Abstract
Background
Most type II restriction-modification (RM) systems have two independent enzymes that act on the same DNA sequence: a modification methyltransferase that protects target sites, and a restriction endonuclease that cleaves unmethylated target sites. When RM genes enter a new cell, methylation must occur before restriction activity appears, or the host's chromosome is digested. Transcriptional mechanisms that delay endonuclease expression have been identified in some RM systems. A substantial subset of those systems is controlled by a family of small transcription activators called C proteins. In the PvuII system, C.PvuII activates transcription of its own gene, along with that of the downstream endonuclease gene. This regulation results in very low R.PvuII mRNA levels early after gene entry, followed by rapid increase due to positive feedback. However, given the lethal consequences of premature REase accumulation, transcriptional control alone might be insufficient. In C-controlled RM systems, there is a ± 20 nt overlap between the C termination codon and the R (endonuclease) initiation codon, suggesting possible translational coupling, and in many cases predicted RNA hairpins could occlude the ribosome binding site for the endonuclease gene.
Results
Expression levels of lacZ translational fusions to pvuIIR or pvuIIC were determined, with the native pvuII promoter having been replaced by one not controlled by C.PvuII. In-frame pvuIIC insertions did not substantially decrease either pvuIIC-lacZ or pvuIIR-lacZ expression (with or without C.PvuII provided in trans). In contrast, a frameshift mutation in pvuIIC decreased expression markedly in both fusions, but mRNA measurements indicated that this decrease could be explained by transcriptional polarity. Expression of pvuIIR-lacZ was unaffected when the pvuIIC stop codon was moved 21 nt downstream from its WT location, or 25 or 40 bp upstream of the pvuIIR initiation codon. Disrupting the putative hairpins had no significant effects.
Conclusions
The initiation of translation of pvuIIR appears to be independent of that for pvuIIC. Direct tests failed to detect regulatory rules for either gene overlap or the putative hairpins. Thus, at least during balanced growth, transcriptional control appears to be sufficiently robust for proper regulation of this RM system.
Background
Bacterial type II restriction-modification (RM) systems are abundant in both the bacterial and the archaeal worlds [1]. Many play important roles in defense against phage [2], but they also appear to modulate horizontal gene transfer [3], and to act as "selfish" toxin-antitoxin addiction modules [4,5]. Type II RM systems generally specify separate DNA methyltransferase (MTase) and restriction endonuclease (REase) proteins [6]. Many type II RM systems are on mobile genetic elements, but even chromosomal RM systems can move into new host cells via transduction, transformation or conjugation [7-10].
PvuII is a plasmid-borne type II RM system from the Gram-negative bacterium Proteus vulgaris [11]. The MTase (M.PvuII) modifies the cognate DNA sequence CAGCTG by methylating the internal cytosine [12], generating N4-methylcytosine [13]; while the REase (R.PvuII) cleaves the central GpC if this sequence is unmethylated [13-15]. The REase and MTase act independently of each other in type II RM systems. As a result, strict regulation is needed after the genes enter a new cell in order to delay REase accumulation until the MTase has had time to protect the new host's DNA. The basis for this regulation is unknown for most RM systems.
A subset of type II RM systems are controlled by a third gene, that was designated as "C" (controller) protein when first discovered in the BamHI and PvuII systems [16,17]. Sequence comparisons quickly identified orthologs in the SmaI and EcoRV systems [17], and since then C proteins have been identified (and in some cases confirmed) in a wide variety of other RM systems [18].
The transcriptional regulation of C-controlled RM systems is understood in outline, from the structure of the C proteins [19-22], through their action at conserved "C boxes" upstream of the C genes [7,9,23-27], to their dual function as activators and repressors [24] and possible interaction with RpoD (σ70) [7,20]. The temporal behavior of one of these systems has also been studied, following its introduction into new host cells [28].
The PvuII genes naturally reside on a mobilizable plasmid [8,12,29]. When these genes enter a new host cell with no pre-existing C.PvuII protein, the MTase gene (pvuIIM) is rapidly transcribed from a pair of C-independent promoters [7,28] (Figure 1A). The REase gene (pvuIIR) has no separate promoter, and depends on two promoters upstream of the C gene pvuIIC [7]. One of these two promoters is weak but independent of C.PvuII. The resulting transcript begins 26 nt upstream of the pvuIIC initiation codon [7] (Figure 1A), so translation relies on a ribosome binding site (RBS, also called a Shine-Dalgarno sequence) [30,31]. Because most of our experiments are carried out in E. coli, it is important that the 3' 60 nt of 16 S rRNA, which includes the "anti-Shine-Dalgarno" sequence complementary to the RBS, is identical in E. coli and P. vulgaris (e.g., compare GenBank accession numbers S000629954 with X07652 or J01874).
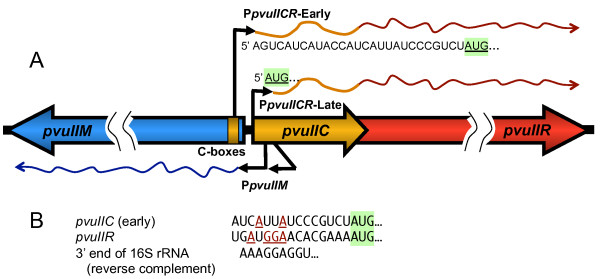
Figure 1.
PvuII transcripts. A. The diagram indicates the expected transcripts early or later after the PvuII genes enter a new host cell. The timing is inferred from in vivo transcript mapping in the presence or absence of active C.PvuII [7], and following synchronous infection of cells with bacteriophage carrying the PvuII genes [28]. "C boxes" are the binding sites for the autogenously-activating C protein. The two pvuIIM (methyltransferase) promoters appear to be constitutive. B. The ribosome-binding (Shine-Dalgarno) sequences are predicted based on location relative to the pvuIIC start codon (shaded green), and comparison to the Logo for ribosome binding sites adapted from [32].
The RBS for pvuIIC is very poor (Figure 1B), based on the very limited similarity to an AGGAGG motif [32]. The expected poor translation initiation would synergize with the lack of transcriptional autogenous activation by C.PvuII [17], by slowing the elongation of any transcripts that did get produced [33]. In contrast, when C.PvuII eventually begins to accumulate, it activates transcription from the second promoter, rapidly boosting transcription of pvuIIC and pvuIIR via a positive feedback loop [7,28]. Furthermore, the resulting C.PvuII-dependent transcript is leaderless [7], beginning at the pvuIIC initiator codon, and is thus independent of the RBS [34-36].
A key area of uncertainty in this model is whether transcription-level regulation is sufficient to protect the cell. Early transcription from the weak C-independent promoter appears to proceed into pvuIIR [28]. If pvuIIR mRNA can be translated well, independent of the rate at which the upstream pvuIIC gene is translated, this might lead to unacceptably rapid early accumulation of restriction activity. Unlike pvuIIC, pvuIIR has an obvious RBS motif (Figure 1B). We accordingly investigated whether there is some form of translational control of pvuIIR.
We were particularly intrigued by two features of the PvuII genes: first, the pvuIIC and pvuIIR genes overlap, and second, pvuIIR is preceded by potential hairpins, one of which would occlude the pvuIIR RBS. The pvuIIC and pvuIIR genes are cotranscribed [9], and many polycistronic transcripts require translation of an upstream gene for efficient translation of a subsequent gene. This phenomenon, resulting from poor loading of free ribosomes onto the internal translation initiator, is called translational coupling, and was first observed in the tryptophan operon of E. coli [37]. Coupling might play an important role in delaying REase accumulation in C-controlled RM systems, relative to that of MTase, after the genes for the system enter a new host cell. The frequent proximity between the C-gene terminators and R-gene initiators in C-protein-controlled RM systems (Figure 2) is suggestive of translational coupling [38,39]. Regarding the hairpins, they were noted in earlier studies, and at least one of them could explain termination products seen in some primer extension experiments [9]. In addition to the overlap and hairpins, in another C-controlled RM system (Eco72I), the C protein itself has an apparent translational regulatory role [25]. Accordingly, we also tested the ability of C.PvuII to affect translation of pvuIICR mRNA in trans.
Figure 2.
Relative Locations of C Terminators and R Initiators of Translation. Selected C.PvuII orthologs that are upstream of known or candidate restriction endonuclease genes were aligned via the endonuclease initiation codons to illustrate the range of relative positions. Names and gene boundaries are available at REBase [1]. Numbers at the left are center-to-center distances between the C gene termination codon and the R gene initiation codon.
Results
Translational lacZ fusions to pvuIIC and pvuIIR
To explore possible translational coupling, we made lacZ translational fusions to the pvuIIC and pvuIIR genes (Figure 3A; Methods). Importantly, in these fusions, transcription is from PlacUV5 and is independent of C.PvuII. Furthermore, the vector's RBS is used, ensuring consistent and efficient translation initiation of pvuIIC.
Figure 3.
pvuII-lacZ transcriptional fusions. A. Sequence from the pvuIIC initiator codon to the pvuIIR-fused lacZ gene. The vector, and source of the polylinker (green) and lacZ gene (blue), is pLex3B (ATCC #87200) [58]. The primers indicated by black arrows and gray shading were used to PCR-amplify pvuIIC and part of pvuIIR; pvuIIR retains its native RBS. The pvuIIC gene includes two unique sites, ClaI and EspI (equivalent to BlpI), at which different null mutations in pvuIIC were generated as previously described [17]. The WT and three different mutants were cloned between the vector XmnI and EcoRI sites such that 'A' of the XmnI site blunt ligated to the 'TG' of the insert on the 5' end to regenerate the pvuIIC initiation codon (under the control of the vector's promoter and RBS); the 3' end ligation used the EcoRI site. In derivatives, synthetic oligonucleotides were inserted between the BglII and EcoRI sites (underlined) to fuse pvuIIC to lacZ or to introduce other changes. The pair of red arrows indicates primers used for mRNA quantitation. B. Oligonucleotides used to alter the pvuIIC-pvuIIR overlap region. A 30 bp oligonucleotide was cloned between the BglII and EcoRI sites (in the sequence shown in Figure 3) to put pvuIIC in-frame with the lacZ gene. In each case, the pvuIIR initiation codon is highlighted in green, and the pvuIIC-frame terminator in red. A unique XbaI site was included to help identify the desired clones and to facilitate shifting the fusion reading frame. A pvuIIC stop codon was introduced in some cases (bottom sequences), to restore it to its native location relative to the pvuIIR initiator. In lines 1 and 2, the pvuIIC-frame terminator is off to the right (not shown).
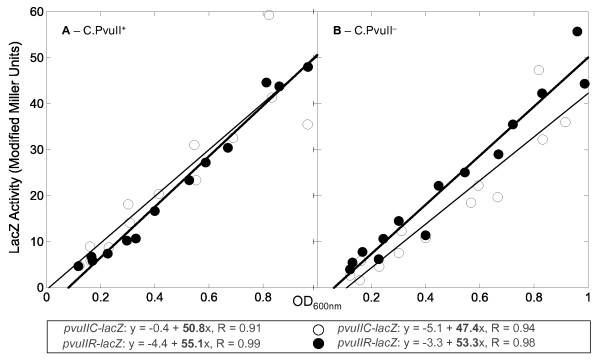
We first measured β-galactosidase activity from WT fusions (see lines 1 and 3 of Figure 3B for junction sequences). Over the course of exponential-phase growth, the plot of activity vs. culture density should be linear if the culture is in balanced growth and the assay is in the linear range. The resulting slopes give a precise measure of relative activity. Using this approach, the level of expression of the WT pvuIIR-lacZ translational fusion was not significantly different from that of the WT pvuIIC-lacZ fusion (Figure 4). Providing WT C.PvuII (or a mutant version) in trans from a second, compatible plasmid had little if any effect on the fusions (Figure 4A versus 4B, open versus closed circles). The four slopes are very similar, giving a combined average ± SE of 51.7 ± 1.7 (i.e., 3.3% SE). This is inconsistent with trans-acting translational effects of C.PvuII protein, unlike the case of C.Eco72I [25].
Figure 4.
Expression of pvuIIC-lacZ and pvuIIR-lacZ translational fusions in the presence and absence of C.PvuII. Cultures of E. coli TOP10 were grown in exponential phase in defined rich medium containing IPTG, and samples were taken at several times for β-galactosidase assays. If the cells are in balanced growth, the plot of activity vs. culture density should be linear. The translational fusions were to pvuIIC (open circles) or pvuIIR (closed circles), and in both cases are transcribed from a C-independent vector promoter. The equations resulting from linear regression are shown. A. A compatible plasmid is providing active C.PvuII in trans. B. As in (A), except the plasmid is providing an inactive version of C.PvuII.
Assessing translational coupling of pvuIIC and pvuIIR
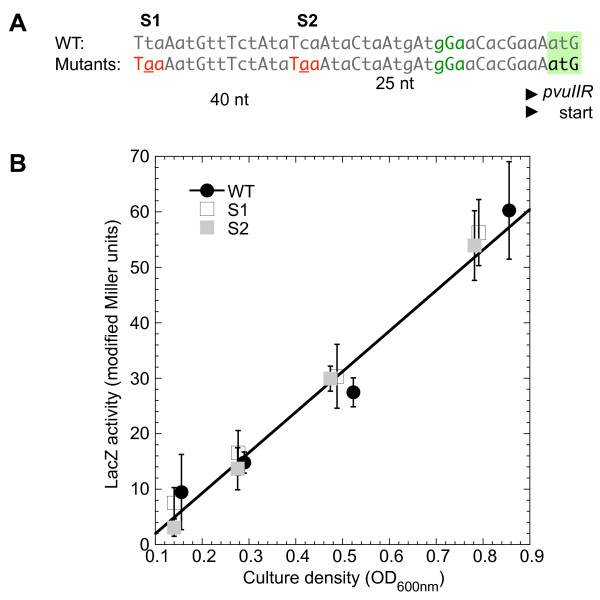
Because coupling is sensitive to the proximity of the termination and start codons of the coupled genes [40], LacZ specific activity was measured in pvuIIR-lacZ fusions with the pvuIIC stop codon at its native site (+20 nt relative to ATG of pvuIIR; Figure 2A) or moved ~20 nt downstream; no effect of this move was seen on translation of pvuIIR-lacZ fusions (not shown). We also generated stop codons 25 or 40 bp upstream of the pvuIIR initiation codon (Figure 5A), in an otherwise-WT pvuIIR-lacZ translational fusion, and measured the LacZ activity (Figure 5B). These stop codons did not detectably affect translation in the pvuIIR reading frame.
Figure 5.
Stop codons created in pvuIIC upstream of pvuIIR-lacZ fusion initiation site. The native pvuIIC translation terminator is downstream of the pvuIIR initiation codon (see top line of Figure 2). A. To test for translational coupling, site-directed mutation was used to introduce pvuIIC terminators farther upstream. The pvuIIR RBS and intiation codon are indicated in green. The two introduced stop codons were in two independent clones. B. lacZ activity of the WT (filled circles) and the mutants (open squares have terminator indicated "S1" in part A; filled squares represent S2). The linear fit is to the WT data.
Nevertheless, previous findings [41] and the results from the WT translational fusions indicate that the translation of pvuIIR is very similar to that of pvuIIC under several conditions, consistent with their translation being coupled. Accordingly we further tested this possibility.
Frameshift mutation in pvuIIC reduces translation of pvuIIR
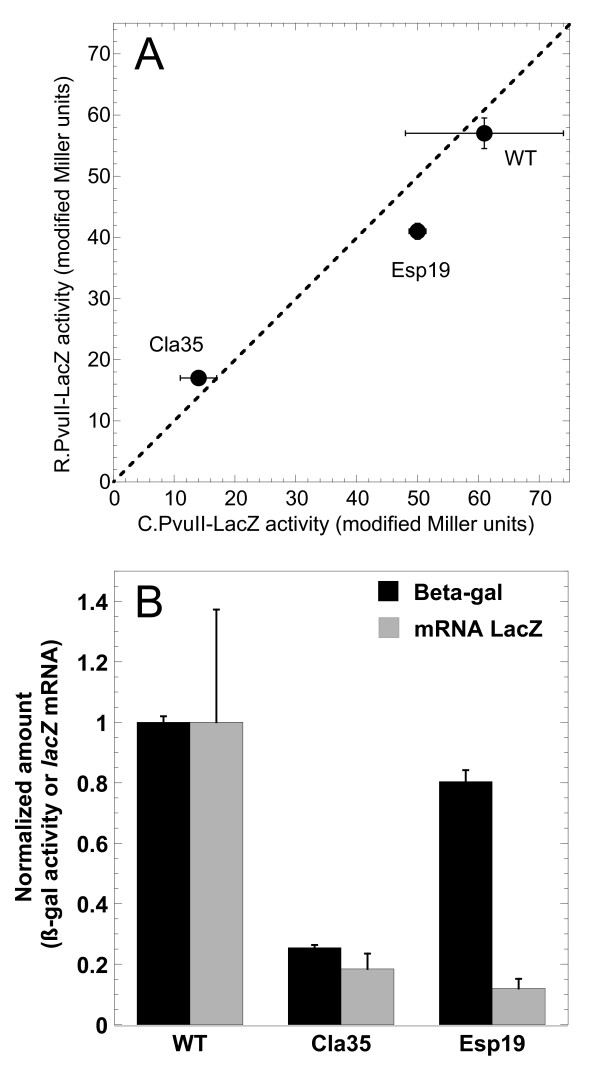
We next determined the effects of mutations within pvuIIC, using our system in which transcription is driven from a promoter unaffected by C.PvuII. WT C.PvuII was supplied from a compatible plasmid in trans. Figure 6A is a correlogram, showing β-galactosidase expression, with the upstream ORF being WT or mutant pvuIIC, and lacZ is fused in the pvuIIC reading frame (x-axis) or in the pvuIIR reading frame (y-axis). The pvuIIC termination codon was, in all cases, in the native location. When pvuIIC was WT (up to the point of the fusion; see Figure 3B), translation was roughly equivalent in both reading frames.
Figure 6.
Effects of pvuIIC mutation. A. Comparison of pvuIIC and pvuIIR translation with WT and mutant variants of pvuIIC. The slopes from triplicate experiments such as that shown in Figure 4 are plotted, showing standard errors (where bars are not visible, errors were smaller than the symbol). In this correlogram, activity from the pvuIIC-lacZ translational fusion is shown on the x-axis, and that from the pvuIIR-lacZ fusion of the same mutant is on the y-axis; if the two fusions for a given variant have equal translation, the point would fall on the dotted line. B. Effects of mutation on mRNA levels and translation activity of pvuIIR-lacZ fusions. Reporter pvuIIR fusions with WT pvuIIC upstream, or with the Cla35 (frameshift) or Esp19 (in-frame) pvuIIC mutations, were grown in triplicate. Real-time RT-PCR was carried out as described in Methods, using the SYBR green method [60] and primers specific to lacZ. Quantitation was based on a standard curve with normalization to recA mRNA. Amounts of mRNA (gray bars) are normalized to the level in the strain carrying WT pvuIIC. The black bars indicate lacZ activity measurements in the same cultures, measured as shown in Figure 4 and normalized to the WT value. Standard errors are shown.
Esp19 is an in-frame insertion of one leucine codon into pvuIIC, within the first helix of the DNA-binding helix-turn-helix motif [41] (see Figure 3A, and Additional file 1, Figure S1). Like the "WT" fusions, the pvuIIC-Esp19 fusions gave similar translational activity in both reading frames. Furthermore, the levels of translation were fairly similar to those when pvuIIC was WT.
Cla35 is a frameshift mutation in pvuIIC that results in a termination codon ~70 nt upstream of the normal stop codon [17] (see Figure 3A, and Additional file 1, Figure S1). The new stop codon is UAG followed by A, which in one in vivo assay system gives a termination rate of about 50% [42]. This mutation led to a roughly fivefold decrease in β-galactosidase expression, relative to the WT fusions, when lacZ was fused in either reading frame. This effect appears to be due to transcriptional polarity (see below), but the important point here is that pvuIIR- and pvuIIC-frame translation changed in parallel.
Quantitative real-time RT-PCR (QRT-PCR) reveals transcriptional polarity
The parallel fivefold drop in pvuIIC and pvuIIR translation in the pvuIIC-Cla35 frameshift mutant (Figure 6A) could reflect premature translation termination leading to premature transcript termination [43,44]. To test this, relative lacZ mRNA levels were determined by QRT-PCR for the WT and both mutant pvuIIR-lacZ fusion strains (see lines 3-5 in Figure 3B). In the case of Cla35, the amount of lacZ mRNA dropped by the same fivefold amount as did translation (Figure 6B), consistent with transcriptional polarity (and, for that matter, with coupling between the rates of pvuIIC and pvuIIR translation).
However, surprisingly, the levels of lacZ mRNA dropped by about the same extent in the strains bearing an in-frame single-codon insertion (Esp19; Figure 6B). The junction sequences of the Cla35 and Esp19 fusions are identical (line 5, Figure 3B). Esp19 only minimally changed the level of pvuIIR-lacZ translation (Figure 6), but this implies that it substantially increased the apparent translational initiation at the downstream pvuIIR initiator, relative to WT, as there is only about an eighth as much mRNA (gray bars in Figure 6B).
Putative hairpins upstream of pvuIIR have no obvious effect on its translation
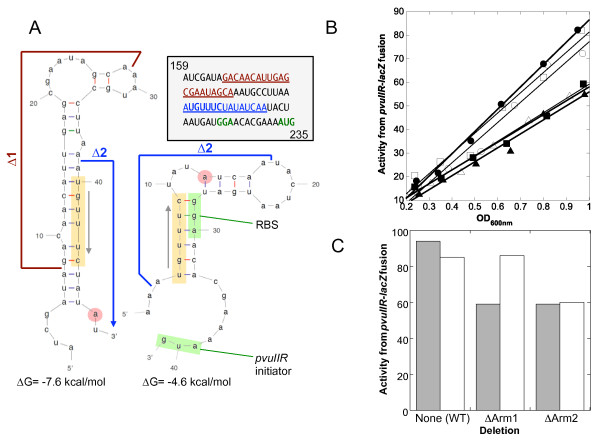
The pvuIIR gene is preceded by two predicted, alternative hairpins ([9]; Figure 7A). Previous primer extension studies [9] were consistent with the presence of the upstream hairpin (left in Figure 7A), as a termination product was seen immediately adjacent to the predicted stem (red circles). These hairpins might regulate translation of pvuIIR, because the downstream hairpin (right side of Figure 7A) would occlude the RBS. Occlusion of an RBS by mRNA secondary structure is a major determinant of translational initiation rates [45-49].
Figure 7.
RNA secondary structure upstream of pvuIIR. A. Putative alternative hairpins in pvuIICR mRNA. The sequence from the pvuIIC gene just upstream of pvuIIR is shown, with numbering corresponding to that in Figure 3A. As previously described [9], the program MFOLD [61] predicts alternative hairpin structures, the downstream one of which would occlude the RBS (structure on right, green highlight). The orange highlighting shows a sequence shared between the two structures, making them mutually exclusive. The red circle indicates the position of a termination product in previous primer extension reactions [7]. The boundaries of two in-frame deletions are indicated. B. Effect of in-frame hairpin arm deletions on translation of pvuIIR. The deletions shown in (A) were introduced into the pvuIIR-lacZ translational fusion, and β-galactosidase activity was measured as described for Figure 4. This was repeated in the presence of WT (closed symbols, gray bars) or null mutant forms of C.PvuII (open symbols, white bars) provided in trans from compatible plasmids. Plot of activity vs. culture density, for WT (circles), arm 1 deletion mutant (squares), or arm 2 deletion mutant (triangles). C. Slopes from (B) are shown to facilitate comparison.
If this model is correct, altering Arm 2 (orange-shaded sequence in Figure 7A) should disrupt both hairpins and thus increase pvuIIR translation, while altering Arm 1 should disrupt the 5' hairpin, promote formation of the 3' hairpin, and thus decrease translation of pvuIIR. We made an in-frame deletion of Arm 2 from the WT construct ("Δ2" in Figure 7A, and Additional file 1, Figure S1) and tested the effects on a pvuIIR-lacZ translational fusion (Figure 7B). Rather than the predicted increase in translation, this deletion reduced expression somewhat. [Supplying WT C.PvuII in trans had no effect (as expected, since a C-independent promoter was being used).] In contrast, the ΔArm1 deletion (Figure 7A, and Additional file 1, Figure S1) was predicted to reduce pvuIIR-lacZ translation, but did so only mildly, and even then only in the presence of WT C.PvuII supplied in trans (Figure 7C).
Discussion
In bacteria such as E coli, genes are often transcribed into polycistronic mRNAs [50]. Ribosomes from an upstream gene can reinitiate translation at the next initiator in a process called translational coupling. The extent of dependence on reinitiation (as opposed to initiation by newly-bound ribosomes) varies, but tends to vary inversely with the distance between the termination and reinitiation codon [51]. In most polycistronic operons, termination codons are close to the initiation codon of the downstream gene; in many cases they even overlap [37], as in the BcnI and AhdI R-M systems (Figure 2A). A high degree of coupling results in coordinated expression of the genes, as illustrated by the gal operon [43,52].
In RM systems that rely on upstream C genes to mediate the delay in REase expression, allowing the MTase time to protect a new host's chromosome [28]; translational coupling could play an important role in the regulatory design. The C and R genes of another C-activated RM system, Esp1396I, are in fact translationally coupled [44]. Furthermore, as shown in Figure 1, translation of pvuIIC from the early pvuIICR transcripts relies on a poor RBS (a feature not shared by most of the C genes). Translational coupling of pvuIIR to pvuIIC would ensure that very little R.PvuII is made from the early transcripts (though in rare cases transcripts lacking RBSs in the leader can still be translated efficiently [53]). Once C.PvuII begins to accumulate, it not only generates a positive feedback loop by activating a second promoter upstream of pvuIIC, but translation of these later transcripts should be much more efficient as they are leaderless [7] and do not rely on the weak RBS.
However our evidence provides no consistent evidence that the translation of pvuIIR is coupled to that of pvuIIC. Most significantly, moving the pvuIIC termination codon in either direction relative to the pvuIIR initiator (Figures 3B, 5) had no apparent effect. In contrast, coupled genes are quite sensitive to the stop-start codon spacing (e.g., see Additional file 1, Figure S2, replotted from data in [40]).
This lack of coupling is consistent with previous work from our laboratory. We had demonstrated a sharp decrease in the expression of pvuIIR in each of four pvuIIC null mutants [17], including two in-frame (EspI) and two frameshift (ClaI) mutants (two of which were used in the current study). These mutants showed greatly-impaired restriction of infection by unmethylated bacteriophage λ, or of transformation by plasmid DNA, and the in vitro R.PvuII activity was ~104-fold less than in strains that carried the intact parental plasmid. Providing pvuIIC in trans had no effect on the WT strain, but providing it to any of the pvuIIC mutants resulted in full restoration of in vivo or in vitro R.PvuII activity. However boosting transcription of frameshift mutants, that terminate translation ~70 nt upstream of the pvuIIR initiation codon (Figure 3A, and Additional file 1, Figure S1), would not have restored a flow of translating ribosomes to the pvuIIR initiator, so these results conflict with coupling.
C proteins themselves can have regulatory effects at the translational level. In the Eco72I RM system [25], providing C.Eco72I in trans to eco72IR-lacZ translational fusions restored REase gene expression even when the upstream eco72IC gene included a frameshift mutation. However, in the case of PvuII, neither pvuIIC-lacZ nor pvuIIR-lacZ translational fusions showed substantial effects of supplying C.PvuII in trans.
Hairpins in the RNA that occlude RBSs can reduce translation initiation [45,47-49], though initiating ribosomes have the capacity to unfold mRNA [54] and may also be prepositioned to slide into place when an RNA hairpin spontaneously unfolds [46]. In-frame deletions in pvuIIC, that would affect predicted alternative hairpins upstream of pvuIIR and thus alter the availability of the pvuIIR RBS (Figure 7A), had no major effect (Figure 7B, C). However, the hairpins might nonetheless reduce the entry of new ribosomes at the pvuIIR RBS, while not affecting progress of translating ribosomes. Searching for potential RBS-occluding hairpins in other C-dependent R-M systems showed that 9/11 had such hairpins with ΔG values ≤ -4.0 kcal/mol (Additional file 1, Figure S3). For comparison, the equivalent PvuII hairpin has a predicted ΔG of -4.6 kcal/mol.
Conclusion
The genes for RM systems are often associated with mobile genetic elements, and temporal control is critical following the entry of these genes into a new cell, to avoid restriction of the host's chromosome. The role of translation-level regulation in this process has not been well studied. In examining the PvuII RM system, we found that translation of the downstream REase gene (pvuIIR) is independent of that of the upstream gene for the autogenous activator/repressor (pvuIIC), at least under the conditions used. This independence was despite the overlapping genes and the presence of putative RNA hairpins involving the pvuIIR RBS. This suggests that the temporal control of PvuII transcription [28], together with the repair capabilities of the bacteria [55-57], is sufficiently robust to protect the new host cell following PvuII gene transfer.
Methods
Cloning and generation of mutants
Table 1 and Additional file 1, Table S1 show, respectively, the plasmids and oligonucleotides that were used in this study. The Cla and Esp mutants of pvuIIC were generated previously by this lab [17]. pPvuRM3.4 contains the WT PvuII R-M system as a 3.4 kb fragment in vector pBR322 [12], and this R-M system includes two unique enzyme sites in the pvuIIC ORF, ClaI and EspI (currently available as the isoschizomer BlpI) (see Figure 3A, and Additional file 1, Figure S1). At the ClaI site, filling in the 5' extensions with Klenow polymerase or digestion using mung bean nuclease, followed by religation, generates frameshift mutations. EspI yields a 3-bp overhang (GC/TNAGC), so fill-in or digestion creates an in-frame mutation [17].
Table 1.
Plasmids used in this study
| Name | Relevant Feature(s) | Reference |
|---|---|---|
| pDK200 | WT pvuIIC gene with its own promoter, ΔpvuIIM, | [7] |
| pDK201 | ΔpvuIIR; TetR, pACYC177 | [7] |
| pDK201-Esp33 | pDK200 with pvuIIC-Esp19 in-frame insertion | This study |
| pDK201-Cla35 | pDK200 with pvuIIC-Esp33 in-frame insertion | This study |
| pPvuIIRM3.4 | pDK200 with pvuIIC-Cla35 frameshift mutation | [12] |
| pPvuIIRM3.4- Esp19 | WT PvuII R-M system; AmpR, pBR322 | [17] |
| pLex3B | pPvuIIRM3.4 with pvuIIC-Esp19 in-frame insertion | [58] |
| Plasmid from ATCC (#87200) used to create translational fusions, to lacZ; AmpR, pMB1 origin |
For this study, DNAs from WT pvuIIC, mutant Esp19 (with insertion of a leucine codon) and mutant Cla35 (frameshift insertion), were purified using the QIAprep plasmid kit (Qiagen, Germantown, MD). Oligonucleotide primers were purchased from Integrated DNA technology company (IDT, Coralville, IA). The amplified region began upstream of the pvuIIC ATG initiator and ended a few codons into pvuIIR (including the entire overlap between pvuIIC and pvuIIR, ~285 bp). High fidelity Pfx polymerase (New England Biolabs, Ipswich, MA) was used to amplify the fragment, which was then purified using a Qiagen gel extraction kit.
The amplified fragments were cloned into TOPO 2.1 vector (Invitrogen, Carlsbad, CA), and ligation products were used to transform TOP-10 chemically competent E. coli (Invitrogen). Transformants were plated onto LB agar with 100 μg/ml ampicillin. Colonies were inoculated into LB broth with appropriate antibiotics, and clones demonstrating the expected restriction pattern were confirmed by sequencing.
LacZ translational fusions
Inserts from sequence confirmed clones in TOPO 2.1 were again amplified using a second set of primers (black arrows in Figure 3A). The secondary amplification began with 'TG' of the ATG initiator of pvuIIC at the 5' end, and extended into pvuIIR (downstream of the pvuIIC termination codon). An EcoRI site was added to the 3' end of the insert. Plasmid pPvuIIRM3.4 carries the WT PvuII RM system in vector pBR322 [12], and pvuIIC contains unique restriction sites (ClaI and EspI/BlpI) that were used to generate mutants [17] (Figure 3A). Wild type and mutant versions of pPvuIIRM3.4 were used as templates. The amplified segments were cloned in-frame with lacZ in vector pLEX3B [58]. PCR amplification used high fidelity Pfx polymerase (New England Biolabs). The EcoRI digested fragment was purified by electrophoresis and the Qiagen gel extraction kit. A similar protocol was followed for all three mutants and the wild type.
We prepared fusions in which the pvuIIC initiation codon was the optimal distance from the RBS of pLEX3B (ATCC # 87200, ATCC, Manassas, VA). Insert DNA was cloned between the XmnI and EcoRI site of the pLEX3B vector, such that on the 5' end 'A' of the XmnI site was blunt ligated to the 'TG' of the insert; the 3' end ligation used the EcoRI site. To facilitate converting these clones from pvuIIC to pvuIIR being in-frame with lacZ, a 30 bp oligonucleotide was acquired with a BglII site at the 5' end (sense strand) and EcoRI at the 3' end (see Figure 3B). In the middle was a unique XbaI site to facilitate identification of the desired clones. This 30 mer duplex was cloned between the BglII and EcoRI sites of the pvuIIR-lacZ fusions (see Figure 3B).
The WT pvuIIC translational fusion contains the full ORF and few codons from pvuIIR in-frame with lacZ. We removed the stop codon from the pvuIIC ORF. In the Cla35 frameshift mutant of pvuIIC, lacZ is in-frame with the shifted pvuIIC reading frame (though there are intervening nonsense codons). Two nucleotides (AG) were deleted to shift the frame, such that pvuIIR was in-frame with lacZ. This shifted the stop codon for pvuIIC by few codons relative to the pvuIIR initiator, so we used site directed mutagenesis to restore the stop codon to its original position by substitution. Primers (IDT) were designed for this purpose, to function with the Stratagene QuickII site-directed mutagenesis protocol and kit (Agilent, Santa Clara, CA).
Ligation products were used for transformation as described above, except that IPTG (1 mM), and X-gal (bromo-chloro-indolyl-galactopyranoside, 40 μg/ml) were also added to the agar. Transformants that turned blue were patched and also inoculated into LB broth with the appropriate antibiotics. Putative lacZ translational fusions were isolated and confirmed by restriction-digestion as well as colony PCR. Clones that demonstrated the desired restriction pattern were confirmed by sequencing.
Hairpin mutants
We prepared in-frame arm1 and arm 2 deletions in the predicted hairpins (Figure 7A) using site-directed mutagenesis kit and protocols as described above. Again this set was confirmed by sequencing (MWG, High Point, NC).
Beta-galactosidase assays
We measured hydrolysis of the substrate ο-nitrophenyl-β-D-galactoside (ONPG). Overnight cultures were grown at 37°C in MOPS rich defined medium (TEKNOVA, Hollister, CA), in the presence of tetracycline (10 μg/ml) and the inducer IPTG (1 mM). This culture was diluted 1:100 into the same medium, without tetracycline, and grown to exponential phase. Samples were collected at regular intervals and the optical density (OD) at 600 nm was measured for every 1 ml sample. Two drops of chloroform and one drop of 0.1% SDS were added, and the sample vortexed for 10 s to open the cells. Samples were placed in a 28°C water bath for 5 min, and 200 μl ONPG (4 mg/ml) were added. Reactions were stopped with 500 μl 1 M Na2CO3, and OD420 nm was determined. The formula of Miller [59] was used to calculate units of β-galactosidase. Miller units were plotted against the culture OD600 nm. The slopes from linear regression indicate relative specific activity; linearity indicates that the culture was in balanced growth.
RNA isolation and quantitation
Cultures were grown as described above, to an OD 600 nm of ~0.3, in the presence of IPTG (1 mM). Samples were taken from the flasks and directly added to two volumes of RNA stabilization buffer (RNA Protect Bacteria Reagent, Qiagen, Valencia, CA). Samples were mixed and left at room temperature for 10 min. The RNeasy miniprep kit (Qiagen) was used to isolate total RNA. Cells in stabilization buffer were harvested by centrifugation at 4°C for 15 min at 5,000 rpm. Pellets were resuspended in 1× TE buffer containing lysozyme (400 μg/ml) after removing the supernatant. Ethyl alcohol was added to precipitate the RNA, which was then dissolved and loaded onto the columns. Following column washing and elution, RNA was treated with RQ1 RNAse-free DNAse (Promega, Madison, WI) to remove DNA. cDNA was synthesized using total RNA as template, random hexamers (Invitrogen), and ImPromII reverse transcriptase (Promega). The random primers were annealed at 25°C for 5 min, and the first strand was then extended at 42°C for 1 h. The reverse transcriptase was then inactivated by heating at 70°C for 1 h.
Red arrows in Figure 3A indicate the primer pair used to quantitate pvuIIC cDNA. Dilutions of cDNA were tested to determine the maximally-efficient concentration for amplification, and also the efficiency of each primer pair. recA expression was also measured, to provide an internal baseline. Cycle threshold (Ct) values were determined using a Roche Light Cycler with SYBR Green. Melting curve analysis was used to confirm the formation of specific products. The standard curve method was used to determine the relative amounts of mRNA, which were normalized to recA [60].
Authors' contributions
MK carried out the strain constructions and experimental analysis, participated in the experimental design, and drafted the manuscript. RB conceived of the study, participated in its design and coordination, helped to draft the manuscript, and generated the figures. Both authors read and approved the final manuscript.
Supplementary Material
Supplemental Information. Figure S1. Location of mutations relative to features of C.PvuII. The amino acid sequence of C.PvuII is shown in single-letter code (GenBank AAA96335.1). The DNA-binding helix-turn-helix motif contains the EspI (BlpI) site; fill-in or resection followed by religation duplicates or removes the leucine in the "SLA" sequence. Orange shading indicates the subunit interface, from comparison to the crystal structure for an orthologous C protein [20]. This interface spans the ClaI site used to generate frameshift mutations. The two in-frame deletions of predicted RNA hairpin elements (see Figure 7A) are indicated as Δ1 and Δ2. The position of the alternate reading frame pvuIIR initiation codon is indicated. Figure S2. Effects of distance between termination and initiation codons on translational reinitiation. This is a replotting of data from [40]. The lacZ gene was introduced into E. coli with varied spacing between the lacZ initiation codon and the terminator for the upstream ORF. Figure S3. Putative RNA hairpins upstream of REase gene initiation codon in selected C-dependent R-M systems. C-dependent R-M systems, from among those shown in Figure 2, were analyzed for potential hairpin structures upstream of the REase gene initiation codon. The 40 nt upstream of each initiator were submitted to MFOLD [61], and the most stable structure returned in each case is shown. The number in parentheses is the predicted ΔG of folding, in kcal/mol. The Esp1396I structure may be further stabilized by its GNRA loop [62].The initiator codon and predicted RBS are highlighted in green. Secondary structures are shown for comparison, whether or not they are likely to be present a significant fraction of the time.
Contributor Information
Meenakshi K Kaw, Email: Meenakshi.Kaw@utoledo.edu.
Robert M Blumenthal, Email: Robert.Blumenthal@utoledo.edu.
Acknowledgements
We thank Drs. Robert Lintner and Iwona Mruk for help with the real-time QRT-PCR, and Dr. Mruk for critically reviewing the manuscript. MKK was supported in part by graduate stipends from the University of Toledo and the Department of Medical Microbiology & Immunology. This material is based upon work supported by the US National Science Foundation under Grant No. MCB-0964728.
References
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010. pp. D234–236. [DOI] [PMC free article] [PubMed]
- Releigh EA, Brooks JE. Restriction Modification systems: where they are and what they do. Chapman & Hall, New York, N.Y; 1998. [Google Scholar]
- Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29(18):3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichige A, Kobayashi I. Stability of EcoRI restriction-modification enzymes in vivo differentiates the EcoRI restriction-modification system from other postsegregational cell killing systems. J Bacteriol. 2005;187(19):6612–6621. doi: 10.1128/JB.187.19.6612-6621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc Natl Acad Sci USA. 1998;95(11):6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K. et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31(7):1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowle D, Lintner RE, Touma YM, Blumenthal RM. Nature of the promoter activated by C.PvuII, an unusual regulatory protein conserved among restriction-modification systems. J Bacteriol. 2005;187(2):488–497. doi: 10.1128/JB.187.2.488-497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderer M, Brust JR, Knowle D, Blumenthal RM. Mobility of a restriction-modification system revealed by its genetic contexts in three hosts. J Bacteriol. 2002;184(9):2411–2419. doi: 10.1128/JB.184.9.2411-2419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijesurier RM, Carlock L, Blumenthal RM, Dunbar JC. Role and mechanism of action of C. PvuII, a regulatory protein conserved among restriction-modification systems. J Bacteriol. 2000;182(2):477–487. doi: 10.1128/JB.182.2.477-487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Abe K, Kobayashi I. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res. 2010;38(7):2428–2443. doi: 10.1093/nar/gkp1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras TR, Greenough L, Schildkraut I, Roberts RJ. Two new restriction endonucleases from Proteus vulgaris. Nucleic Acids Res. 1981;9(18):4525–4536. doi: 10.1093/nar/9.18.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal RM, Gregory SA, Cooperider JS. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J Bacteriol. 1985;164(2):501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus V, Klimasauskas S, Petrauskiene L, Maneliene Z, Lebionka A, Janulaitis A. Interaction of AluI, Cfr6I and PvuII restriction-modification enzymes with substrates containing either N4-methylcytosine or 5-methylcytosine. Biochim Biophys Acta. 1987;909(3):201–207. doi: 10.1016/0167-4781(87)90078-9. [DOI] [PubMed] [Google Scholar]
- Rice MR, Blumenthal RM. Recognition of native DNA methylation by the PvuII restriction endonuclease. Nucleic Acids Res. 2000;28(16):3143–3150. doi: 10.1093/nar/28.16.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice MR, Koons MD, Blumenthal RM. Substrate recognition by the Pvu II endonuclease: binding and cleavage of CAG5mCTG sites. Nucleic Acids Res. 1999;27(4):1032–1038. doi: 10.1093/nar/27.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JE, Nathan PD, Landry D, Sznyter LA, Waite-Rees P, Ives CL, Moran LS, Slatko BE, Benner JS. Characterization of the cloned BamHI restriction modification system: its nucleotide sequence, properties of the methylase, and expression in heterologous hosts. Nucleic Acids Res. 1991;19(4):841–850. doi: 10.1093/nar/19.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J Bacteriol. 1991;173(4):1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin V, Severinov K, Gelfand MS. Systematic prediction of control proteins and their DNA binding sites. Nucleic Acids Res. 2009;37(2):441–451. doi: 10.1093/nar/gkn931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Zhu Z, Mersha F, Chan SH, Dabur R, Xu SY, Balendiran GK. Crystal structure of the restriction-modification system control element C.Bcll and mapping of its binding site. Structure. 2005;13(12):1837–1847. doi: 10.1016/j.str.2005.08.017. [DOI] [PubMed] [Google Scholar]
- McGeehan JE, Streeter SD, Papapanagiotou I, Fox GC, Kneale GG. High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila. J Mol Biol. 2005;346(3):689–701. doi: 10.1016/j.jmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Ball N, Streeter SD, Kneale GG, McGeehan JE. Structure of the restriction-modification controller protein C.Esp1396I. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 9):900–905. doi: 10.1107/S0907444909020514. [DOI] [PubMed] [Google Scholar]
- McGeehan JE, Streeter SD, Thresh SJ, Ball N, Ravelli RB, Kneale GG. Structural analysis of the genetic switch that regulates the expression of restriction-modification genes. Nucleic Acids Res. 2008;36(14):4778–4787. doi: 10.1093/nar/gkn448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeehan JE, Papapanagiotou I, Streeter SD, Kneale GG. Cooperative binding of the C.AhdI controller protein to the C/R promoter and its role in endonuclease gene expression. J Mol Biol. 2006;358(2):523–531. doi: 10.1016/j.jmb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Mruk I, Rajesh P, Blumenthal RM. Regulatory circuit based on autogenous activation-repression: roles of C-boxes and spacer sequences in control of the PvuII restriction-modification system. Nucleic Acids Res. 2007;35(20):6935–6952. doi: 10.1093/nar/gkm837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimseliene R, Vaisvila R, Janulaitis A. The eco72IC gene specifies a trans-acting factor which influences expression of both DNA methyltransferase and endonuclease from the Eco72I restriction-modification system. Gene. 1995;157(1-2):217–219. doi: 10.1016/0378-1119(94)00794-S. [DOI] [PubMed] [Google Scholar]
- Semenova E, Minakhin L, Bogdanova E, Nagornykh M, Vasilov A, Heyduk T, Solonin A, Zakharova M, Severinov K. Transcription regulation of the EcoRV restriction-modification system. Nucleic Acids Res. 2005;33(21):6942–6951. doi: 10.1093/nar/gki998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanova E, Zakharova M, Streeter S, Taylor J, Heyduk T, Kneale G, Severinov K. Transcription regulation of restriction-modification system Esp1396I. Nucleic Acids Res. 2009;37(10):3354–3366. doi: 10.1093/nar/gkp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk I, Blumenthal RM. Real-time kinetics of restriction-modification gene expression after entry into a new host cell. Nucleic Acids Res. 2008;36:2581–2593. doi: 10.1093/nar/gkn097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvin Koons MD, Blumenthal RM. Characterization of pPvu1, the autonomous plasmid from Proteus vulgaris that carries the genes of the PvuII restriction-modification system. Gene. 1995;157(1-2):73–79. doi: 10.1016/0378-1119(94)00618-3. [DOI] [PubMed] [Google Scholar]
- Shultzaberger RK, Bucheimer RE, Rudd KE, Schneider TD. Anatomy of Escherichia coli ribosome binding sites. J Mol Biol. 2001;313(1):215–228. doi: 10.1006/jmbi.2001.5040. [DOI] [PubMed] [Google Scholar]
- Jin H, Zhao Q, Gonzalez de Valdivia EI, Ardell DH, Stenstrom M, Isaksson LA. Influences on gene expression in vivo by a Shine-Dalgarno sequence. Mol Microbiol. 2006;60(2):480–492. doi: 10.1111/j.1365-2958.2006.05110.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Niimura Y, Miura K, Gojobori T. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc Natl Acad Sci USA. 2010;107(14):6382–6387. doi: 10.1073/pnas.1002036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328(5977):504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Hirokawa G, Kiel MC, Kaji A, Blasi U. Translation initiation with 70 S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004;32(11):3354–3363. doi: 10.1093/nar/gkh663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell SM, Janssen GR. Leaderless mRNAs bind 70 S ribosomes more strongly than 30 S ribosomal subunits in Escherichia coli. J Bacteriol. 2002;184(23):6730–6733. doi: 10.1128/JB.184.23.6730-6733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Shimizu Y, Ueda T. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J Biol Chem. 2004;279(10):8539–8546. doi: 10.1074/jbc.M308784200. [DOI] [PubMed] [Google Scholar]
- Oppenheim DS, Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95(4):785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex G, Surin B, Besse G, Schneppe B, McCarthy JE. The mechanism of translational coupling in Escherichia coli. Higher order structure in the atpHA mRNA acts as a conformational switch regulating the access of de novo initiating ribosomes. J Biol Chem. 1994;269(27):18118–18127. [PubMed] [Google Scholar]
- Inokuchi Y, Hirashima A, Sekine Y, Janosi L, Kaji A. Role of ribosome recycling factor (RRF) in translational coupling. EMBO J. 2000;19(14):3788–3798. doi: 10.1093/emboj/19.14.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Guchte M, Kok J, Venema G. Distance-dependent translational coupling and interference in Lactococcus lactis. Mol Gen Genet. 1991;227(1):65–71. doi: 10.1007/BF00260708. [DOI] [PubMed] [Google Scholar]
- Tao T, Blumenthal RM. Sequence and characterization of pvuIIR, the PvuII endonuclease gene, and of pvuIIC, its regulatory gene. J Bacteriol. 1992;174(10):3395–3398. doi: 10.1128/jb.174.10.3395-3398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole ES, Brown CM, Tate WP. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14(1):151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D, Howard BH, Rosenberg M. Efficient expression of Escherichia coli galactokinase gene in mammalian cells. Proc Natl Acad Sci USA. 1982;79(2):257–261. doi: 10.1073/pnas.79.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesnaviciene E, Mitkaite G, Stankevicius K, Janulaitis A, Lubys A. Esp1396I restriction-modification system: structural organization and mode of regulation. Nucleic Acids Res. 2003;31(2):743–749. doi: 10.1093/nar/gkg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA. 1990;87(19):7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit MH, van Duin J. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J Mol Biol. 2003;331(4):737–743. doi: 10.1016/S0022-2836(03)00809-X. [DOI] [PubMed] [Google Scholar]
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324(5924):255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SW, Yang J, Jung GY. Quantitative correlation between mRNA secondary structure around the region downstream of the initiation codon and translational efficiency in Escherichia coli. Biotechnol Bioeng. 2009;104(3):611–616. doi: 10.1002/bit.22431. [DOI] [PubMed] [Google Scholar]
- Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juarez K, Contreras-Moreira B. et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One. 2009;4(10):e7526.. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner BE, Belagaje RM, Schoner RG. Translation of a synthetic two-cistron mRNA in Escherichia coli. Proc Natl Acad Sci USA. 1986;83(22):8506–8510. doi: 10.1073/pnas.83.22.8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D, McKenney K, Sobieski DA, Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982;30(3):865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Skorski P, Leroy P, Fayet O, Dreyfus M, Hermann-Le Denmat S. The highly efficient translation initiation region from the Escherichia coli rpsA gene lacks a shine-dalgarno element. J Bacteriol. 2006;188(17):6277–6285. doi: 10.1128/JB.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer SM, Joseph S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell. 2006;22(1):105–115. doi: 10.1016/j.molcel.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Heitman J, Ivanenko T, Kiss A. DNA nicks inflicted by restriction endonucleases are repaired by a RecA- and RecB-dependent pathway in Escherichia coli. Mol Microbiol. 1999;33(6):1141–1151. doi: 10.1046/j.1365-2958.1999.01556.x. [DOI] [PubMed] [Google Scholar]
- Heitman J, Zinder ND, Model P. Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc Natl Acad Sci USA. 1989;86(7):2281–2285. doi: 10.1073/pnas.86.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayot R, Montagne B, Mazel D, Ricchetti M. An end-joining repair mechanism in Escherichia coli. Proc Natl Acad Sci USA. 2010;107(5):2141–2146. doi: 10.1073/pnas.0906355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederich L, Roth A, Messer W. A versatile plasmid vector system for the regulated expression of genes in Escherichia coli. Biotechniques. 1994;16(5):916–923. [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39(1):75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information. Figure S1. Location of mutations relative to features of C.PvuII. The amino acid sequence of C.PvuII is shown in single-letter code (GenBank AAA96335.1). The DNA-binding helix-turn-helix motif contains the EspI (BlpI) site; fill-in or resection followed by religation duplicates or removes the leucine in the "SLA" sequence. Orange shading indicates the subunit interface, from comparison to the crystal structure for an orthologous C protein [20]. This interface spans the ClaI site used to generate frameshift mutations. The two in-frame deletions of predicted RNA hairpin elements (see Figure 7A) are indicated as Δ1 and Δ2. The position of the alternate reading frame pvuIIR initiation codon is indicated. Figure S2. Effects of distance between termination and initiation codons on translational reinitiation. This is a replotting of data from [40]. The lacZ gene was introduced into E. coli with varied spacing between the lacZ initiation codon and the terminator for the upstream ORF. Figure S3. Putative RNA hairpins upstream of REase gene initiation codon in selected C-dependent R-M systems. C-dependent R-M systems, from among those shown in Figure 2, were analyzed for potential hairpin structures upstream of the REase gene initiation codon. The 40 nt upstream of each initiator were submitted to MFOLD [61], and the most stable structure returned in each case is shown. The number in parentheses is the predicted ΔG of folding, in kcal/mol. The Esp1396I structure may be further stabilized by its GNRA loop [62].The initiator codon and predicted RBS are highlighted in green. Secondary structures are shown for comparison, whether or not they are likely to be present a significant fraction of the time.