriptional repression since all variants were present at similar amounts (Fig. 1C, lanes 1, 5, 9, 17, and 21). Our results indicate that the non-conserved amino acid stretches in the CCID are not required for dPER's repressor activity. With regards to dPER(ΔC3), this deletion encompasses the major DBT binding site on dPER (termed dPDBD), which is required for numerous aspects of dPER metabolism and function, including its ability to repress dCLK-CYC transcriptional activity (Kim et al., 2007; Nawathean et al., 2007). Although we did not perform extensive studies on the dPER(ΔC3) version, it is almost certain that the severely impaired transcriptional repressor function is due to abolishing the dPDBD region.
To examine the role that the C4 region plays in the ability of dPER to function as a transcriptional repressor, we first tested the possibility that C4 is essential for the nuclear localization of dPER. However, ectopically expressed full-length dPER or dPER(ΔC4) did not manifest any significant differences in subcellular localization in S2 cells (supplemental Fig. S1, available at www.jneurosci.org as supplemental material), consistent with previous results indicating that the major nuclear localization sequences on dPER are present in the C3 region (Chang and Reppert, 2003). We also considered the possibility that deletion of C4 might interfere with DBT binding to dPER leading to loss of repressor activity, since dPDBD is adjacent to C4 (Fig. 1A). First, we assayed DBT-induced phosphorylation kinetics with the different dPER deletion variants. As previously shown, the induction of DBT evokes progressive decreases in the mobility of full-length dPER, which are mainly or solely due to differential phosphorylation (Ko et al., 2002) (Fig. 1C). Under our standard conditions, hyperphosphorylated isoforms of dPER are readily observed at 12 h post-dbt induction (Fig. 1C, lane 2) and there is little hypophosphorylated isoforms remaining at 36 h post-dbt induction (lane 4). For dPER(ΔNC3) and dPER(ΔNC4), time-dependent changes in the conversion of hypophosphorylated dPER isoforms to hyperphosphorylated ones were similar to that observed for wild-type dPER (Fig. 1C), indicating that these non-conserved regions play little to no role in the DBT-dependent global phosphorylation of dPER.
Although DBT induction stimulated the time-dependent appearance of slower migrating isoforms of dPER(ΔC3) and dPER(ΔC4), there was a noticeable delay. For example, little to no hyperphosphorylated species of dPER were detected at 12 h post-dbt induction (Fig. 1C, compare lanes 2, 18, and 22; e.g., for the mutant versions, the dPER band is tight and shows little evidence of smearing). In addition, fast-migrating hypophosphorylated versions of dPER(ΔC3) and dPER(ΔC4) were still present even after prolonged incubation with ectopically expressed dbt (Fig. 1C, compare lanes 3, 19 and 23), similar to what was previously shown for dPER(ΔdPDBD) (Fig. 1C, lanes 13–16) (Kim et al., 2007; Nawathean et al., 2007).
To test whether alterations in the ability of DBT to bind dPER are linked to less efficient hyperphosphorylation of dPER(ΔC4), we performed immunoprecipitation assays. Deletion of C3 dramatically decreased the interaction between dPER and DBT (Fig. 1D, top, middle, compare lanes 5 and 3; i.e., more wild-type dPER copurifies with DBT although there is much less total wild-type dPER in the extract compared with the mutant version), as expected based on prior work showing that this region contains the major binding site mediating stable interactions between dPER and DBT (Kim et al., 2007) (Fig. 1D, compare lanes 3 and 4). In sharp contrast, dPER(ΔC4) stably interacts with DBT (Fig. 1D, lane 7). These data strongly suggest that unlike dPDBD's mode-of-action, the attenuation of progressive hyperphosphorylation and inability to block dCLK transcriptional activity by dPER(ΔC4) are not a result of losing the capability to stably interact with DBT. In this regard we also noted that although there is little accumulation of highly phosphorylated isoforms of dPER(ΔC4), this version of dPER still undergoes enhanced degradation with prolonged expression of DBT (Fig. 1E, compare lanes 7 and 8; see also Fig. 1C, compare lanes 24 and 20). Thus, while dPER(ΔC3), dPER(ΔdPDBD) and dPER(ΔC4) all show defects in DBT-dependent global phosphorylation, only the latter retains the ability to undergo significant DBT-mediated decreases in abundance (Fig. 1E, compare lanes 8 and 4, and 6). This further supports previous findings that dPER stability is not strongly linked to global phosphorylation (Chiu et al., 2008) and indicates that the C4 region is required for DBT-dependent hyperphosphorylation in a manner unrelated to promoting strong binding with DBT. Together, our results demonstrate that the ability of dPER to negatively regulate dCLK-CYC-mediated transcription is based on multiple conserved subdomains within the CCID that have distinct biochemical functions.
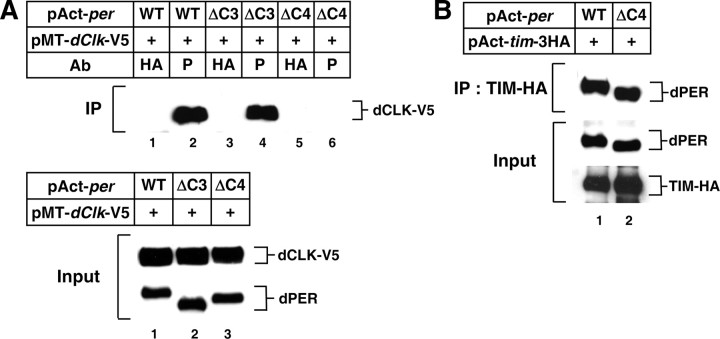
dPER(ΔC4) is highly defective in binding to dCLK
We next considered the possibility that the impaired repressor function of the dPER(ΔC4) variant might be a result of deficient binding with dCLK. Indeed, little to no dPER(ΔC4) copurified with dCLK, in sharp contrast to wild-type dPER or dPER(ΔC3) (Fig. 2A). Importantly, dPER(ΔC4) retains normal ability to bind TIM protein (Fig. 2B). These data together with the finding that dPER(ΔC4) interacts strongly with DBT (Fig. 1D) indicates that the inability of dPER(ΔC4) to stably interact with dCLK is not due to gross conformational changes in dPER resulting from deletion of the C4 region. We conclude that C4 is a critical dCLK binding domain on dPER, although we have not ruled out the possibility that other regions on dPER can function as binding domains for dCLK. Based on these findings we refer to the region between amino acids 926 and 977 as the CBD (for dCLK binding domain). At present it is not clear whether the C4 region on dPER is sufficient to interact with dCLK.
Figure 2.
dPER(ΔC4) interacts with TIM but not with dCLK in S2 cells. A, B, S2 cells were transiently cotransfected with different versions of pAct-dper in combination with pMT-dClk-V5 (A), or pAct-tim-3HA (B). Expression of recombinant dCLK was induced 24 h after transfection by adding 500 μm CuSO4 to the medium. Cells were harvested 24 h after induction (A) or 36 h posttransfection (B), and protein extracts were either directly analyzed by immunoblotting (Input) or following immunoprecipitation (IP). A, IP was performed in the presence of anti-dPER antibodies (P) or a nonspecific antibody (HA), and immune complexes analyzed for dCLK. B, IP was performed in the presence of anti-TIM antibodies and immune complexes analyzed for dPER.
Flies expressing dPER(ΔCBD) display behavioral rhythms with longer periods
To investigate the in vivo significance of the CBD of dPER in clock function, we generated transgenic flies harboring a dper transgene internally deleted for this region, termed p{dper(ΔCBD)}. The parental wild-type dper transgene [herein termed p{dper(WT)}] used to generate the deletion variant has sequences encoding an HA epitope tag followed by a stretch of His residues at the carboxy terminus of the dper open reading frame, facilitating purification and detection of the transgene derived dPER protein (Kim et al., 2007; Chiu et al., 2008). Two independent lines of transgenic flies bearing the p{dper(ΔCBD)} transgene were obtained and evaluated in a per-null wper01 genetic background (Konopka and Benzer, 1971). Locomotor activity rhythms were assayed under standard conditions whereby flies were kept at 25°C for 4 d of 12 h:12 h light:dark (LD) cycle followed by 7 d of complete darkness to determine their free-running periods. As previously shown, transgenic wper01 flies harboring the p{dper(WT)} transgene manifest robust locomotor activity rhythms with ∼23.5 h periods, similar to wild-type flies (Kim et al., 2007) (Table 1). In sharp contrast, although both independent lines of p{dper(ΔCBD)} transgenic flies exhibit strong activity rhythms, the periods are ∼3 h longer than their wild-type control counterparts. The high level of rhythmicity for p{dper(ΔCBD)} flies was surprising given that both dPER(ΔCBD) and dPER(ΔdPDBD) have severely impaired transcriptional repressor functions (Fig. 1B) (Kim et al., 2007) and p{dper(ΔdPDBD)} flies are completely arrhythmic (Kim et al., 2007). This indicates that dPER(ΔCBD) retains some circadian relevant activities and further supports functionally distinct inhibitory domains within the CCID.
Table 1.
Locomotor activity rhythms of p{dper(ΔCBD)} flies and control p{dper(WT)} transgenic fliesa
| Genotypeb | Temperature (°C) | Period ± SEM (h) | Power c | Rhythmicity (%)d | Number e |
|---|---|---|---|---|---|
| w1118 | 18 | 23 ± 0.75 | 41.8 | 50 | 32 |
| wper01;; p{dper(WT)}(M16) | 18 | 23.4 ± 0.42 | 34.6 | 29 | 31 |
| wper01; p{dper((ΔCBD)}(M1) | 18 | 24.3 ± 0.16 | 36.8 | 27.6 | 29 |
| wper01;; p{dper(ΔCBD)}(F3) | 18 | 24.1 ± 0.17 | 60.6 | 93.5 | 31 |
| perS | 18 | 19.2 ± 0.11 | 49.3 | 38.5 | 26 |
| perL | 18 | 27.5 ± 0.38 | 33.9 | 21.9 | 32 |
| w1118 | 25 | 23.3 ± 0.08 | 74.9 | 73.3 | 16 |
| wper01;; p{dper(WT)}(M16) | 25 | 23.3 ± 0.05 | 141.7 | 100 | 32 |
| wper01; p{dper(ΔCBD)}(M1) | 25 | 26.2 ± 0.22 | 69.4 | 68.8 | 16 |
| wper01;; p{dper(ΔCBD)}(F3) | 25 | 26.2 ± 0.15 | 100.1 | 93.1 | 31 |
| perS | 25 | 19.7 ± 0.81 | 108 | 74.2 | 32 |
| perL | 25 | 28.1 ± 0.11 | 116.8 | 89.7 | 32 |
| wper01;; p{dper(WT)}(M16) | 29 | 23.3 ± 0.05 | 106.8 | 100 | 32 |
| wper01; p{dper(ΔCBD)}(M1) | 29 | 28 ± 0.17 | 61.3 | 66.7 | 30 |
| wper01;; p{dper(ΔCBD)}(F3) | 29 | 27.5 ± 0.07 | 122.4 | 100 | 32 |
| perS | 29 | 18.7 ± 0.05 | 74 | 70 | 30 |
| perL | 29 | 29.9 ± 0.08 | 98.9 | 96.9 | 32 |
aFlies were kept at 25°C and exposed to 4 d of 12:12LD followed by 7 d of DD.
bM16, M1, and F3 indicate independent transgenic lines.
cMeasure of the strength or amplitude of the rhythm.
dPercentage of flies with activity rhythms having a power value of ≥10 and a width value of ≥2.
eTotal number of flies that survived until the end of the testing period.
We also determined activity rhythms at different temperatures. As expected, p{dper(WT)} flies exhibit behavioral rhythms with ∼23.5 h periods over a wide range of physiologically relevant temperatures (Table 1). However, behavioral rhythms in p{dper(ΔCBD)} flies lengthen as temperature increases, suggesting the biochemical defect(s) of the dPER(ΔCBD) protein is exacerbated at higher temperatures. Alternatively, this might reveal a more fundamental role for the C4 region on dPER in temperature compensation
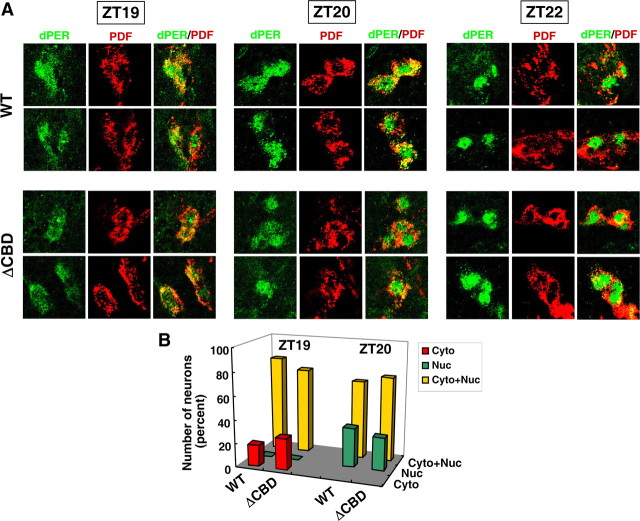
dPER(ΔCBD) exhibits normal timing of nuclear entry in key clock cells
In some cases, mutations in the dper gene that lead to alterations in the period of behavioral rhythms are associated with changes in the timing of dPER nuclear entry in brain clock neurons (Curtin et al., 1995). The circadian system driving adult behavioral rhythms in Drosophila is situated in the brain and comprised of a neural network of several anatomically and functionally distinct clock neuron clusters, wherein the small ventral lateral neurons (s-LNv) are central for the maintenance of locomotor activity rhythms in constant dark conditions (for review, see Nitabach and Taghert, 2008). Pigment dispersing factor (PDF), a circadian relevant neuropeptide, was used as a marker to identify the LNvs and demarcate the cytoplasm (Renn et al., 1999). Wild-type dPER manifests mixed cytoplasmic-nuclear staining at ZT19 and ZT20, and essentially only nuclear staining beginning at ZT22 (Fig. 3). A similar temporal pattern was observed for dPER(ΔCBD), indicating that during a standard light-dark cycle, the C4 region has at best a minor impact on the timing of dPER nuclear entry in key pacemaker neurons (Fig. 3). These findings are consistent with results obtained in cultured S2 cells suggesting the CBD does not have a major impact on dPER subcellular localization (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
Figure 3.
The timing of dPER nuclear entry in the small ventral lateral neurons of p{dper(ΔCBD)} flies is similar to that observed for p{dper(WT)} flies. Adult flies of the indicated genotypes (left of panel) were collected at the indicated times in an LD cycle and processed for immunohistochemistry followed by visualization using confocal microscopy. A, Shown are representative staining patterns obtained for the small ventral lateral neurons (s-LNvs) from at least 5 flies; dPER was visualized with anti-HA (3F10) antibodies labeled with Alexa Fluor 488 (green). PDF was visualized with anti-PDF (C7) antibodies labeled with Alexa Fluor 533 (Cyran et al., 2005). B, The cytoplasmic/nuclear distribution of dPER for s-LNv from each genotype at ZT19 and ZT20 was quantified.
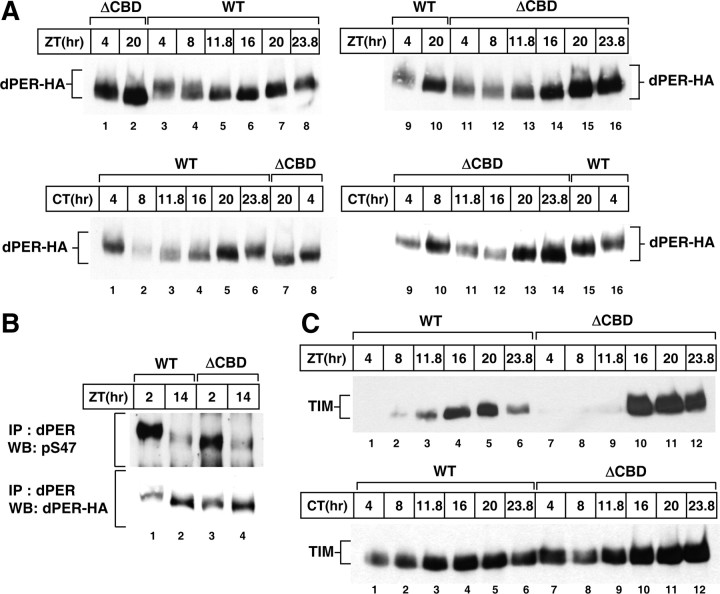
Quasi-normal dPER/TIM biochemical rhythms in p{dper(ΔCBD)} transgenic flies
As a means to more directly probe the central clock mechanism operating in p{dper(ΔCBD)} flies, head extracts were prepared and the daily biochemical cycles in dPER and TIM proteins determined during LD and the first day of DD (Fig. 4). Although dPER(ΔCBD) exhibits daily changes in abundance and phosphorylation, there are several differences when compared with the wild-type control dPER protein. For example, peak levels of dPER(ΔCBD) are 1.5- to 2-fold higher compared with dPER (Fig. 4A, top, compare lanes 2 and 7; Fig. 5A). Also, in light-dark cycles dPER(ΔCBD) attains peak levels between ZT20 and ZT24, while wild-type dPER reaches maximal values by ZT20 (Fig. 4A, top, compare lanes 15 and 16 with 7 and 8). A similar delayed accumulation phase for dPER(ΔCBD) was also observed during the first day of DD (Fig. 4A, bottom, compare lanes 13 and 14 with 5 and 6). These results are consistent with the longer behavioral periods manifested by p{dper(ΔCBD)} flies (Table 1).
Figure 4.
Levels of dPER and TIM proteins undergo robust daily cycles in p{dper(ΔCBD)} flies. A–C, Adult flies of the indicated genotypes [above the panels; WT, wper01;; p{dper(WT)}(M16); ΔCBD, wper01;; p{dper(ΔCBD)}(F3)] were collected at the indicated ZT or CT. Head extracts were prepared and analyzed either directly by immunoblotting (A, C) or following immunoprecipitation (IP) (B). IP was performed in the presence of anti-dPER antibodies, and immune complexes analyzed for either phosphorylated S47 on dPER (pS47) or total dPER. Anti-HA (3F10) or anti-TIM (TR-3) Abs were used to visualize dPER or TIM, respectively.
Figure 5.
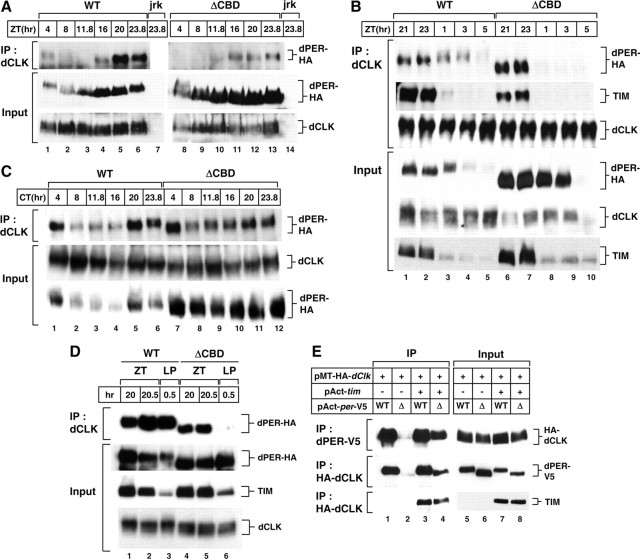
dPER(ΔCBD) can interact with dCLK in the presence of TIM. A–C, Adult flies of the indicated genotypes (above the panels; jrk, dClkjrk) (Allada et al., 1998) were collected at the indicated times during LD cycle (A, B) or first day of DD (C). D, Adult flies of the indicated genotypes were treated with light (LP) or maintained in the dark (ZT) for 30 min starting at ZT20. Head extracts were prepared and either directly analyzed by immunoblotting (Input) or subjected to immunoprecipitation (IP). IP was performed in the presence of anti-dCLK antibodies, and immune complexes analyzed for dPER, TIM, or dCLK, as indicated (right of panels). E, S2 cells were transiently transfected with pAct-dper-V5 or pAct-dperΔCBD-V5 in combination with pMT-HA-dClk. In addition, some cells were additionally transfected with pAct-tim (+). Exogenous expression of dCLK was induced 24 h after transfection by adding 500 μm CuSO4 to the medium. Cells were harvested 24 h after induction, and protein extracts were directly analyzed by immunoblotting (Input) or subjected to immunoprecipitation (IP). The protein targeted by the antibody added during IP is indicated (left of panel) and immune complexes analyzed for the indicated protein (right of panel).
In addition to alterations in the daily abundance cycle, dPER(ΔCBD) exhibits less dramatic shifts in electrophoretic mobility during the late-night/early day when the majority of wild-type dPER is hyperphosphorylated (Fig. 4A, top, compare lanes 1 to 3, and lanes 2 to 7; bottom, compare lanes 1 to 8, and 5 to 7) (see also Fig. 5B). These results are in remarkable agreement with data obtained in cultured S2 cells showing that the DBT-mediated global phosphorylation of dPER(ΔCBD) is impaired (Fig. 1C). Furthermore, although progressive hyperphosphorylation is attenuated, dPER(ΔCBD) still exhibits a robust declining phase in abundance during the early-to-mid day (Fig. 4A, top, compare lanes 11 and 16) (see also Fig. 5B). Thus, as in S2 cells (Fig. 1; supplemental Fig. S2, available at www.jneurosci.org as supplemental material), while the C4 region affects global phosphorylation it has little noticeable effect on the ability of dPER to undergo DBT-mediated degradation. Indeed, prior work showed that although global hyperphosphorylation of dPER moderately enhances its degradation, it is not required for the rapid decline in its levels during the late night/early morning (Chiu et al., 2008). Rather, phosphorylation of Ser47 on dPER during the late night/early day is the key phospho-signal that triggers recognition by the F-box protein SLIMB and ultimately rapid degradation via ubiquitin-proteasome pathway (Ko et al., 2002; Chiu et al., 2008). For example, when dPER is rapidly degraded at ZT2, the intensity of the phosphorylated S47 signal is clearly greater compared with that at ZT14, even though there is less total dPER protein present at ZT2 (Fig. 4B, compare lanes 1 and 2). Although there might be subtle differences in the efficiency of S47 phosphorylation between wild-type dPER and dPER(ΔCBD), the results further support the contention that there is little to no effect of the C4 region on the DBT-dependent mechanism regulating dPER stability. Nonetheless, the attenuated global hyperphosphorylation of dPER(ΔCBD) might contribute to its relatively higher steady-state levels observed during a daily cycle (Fig. 4A). In addition, slightly higher peak levels of dper RNA in p{dper(ΔCBD)} flies (see below; see Fig. 7) might also contribute to the increased overall abundance of the dPER(ΔCBD) protein.
Figure 7.
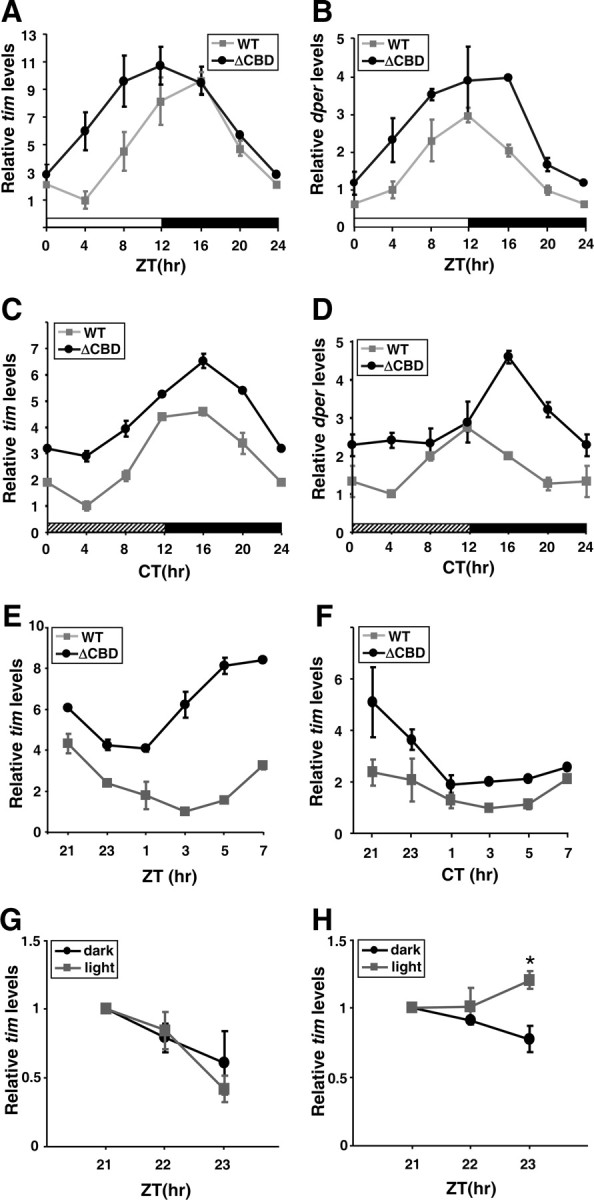
The levels of dper and tim transcripts undergo robust daily cycles in p{dper(ΔCBD)} flies and exhibit rapid increases following light-exposure. A–H, Adult flies of the indicated genotypes were collected at the indicated ZT (A, B, E) or CT (C, D, F). RNA was extracted from fly heads, and quantitative real-time PCR was used to measure the relative levels of total tim (A, C, E, F–H) or dper (B, D) RNAs. G, H, Adult flies of p{dper(WT)} (G) or p{dper(ΔCBD)} (H) flies were collected at the indicated ZT and served as controls (dark). Light exposure was initiated at ZT21, and flies were collected 1 or 2 h after light treatment (light). RNA levels at ZT21 were set to 1 and all other values were normalized. Shown are the average values from three independent experiments. *p < 0.005, error bars denote SEM.
Alterations in the abundance profile of TIM protein from p{dper(ΔCBD)} flies were consistent with those observed for dPER(ΔCBD) protein. Most notably, the overall TIM levels in p{dper(ΔCBD)} flies were 1.5- to 2-fold higher, and the timing in attaining peak levels was delayed (Fig. 4C). TIM protein is essentially only detected during the dark phase in an LD cycle (Fig. 4C, compare top and bottom) (see also Fig. 5B), indicating that its light-mediated degradation is not affected in p{dper(ΔCBD)} flies (Ashmore and Sehgal, 2003).
Light disrupts the interaction between dPER(ΔCBD) and dCLK
To probe for in vivo interactions between dPER(ΔCBD) with dCLK, we prepared head extracts from flies collected at different times during an LD cycle and performed immunoprecipitation assays. In control p{dper(WT)} flies, wild-type dPER interacts with dCLK in a time-dependent manner, with the greatest copurification occurring during the dPER-mediated transcriptional repression phase between ZT20–4, as previously shown (Lee et al., 1998; Bae et al., 2000; Menet et al., 2010) (Fig. 5A, left). To our surprise and unlike the situation when using S2 cells, for which we could not observe interactions between dPER(ΔCBD) and dCLK (Fig. 2), dPER(ΔCBD) stably copurifies with dCLK (Fig. 5A, right). While the staining intensity of dCLK copurifying with dPER (ΔCBD) is less than that observed with the control situation, the overall levels of dCLK in p{dper(ΔCBD)} are also generally lower (Fig. 5A, compare bottom). Nonetheless, it is possible that dPER(ΔCBD) has a weaker association with dCLK compared with wild-type dPER. Intriguingly, the association of dPER(ΔCBD) with dCLK was only observed during the dark phase, unlike wild-type dPER (Fig. 5A, top, compare lanes 1 and 8).
To better understand how light might affect the interaction between dPER(ΔCBD) and dCLK in flies, we examined more time points during the dark-to-light transition. Wild-type dPER is stably associated with dCLK through the late night (i.e., ZT21) and well into the morning (i.e., ZT3) (Fig. 5B, top, lanes 1–4). In sharp contrast, there is a dramatic decrease in the interaction of dPER(ΔCBD) and dCLK following the onset of light (Fig. 5B, top, compare lanes 7 and 8). This striking photic effect on dPER(ΔCBD)-dCLK complex formation cannot be attributed to light-mediated changes in the levels of dPER or dCLK; e.g., in p{dper(ΔCBD)} flies at ZT1, the levels of dPER are higher and those of dCLK comparable to those in p{dper(WT)} flies (Fig. 5B, compare lanes 3 and 8). Together, our data strongly suggest that dPER(ΔCBD) is stably associated in a complex with dCLK during the dark phase, but this interaction is abruptly disengaged upon exposure to photic cues.
What could be causing the photosensitivity underlying the interaction of dPER(ΔCBD) with dCLK? Given that TIM is rapidly degraded upon light stimulation, which is also the case in p{dper(ΔCBD)} flies (Fig. 5B, bottom, compare lanes 2 and 3 with 7 and 8), we reasoned that TIM plays a prominent role in mediating the interaction between dPER(ΔCBD) and dCLK. Indeed, in daily light-dark cycles dPER(ΔCBD) only copurifies with dCLK-containing complexes when TIM is also present in the complex, unlike the wild-type situation where some dPER can interact with dCLK during the early day even in the absence of detectable copurifying TIM (Fig. 5B, lanes 4 and 5). Interactions between dPER(ΔCBD) and dCLK are observed during the “subjective” day in constant darkness (top, compare Fig. 5C, lane 7 to 5B, lane 8), conditions wherein TIM levels do not undergo sharp decreases (Fig. 4C, bottom). Prior work showed that TIM does not require dPER to stably interact with dCLK in vivo (Lee et al., 1998) (Fig. 5B). Moreover, removal of the CBD does not affect the ability of dPER to bind TIM (Fig. 2B). We also investigated the interaction between dPER(ΔCBD) and dCLK in the middle of night, when dPER(ΔCBD) and dCLK is already associated, upon getting rid of TIM by treatment of light. With light pulses of 30 min duration administered on ZT20, rapid TIM degradation is observed (Fig. 5D, third panel, lane 3 and 6). In this condition, while wild-type dPER is strongly associated with dCLK, dPER(ΔCBD) is rapidly dissociated from dCLK (Fig. 5D, first panel, compare lane 3 and 6). Although we did not do a more extensive kinetic analysis, 30 min of light is sufficient to abolish the interaction between dPER(ΔCBD) and dCLK. Together the findings strongly suggest that in vivo TIM is mediating the association between dPER(ΔCBD) and dCLK. As a more direct means to test this possibility we used cultured S2 cells and determined the ability of dPER(ΔCBD) to interact with dCLK in the presence or absence of ectopically expressed TIM. Indeed, TIM strongly enhanced the association between dPER(ΔCBD) and dCLK, whereas it had little to no impact on the interaction between dCLK and wild-type dPER (Fig. 5E).
Direct interaction between dCLK and dPER is not necessary for dPER's repressor function
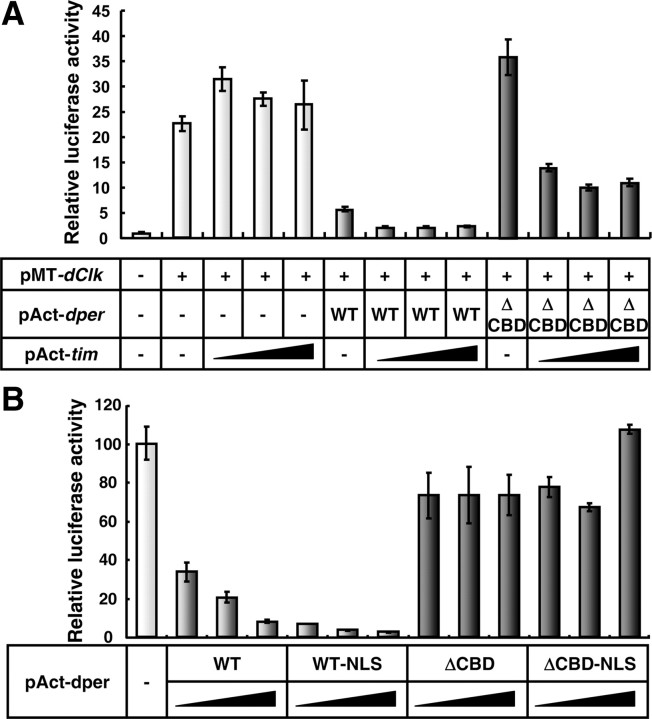
Based on the ability of TIM to mediate the interaction between dPER(ΔCBD) and dCLK in S2 cells (Fig. 5E), we sought to determine whether the presence of TIM could also rescue the ability of dPER(ΔCBD) to inhibit dCLK-CYC-mediated transcription (Fig. 6A). As previously shown, whereas TIM stimulates dPER's function as a transcriptional repressor, TIM manifests little to no repressor activity on its own (Darlington et al., 1998; Rothenfluh et al., 2000; Chang and Reppert, 2003). Remarkably, dPER(ΔCBD) coexpressed with TIM gained the ability to inhibit dCLK-CYC transactivation. The repression exerted by the combination of dPER(ΔCBD) and TIM was ∼50% less compared with that of wild-type dPER, suggesting a partially defective repressor capability. Our findings strongly suggest that TIM can act as a scaffold to bridge the association between dPER and dCLK, enabling dPER to participate in transcriptional repression.
Figure 6.
Ectopic expression of TIM enables dPER(ΔCBD) to inhibit dCLK-dependent transcriptional activity in S2 cells. A, S2 cells were transiently transfected in the presence (+) or absence (−) of pMT-dClk-V5. In addition, 20 ng of pAct-dper (WT) or pAct-dperΔCBD (ΔCBD) were transfected either singly or in combination with increasing amount of pAct-tim (20, 50, 100 ng). Luc activity in the absence of transfecting pMT-dClk-V5 was set to 1, and all other values were normalized. B, S2 cells were transiently transfected with pMT-dClk-V5 in combination with increasing amount of pAct-dper (WT), pAct-dper-NLS (WT-NLS), pAct-dper ΔCBD (ΔCBD), or pAct-dper ΔCBD-NLS (ΔCBD-NLS). Luc activity in the absence of any dper-containing plasmid were set to 100% and all other values normalized. Shown are the average values from three independent experiments for relative luciferase activity(error bars denote SEM).
Nonetheless, it is also possible that TIM stimulates the transcriptional repressor function of dPER(ΔCBD) by increasing its nuclear localization (Saez and Young, 1996; Rothenfluh et al., 2000; Chang and Reppert, 2003). To test this possibility, we sought to augment the nuclear localization of dPER(ΔCBD) by placing a potent nuclear localization signal (NLS) at the C terminus of dPER(ΔCBD) and measured its repressor activity. This strategy was previously shown to enhance dPER's ability to repress dCLK-CYC-mediated transcription in S2 cells (Chang and Reppert, 2003; Nawathean et al., 2007). Despite the increased repressor activity of wild-type dPER containing an ectopic NLS sequence, the presence of a NLS on dPER(ΔCBD) did not enhance its transcriptional inhibition capabilities (Fig. 6B). Thus, the findings clearly indicate that TIM's stimulatory effect on the transcriptional repressor potential of dPER(ΔCBD) is not an “indirect” effect of increased nuclear localization but is almost certainly due to TIM promoting the close interaction between dPER(ΔCBD) and the dCLK-CYC transcription factor.
Light evokes rapid increases in dper/tim RNA levels in p{dper(ΔCBD)} transgenic flies
As a result of the rhythmic inhibition of dCLK-CYC-mediated transcription, the levels of dper and tim mRNAs undergo daily cycles, with peak values attained between ZT12–16 and trough amounts around ZT0–4 (Fig. 7A,B) (Hardin et al., 1990; Sehgal et al., 1994). Although daily oscillations in dper/tim RNA levels were apparent in p{dper(ΔCBD)} flies (Fig. 7A,B), peak values were higher. Higher daily levels of dper/tim transcripts were also observed in constant dark conditions (Fig. 7C,D). These results suggest that repression of dCLK/CYC transactivation via dPER(ΔCBD) is somewhat diminished compared with that of wild-type dPER, in agreement with results obtained in cultured cells (Fig. 6A). However, the quasi-normal daily cycles in dper and tim transcripts indicate that circadian auto-inhibition is quite robust in flies where the sole functional version of dPER is missing the CBD.
Despite quasi-normal daily cycles in dper/tim RNA levels, we noted that in p{dper(ΔCBD)} flies the rising phases are significantly accelerated following lights-on at ZT0, whereas the declining phases were less affected (Fig. 7A,B). Sharp increases in either dper or tim transcripts were not observed for p{dper(ΔCBD)} flies during the subjective day in constant dark conditions (Fig. 7C,D). To better evaluate whether light stimulation affects the daily trajectory in dper/tim transcript accumulation, we measured tim RNA levels in two sets of flies that following entrainment to standard LD cycles were either exposed to light at ZT0 or kept in the dark (Fig. 7E,F). The results clearly indicate that exposure to light dramatically increases the levels of tim during its rising phase, indicating that dPER(ΔCBD)-mediated repression of dCLK/CYC activity is rapidly attenuated by light. We also measured tim RNA levels after introduction of light to p{dper(WT)} and p{dper(ΔCBD)} flies for 2 h starting at ZT21 (Fig. 7G,H). Treatment of light resulted in strong induction of tim RNA in p{dper(ΔCBD)} flies but not in p{dper(WT)} (Fig. 7G,H). These findings are in strong agreement with our demonstration that TIM bridges the interaction between dPER(ΔCBD) and dCLK (Fig. 5), and rescues the transcriptional repressor function of dPER(ΔCBD) (Fig. 6A). Thus, the light-dependent degradation of TIM leads to the rapid dissociation of dPER(ΔCBD) from dCLK-CYC, an event that accelerates the next round of dper/tim expression.
Direct interaction between dPER and dCLK is likely required for maintenance of hyperphosphorylated isoforms of dCLK
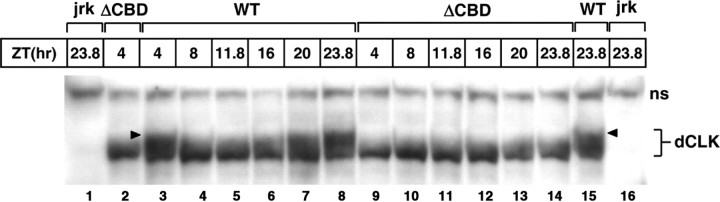
dCLK is phosphorylated in vivo (Lee et al., 1998), with the appearance of hyperphosphorylated isoforms in the late night/early day (Lee et al., 1998; Kim and Edery, 2006; Yu et al., 2006). It is thought that dPER acts as a molecular bridge to deliver kinases such as DBT to dCLK (Kim and Edery, 2006; Yu et al., 2006, 2009; Kim et al., 2007), an event that might be important for repressing dCLK transactivity during the late night/early morning. To determine whether dPER(ΔCBD) might provoke time-dependent hyperphosphorylation of dCLK, we examined biochemical profiles of dCLK in p{dper(WT)} and p{dper(ΔCBD)} flies. In control p{dper(WT)} flies, hyperphosphorylation of dCLK was observed from late night to early morning as previously reported (Kim and Edery, 2006; Yu et al., 2006) (Fig. 8, compare lanes 3 and 4). However, in several independent experiments, we noted that in p{dper(ΔCBD)} flies slowly migrating (highly phosphorylated) dCLK isoforms are either absent or greatly diminished (Fig. 8A, compare lane 2 and 3, and 14 and 15). This result suggests that the late night/early morning-specific hyperphosphorylation of dCLK requires direct interactions between dCLK and dPER. However, we cannot rule out alternative scenarios, such as hyperphosphorylated isoforms of dCLK are less stable in p{dper(ΔCBD)} flies. Nonetheless, our findings clearly indicate that dPER function is critical in regulating the phosphorylated state of dCLK.
Figure 8.
Little to no hyperphosphorylated isoforms of dCLK are detected in p{dper(ΔCBD)} flies. Adult flies of the indicated genotypes (top of the panel) were collected at the indicated ZT. Head extracts were prepared and analyzed by immunoblotting in the presence of anti-dCLK antibodies. ns, Nonspecific bands; arrowheads, hyperphosphorylated isoforms of dCLK.
Discussion
A shared feature of eukaryotic circadian pacemaker mechanisms is that daily cycles in gene expression involve the phase-specific interaction of one or more repressors with core clock transcription factors. Initial findings identified PER proteins in animals and FREQUENCY (FRQ) in Neurospora as key components underlying the main negative feedback loops operating in their respective clocks (Dunlap, 1999). Early models, mostly based on work in Drosophila and Neurospora, suggested that the direct binding of PER or FRQ to their relevant transcription factors was the biochemical mode-of-action underlying these repressors. More recent work is beginning to refine this view and it is now thought that these “repressors” function, at least partly, by acting as molecular bridges to ensure the timely assembly and/or delivery of larger repressor complexes that inhibit elements functioning in the positive arms of circadian transcriptional feedback loops (Cheng et al., 2005; Schafmeier et al., 2005; He et al., 2006; Yu et al., 2006, 2009; Kim et al., 2007; Baker et al., 2009; Chen et al., 2009). Intriguingly, PER and FRQ proteins also share another role in that phosphorylation driven changes in their daily levels are central to setting clock pace (Bae and Edery, 2006; Gallego and Virshup, 2007; Baker et al., 2009). Thus, repressors such as PER and FRQ act as a critical nexus in clock mechanisms by connecting phosphorylation-based biochemical timers to the regulation of transcription, yielding appropriately phased daily cycles in gene expression. How the binding of these period-setting repressors to core clock transcription factors leads to inhibition in transactivation potential is not well understood.
In this study, we identified the C4 region on dPER as the sole or major dCLK binding domain (termed CBD). Despite the inability of dPER(ΔCBD) to bind to dCLK (Fig. 2), p{dper(ΔCBD)} flies manifest quasi-normal feedback circuitry within the core oscillator mechanism (Figs. 4, 7), almost certainly as a result of TIM facilitating the close interaction of dPER with dCLK (Fig. 5), enabling temporal repression of dCLK-CYC-mediated transcription (Figs. 6, 7). Thus, TIM is a bona fide component of the in vivo circadian repressor complex, and presumably plays a role in modulating the interaction between dCLK and dPER. Moreover, we report here that direct binding between dCLK and dPER is not necessary for dPER's repressor activity (Figs. 2, 5, 6), but is likely necessary for normal hyperphosphorylation of dCLK (Fig. 8).
An approach that is providing insights into how these “phospho-timing repressor” clock proteins function is the identity of regions required for promoting transcriptional inhibition. In Drosophila, early work mapped a region on dPER necessary for strong inhibition of dCLK/CYC activity in an S2 cell transcription assay (Chang and Reppert, 2003). This region, termed the CCID domain, contains two highly conserved regions, namely C3 and C4 (Fig. 1A). The C3 region contains the sole or major domain on dPER required for stable interaction with DBT, termed the dPDBD, and is critical not only for hyperphosphorylation of dPER but also for inhibiting dCLK-CYC-mediated transcription, despite the fact that eliminating this region does not abrogate the ability of dPER to stably interact with dCLK in vivo (Kim et al., 2007; Nawathean et al., 2007). On the other hand, the newly identified CBD is required for the physical interaction of dPER with dCLK (Figs. 2, 5). Thus, the CCID is comprised of at least two distinct regions with different biochemical modes-of-action; a region required for physical interaction with dCLK and another that functions as a scaffold to promote binding of regulatory factors that modulate the activity/metabolism of dCLK/CYC.
An unanticipated aspect of our work is that TIM can promote the binding of dPER(ΔCBD) to dCLK in a simplified cell culture system and in flies, an event that rescues dPER's repressor function (Fig. 6). Earlier work suggested that TIM is dispensable for repression of dCLK/CYC transactivity (Rothenfluh et al., 2000; Ashmore et al., 2003; Chang and Reppert, 2003). However, although TIM exhibits little to no repressor activity toward dCLK-CYC-mediated transcription in S2 cells, it enhances that of dPER (Darlington et al., 1998; Rothenfluh et al., 2000; Chang and Reppert, 2003). This enhancement was largely attributed to the fact that TIM stimulates the nuclear localization of dPER (Saez and Young, 1996; Rothenfluh et al., 2000). Our findings suggest a physiological role for TIM in modulating circadian transcription by regulating the interaction between dPER and dCLK. Although dPER can bind dCLK in the absence of TIM (Lee et al., 1998, 1999), it is possible that by interacting with both dPER and dCLK, TIM influences the properties of the repressor complex. For example, TIM might modulate the conformation of dPER, enhancing the assembly/activity of the repressor complex. In support of this view, brief light stimulation in the night leads to subtle but noticeable effects on dper/tim RNA levels that precede significant changes in the abundance of dPER (Lee et al., 1996; Rothenfluh et al., 2000). Of particular interest, dper/tim RNA levels were induced by light exposure in flies expressing a dPER variant (named dPER-ΔC2) missing a short stretch of conserved amino acids (515–568) (Schotland et al., 2000). Although the basis for the impaired function of dPER-ΔC2 was not clear, based on our results it is possible that its interaction with dCLK/CYC is defective in the absence of TIM.
Our findings that TIM has a more pivotal role in transcriptional regulation might also be relevant to a recent study suggesting a two-step mechanism for dPER-mediated inhibition of dCLK-CYC activity, whereby during the first phase of inhibition dPER is bound to dCLK at the chromatin which is followed by a second off-DNA sequestration of dCLK by dPER (Menet et al., 2010). The switch from an on-DNA to an off-DNA mechanism is thought to occur around ZT18, around the time TIM begins to accumulate in the nucleus (Curtin et al., 1995; Menet et al., 2010). We speculate that TIM might regulate progression from an on-DNA to an off-DNA inhibitory mechanism by modulating the interaction between dPER and dCLK/CYC.
Although dPER(ΔCBD) in the presence of TIM can suppress dCLK-CYC-mediated transcription, the efficiency is lower compared with that of wild-type dPER (Figs. 6, 7). Several different scenarios could account for this, including less favorable spatial alignment between dPER(ΔCBD) and dCLK/CYC and/or effects on the ability of dPER(ΔCBD) to bind and/or deliver regulatory factors to dCLK/CYC. Perhaps a more interesting possibility is suggested by the less extensive phosphorylation of dCLK in p{dper(ΔCBD)} flies (Fig. 8). Hyperphosphorylated dCLK is mainly detected in the late night/early day during times when dCLK-CYC transcriptional activity is inhibited, suggesting that highly phosphorylated dCLK is less active (Lee et al., 1998; Kim and Edery, 2006; Yu et al., 2006; Kim et al., 2007). The absence of hyperphosphorylated isoforms of dCLK in p{dper(ΔCBD)} flies suggests that direct association between dPER and dCLK is required to provoke dPER-dependent dCLK phosphorylation. The lack of hyperphosphorylated isoforms of dCLK in p{dper(ΔCBD)} flies might also contribute to the higher overall levels of dper/tim transcripts (Fig. 7). In this context it is noteworthy that FRQ is thought to play a major role in repressing the positive limb of the circadian transcriptional circuits in Neurospora by regulating the phosphorylated state of the WCC complex, the key clock transcription factor driving cyclical gene expression in that system (Schafmeier et al., 2005). Future studies will be required to better understand the biochemical function of dCLK phosphorylation.
In summary, our findings demonstrate that the direct interaction of a key repressor to its target transcription factors is not necessary for its ability to engage in transcriptional inhibition. Moreover, TIM can promote the close association of dPER to dCLK in a manner that sustains dPER's repressor capability, revealing a more direct role for TIM in functional interactions between the negative and positive limbs of the circadian transcriptional feedback circuits operating in Drosophila. The dCLK interaction domain on dPER is situated very close to the dPDBD region that functions, at least partly, by acting as a bridge to promote close interactions between regulatory factors (such as DBT) and the dCLK/CYC complex (Kim et al., 2007). The close spacing on dPER between these two functional regions suggests that complex architectural constraints need to be met to assemble highly efficient repressor complexes. It will be of interest to determine whether other repressors, such as PER proteins in mammals and FRQ in Neursopora, also have similar spatial arrangements. Intriguingly, in mammals the C-terminal region of mPER2, which is downstream of the casein kinase binding (CKB) domain, has been reported to be involved in directly binding to BMAL1 (Chen et al., 2009). Finally, our findings strongly suggest that direct interactions between dPER and dCLK/CYC are required for dCLK hyperphosphorylation. It is possible that some regulatory factors stay tightly bound to key clock repressors (such as DBT to dPER) and thus require very close contact with central clock transcription factors to modulate them, whereas other factors are “delivered” and establishing a high local concentration is sufficient to promote efficient transfer from the repressors to circadian-relevant transcription complexes.
Footnotes
This work was supported by Basic Science Research Program (2008-0058506, NRF-2008-521-C00220) and Chronic Inflammatory Disease Research Center (R13-2003-019) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology. The work was also supported by a National Institutes of Health grant to I.E. (NS03958). We thank Dr. Blau for kindly providing monoclonal anti-PDF antibody.
References
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Ashmore LJ, Sehgal A. A fly's eye view of circadian entrainment. J Biol Rhythms. 2003;18:206–216. doi: 10.1177/0748730403018003003. [DOI] [PubMed] [Google Scholar]
- Ashmore LJ, Sathyanarayanan S, Silvestre DW, Emerson MM, Schotland P, Sehgal A. Novel insights into the regulation of the timeless protein. J Neurosci. 2003;23:7810–7819. doi: 10.1523/JNEUROSCI.23-21-07810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Edery I. Regulating a circadian clock's period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem. 2006;140:609–617. doi: 10.1093/jb/mvj198. [DOI] [PubMed] [Google Scholar]
- Bae K, Lee C, Hardin PE, Edery I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J Neurosci. 2000;20:1746–1753. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell. 2009;34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Más P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Chang DC, Reppert SM. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr Biol. 2003;13:758–762. doi: 10.1016/s0960-9822(03)00286-0. [DOI] [PubMed] [Google Scholar]
- Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Hall JC, Rosbash M. Interspecific comparison of the period gene of Drosophila reveals large blocks of non-conserved coding DNA. EMBO J. 1988;7:3929–3937. doi: 10.1002/j.1460-2075.1988.tb03279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci. 2005;25:5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Edery I. Circadian rhythms in a nutshell. Physiol Genomics. 2000;3:59–74. doi: 10.1152/physiolgenomics.2000.3.2.59. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Hamblen-Coyle MJ, Wheeler DA, Rutila JE, Rosbash M, Hall JC. Behavior of period-altered rhythm mutants of Drosophila in light:dark cycles. J Insect Behav. 1992;5:417–446. [Google Scholar]
- Hardin PE. Essential and expendable features of the circadian timekeeping mechanism. Curr Opin Neurobiol. 2006;16:686–692. doi: 10.1016/j.conb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- He Q, Cha J, He Q, Lee HC, Yang Y, Liu Y. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Edery I. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci U S A. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Ko HW, DiMassa S, Kim EY, Bae K, Edery I. Cis-combination of the classic per(S) and per(L) mutations results in arrhythmic Drosophila with ectopic accumulation of hyperphosphorylated PERIOD protein. J Biol Rhythms. 2007;22:488–501. doi: 10.1177/0748730407306929. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Chen WF, Edery I. Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol. 2004;24:3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Saez L, Young MW. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Price JL, Dembinska ME, Young MW, Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A, Young MW, Saez L. A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron. 2000;26:505–514. doi: 10.1016/s0896-6273(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- Schafmeier T, Haase A, Káldi K, Scholz J, Fuchs M, Brunner M. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Schotland P, Hunter-Ensor M, Lawrence T, Sehgal A. Altered entrainment and feedback loop function effected by a mutant period protein. J Neurosci. 2000;20:958–968. doi: 10.1523/JNEUROSCI.20-03-00958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Sidote D, Majercak J, Parikh V, Edery I. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol. 1998;18:2004–2013. doi: 10.1128/mcb.18.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Price JL, Sehgal A, Saez L, Young MW. Block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Fujii K, Tomioka K. Induction of Drosophila behavioral and molecular circadian rhythms by temperature steps in constant light. J Biol Rhythms. 2007;22:103–114. doi: 10.1177/0748730406298176. [DOI] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]