Abstract
This critical review evaluates progress toward viable point-of-care protein biomarker measurements for cancer detection and diagnostics. The ability to measure panels of specific, selective cancer biomarker proteins in physicians’ surgeries and clinics has the potential to revolutionize cancer detection, monitoring, and therapy. The dream envisions reliable, cheap, automated, technically undemanding devices that can analyze a patient’s serum or saliva in a clinical setting, allowing on-the-spot diagnosis. Existing commercial products for protein assays are reliable in laboratory settings, but have limitations for point-of-care applications. A number of ultrasensitive immunosensors and some arrays have been developed, many based on nanotechnology. Multilabel detection coupled with high capture molecule density in immunosensors and arrays seems to be capable of detecting a wide range of protein concentrations with sensitivity ranging into the sub pg mL−1 level. Multilabel arrays can be designed to detect both high and ultralow abundance proteins in the same sample. However, only a few of the newer ultrasensitive methods have been evaluated with real patient samples, which is key to establishing clinical sensitivity and selectivity.
1. Introduction
Broadly defined, biomarkers for cancer consist of any measurable or observable factors in a patient that indicate normal or disease-related biological processes or responses to therapy.1,2 Biomarkers for disease thus encompass physical symptoms, mutated DNAs and RNAs, secreted proteins, processes such as cell death or proliferation, and serum concentrations of small molecules such as glucose or cholesterol. In this review, we focus on serum levels of proteins as biomarkers that can be used as indicators of the onset, existence or progression of cancer.3 In addition to early detection of cancer, measurement of panels of protein biomarkers holds enormous potential for directing personalized cancer therapy and treatment monitoring.4 However, these applications have yet to be broadly realized in a form that can be readily adapted to point-of-care.5,6 This critical review is aimed mainly at assessing progress toward these goals.
A realizable hope to decrease deaths from cancer and improve therapeutic outcome for patients may be offered by earliest possible detection coupled with new targeted drug delivery therapies featuring personalized biomarker-based monitoring.7,8 Protein biomarkers can be measured in serum and tissue for early cancer detection,9–14 although reliable methodologies have been established clinically for only a handful of biomarkers.1 The poster child of protein cancer biomarkers is prostate specific antigen (PSA),15 which began its career as a clinical biomarker for prostate cancer several decades ago. PSA in serum is the only protein biomarker currently recommended by the American Cancer Society as an early cancer screening tool.16 The danger zone for PSA serum concentration is 4 to 10 ng mL−1, a level indicating the possibility of early stage prostate cancer, while normal levels are typically1 0.5 to 2 ng mL−1. Late stage prostate cancer is characterized by values1 of 10 to 1000 ng mL−1.
It is now apparent that collections or panels of protein cancer biomarkers, as opposed to single biomarkers, will be necessary for reliable cancer detection and monitoring.1,3,10–14 For example, detecting 5 or more biomarkers for a given cancer by liquid chromatography-mass spectrometry (LC-MS) has provided >99% reliable diagnostics.10,12,17,18 On the other hand, single biomarkers often have inadequate predictive value, e.g. about 70% for PSA, which is one of the better single biomarkers.
Increasing predictive power is both a statistical and a biochemical issue. Measurement with equivalent accuracy of elevated levels of 4 to 10 biomarkers for a given cancer should provide a much better statistical basis for successful prediction than measurement of a single biomarker. On the other hand, there are many types of cancers, and even different types of cancers specific to a given organ. A single biomarker expected for a given cancer may for some biochemical reason be poorly expressed in a particular patient, but it would be unlikely that an entire panel of protein biomarkers indicative of that cancer would fail to be expressed. In addition, false positives and negatives may appear quite frequently with a single biomarker, but would be minimized by using a biomarker panel.
Another issue to be considered is the fact that many protein biomarkers are indicative of more than one disease, e.g. serum PSA is elevated in some benign prostate diseases as well as prostate cancer.15 Another biomarker protein, interleukin 6 (IL-6), is overexpressed in oral, prostate, lung, multiple myeloma and renal cell cancers.19 Thus, single cancer biomarkers are often not unique to a specific cancer, but it should be possible to define collections of biomarkers whose elevated individual levels taken together would be reliable predictors for specific cancers.
Detection of panels of biomarkers is further complicated by the fact that ideally both normal and elevated serum levels of biomarkers need to be accurately measured. In addition, concentrations may vary widely for different analyte proteins in the serum. As mentioned for PSA, normal levels are typically 0.5 to 2 ng mL−1 and patents with early stage prostate cancer have levels of 4 to 10 ng mL−1. However, serum IL-6 levels of patients with oral cancer range from ≥20 up to thousands of pg mL−1 compared to <6 pg mL−1 in healthy individuals.19 Thus, the serum concentrations for IL-6 that need to be measured in a given sample may be 1000-fold smaller than those of PSA. In general, collections of cancer biomarkers in serum will have some members at very low concentrations, and other members at much higher concentrations. Also, there are thousands of proteins at significant concentrations in serum, some at much higher than ng mL−1 levels.3 Thus, strategies need to be developed to measure both high and low concentration members of the biomarker panel in the same sample without interference from thousands of other serum proteins, many at higher concentrations than the target analytes.
Owing to low inherent predictive ability of some biomarkers and their overexpression by multiple cancer types, highly reliable prediction for a specific cancer will ultimately require measuring a number of relevant biomarker proteins for each cancer. However, panels are likely to provide accurate prediction and monitoring with a small number of carefully selected members,20 e.g. 4–10. Ideally, measurements need to be done at high accuracy and cheaply at point-of-care, e.g. in a physician’s office or clinic, to reduce costs, minimize sample decomposition, facilitate-on the-spot diagnosis, and alleviate patient stress. Such point-of-care biomarker measurements can also help to guide therapy,4 especially when timely adjustments in treatment become critical. Last but not least, point-of-care biomarker measurements for cancer detection and monitoring will need to be accepted and embraced by the clinical community. Given these realities, development of point-of-care bioanalytical devices to measure multiple cancer biomarkers presents a daunting challenge, but one that nevertheless should be realizable.
Enzyme-linked immunosorbent assays (ELISAs) have served as important methods for clinical protein determinations. Detection limits (DLs) approach 1 pg mL−1 for some protein biomarkers,15,19–22 but ELISA is difficult to develop for point-of-care use and requires significant technical expertise. Classical ELISA methods suffer limitations in analysis time, sample size, equipment cost, and measuring collections of proteins. On the other hand, ELISA-type approaches have been quite useful in assays utilizing immunoarrays, as discussed below.
While modern LC-MS proteomics can achieve multiple biomarker measurements approaching acceptable sensitivity, the current analysis technology appears to be too expensive, often semi-quantitative, time consuming, and technically complex for routine diagnostic point-of-care applications.1–3,20 At present LC-MS is more suited for biomarker discovery research. While vigorous efforts are underway to make LC-MS methods more suitable for point-of-care,1–3,23 arrays for proteins employing optical or electrical detection are also being developed that may have more immediate promise to achieve relatively simple yet highly accurate and sensitive point-of-care measurements at low cost. Unlike gene arrays that are widely available commercially, accurate, simultaneous, sensitive multiple protein arrays are still under development.14,20,24–26 Nevertheless, protein microarrays hold the promise of high selectivity and sensitivity, ease of use, reasonable cost per assay, and a good possibility for future automation. These arrays typically feature a series of analytical spots on a chip with each spot having a selective protein capture agent for a specific biomarker. Following ELISA strategies, the spots in the array may contain primary antibodies or aptamers to capture the desired analyte proteins, and after washing with a cocktail designed to block non-specific binding, a labeled secondary antibody dispersion is added to bind to the analyte proteins (Fig. 1). In the measurement step, the label provides an optical or electrical signal, which can also monitor a reaction catalyzed by an enzyme label. This approach is well suited to early cancer detection and monitoring since it has the potential for relatively rapid high sensitivity determinations of limited panels of biomarkers in serum with good precision and accuracy.20,27
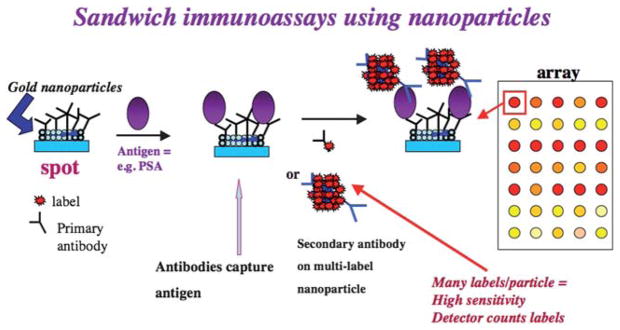
Fig. 1.
Example of an immunoarray strategy designed to detect cancer biomarker proteins (PSA = prostate specific antigen). Gold nanoparticles providing high spot areas are linked to primary antibodies that capture the protein analytes. After washing, a labeled secondary antibody, or as illustrated here a multi-labeled nanoparticle with attached secondary antibody, is added. This species binds to another site on the captured analyte molecules. After additional washing with blocking agents to remove non-specific binding of the labeled species, electrical or optical detection is used to “count” the number of bound labels that is proportional to protein analyte concentration.
This critical review focuses on methodologies for sensitive, specific protein determinations in biomedical samples that we believe have the potential for development for multiplexed point-of-care applications. The paper considers only this limited application, and is not intended as a comprehensive review on protein detection. The next section of this review provides a summary of therapeutic potential of cancer biomarker protein measurements. In subsequent sections we summarize progress in measuring cancer biomarker proteins by optically based, electrochemical, electro-optical and other methods. Our criteria for selection of methodology include the current or future potential for use in multiple protein measurements for cancer detection and monitoring at point-of-care, with all the desired properties of that application as described above. The final section provides future predictions for this field. We have not included LC-MS in this review for the reasons outlined above, although future LC-MS methodology may well reach point-of-care applications. The reader is referred to excellent review articles describing efforts to approach this goal.1–4,23,24
2. Therapeutic value of cancer biomarker proteins
The US National Institutes of Health defines biomarkers more narrowly than above, as “molecules that can be objectively measured and evaluated as indicators of normal or disease processes and pharmacologic responses to therapeutic intervention”.28 By this token, biomarker panels hold value by their remarkable ability to distinguish between two or more biological states. Consequently, the excellent clinical and preclinical applications that these molecules afford have resulted in an area of intense investigation. Significant contributions have recently been made in biomarker identification with the use of powerful discovery and screening technologies such as DNA microarrays and proteomic approaches including mass spectrometry, to essentially accelerate the number of potential biomarker candidates.29–32 Analogous to how automated gel-filled capillaries allowed the early completion of the Human Genome Project, a trend in proteomics that has had a large impact is using capillary electrophoresis separation, other new methods, and sample handling protocols for biomarker protein discovery.33
Most biomarker studies generate data that have the potential of improving our understanding of the underlying disease.34 In the case of human cancers, we now know that progressive changes in cellular behavior from slightly deregulated proliferation to full malignancy are a result of the accumulation of mutations and epigenetic alterations in a sub-set of genes. Identifying their aberrant forms including expressed products such as mRNA or proteins, which may be causal for conversion of normal to malignant tissue, can yield vital information on novel biomarkers for tumor development and progression. Such studies can also suggest candidate drug targets for pharmacological intervention.35 However, a caveat to consider before embarking on detection of cancer biomarkers is that solid tumors generally develop from one or several defective cells, which then proliferate to a self-sustained mass which leads to further growth to a point when patient care is compromised.21 Thus, the single most important goal of a cancer biomarker is the reliable detection of the presence of the smallest number of tumor cells before further growth and when clinical outcome and prognosis are still favorable for the patient.16
To realize the full potential of clinical biomarkers in diagnosis, new bioanalytical technologies are being developed as outlined later in this review. Accompanying the explosive interest in cancer biomarkers is the real potential to improve the accuracy and sensitivity of detection of cancer sub-types as well as markers of prognostic value and predictive of treatment outcome.36 Nevertheless, a limiting factor likely to be encountered is the reliable and accurate detection of cancer biomarkers which is primarily due to insufficient sensitivity of the assays used. Many current methods lack the needed dynamic range to detect proteins of interest that are often expressed as discussed above at levels in the low pg mL−1 range in serum, and in some cases below the detection limit. In contrast, prostate-specific antigen (PSA) levels are normally at a few ng mL−1 or less in healthy subjects, which can be detected by current methodologies including ELISA. The Food and Drug Administration has approved a blood test to measure elevated PSA levels to detect prostate cancer at early or asymptomatic stages of development. However, this test is non-cancer-specific as PSA is also elevated in benign prostate conditions. Thus, at present, higher levels of PSA are not always indicative of disease, but accepted as a biomarker of disease when combined with clinical examination. Unfortunately prostate cancer can still only be definitively diagnosed with a positive biopsy.37 In this regard, the use of new technologies is essential to simultaneously and accurately measure biomarkers that are inherently elevated in cancer and as members of panels of multiple biomarkers. This approach can then result in early cancer detection, as well as in improvement in patient prognosis and care.
Validated biomarkers of cancer development and progression thus need to have excellent specificity, which can be defined as the ability of the assay to rule out a condition when it is absent and, sensitivity, defined as the ability of the assay to identify a condition when it is present. These clinical specificity and sensitivity parameters are closely linked to method used for measurement, and need to be high (e.g. >90%) to avoid false positives and false negatives, which is crucial to avoid misdiagnosis.38 Notably, it is during the validation step where low specificity of selected biomarkers often fails to move them to the next stage.
However, with well designed experimental systems using large collections of clinical samples, the chances of validating potential biomarkers increase, together with our knowledge of the biology of the underlying disease condition.39 In this regard, immunohistochemistry remains an important technique in biomarker validation in cancer tissues. Combined with the availability of using multiple antibodies against potential cancer biomarker proteins using tissue microarrays (TMAs) consisting of hundreds of normal and tumor tissues on a single slide, the expression and activity status of relevant molecules can be explored and validated simultaneously in large sample collections from a single immunostaining procedure. This provides valuable information about suitable biomarkers and drug targets, for example for the development and testing of novel therapies.40
Currently, fully validated biomarkers hold great promise for use as diagnostic, prognostic, and predictive markers for neoplastic diseases, especially when used in panels. Diagnostic markers are used to detect early stage disease where intervention can provide a favorable outcome under the current standard of care. The success of improving patient prognosis is dependent on the existence of evidence establishing the advantage of early intervention, as is clearly the case in cancer.41 Similarly, diagnosis can be applied to histopathological tumor classification, primarily to understand the biological complexity underlining the disease. When metastasis is likely, an aggressive adjuvant therapy can be often prescribed. For example, a sub-set of breast cancer patients correctly diagnosed with early stage and high-risk tumors can benefit from appropriate personalized treatment. Similarly, if biomarker detection suggests that metastasis is likely in a patient, then an aggressive adjuvant therapy can be prescribed.42 On the other hand, biomarker levels indicative of disease progression can be used to identify those patients who are likely to benefit from a specific targeted treatment, and may also provide molecular endpoints that can be used to predict and monitor treatment efficacy.43,44 Appropriate biomarkers may constitute components of dysregulated signaling pathways and can be effectively screened with increased sensitivity and specificity by using available antibodies and TMAs. Ultimately, this knowledge may provide tremendous opportunities not only for the diagnosis of disease, but also for the development of novel molecular-targeted strategies. Realization of these opportunities depends on the development of accurate, sensitive, cheap, automated devices for the detection of multiple protein biomarkers in patient samples, preferably at point-of-care.
3. Optical methods and arrays
3.1. Introduction
Array-based optical methods, fluorescence spectroscopy in particular, are important approaches for the multiplexed analysis of important cancer biomarkers with good sensitivity and low detection limits.25 There are two distinct types of goals that need to be addressed by these methods. One is the determination of a small panel of specific protein biomarkers in multiple samples that are obtained at different stages of cancer diagnosis, prognosis, treatment and monitoring. Success with this goal is critical for personalized medicine where each individual patient is cared-for in an appropriate manner. The other major goal is the determination of specific biomarker panels in samples obtained from a group of patients for the early detection of cancer or other diseases. These studies are directed toward validation or discovery of biomarkers. Progress in this second area is expected to accelerate early screening of high risk populations at point-of-care.45
To achieve these two distinct goals, arrays of antibodies,46 nucleic acid aptamers,47 peptide-like molecules called peptoids,48 peptide aptamers,49 and small molecules,50 are being used to determine the concentrations of proteins in a variety of biological samples that include serum, saliva, sputum, tears, tissue lysates, and urine.25 The following sections of the article focus mainly on frequently used antibody arrays for detection of single or multiple analytes by optical methods. The reader may consult excellent articles, suggested above, for progress on other types of arrays and detection methods. Next, we focus on fluorescence-based detection by using the sandwich immunoassay approach.
3.2. Fluorescent arrays
Strategies for high sensitivity fluorescence detection of biomarkers by sandwich immunoassay on an antibody array involve the following steps: (1) capturing the analyte on spots of the array using high affinity antibodies, called the capture anti-bodies (Ab1), which are immobilized on a solid support without diminishing its bioactivity;51 (2) blocking or minimizing the non-specific binding (NSB) of the analyte to the solid support, usually with a protein such as bovine serum albumin and a detergent; (3) exposure of the array to the sample for capture of the analytes by Ab1; (4) tagging the immobilized analytes with secondary antibodies (Ab2) labeled with a high efficiency fluorophore, while blocking NSB of this species; (5) imaging the array by exciting the fluorophore at particular wavelengths and monitoring the fluorescence such that the signal is highly selective for the fluorophore label while maintaining low background levels; and (6) quantitation of fluorescence from specific spots of the array in terms of the corresponding analyte concentrations.
Detection limits for fluorescence immunoassays, in general, are in the range of mid- to lower picomolar (ng mL−1) concentrations.25 However, cancer biomarker detection in patient samples such as serum often requires much lower detection limits, in the range of femtomolar (pg mL−1) concentrations or lower. The need for low detection limits is essential for early detection of cancer, since a large number of important cancer biomarkers exist at very low levels at early stages of cancer and at normal levels. Ultralow detection levels are also essential for the prompt detection of relapse after cancer treatment and therapy to ensure positive therapeutic outcomes. An additional challenge is that the real samples consist of complex mixtures of proteins, lipids and carbohydrates where achieving high specificity and low levels of NSB are major challenges. NSB to un-intended targets, where the analyte of interest is in very low concentrations and many other components are in high concentrations, can severely interfere with the assay. To achieve desired detection limits for biomedical applications, it is almost always essential to integrate specific signal amplification strategies by biochemical52,53 electrochemical (see below), chemiluminescent,54 or nanoparticle-based amplification strategies.55,56 These amplification strategies need to be compatible with biomarker arrays, while providing high sensitivity and low background signals. One major challenge for signal amplification is that the amplification agent that results in the enhanced signal should not migrate from spot to spot within the array such that spatial resolution of the signal is maintained.
Fluorescence-based multiplexed detection methods are still in developmental stages, and a representative set of examples is given in Table 1 to illustrate the type of antigens that have been successfully detected with corresponding detection limits. These methods illustrate the detection of cytokines, toxins, antigen/antibody pairs and other proteins of clinical importance at levels that are biologically significant. These examples demonstrate the feasibility of the antibody array approach for the detection of biomarkers at physiologically relevant levels in serum, tissue and other biological media. While these examples of biomarker detection are impressive, significantly improved sensitivity and detection limits are still desirable for practical applications. While many studies focused on single analyte measurements in buffer or serum, simultaneous analysis of multiple biomarkers in relevant biological fluids, with little or no pre-treatment, will be of great significance in cancer diagnosis, treatment and management. Such an approach is particularly relevant for point-of-care (POC) applications. Currently, there are no commercial multiplexed biomarker detection systems specifically designed for POC applications. Such devices are highly desirable for improved treatment and lowering health care costs, and have the potential to provide diagnostic cancer testing for millions of patients worldwide.
Table 1.
Biomarker detection by fluorescence methods
| Array | Detection | Amplification | Assay type | Detection limits | Ref. |
|---|---|---|---|---|---|
| Microsphere | Fluorescence | RCAa | Sandwich immunoassay | 10 fM IL-6, IL-8 | 57 |
| DNA barcode | Fluorescence | PCRb | Sandwich immunoassay | 500 aM PSA | 58 |
| Microspheres | Fluorescence | None | Sandwich immunoassay | 0.3 pg mL−1, toxins | 59 |
| Microspheres (Luminex) | Flow cytometry | None | Sandwich immunoassays | Approaching 10 pg mL−1 for multiplexed proteins | 60 |
| None | Fluorescence | None | Sandwich immunoassay | 1 ng mL−1 C-reactive protein | 61 |
| Au | Scattering | Au-deposition | 1 fM PSA | 62 | |
| Antibody array | Fluorescence | RCAa | Sandwich immunoassay | 1–0.5 pg mL−1, 71 cytokines | 63 |
| Antibody array | Fluorescence | RCAa | 10 fM various proteins | 57 | |
| Antibody array | Fluorescence | None | Dye-labeled | 100 pg mL−1, 115 proteins | 46 |
Rolling cycle amplification.
Polymerase chain reaction.
The xMAP technology marketed by Luminex employs 5.6 μm beads labeled with two fluorescent dyes at different levels for identification of multiple beads addressing different analytes. The beads in the assay are identified and counted by laser flow cytometry. This approach can be used for multiplexed fluorescent detection of proteins employing commercial kits for up to 100 or more proteins.64 The sandwich immunoassays employ a bead with a secondary antibody attached to a third fluorescent dye label. The analytical label and two-level identification dyes are read for each bead simultaneously by flow cytometry. While this approach is appropriate for research and hospital laboratory use, it has not yet been developed for point-of-care applications. Flow cytometry is generally expensive and somewhat technically demanding. However, future breakthroughs in this field may overcome current limitations.
3.3. Chemiluminescence based arrays
One approach to improve upon the sensitivity of fluorescence-based arrays is to use chemiluminescence to generate the signal while optimizing the method such that the signal is proportional to analyte concentration over a wide dynamic range. ELISA-like arrays have been used to detect cytokines from patient sera using chemiluminescence detection.65,66 Cytokines were captured on an IgG array supported on polyvinylidene difluoride membrane, and captured ligands were detected by exposure to biotin-conjugated antibodies, which also carried reagents required for chemiluminescence. Binding of the ligand resulted in enhanced chemiluminescence, thereby providing on-chip amplification of the emitted light signal by several orders of magnitude.67
3.4. Surface plasmon resonance (SPR)
Introduction
Surface plasmon resonance (SPR) is a powerful platform68 to investigate biomolecular interactions with a high degree of accuracy, precision, information content, and sensitivity. Key biomedical applications of SPR include, but are not limited to, antigen–antibody interactions, protein–ligand binding, DNA-hybridization and high sensitivity detection of a variety of biomarkers.69 SPR is a phenomenon that occurs when light propagates from a high-refractive-index medium (Fig. 2, prism or grating) toward an interface containing a low-refractive-index material (sample solution). When a thin Au or Ag film is placed at the glass–liquid interface, incident photons in the evanescent wave generated at the interface by rear illumination couple with the electrons in the metal (plasmons). This coupling occurs only when the conditions of resonance are established. Experimental variables that control resonance include the polarization of the incident light with respect to the metal film, the angle of incidence, and the wavelength of incident light, for a given pair of low and high refractive index materials. Resonance coupling between the incident photons and the plasmons of the metal film results in the exchange of momentum between the two entities, and this interaction decreases the intensity of the reflected light from the metal–glass interface. Minimum reflectivity, therefore, occurs at an angle called the SPR angle at a specific wavelength, or at a specific incident angle at the resonant wavelength. Therefore, SPR signals can be monitored as a function of incidence angle or incidence wavelength, and the reflectivity changes monitored with a sensitive optical detector.
Fig. 2.
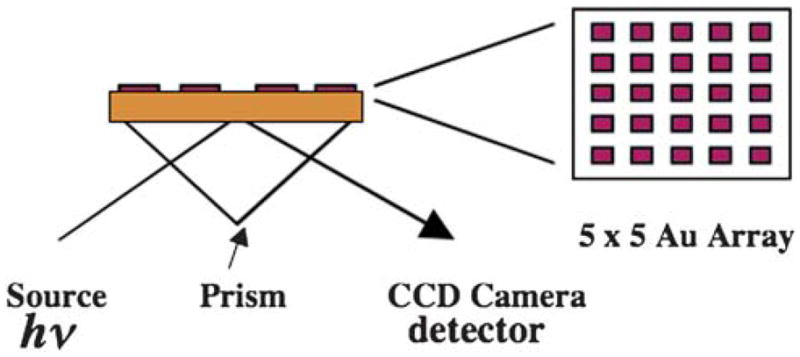
Illustration of SPR arrays on gold films that can be used for measuring multiple protein biomarkers shown as a side view of an array that is coupled to a CCD camera for imaging. On the right is the top view of a 5 × 5 array.
When biomolecules bind to the external surface of the metal film, the resulting change in the dielectric properties of the medium adjacent to the metal film results in changes in the resonance conditions, and hence, modulates the intensity of the reflected light. Thus, SPR can serve as a label-free detection technique to monitor the refractive index changes of the medium adjacent to the metal film brought about by a variety of events. The SPR signal is extremely sensitive to the dielectric constant as well as the mass of the material in contact with the Au film.70 For example, binding of a ligand to a receptor attached to the metal surface will change the intensity of the reflected light, and hence, generate an SPR signal that can be monitored in real time. Kinetics of biomolecular interactions or concentrations of specific analytes such as proteins that bind to molecules attached to the sensor surface can be monitored during the SPR experiment.
Changes in refractive index of the medium up to ~300 nm above the Au film surface influence the SPR signal. Thus, almost any biological binding event can be detected by SPR when one of the binding partners is immobilized on the gold sensor within this distance. SPR can also be used to monitor protein conformational changes, an application that we pioneered to study protein folding thermodynamics on surfaces.71,72 Angular-dependent SPR is convenient for single analyte samples, optimization and calibration, measuring on and off rates of binding events, and affinity constants. SPR has been extended to array format25,69 and a number of analytes can be detected simultaneously (Fig. 2). The general strategy has been to spot multiple antibodies, each specific to a particular antigen, and use the array to simultaneously record SPR signals from each spot after the array is treated with a mixture of antigens.
SPR is a high sensitivity, real-time, label-free method for the detection of virtually any biomarker. Recent innovations have resulted in specific signal amplification methods and decreased detection limits (DLs). The DLs can vary depending on the protocol used for the SPR detection, and they are in the range of a few ng mL−1 when the SPR signal is not amplified.73 However, DLs are improved to pg mL−1 levels or better by amplification strategies such as labeling with nanoparticles or DNA-based amplification.25 While the label-free designation is lost by this improvement, the gain can be exquisite sensitivity. Specifically, the use of secondary antibodies labeled with Au nanoparticles, nanorods or magnetic nanoparticles, as well as enzyme-mediated precipitation reactions, provides excellent methods to amplify SPR signals and improve DLs by as much as 10 000 times with respect to un-amplified signals.74
SPR detection of single biomarkers
Vascular endothelial growth factor (VEGF), which is a cancer biomarker for breast cancer, lung cancer, and colorectal cancer, as well as rheumatoid arthritis, was detected by SPR at biologically relevant levels of 45 pg mL−1 (1 pM).53 In healthy humans, VEGF levels are in the range of 1–2 pM but in cancer patients VEGF levels can be considerably higher.80 A novel aspect of this approach was that RNA aptamers were used to capture VEGF as an aptamer–protein–Ab2 sandwich. RNA aptamers are short stretches of RNA sequences that show unusually high affinities and selectivities for specific antigens, and these are typically obtained by in vitro-selection and amplification. A number of aptamers specific for corresponding ligands can be purchased from commercial sources, although there are far less aptamers available than antibodies. In the above study, Ab2 was labeled with horseradish peroxidase (HRP), which catalyzed a precipitation reaction with tetramethylbenzidine at the sensor surface. Precipitation of the solid amplified the SPR signals by 10 000-fold when compared to the unamplified signals.
SPR was also used for the detection of prostate specific antigen (PSA) in human serum at nanogram levels75 using a sandwich immunoassay. Capture antibody Ab1 was first immobilized onto the SPR sensor surface, and binding of PSA to Ab1 was detected as the Ab1–PSA–Ab2 complex, where Ab2 is the secondary antibody, which was labeled with biotin. Biotin present on Ab2, in turn, was complexed with streptavidin-coated Au nanoparticles (20 nm) to enhance the SPR signals. The signal amplification was on the order of several-hundred fold due to the binding of the streptavidin-coated nanoparticle to Ab2, and no binding occurred without PSA. Thus, the Ab1–PSA–Ab2–biotin–streptavidin–NP configuration facilitated sensitive detection (DLs of 1 ng mL−1) of PSA in human serum samples. Note that PSA levels in healthy humans (males) are 0.5–2 ng mL−1 and the SPR approach was sensitive enough to detect elevated PSA levels of 4–10 ng mL−1, which are indicative of the onset of prostate cancer. Preliminary results from our laboratory showed that SPR can be used to monitor PSA in serum at levels into the low fg mL−1 range. The SPR signals were amplified by using magnetic nanoparticles (MNPs) coated with Ab2 for off-line capture of PSA. This type of SPR protocol, and other innovative approaches (Table 2), need to be extended for the detection and analysis of multiple biomarkers with improved DLs and reliability.
Table 2.
Selected examples of SPR-based detection of cancer biomarkers
| Biomarker | Amplification | Assay type | DL | Ref. |
|---|---|---|---|---|
| VEGF | Precipitation | RNA–aptamer microarrays | 45 pg mL−1 | 53 |
| PSA | Au-NP | Ab1/PSA/Ab2–biotin/streptavidin–AuNP | 1 ng mL−1 | 75 |
| p53 | None | SPR binding assay | 0.2–0.5 ng mL−1 | 76 |
| IL-6 | None | Protein A/Ab1/IL6/IL6 receptor | 3.75 ng mL−1 | 77 |
| IL-8 | None | Ab1/IL8/Ab2 | <2 ng mL−1 | 78 |
| PSA | DNA barcode | Ab1–NP/Ab2–magnetic microparticles | 30 aM PSA | 79 |
Protein 53 (p53) is a transcription factor and it regulates the cell cycle and functions as a tumor suppressor. For this reason p53 is considered to be “the guardian of the genome”. Mutation of p53 is one of the most common molecular markers of the squamous cell carcinoma of the head and neck.81 Recently, SPR was used to monitor the binding of p53 to its target double stranded DNA82 as well as other biological partners. Interactions between the HPV E6 protein, p53 and ubiquitin ligase E6AP were monitored successfully by SPR.76
The analysis required 10 nL of 5–10 μM p53 solution and demonstrated the viability of p53 detection by SPR with high sensitivity, prior to signal amplification, but left room for considerable improvement. The method was neither optimized nor validated but we estimate a detection limit of 2–5 pg in the 10 nL sample, based on data presented in the article. It is likely that the DLs can be further improved with appropriate SPR amplification steps and that the method can be extended for the detection of native p53 as well as mutants.
Recombinant interleukin-6 (IL-6) was detected with good DLs by SPR using the immobilized antibody assay. IL-6 is a multi-functional cytokine and it is implicated in growth regulation and differentiation of cells.77 IL-6 is a 20 kDa protein with two N-glycosylation sites, the human form has been cloned, and the recombinant IL-6 is commercially available. In order to monitor the activities of different batches of recombinant IL-6, SPR methods were developed, calibration graphs constructed and the procedure validated.77 The binding of IL-6 to its antibody was monitored, which was followed by the binding of soluble IL-6 receptor to the Ab1–IL-6 complex (Ab1–IL-6–IL-6 receptor). Optimal results were obtained when Protein A was first immobilized on the sensor surface, followed by the binding of anti-mouse antibody and then IL-6 antibody. This approach gave a DL of 4 ng mL−1 of IL-6, which was highly reproducible from day to day, sample to sample, and did not involve any amplification steps. These data clearly support reliable SPR-based detection of IL-6, but at concentrations above those that are clinically useful. Sensitivity and DL may be improved by introducing specific amplification strategies.
Interleukin-8 (IL-8) is another cytokine which plays an important role in human cancers including breast, prostate, and Hodgkin’s disease.83 Expression levels of IL-8 vary with respect to the cancer type, and there is a significant difference in the salivary levels of IL-8 between healthy individuals (30 pM or 250 pg mL−1) and patients with oropharyngeal squamous cell carcinoma (OSCC) (86 pM or 720 pg mL−1).84 Inflammation of the oral cavity increases salivary cytokine levels but the above differences in IL-8 levels are significant when compared to the corresponding IL-6 levels. Elevation of IL-8 levels in the saliva of the cancer patients is elevated over and above levels due to inflammation of the oral cavity. SPR was used to determine the levels of IL-8 in human saliva of oral cancer patients using a sandwich immunoassay. The assay had a detection limit of 184 pM (<2 ng mL−1) which is greater than the IL-8 levels in patients or healthy individuals. Therefore, ten-fold pre-concentration of the initial sample was suggested in order to determine IL-8 levels in patients.78
Virus-like particles (VLPs) consisting of the human immuno-deficiency virus (HIV) oncoproteins were used in an ELISA format85 to detect human IgG antibodies against human papillomavirus HPV-16 infection in cancer patients, a known cause of cervical and oral cancer.86 Subsequent studies showed that ELISA detection of infection by HPV-16 can be improved by the use of full length recombinant biotin labeled E6 and E7 proteins as antigens instead of VLPs.87,88 Serum samples of patients of head and neck squamous cell carcinoma (HNSCC) (240 HNSCC cases, and 322 matched control subjects without cancer) have been analyzed in this way. These serology studies when coupled with DNA sequencing and histological studies indicated that HPV-16-positive and HPV-16-negative oral cancers in HNSCC patients are distinct and that these should be considered as separate cancers. Such approaches should also be valuable in detection by SPR.
Overall, amplified SPR detection of cancer biomarkers is promising for early diagnosis of cancer, and such technologies may eventually find their way into POC applications. One problem remains the expense of optical detection, and the need for a robust, stable, portable SPR device. Cost might be decreased for POC using simple optics in conjunction with CCD camera detection. Such devices are not currently available for clinical biomarker panel detection.
4. Electrochemical immunoassays and arrays
4.1. History
Heineman and Halsall pioneered enzyme-linked electrochemical immunoassays for small molecules and proteins several decades ago.89 They have continued to develop their approach, in which the binding of antigens to capture antibodies is separated in space and time from enzyme-label detection.90,91 Their enzyme label alkaline phosphatase produces electroactive products that are transported by a chromatographic or fluidic system to an electrode detector. They have obtained excellent detection limits (DLs) in the pg mL−1 to ng mL−1 range for small molecules and proteins depending on the design characteristics of the system and the nature of the detector. Interdigitated electrodes have provided the highest sensitivities. For automation and miniaturization, Heinemann’s team was among the first to employ microfluidics in electrochemical immunoassays for measurements of proteins.92
4.2. Single protein electrochemical immunosensors
In the 1990s, significant effort began to be directed toward developing self-contained, single analyte enzyme-linked electrochemical immunosensors.26,93–97 Most approaches feature antibodies (Ab) attached to the sensor surface, so that antigen capture, enzyme-labeled secondary antibody binding, and detection are all done on the same surface. The most selective and sensitive electrochemical immunoassays employ the sandwich format (cf. Fig. 1).
In addition to alkaline phosphatase and glucose oxidase, horseradish peroxidase (HRP) is a suitable label. It can be activated by hydrogen peroxide, which oxidizes the iron heme HRP enzyme to the FeIV=O form that is reduced to produce electro-chemical signals.98 HRP-labeled Ab2 is a good choice for inclusion into arrays since immobilization of the electroactive enzyme label on the electrode minimizes diffusional crosstalk between array elements, which may complicate detecting soluble electroactive products, e.g. from alkaline phosphatase.99 Many other types of labels have been used, including electroactive metal ions and complexes, nanoparticles,100,101 conductive polymers, and liposomes loaded with electroactive compounds.97 Savéant and co-workers provided theoretical analyses of amperometric and voltammetric enzyme-labeled immunosensor responses, and identified key factors in sensitivity.102–104
A critical issue in an immunoassay is minimization of NSB of the labeled-Ab2 that arises when this species is bound to non-antigen sites on the sensor. In NSB, labeled-Ab2 still gives a signal, but it is not proportional to the analyte protein concentration, and can raise the detection limit and degrade sensitivity. NSB can be minimized by washing with casein or bovine serum albumin along with detergents in NSB-blocking steps in the assay, as discussed earlier. The literature on protein adsorption onto solid surfaces suggests that tailoring the sensor surface with appropriate chemical groups may retard protein adsorption, with one of the most effective surfaces featuring polyethylene glycol (PEG) moieties.97,105,106 However, most studies on which these predictions are based have measured protein adsorption in the absence of a sensitive analytical target, and few have addressed the trace levels of adsorbed proteins relevant to immunoassays. Such functionalized surfaces may still be subject to small amounts of NSB of labeled species that could significantly increase background in pg to ng mL−1 analyte concentration ranges.
More recently, the advantages of labeled vs. unlabeled immunoassays have been argued. Unlabeled immunoassays can be achieved with SPR, atomic force cantilevers, and quartz crystal microbalance (QCM), impedance/capacitance, field-effect transistors (FETs) or nanowire transistors.107,108 While most of these techniques can provide excellent sensitivity, some like SPR (see above) and QCM still require labels to reach sensitivity levels needed for clinical cancer biomarker detection. Label-free methods such as AFM and ultrasensitive nanowire transistors require significant future development and cost reductions before they are able to compete with more accessible, lower cost procedures. In this context, a less-sensitive nanoribbon sensor was recently coupled with a microfluidic sample preparation device for the detection of PSA and a breast cancer biomarker in an approach that shows considerable promise.109
Proponents of unlabeled approaches argue that labeling of antibodies is a troublesome and perhaps unnecessary procedure, and adding labeled Ab2 requires an additional step in sandwich assays. These are, of course, relevant issues only when high sensitivity can be achieved without labels. In addition, there is a distinct difference in uniformly labeling all the analyte proteins, which is difficult to achieve, and using a labeled Ab2 for detection that can be easily prepared in batches and stored prior to the assay. The latter approach shares many of the attributes of true non-labeled methods, but fewer of the problems.108 Further, NSB of the large number of non-analyte serum proteins in non-specific label-free methods such as SPR, nanowire transistor, and impedance/capacitance can be significant, since any adsorbed molecule will contribute to the NSB background. Proponents of labeled approaches also point out that large numbers of labeled secondary antibodies for sandwich immunoassays are available commercially, and that multi-label strategies using nanoparticles have already achieved exquisite sensitivities and detection limits for protein biomarkers, as well as good accuracy in real biomedical samples.100,101,105,106,110,111 In addition, predominantly NSB of labeled Ab2 will contribute to background signals and presents the main NSB problem to be solved.
4.3. Ultrasensitive electrochemical immunosensors
As suggested above, reliable early cancer detection via protein biomarker measurements in serum requires methods with detection limits below that of the normal patient concentration of the least abundant protein, and adequate sensitivity for all the protein biomarkers at normal and elevated levels in serum. These considerations suggest that detection limits below the pg mL−1 level and good sensitivity up to hundreds of ng mL−1 will be necessary. Considerable effort has been directed toward fabrication of high sensitivity electrochemical immunosensors for proteins with ultralow detection limits (DLs) and excellent sensitivities. An added benefit may be that unknown biomarker proteins with very low serum concentrations could emerge once very high sensitivity can be achieved routinely.
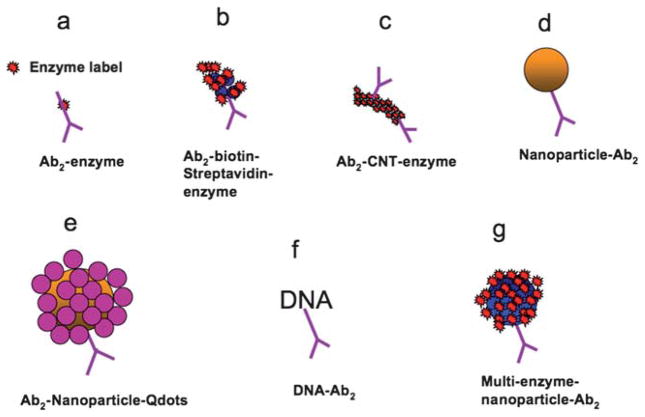
Promising approaches utilizing secondary antibodies (Ab2) labeled with nanoparticles in sandwich immunosensors have employed strategies including dissolvable nanoparticles, and nanoparticles with multiple enzyme labels (Fig. 3) or multiple redox probes.100,101,105–115 These approaches yield high signal amplification by providing many signal generating electrochemical events for each analyte bound to the sensor.
Fig. 3.
Amplification strategies for electrochemical immunosensors using nanoparticles or other moieties attached to secondary antibody Ab2.
A recent review by Wang100 summarizes the various approaches using electrochemically detectable metal nano-particles as labels in protein immunosensors. Dequaire et al. provided proof of concept for the basic approach,116 in which the metal labels are dissolved in acid after Ab2 binding and NSB blocking steps. They used gold nanoparticle–Ab2 labels, and detected gold ions released after acid dissolution by anodic stripping voltammetry to achieve a 3 pM DL for IgG protein in buffer. Wang developed several strategies to amplify the sensitivity even further,100 including a cyclic accumulation of gold nanoparticles and the use of these particles to catalyze the subsequent precipitation of Ag. Both approaches provide a large number of metal ions to be detected in the electrochemical stripping measurement step. The Ag-deposition approach was used to achieve a 0.5 ng mL−1 (22 pM) DL for cardiac troponin I.117 Multiple gold nanoparticles have also been attached to larger Au spheres and used for Ag-deposition enhancement.100 CdS quantum dots (Qdots) on magnetic particles have also been used as labels, with magnetic collection and dissolution of the Qdot and electrochemical stripping detection of Cd, which was further enhanced by Cd-deposition.100 Ag-deposition has also been used in high sensitivity conductivity immunoassays of human IgG in buffer.118 Other strategies as extensively reviewed100,101,105,112–114 have included loading Ab2-nanoparticles or Ab2–polymer beads with electroactive labels such as ferrocene derivatives, and releasing these redox probes for electrochemical detection following the binding steps. Multiplexing strategies using the above approaches are discussed in a later section.
Wang’s team also pioneered the use of multilabel enzyme nanoparticles with attached Ab2, while first demonstrating this approach for ultrasensitive detection of DNA.119 Multi-wall carbon nanotubes (MWCNTs) were used to carry thousands of alkaline phosphatase enzyme molecules per CNT and secondary antibodies to achieve a fM detection limit for proteins in buffer. The carbon nanotubes also served to preconcentrate the enzyme reaction product α-naphthol by adsorptive accumulation. A similar enhancement strategy using layer-by-layer film deposition of alkaline phosphatase with oppositely charged polyions on MWNTs achieved a remarkably low DL of ~70 aM for IgG in buffer.120
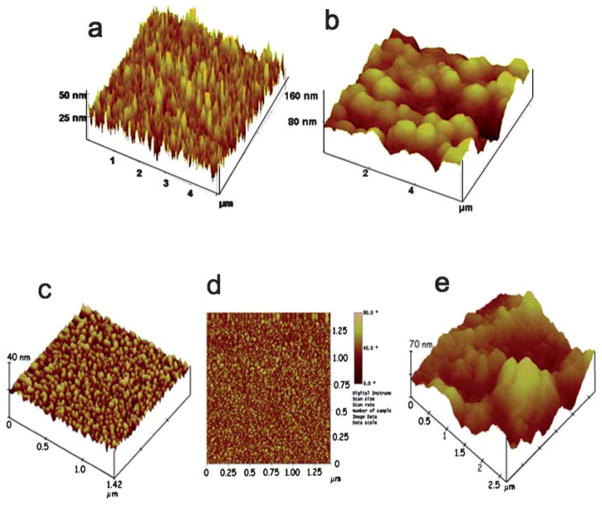
Our research team has exploited the multi-enzyme label strategy in designing immunosensor protocols for PSA, IL-6, and other prostate cancer biomarkers.106,112,121–123 Sensitivity and detection limits were further improved by using conductive nanostructured electrode platforms, e.g. films of oxidatively shortened upright single wall carbon nanotube (SWCNT) forests110 or 5 nm glutathione-decorated gold nanoparticles, both of which contain carboxylate groups ready for attachment of large amounts of capture antibodies by amidization.111 AFM images of these sensor surfaces confirm their large surface areas (Fig. 4).
Fig. 4.
Tapping mode atomic force microscopy (AFM) images of: (a) SWNT forest on smooth silicon and (b) anti-biotin antibody functionalized SWNT on smooth silicon. (c) PDDA/gold (5 nm) nanoparticle bilayer on a smooth mica surface suggesting a densely packed nanoparticle layer; (d) phase contrast image of gold nanoparticle electrode on mica, again suggestive of full coverage with the nanoparticles; and (e) gold nanoparticle electrode as in “c” after covalent linkage of the antibody anti-PSA onto the surface. Images a and b reproduced with permission from X. Yu, S. Kim, F. Papadimitrakopoulos, J. F. Rusling, Mol. BioSyst., 2005, 1, 70–78. Copyright Royal Society of Chemistry (UK), 2005. Images c, d, and e reproduced with permission from ref. 111, Copyright American Chemical Society, 2009.
SWCNT and AuNP nanostructured sensor surfaces were first used to fabricate sandwich immunoassays for prostate cancer biomarker PSA.106,110,111,121 Two detection protocols were evaluated. As in Fig. 3a, conventional secondary antibodies (Ab2) conjugated with enzyme label HRP were used, as well as a higher sensitivity protocol using oxidized multiwall carbon nanotubes (CNTs) conjugated with HRP and Ab2 (CNT–HRP–Ab2) at high HRP/Ab2 ratios (Fig. 3c). These CNT–HRP–Ab2 with 170 HRPs per 100 nm length124 can replace the conventional HRP–Ab2 in the immunoassay procedure to greatly enhance sensitivity.
Fig. 5A shows amperometric SWNT immunosensor responses to PSA in calf serum using CNT–HRP–Ab2. A PSA detection limit (DL) as a signal 3 times the noise above the zero PSA control was 4 pg mL−1 (150 fM). Controls (b) and (c) in Fig. 5A show that SWNT forests provided a significant gain in sensitivity over immunosensors without nanotubes, presumably because of an increased density of Ab1. Recent unpublished results show that such SWCNT forests can achieve a 10 to 15-fold increase in the number of surface antibodies compared to a flat immunosensor.
Fig. 5.
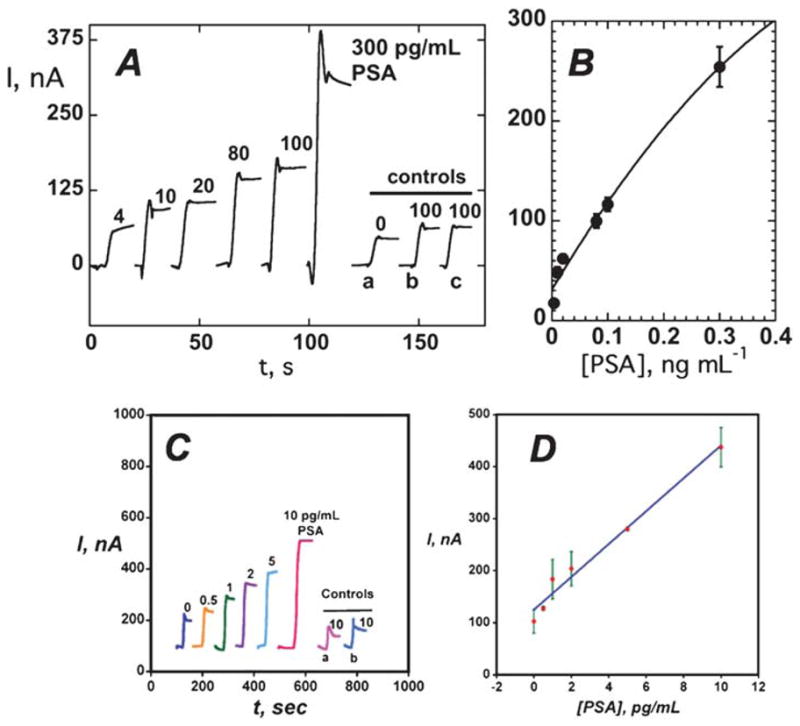
Amperometric response for nanostructured immunosensors incubated with PSA in 10 μL serum in buffer containing 1 mM hydroquinone after injecting 0.04 mM H2O2 to develop the signal (A) current for SWNT immunosensors at −0.3 V and 3000 rpm using the Ab2–CNT–HRP bioconjugate with 170 labels per 100 nm at PSA concentrations shown: controls: (a) full SWNT immunosensor omitting addition of PSA, (b) immunosensor built on bare PG surface for 100 pg mL−1 PSA, (c) immunosensor built on Nafion–iron oxide-coated PG electrode for 100 pg mL−1 PSA. (B) Influence of PSA concentration on steady state current for SWNT immunosensor using Ab2–CNT–HRP bioconjugate. (C) Current at −0.3 V and 3000 rpm for AuNP immunosensors using Ab2–magnetic bead–HRP with 7500 labels/bead at PSA concentrations shown. Controls: (a) immunosensors built on bare PG at 10 pg mL−1 PSA; (b) immunosensors built on PDDA coated PG surface at 10 pg mL−1 PSA; (D) influence of PSA concentration on steady state current for AuNP immunosensor using multi-label Ab2–magnetic bead–HRP. A and B reproduced with permission from ref. 110, copyright American Chemical Society 2006. C and D reproduced with permission from ref. 111, Copyright American Chemical Society, 2009.
Our AuNP electrodes were fabricated by depositing a layer of 5 nm glutathione-decorated AuNPs onto a previously deposited 0.5 nm polycation layer on PG. High sensitivity was obtained by using 1 μm magnetic bead–Ab2–HRP bioconjugates with ~7500 HRPs per bead.111 Using these multiply labeled magnetic beads with AuNP electrodes (Fig. 4C), detection limit at 0.5 pg mL−1 (20 fM) PSA was eight fold better, and sensitivity was 4-fold better than SWCNT forest immunosensors featuring the multi-label CNT–HRP–Ab2. Controls (a) and (b) in Fig. 5C show that AuNPs also provided enhanced sensitivity over immunosensors without AuNPs. With both SWNT forest and AuNP immuno-sensors, excellent correlations were found with a standard ELISA for determination of PSA in serum of human cancer patients.110,111
A head-to-head comparison was made between SWCNT and AuNP immunosensors for determination of IL-6 in serum.122 Immunoassays employed HRP labels on streptavadin bound to biotinylated Ab2, providing 14–16 labels per analyte (Fig. 3b). The AuNP immunosensor gave DL of 10 pg mL−1 IL-6 (500 fM) in 10 μL calf serum, 3-fold better than 30 pg mL−1 found for the SWCNT forest immunosensor. The AuNP sensors provided a linear dynamic range of 20–4000 pg mL−1, covering most of the clinically relevant range, and was much larger than the SWNT immunosensors (40–150 pg mL−1).122
While there has been considerable development of label-free electrochemical impedance immunosensors, a recent review concluded that in general these methods may not be able to provide ultrasensitive protein analyses without some mode of amplification or other improvements in sensitivity.125 Significant amplification and high sensitivities have been obtained, for example, using metal nanoparticle labels or AuNP labels that catalyze subsequent silver deposition. Furthermore, several carefully designed label-free impedance/conductance methods have been able to achieve notably high sensitivity for proteins. A capacitance method using a ferri/ferrocyanide probe and a potentiostatic step gave a DL of 10 pg mL−1 (500 fM) for IL-6 in buffer.126 Optimization of experimental parameters and protocols using flow injection electrochemical impedance spectroscopy led to sensitivity in the low aM range for interferon-γ in buffer.127 Such approaches appear promising for future point-of-care applications provided NSB from the numerous serum proteins can be minimized. A recent review by Tkac and Davis discussed these high sensitivity approaches in detail.108
4.4. Multiplexed protein arrays
Two major approaches have been pursued for ultrasensitive multiplexed electrochemical protein measurements. In one, a “barcode” approach is used to label secondary antibodies with distinct nanoparticles having different electrochemical characteristics, e.g. different dissolvable metals or quantum dots (Qdots) that can be detected by stripping voltammetry.103 In the second approach, multi-electrode microelectronic arrays are used in which each electrode has a different antibody attached. Conceivably these approaches could also be combined.103
As an example of the first approach, cadmium sulfide, zinc sulfide, copper sulfide and lead sulfide quantum dots (Qdots) were attached to four different secondary antibodies to detect four different proteins by sandwich immunoassay.128 The four metal ions, each attached to a specific antibody for a different protein, were measured after binding of the antigen by stripping voltammetry and dissolution of the particle. Multiple-metal spheres, striped rods or alloy rods have also been used as labels. The rods can be capped with a gold end for attachment to Ab2. Upon dissolution, these materials give a series of stripping peaks whose peak potentials and relative intensities are characteristic of a single analyte protein.100 In principle, such “barcode” labels can be employed for the determination of thousands of proteins, with computer deconvolution of the identities and concentrations.
Pairs of iridium oxide electrodes to which capture antibodies for two proteins were covalently attached have been used to determine proteins.129 Separation of the electrodes by 2.5 mm to eliminate cross-talk between the electrodes enabled simultaneous electrochemical immunoassays using alkaline phosphatase-labeled Ab2 and detection of the electroactive enzyme product hydroquinone. Detection limits were ~1 ng mL−1 for goat IgG, mouse IgG, and cancer biomarkers carcinoembryonic antigen (CEA) and α-fetoprotein (AFP). This concept was extended to an 8-electrode array and an electrochemical immunoassay was developed for simultaneous detection130 of goat IgG, mouse IgG, human IgG, and chicken IgY. DLs of ~3 ng mL−1 were achieved for these analytes, and the method was validated vs. a referee method using synthetic sera containing the 4 proteins. Eight-electrode iridium oxide arrays designed into each well of a 12-well plate were used to simultaneously measure131 seven cancer biomarkers: AFP, ferritin, CEA, hCG-β, CA 15–3, CA 125, and CA 19–9 with DLs of ~2 ng mL−1. The method showed good correlation for a standard serum sample against commercial ELISA methods. Unfortunately, none of these systems were useful in the pg mL−1 range.
CombiMatrix VSLI chips with over 1000 electrodes cm−2 have been used for sandwich immunoassays of proteins.132 Oligonucleotides were synthesized on the 100 μm diameter electrodes and antibodies were outfitted with complementary oligonucleotide strands so that they could bind to specific electrodes. This method was used to determine human α1 acid glycoprotein (APG), ricin, M13 phage, Bacillus globigii spores, and fluorescein with a 5 pg mL−1 DL for the protein APG. A similar approach was used with two or three different enzyme labels on the secondary antibodies for multianalyte detection at several different measuring potentials.133 While these chips have the potential for massively parallel array detection of thousands of proteins, this has yet to be realized. The approach has been commercialized by CombiMatrix for DNA and RNA arrays. The bottleneck for protein arrays would seem to be attachment of the oligonucleotide affinity strands to the large number of antibodies that would be necessary for such a chip.
Wong et al. reported several approaches to arrays to detect the oral cancer biomarkers IL-8 protein and RNA in the same sample. In 2007, they reported a prototype microfluidic array with detection capabilities of ~50 pg mL−1.134 More recently, they reported a 16-sensor electrochemical chip with each sensor having its own reference and counter electrode.135 Individual sensors were coated with a DNA dendrimer/conducting polymer film decorated with binding sites for capture antibodies. Two oral cancer protein markers (IL-8 and IL-1b) and IL-8 mRNA were measured in buffer with LD of 100–200 fg mL−1 for the proteins and 10 aM for IL-8 mRNA in 4 μL samples, which was 1000-fold better than without the DNA dendrimer/conducting polymer. This system was applied to measurements in saliva, with considerably higher DL for IL-8 mRNA (~4 fM) and for IL-8 (7.4 pg mL−1).136 Using these arrays to analyze a collection of oral cancer saliva samples and controls gave ~90% clinical sensitivity and specificity for both IL-8 mRNA and IL-8 protein. These studies illustrate the critical importance of testing arrays in real biomedical samples, as they show considerable degradation of DL for both analytes when moving from buffer to saliva. In addition, the last study is an excellent example of essential method validation and analytical performance evaluation using real patient samples.
Our research team recently used a 4-unit electrochemical immunoarray equipped with single-wall carbon nanotube forest electrodes to simultaneously measure multiple prostate cancer biomarkers in cancer patient serum.137 The proteins were PSA, prostate specific membrane antigen (PSMA), platelet factor-4 (PF-4) and IL-6. Method sensitivity was tailored to analytical requirements of each protein by combining single and multiply labeled strategies. Biotinylated secondary antibodies (Ab2) that bind specifically to streptavidin–HRP conjugates to provide 14–16 labels per antibody (see Fig. 2b) were used in a sandwich immunoassay format to give the necessary sensitivity to detect PF-4 (DL ≈ 1 ng mL−1) and IL-6 (DL ≈ 30 pg mL−1). Singly labeled Ab2–HRP (Fig. 2a) was used for PSA (DL ≈ 1 ng mL−1) and PSMA (DL ≈ 10 ng mL−1) in the ng mL−1 range. Immunoarray determinations of the four proteins in serum samples of prostate cancer patients and controls gave excellent correlations to standard ELISA assays.137
Electrochemiluminescence (ECL) detection is also promising for multiple protein assays. Tris(2,2′-bipyridyl)ruthenium(II), [Ru-(bpy)3]2+, or RuBPY, initiates ECL when its oxidized form reacts with a suitable sacrificial reductant, and can be used as a label in immunoassays.138 The result is light emission with electrical rather than optical excitation, thus simplifying instrumentation. Magnetic bead methods have been most successful with ECL detection, and are available from several companies.139,140 A typical ECL magnetic bead immunoassay features antibody–streptavidin-coated magnetic beads that capture the protein analyte from the sample. Then, RuBPY-labeled bio-tinylated secondary antibody is added and the beads are washed to remove non-specific binding. The magnetic beads are transported to an electrochemical cell where they are magnetically captured onto an electrode for ECL measurement.140 Manual and automated systems are available along with kits for up to 10 selected proteins, although most applications measure one or two proteins per sample. Current automated ECL bead technology is relatively expensive and requires a high level of technical expertise and maintenance. While ECL-bead protein detection is making inroads in research and some hospital clinical laboratories, it will require considerable development and cost reduction for widespread point-of-care applications. A recent innovation involved Ab2–ECL–RuBPY beads with a gold-coated fiber-optic array used to demonstrate detection of 3 proteins in sandwich immunoassays.141 By using 3 different levels of fluorescent labels, single beads representative of 3 different protein analytes were identified and imaged using ECL for quantitation. While promising, this method is in very early development.
5. Outlook for the future
Optical and electrochemical approaches have been developed that are capable of detecting proteins at levels at or below a few pg mL−1, at concentration at or below the normal levels of many cancer biomarker proteins. Many new approaches using nano-technology have been designed to detect proteins at even higher sensitivity. While only a few of these approaches have been validated against cancer patient samples, they have demonstrated good accuracy and sensitivity for clinically important levels of biomarker proteins. However, cancer detection and diagnostics using protein biomarkers will require accurate detection of panels of an estimated 4–10 biomarkers for each cancer, and much less progress has been made on this front. Extensive testing will be necessary on patient samples such as serum or saliva to establish analytical reliability, and clinical sensitivity and selectivity.
Currently no devices or methodologies for multiplexed cancer biomarker protein detection are in widespread POC use, although some commercial bead methods utilizing ECL and fluorescence are appropriate and reliable for laboratory based sample analyses. POC devices will require simple, low cost, technically undemanding methodology. Electrochemical approaches are inherently inexpensive; optical methods will need technical advances to produce lower cost detection. We can distinguish between methodology where binding events and detection are done simply and directly on a sensor or array surface vs. those that require further processing or dissolving of labels and detection in solution. Clearly the simpler approaches have the current advantage. Finally, the marriage of sensitive reliable, simple detection protocols with microfluidic or other sample handling technology seems to be a necessary future step to reach POC. However, the payoff in future cancer diagnostics and therapy is large, and significant efforts and funding are being committed with the hope of solving these formidable analytical challenges in the relatively near future.
Acknowledgments
Preparation of this review was supported jointly by PHS grant ES013557 from NIEHS/NIH, by the National Science Foundation (DMR-0604815), by the Intramural Research Program, NIDCR/NIH, and by a Walton Research Fellowship awarded by Science Foundation Ireland to JFR.
Biographies

James Rusling obtained his BSc in Chemistry from Drexel University and PhD from Clarkson University. He is Professor of Chemistry at University of Connecticut and Professor of Cell Biology at University of Connecticut Health Center. Current research includes electrochemical arrays for toxicity prediction and early cancer detection, and fundamental bioelectrochemistry. He has authored over 300 research papers, several books, and is also a musician interested in Irish and American folk styles.

Challa Vijaya Kumar obtained his PhD from the Indian Institute of Technology Kanpur, India in 1982 and worked as research associate at the University of Notre Dame and Columbia University. He joined the University of Connecticut in 1988 as assistant professor of Chemistry and he is currently Professor of Biological and Physical Chemistry, and Head, the Division of Physical Chemistry. His research interests include DNA/RNA binders, Biocatalytic Nanomaterials, and Protein Biomarkers and Photoscissors.

J. Silvio Gutkind received his PhD in Pharmacy and Biochemistry from the University of Buenos Aires, Argentina. He was trained as a post-doc at NIMH and NCI, and joined the NIDCR, NIH, in 1988, where he is currently the Chief of the Oral and Pharyngeal Cancer Branch. His research team studies the molecular basis of cancer, with emphasis on basic mechanisms of signal transduction and cell growth control, and their dysregulation in oral squamous cell carcinogenesis and AIDS-malignancies. Dr Gutkind has published over 320 scientific manuscripts, edited three books, and is currently leading a multi-institutional effort aimed at exploring novel molecular-targeted therapies for oral cancer patients.

Vyomesh Patel received his PhD from the University of Bristol in 1993. After Post-Doctoral training at the NIH, in Bethesda, USA, he is currently a Staff Scientist. His research interests are in understanding the molecular basis of squamous cancer development and progression, and in the use of technology to drive some of these research areas, to potentially identify biomarkers, that may have use for example at point-of-care, for diagnostic and prognostic application.
References
- 1.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 2.Hawkridge AM, Muddiman DC. Mass spectrometry-based biomarker discovery: toward a global proteome index of individuality. Annu Rev Anal Chem. 2009;2:265–277. doi: 10.1146/annurev.anchem.1.031207.112942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 5.Wang J. Electrochemical biosensors: towards point-of-care cancer diagnostics. Biosens Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Giljohan DA, Mirkin CA. Drivers of biodiagnostic development. Nature. 2009;426:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinstein B, Joe AK. Mechanisms of disease: oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–547. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 8.Le Tourneaua C, Faivrea S, Siu LL. Molecular targeted therapy of head and neck cancer: review and clinical development challenges. Eur J Cancer. 2007;43:2457–2466. doi: 10.1016/j.ejca.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Xiao Z, Prieto D, Conrads TP, Veenstra TD, Issaq HJ. Proteomics patterns: their potential for disease diagnosis. Mol Cell Endocrinol. 2005;23:95–106. doi: 10.1016/j.mce.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Ebert MPA, Korc M, Malfertheiner P, Röcken C. Advances, challenges, and limitations in serum-proteome-based cancer diagnosis. J Proteome Res. 2006;5:19–25. doi: 10.1021/pr050271e. [DOI] [PubMed] [Google Scholar]
- 11.Weston AD, Hood L. Systems biology, proteomics and future of health care: toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 12.Tothill IE. Biosensors for cancer markers diagnosis. Semin Cell Dev Biol. 2009;20:55–62. doi: 10.1016/j.semcdb.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Stevens EV, Liotta LA, Kohn EC. Proteomic analysis for early detection of ovarian cancer. Int J Gynecol Cancer. 2003;13:133–139. doi: 10.1111/j.1525-1438.2003.13358.x. [DOI] [PubMed] [Google Scholar]
- 14.Wilson DS, Nock S. Recent developments in protein microarray technology. Angew Chem, Int Ed. 2003;42:494–500. doi: 10.1002/anie.200390150. [DOI] [PubMed] [Google Scholar]
- 15.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 16.Smith RA, Cokkinides V, Brawley OW. Cancer screening in the United States, 2008: a review of current American cancer society guidelines and cancer screening issues. Ca–Cancer J Clin. 2008;58:161–179. doi: 10.3322/CA.2007.0017. [DOI] [PubMed] [Google Scholar]
- 17.Wagner PD, Verma M, Srivastava S. Challenges for biomarkers in cancer detection. Ann N Y Acad Sci. 2004;1022:9–16. doi: 10.1196/annals.1318.003. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 19.Riedel F, Zaiss I, Herzog D, Götte K, Naim R, Hörman K. Serum levels of interleukin 6 in patients with primary head and neck squamous cell carcinoma. Anticancer Res. 2005;25:2761–2766. [PubMed] [Google Scholar]
- 20.Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discovery. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams TI, Toups KL, Saggese DA, Kalli KR, Cliby WA, Muddiman DC. Epithelial ovarian cancer: disease etiology, treatment, detection, and investigational gene, metabolite, and protein biomarkers. J Proteome Res. 2007;6:2936–2962. doi: 10.1021/pr070041v. [DOI] [PubMed] [Google Scholar]
- 22.Ward MA, Catto JWF, Hamdy FC. Prostate specific antigen: biology, biochemistry and available commercial assays. Ann Clin Biochem. 2001;38:633–651. doi: 10.1258/0004563011901055. [DOI] [PubMed] [Google Scholar]
- 23.Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 24.Bensmail H, Haoudi A. Postgenomics: proteomics and Bioinformatics in cancer research. J Biomed Biotechnol. 2003;4:217–230. doi: 10.1155/S1110724303209207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HJ, Wark AW, Corn RM. Microarray methods for protein biomarker detection. Analyst. 2008;133:975–983. doi: 10.1039/b717527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warsinke A, Stocklein W, Leupold E, Micheel E, Scheller FW. Electrochemical Immunosensors on the route to proteomic chips. In: Palacek E, Scheller F, Wang J, editors. Electrochemistry of Nucleic Acids and Proteins (Perspectives in Bioanalysis) Vol. 1. Elsevier B. V; Amsterdam: 2007. pp. 451–483. [Google Scholar]
- 27.Rasooly A, Jacobson J. Development of biosensors for cancer clinical testing. Biosens Bioelectron. 2006;21:1851–1858. doi: 10.1016/j.bios.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Manne U, Srivastava RG, Srivastava S. Recent advances in biomarkers for cancer diagnosis and treatment. Drug Discovery Today. 2005;10:965–976. doi: 10.1016/S1359-6446(05)03487-2. [DOI] [PubMed] [Google Scholar]
- 29.Lonergan KM, Chari R, Coe BP, Wilson IM, Tsao MS, Ng RT, Macaulay C, Lam S, Lam WL. Transcriptome profiles of carcinoma—in-situ and invasive non-small cell lung cancer as revealed by SAGE. PLoS One. 2010;5:e9162. doi: 10.1371/journal.pone.0009162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishizuka S, Chen ST, Gwadry FG, Alexander J, Major SM, Scherf U, Reinhold WC, Waltham M, Charboneau L, Young L, Bussey KJ, Kim S, Lababidi S, Lee JK, Pittaluga S, Scudiero DA, Sausville EA, Munson PJ, Petricoin EF, Liotta LA, Hewitt SM, Raffeld M, Weinstein JN. Diagnostic markers that distinguish colon and ovarian adenocarcinomas: identification by genomic, proteomic, and tissue array profiling. Cancer Res. 2003;63:5243–5250. [PubMed] [Google Scholar]
- 31.Patel V, Hood BL, Molinolo AA, Lee NH, Conrads TP, Braisted JC, Krizman DB, Veenstra TD, Gutkind JS. Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin Cancer Res. 2008;14:1002–1014. doi: 10.1158/1078-0432.CCR-07-1497. [DOI] [PubMed] [Google Scholar]
- 32.Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M, Gutkind JS. Oncogene. 2000;19:3220–3224. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]
- 33.Fang X, Balgley BM, Lee CS. Recent advances in capillary electrophoresis-based proteomic techniques for biomarker discovery. Electrophoresis. 2009;30:3998–4007. doi: 10.1002/elps.200900219. [DOI] [PubMed] [Google Scholar]
- 34.Dalton WS, Friend SH. Cancer biomarkers—an invitation to the table. Science. 2006;312:1165–1168. doi: 10.1126/science.1125948. [DOI] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 36.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 37.Ploussard G, de la Taille A. Urine biomarkers in prostate cancer. Nat Rev Urol. 2010;7:101–109. doi: 10.1038/nrurol.2009.261. [DOI] [PubMed] [Google Scholar]
- 38.Moncada V, Srivastava S. Biomarkers in oncology research and treatment: early detection research network: a collaborative approach. Biomarkers Med. 2008;2:181–191. doi: 10.2217/17520363.2.2.181. [DOI] [PubMed] [Google Scholar]
- 39.Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: theoretical considerations and practical challenges. J Clin Oncol. 2009;27:4027–4034. doi: 10.1200/JCO.2009.22.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hewitt SM. Tissue microarrays as a tool in the discovery and validation of tumor markers. Methods Mol Biol (Totowa, NJ, U S) 2009;520:151–161. doi: 10.1007/978-1-60327-811-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 42.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein-Lønning P, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 44.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 45.Paweletz CP, Charboneau L, Bichsel VE, Chen SNLT, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin IE, Liotta LA. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 46.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2:1–5. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bock C, Coleman M, Collins BD, Davis J, Foulds G, Gold L, Greef C, Heil J, Heilig JS, Hicke B, Hurst MN, Miller D, Ostroff R, Petach H, Schneider D, Vant-Hull B, Waugh S, Weiss A, Wilcox SK, Zichi D. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics. 2004;4:609–618. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]
- 48.Reddy MM, Kodadek T. Protein “fingerprinting” in complex mixtures with peptoid microarrays. Proc Natl Acad Sci U S A. 2005;102:12672–12677. doi: 10.1073/pnas.0501208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray E, McKenna EO, Burch LR, Dillon J, Langridge-Smith P, Kolch W, Pitt A, Hupp TR. Microarray-formatted clinical biomarker assay development using peptide aptamers to anterior gradient-2. Biochemistry. 2007;46:13742–13751. doi: 10.1021/bi7008739. [DOI] [PubMed] [Google Scholar]
- 50.Panicker RC, Huang X, Yao SQ. Recent advances in peptide-based microarray technologies. Comb Chem High Throughput Screening. 2004;7:547–556. doi: 10.2174/1386207043328517. [DOI] [PubMed] [Google Scholar]
- 51.Angenendt P. Progress in protein and antibody microarray technology. Drug Discovery Today. 2005;10:503–511. doi: 10.1016/S1359-6446(05)03392-1. [DOI] [PubMed] [Google Scholar]
- 52.Orchekowski R, Hamelinck D, Li L, Gliwa E, VanBrocklin M, Marrero JA, Vande Woude GF, Feng Z, Brand R, Haab BB. Antibody microarray profiling reveals individual and combined serum proteins associated with pancreatic cancer. Cancer Res. 2005;65:11193–11202. doi: 10.1158/0008-5472.CAN-05-1436. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Lee HJ, Corn RM. Detection of protein biomarkers using RNA aptamer microarrays and enzymatically amplified surface plasmon resonance imaging. Anal Chem. 2007;79:1082–1088. doi: 10.1021/ac061849m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H, Bouwman K, Schotanus M, Verweij C, Marrero JA, Dillon D, Costa J, Lizardi P, Haab BB. Two-color, rolling-circle amplification on antibody microarrays for sensitive multiplexed serum–protein measurements. Genome Biol. 2004;5:R28. doi: 10.1186/gb-2004-5-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoeva SI, Lee J-S, Smith JE, Rosen ST, Mirkin CA. Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. J Am Chem Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 56.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 57.Konry T, Hayman RB, Walt DR. Microsphere-based rolling circle amplification microarray for the detection of DNA and proteins in a single assay. Anal Chem. 2009;81:5777–5782. doi: 10.1021/ac900694y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goluch ED, Nam JM, Chiesl TN, Shaikh KA, Ryu KS, Barron AE, Mirkin CA, Liu C. Development of a Microfluidic Device using Nanoparticle Based Bio-Barcode for Ultra-Sensitive Detection of Proteins. 8th International Conference on Miniaturized Systems for Chemistry and Life Sciences; September 26–30, 2004; Malmo. Sweden. [Google Scholar]
- 59.Pauly D, Kirchner S, Stoermann B, Schreiber T, Kaulfuss S, Schade R, Zbinden R, Avondet M-A, Dorner MB, Dorner BG. Simultaneous quantification of five bacterial and plant toxins from complex matrices using a multiplexed fluorescent magnetic suspension assay. Analyst. 2009;134:2028–2039. doi: 10.1039/b911525k. [DOI] [PubMed] [Google Scholar]
- 60.Luminex Corp. http://www.luminexcorp.com/technology/index.html.
- 61.Gervaisab L, Delamarche E. Toward one-step point-of-care immunodiagnostics using capillary-driven microfluidics and PDMS substrates. Lab Chip. 2009;9:3330–3337. doi: 10.1039/b906523g. [DOI] [PubMed] [Google Scholar]
- 62.Kim D, Daniel WL, Mirkin CA. Microarray-based multiplexed scanometric immunoassay for protein cancer markers using gold nanoparticle probes. Anal Chem. 2009;81:9183–9187. doi: 10.1021/ac9018389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, Shu Q, Laroche I, Zhou Z, Tchernev VT, Christiansen J, Veleeca M, Kingsmore SF. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat Biotechnol. 2002;20:359. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rules Based Medicine Inc. http://www.rulesbasedmedicine.com/
- 65.Huang RP. Simultaneous detection of multiple proteins with an array-based enzyme-linked immunosorbent assay (ELISA) and enhanced chemiluminescence (ECL) Clin Chem Lab Med. 2001;39:209–214. doi: 10.1515/CCLM.2001.032. [DOI] [PubMed] [Google Scholar]
- 66.Lin Y, Huang R, Cao X, Wang SM, Shi Q, Huang RP. Detection of multiple cytokines by protein arrays from cell lysate and tissue lysate. Clin Chem Lab Med. 2003;41:139–145. doi: 10.1515/CCLM.2003.023. [DOI] [PubMed] [Google Scholar]
- 67.Huang RP, Yang W, Yang D, Flowers L, Horowitz IR, Cao X, Huang R. The promise of cytokine antibody arrays in the drug discovery process. Expert Opin Ther Targets. 2005;9:601–615. doi: 10.1517/14728222.9.3.601. [DOI] [PubMed] [Google Scholar]
- 68.Earp RL, Dessy RE. Surface Plasmon Resonance. In: Ramsay G, editor. Commercial Biosensors. Wiley; NY: 1998. pp. 99–164. [Google Scholar]
- 69.Shumaker-Parry JS, Aebersold R, Campbell CT. Parallel, quantitative measurement of protein binding to a 120-element double-stranded DNA array in real time using surface plasmon resonance microscopy. Anal Chem. 2004;76:2071–2082. doi: 10.1021/ac035159j. [DOI] [PubMed] [Google Scholar]
- 70.Lahiri J, Isaacs JT, Whitesides GM. A strategy for the generation of surfaces presenting ligands for studies of binding based on an active ester as a common reactive intermediate: a surface plasmon resonance study. Anal Chem. 1999;71:777–790. doi: 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- 71.Chah S, Kumar CV, Hammond MR, Zare RN. Denaturation and renaturation of self-assembled yeast iso-1-cytochrome c. Anal Chem. 2004;76:2112. doi: 10.1021/ac035416k. [DOI] [PubMed] [Google Scholar]
- 72.Bhambhani A, Chah S, Hvastkovs EG, Jensen GC, Rusling JF, Zare RN, Kumar CV. Folding control of yeast iso-1-cytochrome c sandwiched between Au and layered zirconium materials monitored by surface plasmon resonance. J Phys Chem B. 2008;112:9201–9208. doi: 10.1021/jp7121642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee H-J, Nedelkov D, Corn RM. Surface plasmon resonance imaging measurements of antibody arrays for the multiplexed detection of low molecular weight protein biomarkers. Anal Chem. 2006;78:6504–6510. doi: 10.1021/ac060881d. [DOI] [PubMed] [Google Scholar]
- 74.Teramura Y, Arima Y, Iwata H. Surface plasmon resonance-based highly sensitive immunosensing of brain natriuretic peptide using nanobeads for signal amplification. Anal Biochem. 2006;357:208–215. doi: 10.1016/j.ab.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 75.Choi J-W, Kanga D-Y, Jang Y-H, Kima H-H, Mind J, Oh B-K. Ultra-sensitive surface plasmon resonance based immunosensor for prostate-specific antigen using gold nanoparticle–antibody complex. Colloids Surf, A. 2008:313–314. 655–659. [Google Scholar]
- 76.Ro H-S, B-Koh H, Jung SO, Park HK, Shin Y-B, Kim M-G, Chung BH. Surface plasmon resonance imaging of protein arrays for analysis of triple protein interactions of HPV, E6, E6AP, and p53. Proteomics. 2006;6:2108–2111. doi: 10.1002/pmic.200500635. [DOI] [PubMed] [Google Scholar]
- 77.Deckert F, Legay F. Development and validation of an IL-6 immuno-receptor assay based on surface plasmon resonance. J Pharm Biomed Anal. 2000;23:403–412. doi: 10.1016/s0731-7085(00)00313-7. [DOI] [PubMed] [Google Scholar]
- 78.Yang C-Y, Brooks E, Li Y, Denny P, Ho C-M, Qi F, Shi W, Wolinsky L, Wu B, Wong DTW, Montemagno CD. Detection of picomolar levels of interleukin-8 in human saliva by SPR. Lab chip. 2005;5:1017–1023. doi: 10.1039/b504737d. [DOI] [PubMed] [Google Scholar]
- 79.Nam J-M, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2005;301:1884. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 80.Adams J, Parder PJ, Downey S, Forbes MA, MacLennan K, Allagar V, Kaufman S, Hallam S, Bicknell R, Walker JJ, Cairnduff F, Selby PJ, Perren TJ, Landsdown M, Banks R. Vascular endothelial growth factor (VEGF) in breast cancer: comparison of plasma, serum, and tissue VEGF and microvessel density and effects of tamoxifen. Cancer Res. 2000;60:2898–2905. [PubMed] [Google Scholar]
- 81.Gasco M, Crook T. The p53 network in head and neck cancer. Oral Oncol. 2003;39:222–231. doi: 10.1016/s1368-8375(02)00163-x. [DOI] [PubMed] [Google Scholar]
- 82.Campagnolo C, Ryan TE, Atkinson RC, Batt CA. Real-time, label-free monitoring of the binding of human sera antibodies to tumor antigens. 58th Northwest Regional Meeting of the ACS; Bozeman, MT. 2003. [Google Scholar]
- 83.Xie K. Interleukin-8 and human cancer biology. Cytokine Growth Factor Rev. 2001;12:375–391. doi: 10.1016/s1359-6101(01)00016-8. [DOI] [PubMed] [Google Scholar]
- 84.St John MAR, Li Y, Zhou X, Denny P, Ho C-M, Montemagno C, Shi W, Qi F, Wu B, Sinha BU, Jordan R, Wolinsky L, Park N-H, Liu H, Abemayor E, Wong E, David TW. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130:929–935. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- 85.Viscidi RP, Ahdieh-Grant L, Clayman B, Fox K, Massad LS, Cu-Uvin S, Shah KV, Anastos KM, Squires KE, Duerr A, Jamieson DJ, Burk RD, Klein RS, Minkoff H, Palefsky J, Strickler H, Schuman P, Piessens E, Miotti P. Serum immunoglobulin G response to human papillomavirus type 16 virus-like particles in human immunodeficiency virus (HIV)-positive and risk-matched HIV-negative women. J Infect Dis. 2003;187:194–205. doi: 10.1086/346052. [DOI] [PubMed] [Google Scholar]
- 86.Psyrri A, DiMaio D. Human papillomavirus in cervical and head-and-neck cancer. Nat Clin Pract Oncol. 2008;5:24–31. doi: 10.1038/ncponc0984. [DOI] [PubMed] [Google Scholar]
- 87.Ravaggi A, Romani C, Pasinetti B, Tassi RA, Bignotti E, Bandiera E, Odicino FE, Ragnoli M, Donzelli C, Falchetti M, Calza S, Santin AD, Pecorelli S. Correlation between serological immune response analyzed by a new ELISA for HPV-16/18 E7 oncoprotein and clinical characteristics of cervical cancer patients. Arch Virol. 2006;151:1899–1916. doi: 10.1007/s00705-006-0787-y. [DOI] [PubMed] [Google Scholar]
- 88.Sehr P, Zumbach K, Pawlita M. A generic capture ELISA for recombinant proteins fused to glutathione S-transferase: validation for HPV serology. J Immunol Methods. 2001;253:153–162. doi: 10.1016/s0022-1759(01)00376-3. [DOI] [PubMed] [Google Scholar]
- 89.Heineman WR, Halsall HB. Strategies for electrochemical immunoassay. Anal Chem. 1985;75:1321A–1331A. doi: 10.1021/ac00289a804. [DOI] [PubMed] [Google Scholar]
- 90.Vijayawardhana CA, Halsall HB, Heineman WR. Milestones of Electrochemical Immunoassay at Cincinnati. In: Chambers JQ, Bratjer-Toth A, editors. Electroanalytical Methods for Biological Materials. Marcel Dekker; NY: 2002. pp. 195–231. [Google Scholar]
- 91.Ronkainen-Matsuno NJ, Thomas JH, Halsall HB, Heineman WR. Electrochemical immunoassay moving into the fast lane. TrAC Trends Anal Chem. 2002;21:213–225. [Google Scholar]
- 92.Bange A, Halsall HB, Heineman WR. Microfluidic immunosensor systems. Biosens Bioelectron. 2005;20:2488–2503. doi: 10.1016/j.bios.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 93.Lu B, Smyth MR, O’Kennedy R. Immunological activities of IgG antibody on pre-coated Fc receptor surfaces. Anal Chim Acta. 1996;331:97–102. [Google Scholar]
- 94.Carter RM, Poli MA, Pesavento M, Sibley DET, Lubrano GJ, Guilbault GG. Immunoelectrochemical biosensors for detection of saxitoxin and brevetoxin. ImmunoMethods. 1993;3:128–133. [Google Scholar]
- 95.Warsinke A, Benkert A, Scheller FW. Electrochemical immunoassays. Fresenius’ J Anal Chem. 2000;366:622–634. doi: 10.1007/s002160051557. [DOI] [PubMed] [Google Scholar]
- 96.Lu B, Smyth MR, O’Kennedy R, Moulds J, Frame T. Development of an amperometric immunosensor based on flow-injection analysis for the detection of red blood cells. Anal Chim Acta. 1997;340:175–180. [Google Scholar]
- 97.Yakovleva J, Emneus J. Electrochemical Immunoassays. In: Bartlett PN, editor. Handbook of Bioelectrochemistry. John Wiley; NY: 2008. pp. 377–410. [Google Scholar]
- 98.Ruzgas T, Lindgren A, Gorton L, Hecht H-J, Reichelt J, Bilitewski U. Electrochemistry of Peroxidases. In: Chambers JQ, Brajter-Toth A, editors. Electroanalytical Methods for Biological Materials. Marcel Dekker; New York: 2002. pp. 233–254. [Google Scholar]
- 99.Kojima K, Hiratsuka A, Suzuki H, Yano K, Ikebukuro K, Karube I. Electrochemical protein chip arrayed immunosensors with antibodies immobilized in a plasma-polymerized film. Anal Chem. 2003;75:1116–1122. doi: 10.1021/ac0257391. [DOI] [PubMed] [Google Scholar]
- 100.Wang J. Nanoparticle-based electrochemical bioassays of proteins nanoparticle-based electrochemical bioassays of proteins. Electroanalysis. 2007;19:769–776. [Google Scholar]
- 101.Luo X, Morrin A, Killard AJ, Smyth MR. Application of nanoparticles in electrochemical sensors and biosensors. Electroanalysis. 2006;18:319–326. [Google Scholar]
- 102.Bourdillon C, Demaille C, Moiroux J, Savéant J-M. Activation and diffusion in the kinetics of adsorption and molecular recognition on surfaces. Enzyme-amplified electrochemical approach to biorecognition dynamics illustrated by the binding of antibodies to immobilized antigens. J Am Chem Soc. 1999;121:2401–2408. [Google Scholar]
- 103.Limoges B, Marchal D, Mavré F, Savéant J-M, Schöllhorn B. Theory and practice of enzyme bioaffinity electrodes. Direct electrochemical product detection. J Am Chem Soc. 2008;103:7259–7275. doi: 10.1021/ja7102845. [DOI] [PubMed] [Google Scholar]
- 104.Limoges B, Marchal D, Mavré F, Savéant J-M. Theory and practice of enzyme bioaffinity electrodes. Chemical, enzymatic, and electrochemical amplification of in situ product detection. J Am Chem Soc. 2008;103:7276–7285. doi: 10.1021/ja7102873. [DOI] [PubMed] [Google Scholar]
- 105.Veetil JV, Ye K. Development of immunosensors using carbon nanotubes. Biotechnol Prog. 2007;23:517–531. doi: 10.1021/bp0602395. [DOI] [PubMed] [Google Scholar]
- 106.Kim S-N, Rusling JF, Papadimitrakopolous F. Carbon nanotubes in electronic and electrochemical detection of biomolecules. Adv Mater. 2007;19:3214–3228. doi: 10.1002/adma.200700665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Patolsky F, Zheng G, Lieber CM. Nanowire-based biosensors. Anal Chem. 2006;78:4260–4269. doi: 10.1021/ac069419j. [DOI] [PubMed] [Google Scholar]
- 108.Tkac J, Davis JJ. Label-Free Field Effect Protein Sensing. In: Davis JJ, editor. Engineering the Bioelectronic Interface. Royal Society of Chemistry; UK: 2009. pp. 193–224. [Google Scholar]
- 109.Stern E, Vacic A, Rajan NK, Criscione JM, Park J, Ilic BR, Mooney DJ, Reed MA, Fahmy TM. Label-free biomarker detection from whole blood. Nat Nanotechnol. 2009;5:138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu X, Munge B, Patel V, Jensen G, Bhirde A, Gong J, Kim S, Gillespie J, Gutkind S, Papadimitrakopolous F, Rusling JF. Carbon nanotube amplification strategies for highly sensitive immunosensing of cancer biomarkers in serum and tissue. J Am Chem Soc. 2006;128:11199–11205. doi: 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mani V, Chikkaveeraiah BV, Patel V, Gutkind JS, Rusling JF. Ultrasensitive immunosensor for cancer biomarker proteins using gold nanoparticle film electrodes and Multienzyme-particle amplification. ACS Nano. 2009;3:585–594. doi: 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang H, Zhao Q, Li X-F, Le XC. Ultrasensitive assays for proteins. Analyst. 2007;132:724–737. doi: 10.1039/b704256f. [DOI] [PubMed] [Google Scholar]
- 113.Wang J. Nanomaterial-based electrochemical biosensors. Analyst. 2005;130:421–426. doi: 10.1039/b414248a. [DOI] [PubMed] [Google Scholar]
- 114.Wang J. Nanomaterial-based amplified transduction of biomolecular interactions. Small. 2005;1:1036–1043. doi: 10.1002/smll.200500214. [DOI] [PubMed] [Google Scholar]
- 115.Rusling JF, Yu X, Munge BS, Kim SN, Papadimitrakopoulos F. In: Engineering the Bioelectronic Interface. Davis J, editor. Royal Society of Chemistry; UK: 2009. pp. 94–118. [Google Scholar]
- 116.Dequaire M, Degrand C, Limoges B. An electrochemical metalloimmunoassay based on a colloidal gold label. Anal Chem. 2000;72:5521. doi: 10.1021/ac000781m. [DOI] [PubMed] [Google Scholar]
- 117.Guo H, He N, Ge S, Yang D, Zhang J. MCM-41 mesoporous material modified carbon paste electrode for the determination of cardiac troponin I by anodic stripping voltammetry. Talanta. 2005;68:61. doi: 10.1016/j.talanta.2005.04.067. [DOI] [PubMed] [Google Scholar]
- 118.Velev OD, Kaler EW. In situ assembly of colloidal particles into miniaturized biosensors. Langmuir. 1999;15:3693. [Google Scholar]
- 119.Wang J, Liu G, Jan MR. Ultrasensitive electrical biosensing of proteins and DNA: carbon-nanotube derived amplification of the recognition and transduction events. J Am Chem Soc. 2004;126:3010–3011. doi: 10.1021/ja031723w. [DOI] [PubMed] [Google Scholar]
- 120.Munge B, Liu G, Collins G, Wang J. Multiple enzyme layers on carbon nanotubes for electrochemical detection down to 80 DNA copies. Anal Chem. 2005;77:4662–4666. doi: 10.1021/ac050132g. [DOI] [PubMed] [Google Scholar]
- 121.Rusling JF, Sotzing G, Papadimitrakopoulos F. Designing nanomaterials-enhanced electrochemical immunosensors for cancer biomarker proteins. Bioelectrochemistry. 2009;76:189–194. doi: 10.1016/j.bioelechem.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Munge BS, Krause CE, Malhotra R, Patel V, Gutkind JS, Rusling JF. Electrochemical immunosensors for interleukin-6. Comparison of carbon nanotube forest and gold nanoparticle platforms. Electrochem Commun. 2009;11:1009–1012. doi: 10.1016/j.elecom.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Malhotra R, Patel V, Vaqué JP, Gutkind JS, Rusling JF. Ultrasensitive electrochemical immunosensor for oral cancer biomarker IL-6 using carbon nanotube forest electrodes and multilabel amplification. Anal Chem. 2010;82:3118–3123. doi: 10.1021/ac902802b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jensen GC, Yu X, Munge B, Bhirde A, Gong JD, Kim SN, Papadimitrakopoulos F, Rusling JF. Characterization of multienzyme–antibody–carbon nanotube bioconjugates for immunosensors. J Nanosci Nanotechnol. 2009;9:249–255. doi: 10.1166/jnn.2008.J016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Daniels JS, Pourmanda N. Label-free impedance biosensors: opportunities and challenges. Electroanalysis. 2007;19:1239–1257. doi: 10.1002/elan.200603855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Berggren C, Bjarnason B, Johansson G. An immunological interleukine-6 capacitive biosensor using perturbation with a potentiostatic step. Biosens Bioelectron. 1998;13:1061–1068. doi: 10.1016/s0956-5663(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 127.Bart M, Stigter ECA, Stapert HR, de Jong GJ, van Bennekom WP. On the response of a label-free interferon-γ immunosensor utilizing electrochemical impedance spectroscopy. Biosens Bioelectron. 2005;21:49–59. doi: 10.1016/j.bios.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 128.Liu G, Wang J, Kim J, Jan M, Collins G. Electrochemical coding for multiplexed immunoassays of proteins. Anal Chem. 2004;76:7126–7130. doi: 10.1021/ac049107l. [DOI] [PubMed] [Google Scholar]
- 129.Wilson MS. Electrochemical immunosensors for the simultaneous detection of two tumor markers. Anal Chem. 2005;77:1496–1502. doi: 10.1021/ac0485278. [DOI] [PubMed] [Google Scholar]
- 130.Wilson MS, Nie W. Electrochemical multianalyte immunoassays using an array-based sensor. Anal Chem. 2006;78:2507–2513. doi: 10.1021/ac0518452. [DOI] [PubMed] [Google Scholar]
- 131.Wilson MS, Nie W. Multiplex measurement of seven tumor markers using an electrochemical protein chip. Anal Chem. 2006;78:6476–6483. doi: 10.1021/ac060843u. [DOI] [PubMed] [Google Scholar]
- 132.Dill K, Montgomery DD, Ghindilis AL, Schwarzkopf KR, Ragsdale SR, Oleinikov AV. Biosens Bioelectron. 2004;20:736–742. doi: 10.1016/j.bios.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 133.Dill K, Ghindilis A, Schwarzkopf K. Multiplexed analyte and oligonucleotide detection on microarrays using several redox enzymes in conjunction with electrochemical detection. Lab Chip. 2006;6:1052–1055. doi: 10.1039/b600126b. [DOI] [PubMed] [Google Scholar]
- 134.Gau V, Wong D. Oral fluid Nanosensor test (OFNASET) with Advanced electrochemical-based molecular analysis platform. Ann N Y Acad Sci. 2007;1098:401–410. doi: 10.1196/annals.1384.005. [DOI] [PubMed] [Google Scholar]
- 135.Wei F, Liao W, Xu Z, Yang Y, Wong DT, Ho C-M. Bio/abiotic interface constructed from nanoscale DNA dendrimer and conducting polymer for ultrasensitive biomolecular diagnosis. Small. 2009;5:1784–1790. doi: 10.1002/smll.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wei F, Patel P, Liao W, Chaudhry K, Zhang L, Arellano-Garcia M, Hu S, Elashoff D, Zhou H, Shukla S, Shah F, Ho C-M, Wong DT. Electrochemical sensor for multiplex biomarkers detection. Clin Cancer Res. 2009;15:4446–4452. doi: 10.1158/1078-0432.CCR-09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chikkaveeriah BV, Bhirde A, Malhotra R, Patel V, Gutkind JS, Rusling JF. Single-wall carbon nanotube forest immunoarrays for electrochemical measurement of 4 protein biomarkers for prostate cancer. Anal Chem. 2009;81:9129–9134. doi: 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Debad JB, Glezer EN, Leland JK, Sigal GB, Wholstadter J. In: Electrogenerated Chemiluminescence. Bard AJ, editor. Marcel Dekker; NY: 2004. pp. 359–396. [Google Scholar]
- 139.http://rochediagnostics.ca/lab/solutions/e2010.php.
- 140.http://www.meso-scale.com/CatalogSystemWeb/WebRoot/technology/ecl.htm.
- 141.Deiss F, LaFratta CN, Symer M, Blicharz TM, Sojic N, Walt DR. Multiplexed sandwich immunoassays using electrochemiluminescence imaging resolved at the single bead level. J Am Chem Soc. 2009;131:6088–6089. doi: 10.1021/ja901876z. [DOI] [PMC free article] [PubMed] [Google Scholar]