Abstract
The GLUT4 gene is subject to complex tissue-specific and metabolic regulation, with a profound impact on insulin-mediated glucose disposal. We have shown, by using transgenic mice, that the human GLUT4 promoter is regulated through the cooperative function of two distinct regulatory elements, domain 1 and the myocyte enhancer factor 2 (MEF2) domain. The MEF2 domain binds transcription factors MEF2A and MEF2D in vivo. Domain I binds a transcription factor, GLUT4 enhancer factor (GEF). In this report, we show a restricted pattern of GEF expression in human tissues, which overlaps with MEF2A only in tissues expressing high levels of GLUT4, suggesting the hypothesis that GEF and MEF2A function together to activate GLUT4 transcription. Data obtained from transiently transfected cells support this hypothesis. Neither GEF nor MEF2A alone significantly activated GLUT4 promoter activity, but increased promoter activity 4- to 5-fold when expressed together. Deletion of the GEF-binding domain (domain I) and the MEF2-binding domain prevented activation, strengthening the conclusion that promoter regulation occurs through these elements. GEF and MEF2A, isolated from nuclei of transfected cells, bound domain I and the MEF2 domain, respectively, which is consistent with activation through these regulatory elements. Finally, GEF and MEF2A coimmunoprecipitated in vivo, strongly supporting a mechanism of GLUT4 transcription activation that depends on this protein–protein interaction.
GLUT4 is one member of a family of glucose transport proteins and is principally responsible for insulin-mediated glucose uptake in muscle and adipose tissue. Manipulation of GLUT4 levels in transgenic mice has revealed that glucose homeostasis is highly sensitive to the level of GLUT4 expression (1–3). Furthermore, It has been demonstrated in transgenic mice that a diabetic phenotype can be alleviated by increasing expression of GLUT4 (4–6). These data suggest that regulation of the GLUT4 gene may serve as a potential intervention point for treatment of type II diabetes. To this end, it is important to clarify the molecular basis underlying GLUT4 gene regulation.
The GLUT4 gene undergoes a complex program of gene regulation in vivo, being subject to both tissue-specific and hormonal/metabolic regulation. GLUT4 mRNA is largely restricted to brown and white adipose tissue, and skeletal and cardiac muscle. Small amounts ofGLUT4 mRNA and protein have been detected in specialized cell types of other tissues. Changes in GLUT4 expression are observed in physiologic states of altered glucose homeostasis. In general, GLUT4 mRNA expression is down-regulated in states of relative insulin deficiency such as streptozotocin (STZ)-induced diabetes (7). GLUT4 gene expression varies in a tissue-specific manner. For example, GLUT4 mRNA expression levels change more rapidly in adipose tissue compared with skeletal muscle in response to STZ-induced diabetes (8). In addition, GLUT4 mRNA increases with exercise training but decreases during insulin deficiency (9–11), and these changes are due to alterations in the transcription rate (12, 13). Understanding the regulation of GLUT4 transcription may lead to new insights into the control of genes expressed in other highly differentiated tissues.
By using transgenic mice, we have shown that cis-acting elements regulating transcription of the human Glut4 promoter are contained within 895 base pairs upstream of the initiation site (14–16). This region contains two nonoverlapping regulatory domains, each required for maximum transcription from the human GLUT4 promoter. The region referred to as the myocyte enhancer factor 2 (MEF2) domain binds isoforms of the MEF2 family of transcription factors (15, 17). A second region, referred to as domain I, binds a transcriptional activator recently cloned in our laboratory, and is named GLUT4 enhancer factor (GEF) (14). Data obtained in our laboratory strongly suggest that both tissue specificity and down-regulation of the GLUT4 gene during STZ-induced diabetes are controlled through these two elements (14, 15). In addition, transcriptional activation of the GLUT4 gene after chronic activation of AMP-kinase requires these two regulatory elements (18). The mechanism of regulation of the GLUT4 gene may have implications for transcription promoters of other genes responding to complex physiologic stimuli.
In this report, we provide further evidence implicating GEF in GLUT4 transcription. Our experiments confirm and extend, in a cell-culture model, previous results observed in transgenic animals. In addition, experiments in transiently transfected cell cultures demonstrate GEF and MEF2A function together to activate GLUT4 transcription. A physical interaction between GEF and MEF2A is demonstrated by coimmunoprecipitation of GEF and MEF2A proteins expressed cell culture. Finally, we characterize GEF with respect to its tissue distribution, and propose a model in which both GEF and MEF2A are required for efficient expression of GLUT4.
Materials and Methods
Northern Blot Analysis. A commercially prepared multiple tissue Northern blot filter (Clontech) loaded with 2 μg of poly(A) RNA per lane was probed with a random-primed cDNA probe (Invitrogen) corresponding to the 3′ end of GEF (GEFdb), according to manufacturer's specifications. The blot was then stripped and reprobed by using a random-primed cDNA probe corresponding to actin.
Immunoprecipitation and Western Blot. Cells and tissues were extracted with a total protein extraction reagent (T-PER, Pierce) and were quantitated by using Coomassie plus protein assay reagent (Pierce). Total lysate (50 μg) was fractionated by SDS/PAGE using 10% acrylamide gels, transferred to PVDF membranes, and labeled with antibodies as indicated. Antiserum against the GST-GEF (amino acids 1–161) fusion protein was prepared commercially in rabbits (Cocalico Biologicals, Reamstown, PA). IgG proteins were purified from preimmune rabbit serum and GST-GEF antiserum by protein G affinity chromatography (Pierce). α-MEF2A monoclonal and polyclonal antibodies (Santa Cruz Biotechnology) and α-MEFD antibody (Transduction Laboratories, Lexington, KY) were purchased. Immunoreactive proteins were visualized by enhanced chemiluminescence (SuperSignal, Pierce).
Preparation of Nuclear Extracts by Electrophoretic-Mobility Shift Assay (EMSA). Nuclear extract-transfected Cos 7 cells were prepared by using a nuclear protein extraction kit (NE-PER, Pierce). Total protein was measured by using Coomassie Plus protein assay reagent (Pierce) and aliquots were stored at –70°C. EMSA was performed as described (14).
Preparation of Fusion Proteins and Coprecipitations. GST fusion proteins were expressed in Escherichia coli strain, BL-21, which contained plasmids encoding fusion proteins. Cultures expressing GST-GEF were incubated with 100 μM Zn acetate. Affinity purification of fusion proteins was carried out by using a glutathione-Sepharose columm (Amersham Bioscience, Piscataway, NJ) for GST fusion proteins. Purity of the fusion proteins was assessed by SDS/PAGE and Coomassie staining.
For pull-down experiments, equimolar amounts (35 pmol each) of GST and full-length GST-GEF protein (attached to glutathione agarose beads) were mixed with nuclear extracts (200 μg total protein) from Cos cells overexpressing MEF2A or MEF2D in 1 ml of binding buffer (25 mM Tris·HCl, pH 7.5/20 μM Zn acetate/1 mM DTT/1 mM MgCl2/40 mM KCl) and rocked end over end for 1 h at 4C. The beads were washed three times in TBS containing 0.5% Nonidet P-40 and two times in TBS. Protein complexes were eluted in Laemmli buffer, fractionated by SDS/PAGE, and analyzed by Western blot.
Coimmunoprecipitation experiments were performed in nuclear extracts (1 mg of protein) prepared from Cos cells overexpressing both GEF and either MEF2A or MEF2D by using either nonimmune rabbit IgG or affinity-purified anti-GEF antibody, as described above. Extracts were incubated with 10 μg of antibody in 1 ml of binding buffer (25 mM Tris·HCl, pH 7.5/20 μM Zn acetate/1 mM MgCl2/40 mM KCl) and rocked end over end for 15 h at 4C. Antigen–antibody complexes were captured on protein AG-Plus agarose beads (Santa Cruz Biotechnology). The beads were washed three times in TBS. Protein complexes were eluted in Laemmli buffer, fractionated by SDS/PAGE, and analyzed by Western blot.
Transient Transfections and Reporter Assays. Transient transfections were performed on subconfluent COS cells by using FuGENE 6 (Roche Applied Science), according to manufacturer's specifications. Total transfected DNA was held constant at 2 μg by the addition of plasmid vector (pCDNA). Human GLUT4 promoter expression was measured as firefly luciferase activity by using a Femtomaster FB 12 luciferometer (Zylux, Maryville, TN). Human GLUT4 reporter constructs were cotransfected with a plasmid control for transfection efficiency containing Renilla luciferase expressed from the HSV-TK promoter (pRL-TKluc, Promega). Additional plasmids encoding potential transactivators were added to cotransfections, as described in the legends to Figs. 3, 4, and 6. Genes encoding GEF, MEF2A, MEF2C, and MEF2D were cloned into plasmid vectors downstream of the human cytomegalovirus immediate early promoter. Expression plasmids encoding MEF2 isoforms were a generous gift from E. Olson (University of Texas Southwestern Medical Center, Dallas). Dual luciferase assays were performed according to the manufacturer's specifications (Promega).
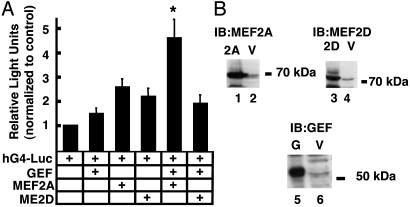
Fig. 3.
Transcription from the human GLUT4 promoter in a cell-culture system. (A) COS 7 cells were transfected according to Materials and Methods. Each transfection included 500 ng of the human GLUT4 promoter fused to firefly luciferase (hG4-luc), 100 ng of a plasmid used to control transfection efficiency (pRLTk-luc), and 500 ng of various combinations of plasmids encoding transcription factors GEF and/or members of the MEF2 family. Light units expressed from the firefly luciferase gene (GLUT 4 promoter) were corrected for light units expressed from the sea pansy luciferase gene (transfection efficiency reporter, pRLTk-luc). Data were analyzed by using two-way ANOVA. *, statistically significant (P < 0.05) interaction between factors (GEF and MEF2A). (B) Immunoblots of lysates obtained from COS 7 cells transfected with plasmids encoding transcription factors GEF (G), MEF2A (2A), MEF2D (2D), or empty vector (V). Transfected cell lysates were probed by using corresponding antibodies specific for MEF2A, MEF2D, and GEF, respectively.
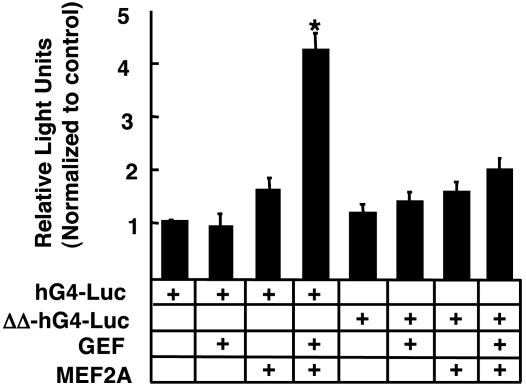
Fig. 4.
Transcription of human GLUT4 reporter (hG4-luc) or mutant GLUT4 promoter (ΔΔ-hG4-luc) containing deletions of the GEF- and MEF2-binding sites. Transfections were carried out as described in Materials and Methods and Fig. 3. Differences between the reporter constructs were determined with a Student's t test. *, significant difference (P < 0.01).
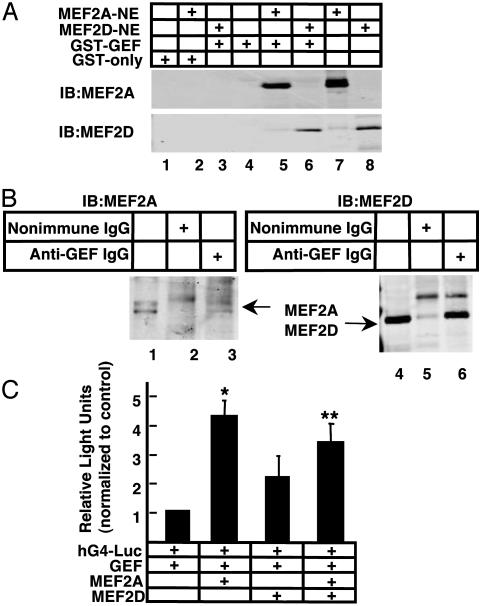
Fig. 6.
Coprecipitation of GEF and MEF2A in vitro and in vivo. (A) Purified GST-GEF or GST alone were incubated with nuclear extracts from Cos cells expressing either MEF2A or MEF2D. GST fusion proteins were captured by using glutathione agarose beads and washed, and the associated proteins were fractionated by SDS/10% PAGE and Western blot analysis by using anti-MEF2A polyclonal or anti-MEF2D antibody. Lane 7 is 25% of input of the MEF2A-containing lysate and lane 8 is 10% input of the MEF2D-containing lysate. (B) Coimmunoprecipitation of MEF2A and MEF2D using anti-GEF IgG in nuclear extracts expressing exogenous GEF and MEF2A or GEF and MEF2D. As a control for specificity, lysates were precipitated with nonimmune IgG (lanes 2 and 5). Antigen–antibody complexes were captured by using protein A/G plus agarose, washed, and fractionated on SDS/10% PAGE. A sample of each lysate representing 10% of the sample was loaded in lanes 1 and 4 of each Western blot. Blots were labeled with mouse monoclonal antibodies specific for MEF2A or MEF2D. (C) COS 7 cells were transfected as described in Fig. 3. Plasmids encoding GEF, MEF2A, or MEF2D were transfected at a combined total of 500 ng. The data were analyzed by two-way ANOVA. **, the contribution of MEF2A (P < 0.0001); *, significance of the interaction term (MEF2A and MEF2D) (P = 0.0186).
Cell Fixation and Confocal Microscopy. Full-length GEF cDNA was inserted in-frame in pEGFP-C1 expression plasmid (Clontech) to generate a fusion protein with GEF at the C terminus of the enhanced green fluorescent protein (GEF-EGFP). Cos cells were transfected with plasmid DNA GEF-GFP or GFP only. After 24 h, cells fixed 10 min at room temperature (RT) in a solution containing 2% paraformaldehyde, 0.1% gluteraldehyde, 2 mM MgCl2, 0.1 M Pipes, pH 6.8, and 20 mM EGTA. Fixed cells were washed again three times for 5 min with PBS and permeabilized with a solution of 0.5% Triton X-100 in PBS for 10 min at RT. Three washes of 10 min each in 2.5 mg/ml NaBH4 dissolved in 50% ethanol were performed to quench free aldehyde groups. Cells were washed again three times for 5 min with PBS and the nuclei were stained by incubation for 15 min at RT in a solution of 2 μM ToPro 3 (Molecular Probes) diluted in PBS. Cells were washed again as before, rinsed briefly with water, and mounted under coverslips by using ProLong antifade reagent (Molecular Probes). Z sections (12–15 per sample) of ≈0.25 m were obtained for each sample by using a Leica TNS confocal microscope. Images were compiled and analyzed by using image j or leica lcs lite software.
Results
In a previous article (14), we reported cloning the DNA-binding domain of a novel human GLUT4 transcriptional activating protein, named GEF. To obtain the full-length GEFcDNA, we screened 2 × 106 phage containing fragments of a human skeletal muscle library (Stratagene), by using a probe derived from the original GEF clone (GEFdb). Five clones that contained at least 300 base pairs of 5′ end absent in the original GEFbd clone were isolated and sequenced. To confirm that the newly cloned 5′ end was contiguous with the original cDNA, we performed Southern blot analysis by using human genomic DNA (Clontech) digested with either EcoRI or BamHI. Probes from the GEFdb cDNA and the newly cloned 5′ region hybridized with identical restriction fragments of human genomic DNA on Southern blots, indicating that the previously isolated 3′ end and the newly cloned 5′ end resided on the same gene (see Fig. 7, which is published as supporting information on the PNAS web site). The sequence of the full-length GEF is available from GenBank (accession no. AF249267).
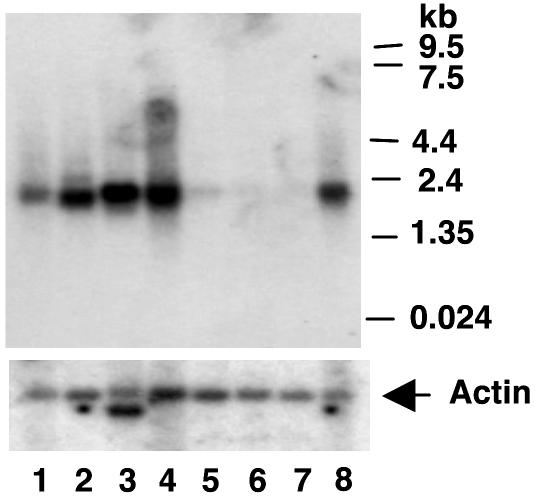
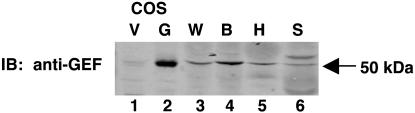
The human tissue distribution and size of the full-length mRNA encoding GEF were determined by using a GEF cDNA probe (Fig. 1). The GEFdb cDNA probe hybridized to a single band of RNA corresponding to ≈1.9 kb. The pattern of GEF mRNA expression correlated with the pattern of GEF protein binding to domain I by using EMSA analysis (14), and was restricted to heart, skeletal muscle, liver, kidney, and pancreas (Fig. 1). No GEF mRNA was evident in lung, placenta, or brain. Results in Fig. 1 were confirmed and extended by Western blot analysis of GEF protein in mouse tissues (Fig. 2). GEF protein was detected in heart and skeletal muscle, which was consistent with the RNA data. In addition, GEF was detected in white and brown adipose tissues (Fig. 2). The combined data from Figs. 1 and 2 demonstrate GEF expression in all insulin-responsive tissues. The tissue distribution of MEF2A is distinct from, but overlaps with, expression of GEF. The overlap between MEF2A and GEF in the major GLUT4-expressing tissues (15, 19).
Fig. 1.
Distribution of GEF mRNA using a human multitissue Northern blot probed with a radiolabeled GEFdb cDNA. The blot was stripped and reprobed with an actin probe to control for loading. The commercially obtained Northern blot (Clontech) was loaded with mRNA from the following tissues: lane 1, pancreas; lane 2, kidney; lane 3, skeletal muscle; lane 4, liver; lane 5, lung; lane 6, placenta; lane 7, brain; and lane 8, heart.
Fig. 2.
GEF protein distribution in mouse. Fifty micrograms of mouse white adipose tissue (W), brown adipose tissue (B), heart (H), and skeletal muscle (S) lysates were immunoblotted by using antiserum raised against the N-terminal 161 amino acids of GEF. Control lanes 1 and 2 were obtained from COS 7 cells transfected with vector plasmid (V) or full length GEF cDNA (G).
Evidence obtained from transgenic mice has demonstrated two critical elements within the human GLUT4 promoter conferring regulation of transcription. These results suggested the possibility of recapitulating GLUT4 transcriptional activation in a cell-culture system by focusing on these two regions and their cognate binding proteins, GEF and MEF2 isoforms. To this end, we constructed a firefly luciferase reporter expressed from the full-length human GLUT4 promoter (hG4-luc). COS 7 cells were transfected with hG4-Luc in conjunction with plasmids expressing GEF, MEF2 isoforms, or combinations of both. A control plasmid expressing the sea pansy luciferase was included to correct for efficiency of transfection. Light units obtained by using lysates from cells transfected with hG4-Luc alone were corrected for efficiency of transfection and assigned a value of one, providing a comparison point for other experimental conditions. Fig. 3A summarizes the results of these experiments. Values shown in Fig. 3 represent a minimum of five independent transfections for each experimental condition. Cotransfection of hG4-Luc with plasmids expressing GEF, MEF2A, or MEF2D alone resulted in slight increases in luciferase expression of 1.5-, 2.6-, and 2.2-fold, respectively, indicating that each of these proteins is capable of transactivating the GLUT4 promoter, albeit at low levels. Cotransfections of plasmids encoding both MEF2A and GEF resulted transactivation of the human GLUT4 promoter at a level significantly greater than that for either factor alone (P > 0.05, by using two-way ANOVA for interaction). In contrast, cotransfection of GEF and MEF2D did not significantly increase GLUT4 promoter function, indicating that this effect is specific for the MEF2A isoform. Results similar to those observed for MEF2D were obtained with MEF2C (data not shown).
To confirm that plasmids encoding the transactivators were expressing proteins, particularly in cases where cotransfection of the plasmid had no effect on transcription from hG4-Luc, Western blotting of transfected whole-cell lysates was performed (Fig. 3B). These results demonstrate that plasmids encoding GEF, MEF2A, and MEF2D all expressed proteins of the expected size, reacting with their cognate antibodies.
The implication of the experiments described above is that transactivation of the GLUT4 promoter by GEF and MEF2A occurs through the DNA elements first identified in transgenic mice. To directly address this issue, we deleted the GEF- and MEF-binding sites from the GLUT4 promoter (ΔΔ-hG4-Luc), and performed cotransfections by using this reporter in conjunction with plasmids encoding GEF and MEF2A. Experiments summarized in Fig. 4 demonstrate that deletion of the two DNA elements identified in mice results in a GLUT4 promoter that is severely deficient in its ability to be transactivated by GEF plus MEF2A.
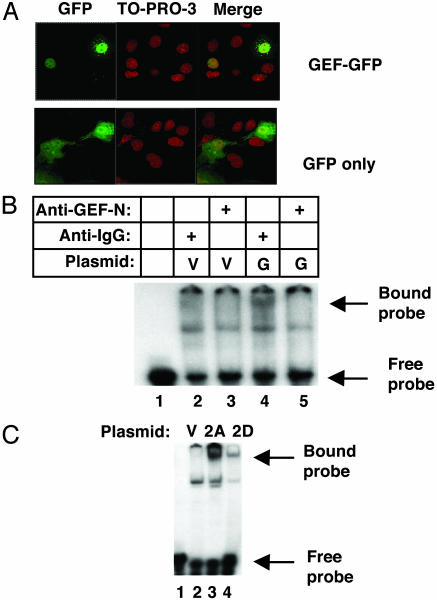
Consistent with a role for GEF as an activator of transcription, we observed nuclear localization of GEF by two different methods. First, we transfected a DNA construct in which GFP was fused in-frame with the N terminus of GEF (pGEF-GFP). Fig. 5A depicts images of COS7 cells transfected with either pGEF-GFP or a plasmid expressing GFP only. Cells were fixed and treated with ToPro3 (Molecular Probes) to delineate the boundary of the nucleus. These experiments show clearly that the GEF-GFP chimera is concentrated within the nucleus, which is in contrast to the virtually uniform distribution of GFP throughout the cell. This result is not unexpected, because the deduced amino acid sequence of GEF contains a putative nuclear localization signal. Experiments using a biochemical approach confirm and extend immunofluorescence experiments, demonstrating nuclear localization of GEF. Fig. 5 B and C depict data obtained from EMSA, using protein samples isolated from nuclei of COS7 cells transfected with plasmids encoding GEF and MEF isoforms (the same constructs used in Fig. 3). Nuclear protein extracts were incubated with radiolabeled probes consisting of the GEF- or the MEF2-binding sites, and followed by electrophoresis on nondenaturing gels. Comparison of cells transfected with GEF cDNA (G) versus the parent cloning vector (V) control revealed a labeled band corresponding to the protein-bound GEF probe, which is visible near the top of the gel (Fig. 5B, lanes 2 and 4). To confirm that this band corresponds to a nuclear protein complex containing GEF, control and GEF-expressing extracts were preincubated with either nonimmune IgG or anti-GEF IgG. Bound anti-GEF antibody inhibited the GEF–DNA complex from entering the gel (Fig. 5B, compare lanes 4 and 5). These results demonstrate a direct interaction between the exogenous protein and the DNA motif known to bind GEF, as well as to confirm the previous finding that exogenous GEF is localized to the nucleus.
Fig. 5.
Nuclear localization of transfected GEF and MEF2. A plasmid encoding GFP-GEF or GFP alone was transfected into COS 7 cells. Fixed cells expressing the protein were scanned by using confocal microscopy, and Z sections were compiled to obtain the images shown in A. To-Pro-3 (Molecular Probes) was included during staining to demarcate the nucleus. EMSA of lysates obtained from cells transfected with plasmids expressing vector only (V), GEF (G), MEF2A (2A), and MEF2D (2D) were performed with radiolabeled oligonucleotides corresponding to the domain I-binding site (B) or GLUT4 MEF2-binding site (C). Lysates in B were incubated with anti-GEF IgG (lanes 3 and 5), or with a nonspecific IgG (lanes 2 and 4).
Similar gel shift experiments were carried out by using the MEF2 site. In these experiments, exogenously expressed MEF2A and MEF2D extracted from nuclei were able to bind a MEF2-specific probe (Fig. 5C). Thus, the recombinant proteins were capable of entering the nucleus and of binding to their specific promoter element.
Our data from transgenic mice (14, 15) and from the transient transfections in tissue culture cells (Figs. 3 and 4) demonstrate that GEF and MEF2A cooperate in the regulation of GLUT4 transcription, but the nature of their interaction is unknown. The simplest mechanism to explain cooperation is direct protein–protein interaction resulting in formation of a GEF–MEF2A complex required for transcription activation. To begin to test this hypothesis, we expressed GEF in bacterial cells and determined whether or not the purified proteins could directly interact with MEF2A or MEF2D in vitro. GEF, fused in-frame to GST (GST-GEF), attached to glutathione agarose beads, was incubated with nuclear extracts from Cos cells overexpressing either MEF2A or MEF2D. Agarose-bound GEF and interacting proteins were immunoblotted by using anti-MEF2A or anti-MEF2D antibody. Results of these coprecipitation experiments are shown in Fig. 6A. The position of the MEF2A protein on the blot is visible in lane 7, which shows 25% of total input of nuclear extract used in the coprecipitation. The position of MEF2D protein on the blot is visible in lane 8, which represents 10% of input of nuclear extract used in the coprecipitation. GST alone did not result in binding of any MEF2-immunoreactive material (Fig. 6A, lanes 2 and 3). In contrast, binding of MEF2A and MEF2D to GST-GEF was observed (Fig. 6A, lanes 5 and 6). These results indicate that GEF binds to both MEF2A and MEF2D in vitro, and strongly suggests potential interactions between GEF and MEF2A and/or MEF2D in vivo.
To determine whether MEF2A and MEF2D isoforms interact with GEF in vivo, we performed coimmunoprecipitation assays by using an affinity-purified antibody raised against the N terminus of GEF with extracts from Cos cells expressing GEF and MEF2A, or GEF and MEF2D (Fig. 6B). Anti-GEF IgG was able to coimmunoprecipitate both MEF2A (Fig. 6B, lane 3) and MEF2D (Fig. 6B, lane 6). Rabbit IgG did not coimmunoprecipitate either MEF2 isoform (Fig. 6B, lanes 2 and 5). An aliquot of lysate (10% of input) was loaded in lanes 1 and 4 (Fig. 6B) to show the position in the gel.
Because MEF2D does not activate transcriptional activity of the human GLUT4 promoter, but can bind the MEF2-binding site, we carried out cotransfection experiments to determine whether MEF2D can affect transcriptional activity mediated by GEF and MEF2A (Fig. 6C). In these experiments, hG4-Luc was cotransfected with a plasmid expressing GEF and plasmids expressing MEF2A, MEF2D, or both. As before, expression of MEF2A, together with GEF, resulted in a 4.3-fold increase (P < 0.0001, two-way ANOVA) in luciferase reporter activity, whereas expression of MEF2D plus GEF increased reporter activity 2.2-fold (a value identical to MEF2D alone, as shown in Fig. 3). When MEF2D and MEF2A were cotransfected together with GEF, reporter activity was increased only 3.4-fold over hG4-Luc alone. This result was significantly reduced, compared with activation by MEF2A plus GEF (P = 0.0186, using two-way ANOVA for interaction). In this experimental system, MEF2D interferes with the transcriptional activation promoted by the cooperative function between MEF2A and GEF.
Discussion
This article provides evidence supporting the hypothesis that GEF is a bona fide transcription factor, regulating expression of the human GLUT4 gene, in cooperation with MEF2A. An interaction between GEF and MEF2 was first suggested by experiments in transgenic mice, where deletion of either the MEF2-binding site or the binding site of an unknown protein (later identified as GEF) ablated mRNA expression from the human GLUT4 promoter (14, 15, 19). The distributions of GEF and MEF2 mRNA and protein, and their comparison with GLUT4 expression, led us to hypothesize that both GEF and MEF2 are required for expression of high levels of GLUT4, and that neither transcription factor alone is sufficient to support GLUT4 transcription at maximum levels.
Our data are consistent with the hypothesis that GEF and MEF2 are the primary proteins regulating GLUT4 transcription: both are necessary and neither alone is sufficient for optimal transactivation of GLUT4. This hypothesis presupposes expression of both GEF and MEF2 in all major GLUT4-expressing tissues. The major GLUT4-expressing tissues are heart, skeletal muscle, brown adipose tissue, and white adipose tissue (20–22). We have demonstrated the presence of GEF mRNA in human heart and skeletal muscle, as well as liver, pancreas, and kidney, and have detected GEF protein in mouse heart, skeletal muscle, brown adipose tissue, and a small amount in white adipose tissue. Ubiquitously expressed MEF2-related mRNAs accumulate preferentially in human skeletal muscle, heart, and brain, but have been detected at low levels in placenta, lung, and kidney, by using a probe specific for the conserved MADS domain (19). This finding is notable, because kidney represents the only example we are aware of, in which mRNAs coding for both GEF and MEF2 can be detected in a tissue that does not express high levels of GLUT4 (small amounts of GLUT4 have, however, been detected in kidney; ref. 23). Kidney represents a nonhomogeneous tissue type, which is composed of a diverse cell population. It is difficult, therefore, to determine conclusively whether a subpopulation of kidney cells is expressing GEF, MEF2, and high levels of GLUT4, whereas the majority of cells in the tissue are not. MEF2A mRNA and protein have both been detected in heart, skeletal muscle, and white and brown adipose tissue, whereas MEF2C mRNA has been detected in primarily in skeletal muscle and brain (24). The MEF2D has been shown to have a ubiquitous distribution, whereas the distribution of MEF2C mRNA does not include adult heart or adipose tissue. These findings led us to focus on the 2A and 2D isoforms in transient transfection studies. By using nuclear extracts from skeletal muscle tissue, MEF2C did not form a complex with either MEF2A or MEF2D and did not bind to the MEF2-binding site found in the human GLUT4 promoter (15, 17). MEF2C was shown to transactivate a GLUT4 promoter/reporter construct when overexpressed with the transcriptional coactivator, PGC-1 (25). The physiologic significance of this result has been called into question because overexpression of PGC-1 did not upregulate GLUT4 expression in transgenic mice (26).
Our experiments demonstrating transcription activation of the human GLUT4 promoter in a cell type not normally expressing GLUT4, and the observation that the highest levels of activation occur only in the presence of both GEF and MEF2A, provide the strongest evidence in support of our hypothesis. In transfected COS7 cells, GEF or MEF2 isoforms were able to stimulate transcription only slightly or not at all, whereas GEF in conjunction with MEF2A, but not MEF2C or MEF2D, significantly increased expression from the human GLUT4 promoter (Figs. 3 and 4). This increase was greater than simply additive, suggesting that some type of synergism is occurring between the two proteins. These results strongly suggest that the 2A isoform of MEF cooperates with GEF to regulate transcription of GLUT4, but does not conclusively rule out the participation of other currently unidentified factors. Identification of these factors, if any, awaits further experimentation.
Results from our studies indicate that when MEF2D is cotransfected together with MEF2A and GEF, GLUT4 transcriptional activity is reduced (Fig. 6C); however, when MEF2D and GEF are cotransfected together, there is a small increase in GLUT4 transcriptional activity (Fig. 3). These two observations are consistent with a model in which MEF2D competes with MEF2A in activation of the GLUT4 promoter. Because MEF2D binds the GLUT4 MEF2-binding site (Fig. 5C), and binds GEF (Fig. 6B), MEF2D appears to be able to attenuate the formation of a functional complex composed of MEF2A, GEF, and their respective DNA-binding sites. This finding may have important implications for the regulation of the human GLUT4 promoter in vivo. We and others (15, 27) have shown that levels MEF2A, but not MEF2D, are decreased in STZ-induced diabetes. In the diabetic state, the ratio of MEF2D to MEF2A is higher than normal, and formation of a less active transcription complex consisting mainly of GEF and MEF2D may be responsible in part for decreased transcription of GLUT4 observed under this physiologic state. Further studies to investigate this model are warranted.
GLUT4 gene transcription increases with exercise (13). Exercise has been shown to increase MEF2 transcriptional activity (28), raising the possibility that MEF2A transcriptional activation of the GLUT4 promoter occurs via a calcium-dependent pathway. An increase in intracellular Ca levels in skeletal muscle correlates with increases in MEF2A and MEF2D, and GLUT4 protein levels (29); however, a direct effect of Ca on the GLUT4 promoter activation has not been made. Chronic activation of AMP kinase does increase GLUT4 MEF2-binding activity in skeletal muscle (18, 30), which is consistent with the idea that MEF2A is involved in regulating GLUT4 gene expression.
Studies of other gene promoters with functional MEF2-binding domains indicate that, in many cases, MEF2 isoforms require at least one additional transcription factor to regulate transcription. For example, the desmin gene has an MEF2-binding site that is necessary for cardiac and skeletal muscle transcription (31). However, when this MEF2 site is used to promote expression of a reporter gene in transgenic mice, the reporter is detected in skeletal muscle, but not cardiac tissue, indicating that the MEF2-binding site of the desmin promoter is necessary, but is not sufficient, for expression in adult cardiac tissue (32). This finding is analogous to our observations that the MEF2 site will support transcription of a GLUT4 reporter in skeletal muscle, but it is not sufficient to promote transcription in heart or adipose tissue (16). The functional GLUT4 promoter requires not only the MEF2 site but also the domain I-binding site as well, suggesting that the transcription factors binding to these elements have a functional interaction (14, 15).
Direct interaction between GEF and MEF2A, resulting in recruitment of RNA polymerase activity, is the simplest model available to begin to explain their cooperative effect. Consistent with this idea, we demonstrated a direct interaction between GEF and both MEF2A and MEF2D, in an in vitro-binding assay and coimmunoprecipitation (Fig. 6). Direct MEF2A–GEF interaction does not preclude involvement of other factors, or rule out activities such as DNA unwinding by GEF, MEF2A, or unidentified factors in the transcription complex. Other proteins found in differentiated 3T3-L1 adipocyte nuclear extracts including NF1 and Olf-1/early B cell factor have been found to bind to domain I of the mouse GLUT4 promoter (33, 34). Unlike GEF, these proteins have not been shown to transactivate the GLUT4 promoter, but appear instead to mediate the downregulation of GLUT4 promoter activity observed in that cell line (33, 34). Transactivation of the rat GLUT4 promoter in 3T3-L1 adipocytes and C2C12 myoblasts has been shown occur by overexpression of the Kruppel-like factor, KLF15; however, it is not clear whether this factor directly binds to the GLUT4 promoter, and its mechanism of action is unknown (35).
In summary, we have demonstrated that a protein called GEF, obtained by screening a human library for proteins binding to domain I of the human GLUT4 promoter, transactivates the GLUT4 promoter in vitro. We show that this transcription factor, when transiently expressed in cultured cells, binds to the domain 1-regulatory region of the human GLUT4 promoter. Furthermore, GEF functions cooperatively with MEF2A to stimulate transcription, recapitulating the regulation of the human GLUT4 promoter observed in transgenic mice. Finally, we show that the MEF2A proteins and GEF can be specifically coprecipitated in vitro, strongly suggesting that the mechanism for cooperative function between these transcription factors results from direct protein–protein interaction. The nature of the cooperation between GEF and MEF2A in regulation of GLUT4 transcription, particularly with respect to their DNA- and protein-binding domains, are areas of investigation for the future. Comparison of data from the in vitro cell-culture system with data from transgenic mice is likely to be a productive avenue of investigation into the mechanism of GLUT4 promoter regulation. It is likely that a greater understanding of GLUT4 transcription will lead to a better understanding of diabetes and may lead to a useful therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Quwanza S. Duggins and Pablo Ceres for excellent technical assistance. This work was supported by grants from the Juvenile Diabetes Foundation and National Institutes of Health Grant DK62341 (to A.L.O.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GEF, GLUT4 enhancer factor; MEF, myocyte enhancer factor; EMSA, electrophoretic mobility-shift assay.
References
- 1.Tsao, T. S., Burcelin, R., Katz, E. B., Huang, L. & Charron, M. J. (1996) Diabetes 45, 28–36. [DOI] [PubMed] [Google Scholar]
- 2.Treadway, J. L., Hargrove, D. M., Nardone, N. A., McPherson, R. K., Russo, J. F., Milici, A. J., Stukenbrok, H. A., Gibbs, E. M., Stevenson, R. W. & Pessin, J. E. (1994) J. Biol. Chem. 269, 29956–29961. [PubMed] [Google Scholar]
- 3.Shepherd, P. R., Gnudi, L., Tozzo, E., Yang, H., Leach, F. & Kahn, B. B. (1993) J. Biol. Chem. 268, 22243–22246. [PubMed] [Google Scholar]
- 4.Gibbs, E. M., Stock, J. L., McCoid, S. C., Stukenbrok, H. A., Pessin, J. E., Stevenson, R. W., Milici, A. J. & McNeish, J. D. (1995) J. Clin. Invest. 95, 1512–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leturque, A., Loizeau, M., Vaulont, S., Salminen, M. & Girard, J. (1996) Diabetes 45, 23–27. [DOI] [PubMed] [Google Scholar]
- 6.Tozzo, E., Gnudi, L. & Kahn, B. B. (1997) Endocrinology 138, 1604–1611. [DOI] [PubMed] [Google Scholar]
- 7.Olson, A. L. & Knight, J. B. (2003) Front. Biosci. 8, 5401–5409. [DOI] [PubMed] [Google Scholar]
- 8.Richardson, J. M., Balon, T. W., Treadway, J. L. & Pessin, J. E. (1991) J. Biol. Chem. 266, 12690–12694. [PubMed] [Google Scholar]
- 9.Ren, J. M., Semenkovich, C. F., Gulve, E. A., Gao, J. & Holloszy, J. O. (1994) J. Biol. Chem. 269, 14396–14401. [PubMed] [Google Scholar]
- 10.Kawanaka, K., Tabata, I., Katsuta, S. & Higuchi, M. (1997) J. Appl. Physiol. 83, 2043–2047. [DOI] [PubMed] [Google Scholar]
- 11.Host, H. H., Hansen, P. A., Nolte, L. A., Chen, M. M. & Holloszy, J. O. (1998) J. Appl. Physiol. 84, 798–802. [DOI] [PubMed] [Google Scholar]
- 12.Gerrits, P. M., Olson, A. L. & Pessin, J. E. (1993) J. Biol. Chem. 268, 640–644. [PubMed] [Google Scholar]
- 13.Neufer, P. D., Carey, J. O. & Dohm, G. L. (1993) J. Biol. Chem. 268, 13824–13829. [PubMed] [Google Scholar]
- 14.Oshel, K. M., Knight, J. B., Cao, K. T., Thai, M. V. & Olson, A. L. (2000) J. Biol. Chem. 275, 23666–23673. [DOI] [PubMed] [Google Scholar]
- 15.Thai, M. V., Guruswamy, S., Cao, K. T., Pessin, J. E. & Olson, A. L. (1998) J. Biol. Chem. 273, 14285–14292. [DOI] [PubMed] [Google Scholar]
- 16.Olson, A. L. & Pessin, J. E. (1995) J. Biol. Chem. 270, 23491–23495. [DOI] [PubMed] [Google Scholar]
- 17.Mora, S. & Pessin, J. E. (2000) J. Biol. Chem. 275, 16323–16328. [DOI] [PubMed] [Google Scholar]
- 18.Zheng, D., MacLean, P. S., Pohnert, S. C., Knight, J. B., Olson, A. L., Winder, W. W. & Dohm, G. L. (2001) J. Appl. Physiol. 91, 1073–1083. [DOI] [PubMed] [Google Scholar]
- 19.Yu, Y. T., Breitbart, R. E., Smoot, L. B., Lee, Y., Mahdavi, V. & Nadal-Ginard, B. (1992) Genes Dev. 6, 1783–1798. [DOI] [PubMed] [Google Scholar]
- 20.James, D. E., Strube, M. & Mueckler, M. (1989) Nature 338, 83–87. [DOI] [PubMed] [Google Scholar]
- 21.Charron, M. J., Brosius, F. C., III, Alper, S. L. & Lodish, H. F. (1989) Proc. Natl. Acad. Sci. USA 86, 2535–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birnbaum, M. J. (1989) Cell 57, 305–315. [DOI] [PubMed] [Google Scholar]
- 23.Marcus, R. G., England, R., Nguyen, K., Charron, M. J., Briggs, J. P. & Brosius, F. C., III (1994) Am. J. Physiol. 267, F816–F824. [DOI] [PubMed] [Google Scholar]
- 24.Martin, J. F., Schwarz, J. J. & Olson, E. N. (1993) Proc. Natl. Acad. Sci. USA 90, 5282–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michael, L. F., Wu, Z., Cheatham, R. B., Puigserver, P., Adelmant, G., Lehman, J. J., Kelly, D. P. & Spiegelman, B. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3820–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, J., Wu, H., Tarr, P. T., Zhang, C.-Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., et al. (2002) Nature 418, 797–801. [DOI] [PubMed] [Google Scholar]
- 27.Mora, S., Yang, C., Ryder, J. W., Boeglin, D. & Pessin, J. E. (2001) Endocrinology 142, 1999–2004. [DOI] [PubMed] [Google Scholar]
- 28.Wu, H., Rothermal, B., Kanatous, S., Rosenberg, P., Naya, F. J., Shelton, J. M., Hutcheson, K. A., DiMaio, J. M., Olson, E. N., Bassel-Duby, R. & Williams, R. S. (2001) EMBO J. 20, 6414–6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ojuka, E. O., Jones, T. E., Nolte, L. A., Chen, M., Wamhoff, B. R., Sturek, M. & Holloszy, J. O. (2002) Am. J. Physiol. 282, E1008–E1013. [DOI] [PubMed] [Google Scholar]
- 30.Song, X. M., Fielder, M., Galuska, D., Ryder, J. W., Fernstrom, M., Chibalin, A. V., Wallber-Henriksson, H. & Zierath, J. R. (2002) Diabetologia 45, 56–65. [DOI] [PubMed] [Google Scholar]
- 31.Kusik, I. R., Li, H., Tran, D. & Capetanaki, Y. (1996) Dev. Biol. 174, 1–13. [DOI] [PubMed] [Google Scholar]
- 32.Naya, F. J., Wu, C., Richardon, J. A., Overbeek, P. & Olson, E. N. (1999) Development (Cambridge, U.K.) 126, 2045–2052. [DOI] [PubMed] [Google Scholar]
- 33.Cooke, D. W. & Lane, M. D. (1999) J. Biol. Chem. 274, 12917–12924. [DOI] [PubMed] [Google Scholar]
- 34.Dowell, P. & Cooke, D. W. (2002) J. Biol. Chem. 277, 1712–1718. [DOI] [PubMed] [Google Scholar]
- 35.Gray, S., Feinberg, M. W., Hull, S., Kuo, C. T., Watanabe, M., Banerjee, S. S., DePina, A., Haspel, R. & Jain, M. K. (2002) J. Biol. Chem. 277, 34322–34328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.