Abstract
Constitutive androstane receptor (CAR) and pregnane X receptor (PXR) are closely related orphan nuclear receptor proteins that share several ligands and target overlapping sets of genes involved in homeostasis and all phases of drug metabolism. CAR and PXR are involved in the development of certain diseases, including diabetes, metabolic syndrome and obesity. Ligand screens for these receptors so far have typically focused on steroid hormone analogs with pharmacophore-based approaches, only to find relatively few new hits. Multiple CAR isoforms have been detected in human liver, with the most abundant being the constitutively active reference, CAR1, and the ligand-dependent isoform CAR3. It has been assumed that any compound that binds CAR1 should also activate CAR3, and so CAR3 can be used as a ligand-activated surrogate for CAR1 studies. The possibility of CAR3-specific ligands has not, so far, been addressed. To investigate the differences between CAR1, CAR3 and PXR, and to look for more CAR ligands that may be of use in quantitative structure-activity relationship (QSAR) studies, we performed a luciferase transactivation assay screen of 60 mostly non-steroid compounds. Known active compounds with different core chemistries were chosen as starting points and structural variants were rationally selected for screening. Distinct differences in agonist versus inverse agonist/antagonist effects were seen in 49 compounds that had some ligand effect on at least one receptor and 18 that had effects on all three receptors; eight were CAR1 ligands only, three were CAR3 only ligands and four affected PXR only. This work provides evidence for new CAR ligands, some of which have CAR3-specific effects, and provides observational data on CAR and PXR ligands with which to inform in silico strategies. Compounds that demonstrated unique activity on any one receptor are potentially valuable diagnostic tools for the investigation of in vivo molecular targets.
Keywords: CAR, PXR, QSAR, nuclear receptor, isoform-specific activity, mid-throughput screening
1. Introduction
Constitutive androstane receptor (CAR, NR1I3) and pregnane X receptor (PXR, NR1I2, also known as steroid and xenobiotic receptor (SXR)) are orphan nuclear receptor (NR) proteins, closely related in structure and function, which regulate the transcription of genes involved in all phases of endo- and xeno-biotic metabolism, including various cytochrome P450 genes [1]. Animal studies have uncovered key roles for CAR and PXR in the regulation of hepatic energy metabolism through complex cross-talk involving insulin and thyroid hormone and transcription factors including peroxisome proliferator-activated receptor (PPAR)-α, PPAR-γ coactivator 1-α (PGC1-α) and forkhead box O1 (FOXO1) [2,3,4,5,6,7]. CAR plays varying roles in weight loss. CAR null mice show increased weight loss compared to wild type controls when under caloric restriction [3]. Wild type mice with high-fat diet induced obesity and the leptin deficient ob/ob mice show weight loss when treated with mouse CAR selective activator 1, 4- Bis[2-(3, 5- dichloropyridyloxy)]benzene (TCPOBOP) [8]; however CAR null mice on a high-fat diet show no response to TCPOBOP [7]. While there are significant differences between human and mouse CAR ligands, the metabolic pathways in which the nuclear receptors themselves are involved show more similarity. The activation of human CAR and/or PXR by exposure to environmental contaminants, pharmaceutics and certain natural products therefore has implications for human health, in particular for metabolic syndrome, diabetes and obesity.
Multiple CAR mRNA isoforms have been detected in human liver, reviewed in [1], at least 15 of which have NCBI reference sequences (http://www.ncbi.nlm.nih.gov/gene/9970). The two most abundant hepatic CAR isoforms are wild type reference CAR (CAR1) and the CAR3 splice variant (also known as SV2) [9] that contains a 5 amino acid (APYLT) insertion between exons 7 and 8 in the ligand binding domain (LBD). The third most common CAR variant, CAR2 (SV1), has a four amino acid (SPTV) insertion in exon 7. As the name suggests, CAR1 has constitutively high basal activity on the promoters of target genes in the absence of ligand, however, splice variants CAR3 and CAR2, are ligand-activated like PXR. Since many CAR ligands also activate PXR [10,11,12] (reviewed in [1]), few CAR-specific ligands have been discovered and CAR splice variant-specific ligands have not been fully investigated. A recent study by DeKeyser et al. (2009) characterized the plasticizer di(2-ethylhexyl)phthalate as a potent specific activator of CAR2 [13]. It is therefore feasible that CAR3 may also have specific ligands and that CAR1 ligands may have different activities with CAR3.
There are many structural similarities between PXR and CAR, however PXR has a larger, more flexible, LBD (1280–1544 Å3 [14]) than CAR1 (675 Å3 [15]) and has a greater number of known activators that vary considerably in size and shape. There are currently no crystal structures available for the CAR splice variants. CAR proteins from different species also share a high degree of sequence and structural homology but with significant variations in the LBDs which correlate with species-specific effects [16]. There are relatively few ligands of both mouse and human CAR, and of these, most mouse CAR ligands have inverse agonist effects on human CAR making it difficult to extrapolate from mouse models to effects on human conditions. Both CAR and PXR form heterodimers with retinoid X receptor α (RXRα, NR2B1), in which the DNA binding domains (DBD) of each protein bind to one half of various nuclear receptor binding site motifs in the promoter regions of target genes (including direct repeats (DR1 [17], DR3, DR4 [18] and DR5 [19]), and everted repeat-8 (ER8) elements [18]). DR4 response elements are especially optimal for human CAR responses [18] whereas PXR favors ER6 motifs. CAR and PXR also interact with an overlapping set of accessory proteins to activate or repress transcription. Reporter plasmids containing optimized consensus DR4 sequence or ER6 motifs upstream of firefly luciferase can be used in transactivation assays to screen compounds for specific CAR or PXR effects, respectively. CAR1 has constitutive high basal activity on the promoters of downstream targets in the absence of ligand. CAR1 ligands can therefore have agonist or inverse agonist effects on the transcription of target genes via the recruitment of coactivators or corepressors, respectively. Conversely, CAR3 and PXR have little or no basal activity and require ligand binding to initiate activation or repression of downstream target genes. CAR can also be activated independently of ligand binding by compounds including phenobarbital, phenytoin and bilirubin [20,21,22], treatment with which causes cytoplasmic CAR1 to translocate to the nucleus and up-regulate CYP450 2B6. PXR is ligand-activated but additional ligand binding sites have been described for PXR outside of the LBD pocket [23], adding another layer of complexity to PXR bio-sensing and control.
Given the promiscuous nature of CAR and its pivotal role in detoxification and the maintenance of metabolic homeostasis, it is perhaps surprising that relatively few human CAR ligands have been reported so far, reviewed in [1]. Some pharmaceutical agents are known ligands of human CAR, including the antihistamine meclizine, which is an inverse agonist [24], and the antifungal compound clotrimazole, which has been reported variously as an agonist and an inverse agonist [25,10]. By far the strongest reported CAR ligand is the wholly synthetic compound 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) with an EC50 of 49 nM [11]. Experience with CITCO however is far from uniform and problems with stability have been noted [26]. We have experienced similar variability with CITCO over time and believe the discovery of more reliable positive controls for in vitro studies would be beneficial.
Off-target activation of CAR and PXR by pharmaceutics can lead to adverse drug-drug interactions [27]. There is a need to define clear structure-activity relationships among CAR and PXR ligands in order to optimize drug therapies and to better understand the possible impacts of environmental contaminants and dietary components on health and disease. A recent review of CAR ligand searches from in silico quantitative structure-activity relationship (QSAR) and pharmacophore modeling approaches demonstrated that, despite crystal structures of human CAR now being available, true CAR and PXR ligands are still hard to find using even the most complex of models [26]. One approach to rational drug discovery uses known active structures as starting points for modification. We hypothesized that subtle differences in the structural components of small molecules define CAR1, CAR3 and/or PXR binding preference and determine agonist, antagonist or inverse agonist activity. We performed a rational QSAR (RQSAR), a mid-throughput targeted screen of 60 compounds in transient transfection luciferase assays in human hepatoma cell line, HuH-7, using human CAR1, CAR3 and PXR expression plasmids to look for differences in ligand behavior. We were particularly interested in looking for structural differences in the types of compounds that act as agonists and inverse agonists of CAR1, and whether or not the same compounds were also ligands of CAR3. Steroid-based structures have been more fully investigated elsewhere, so this study focused on exogenous, non-steroidal groups of compounds. Structurally similar compounds that have opposite effects on the two CAR isoforms may provide important clues as to the nature of characteristics important for ligand binding. The results presented here provide more detail on the molecular features that do and do not contribute to agonist and antagonist activity, of utility for pharmacophore development.
2. Methods
2.1 Reagents
Chemical compounds were sourced from Acros Organics (Morris Plains, NJ), Alexis Biochemicals (part of Enzo Life Sciences, Farmingdale, NY), Fisher Scientific (Pittsburgh, PA), LKT Laboratories, Inc. (St. Paul, MN), MP Biomedicals (Solon, OH), Sigma-Aldrich (St. Louis, MO), Steraloids (Newport, RI), Tocris Bioscience (Ellisville, MO) and Ultra (N. Kingstown, RI). See supplementary table S1 for individual names, suppliers and CAS numbers. All compounds were dissolved in dimethylsulfoxide (DMSO) (Fisher Scientific) to 1000 × final concentration so that solvent concentration was not more than 0.2% of the final volume.
2.2 Cell culture
Human hepatoma HuH-7 cells (JCRB0403) were maintained in a humidified incubator at 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 5% fetal bovine serum (v/v), 1.5 g/L sodium bicarbonate, 1 µM sodium pyruvate, 10 mM HEPES, 2 mM L-glutamine, 100 units/mL penicillin 100 µg/mL streptomycin, 100 µM non-essential amino acids. Tissue culture media and additives were from Invitrogen/GIBCO Corp. (Carlsbad, CA) or Lonza (Hopkinton, MA).
2.3 Rational selection of compounds for mid-throughput QSAR screen
Compounds were selected to represent different structural groups, to expand sets of known ligands and to act as internal controls for key structural features. Known small-molecule ligands of CAR and/or PXR were used as starting points and, where possible, compounds were selected to differ from the next by just one substituent. Group A (triphenyls) centers on clotrimazole. Group B (phthaleins) is a logical expansion of the triphenyl set A. Group C is a group of alkylphenols. Group D contains artificial estrogen stilbenes and related natural plant derivative stilbenoids. The diphenyl group (E) shares similarities with group D but are centered on the known endocrine-disruptor bisphenol A (BPA). Group F (antihistamines and antifungals) are partly an extension of the diphenyl set, from two benzene rings to multi-ring structures, including indoles and azoles; this set in particular includes compounds with larger, more flexible molecular structures and are also licensed pharmaceutics. Groups G and H contain pairs of related and singlet small molecules of interest that do not naturally fit with the other groups.
2.4 Transcriptional activation assays
2.4.1 Plasmids
The CAR-responsive reporter plasmid was pFR-Luc, which contains five copies of the yeast GAL4 binding element upstream activation sequence (UAS), upstream of firefly luciferase; the PXR-responsive plasmid was pCYP3A4-XREM-TK-Luc which contains the proximal ER6 and distal XREM elements of human CYP3A4 promoter upstream of thymidine kinase (TK) enhancer element and firefly luciferase. CAR expression plasmids were pM-CAR1 (GAL4 DBD upstream of full length reference CAR) and splice variant pM-CAR3 (GAL4 DBD-full length reference CAR with APYLT insertion in the LBD). The PXR expression plasmid p3XFLAG-PXR (3X FLAG is an amino-terminal fusion of three copies of Sigma Company flu antigen-derived epitope) was cotransfected with dimerization partner p3XFLAG-RXRα. Green fluorescent protein plasmid, pEGFP-C1 (Clontech, TAKARA Bio Group, Mountain View, CA), and Renilla luciferase plasmid, pRL-CMV (Promega, Madison, WI), were co-transfected for quality control and data normalization, respectively. All expression and reporter plasmids used in this study were from human gene variants. Original cloning was performed in the laboratory of Dr Curtis Omiecinski, The Pennsylvania State University, University Park, PA [28,29].
2.4.2 Transient transfections
Polyethylenimine (PEI, 25kD 1 mg/ml): plasmid DNA complexes (1 µg plasmid: 4 µL PEI) were formed in aqueous solution (20 mM HEPES, pH 7) and added to HuH-7 cells in serum-free α-minimal essential medium (α-MEM). Each near-confluent T25 flask HuH-7 cells was transfected with 5 µg reporter plasmid combined with 0.5 µg pRL-CMV, and 1.25 µg each of CAR or PXR and RXRα expression plasmid and pEGFP-C1.
2.4.3 Treatments
After 20 h transfection, cells were trypsinized (0.25% Trypsin/0.038% EDTA) and transferred to white-walled, clear-bottomed 96-well plates in fully supplemented DMEM. After 2–4 h attachment, chemical treatments were added at 10 µM final concentration. CAR1 transfected cells were cotreated with 10 µM 1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide (PK11195), a known CAR1 inverse agonist [12], to lower basal activity, widening the dynamic range of the assay to show any agonist effects more clearly. Similarly, PXR plates were cotreated with known PXR agonist, rifampicin (25 µM) [30], to raise PXR activity in order to show any antagonist effects of screening compounds.
2.4.4 Data collection and analysis
After 20 h incubation at 37°C, cells were passively lysed and read in situ with Stop-N-Glo reagent (Promega, Madison, WI) on a Lumicount luminometer. Firefly luciferase values were normalized against Renilla luciferase, per well. Replicates were subject to Grubbs’ outlier detection and any significant outliers (z > 1.15) were removed prior to mean calculation. The number of replicate wells per plate (n) equals four unless otherwise stated (see individual figure captions). Student’s unpaired t-tests were performed within each plate and significance at p < 0.05, 0.01 or 0.001 was determined. Fold change relative to within-plate DMSO control was calculated prior to between-plate comparison. For graphical display, a median DMSO control value was selected from among plates for the PK11195 and rifampicin cotreatments. In order to give equal visual weight to agonist and inverse agonist/antagonist effects, CAR1 and PXR data are displayed as log2 fold change, and CAR3 data are displayed using log10 scale, +/− SEM corrected for the appropriate log scale. SEM provides a measure of the variability surrounding the mean, while taking into account the number of replicates.
2.5 In silico molecular properties calculation
Molecular properties were determined using Discovery Studio Client (Accelrys, San Diego, CA) general protocols. Input MDL.mol files were drawn in 2D using MarvinSketch (www.chemaxon.com) and converted to 3D format in Discovery Studio. See supplementary data Excel file S2.
3. Results and Discussion
3.1 Group A – Triphenyls (triphenylmethane derivatives), Figure 1, Table 1
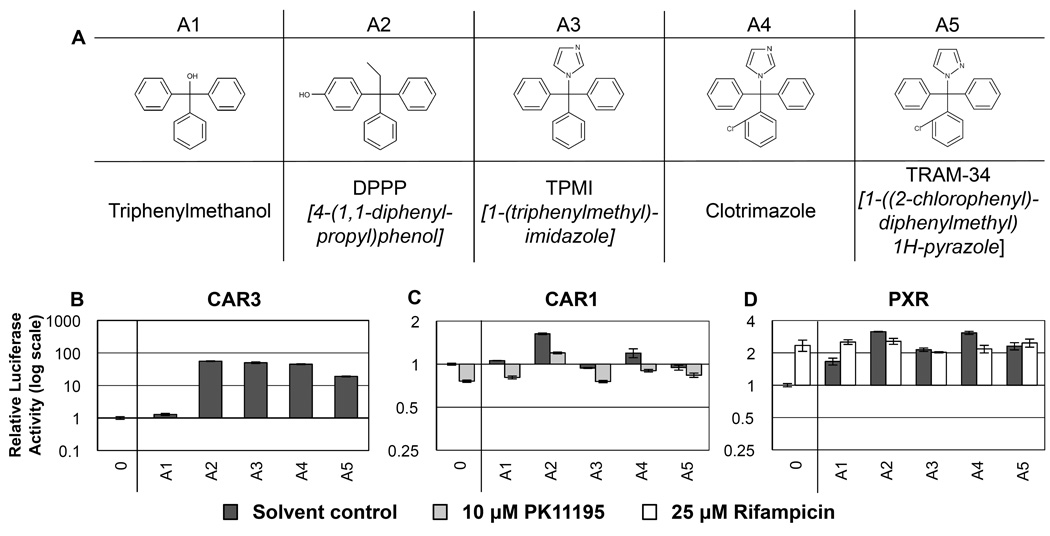
Figure 1. RQSAR for group A compounds - Triphenyls.
(A) 2D structures for group A triphenyls. (B) CAR3-driven relative luciferase activity, (C) CAR1, and (D) PXR treated with 10 µM test compound, plotted using log scales. 0 = DMSO solvent control. Cotreatments were PK11195 (10 µM) for CAR1 and rifampicin (25µM) for PXR. (n=4 except for B: A2, A4 and A5 n=8. C: A1 n=3).
Table 1.
Relative luciferase activity values for CAR1, CAR3 and PXR treated with group A compounds (Triphenyls).
| CAR3 | CAR1 | PXR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PK11195 | Control | Rifampicin | ||||||||
| ID | Chemical Name | Value | Siga | Value | Sig | Value | Sig | Value | Sig | Value | Sig |
| 0 | Solvent control | 1.00 | - | 1.00 | - | 0.76 | - | 1.00 | - | 2.35 | - |
| A1 | Triphenylmethanol | 1.29 | - | 1.06 | ** | 0.81 | - | 1.66 | ** | 2.52 | - |
| A2 | DPPP [4-(1,1-diphenylpropyl)phenol] | 55.86 | *** | 1.63 | *** | 1.20 | *** | 3.15 | *** | 2.57 | - |
| A3 | TPMI [1-(triphenylmethyl) imidazole] | 50.10 | *** | 0.94 | ** | 0.76 | - | 2.14 | *** | 2.03 | - |
| A4 | Clotrimazole | 45.18 | *** | 1.20 | - | 0.90 | - | 3.07 | *** | 2.18 | - |
| A5 | TRAM-34 [1-((2-chlorophenyl) diphenylmethyl)-1H-pyrazole] | 19.02 | *** | 0.95 | - | 0.84 | - | 2.31 | * | 2.47 | - |
Sig = significance in unpaired t-test
 Significant agonist effect
Significant agonist effect
 Significant inverse agonist/antagonist effect
Significant inverse agonist/antagonist effect
0.01 < p < 0.05;
0.001 < p < 0.01;
p < 0.001
The triphenyl anti-fungal compound clotrimazole (A4), a strong human PXR agonist, has been reported variously to have human CAR agonist and inverse agonist response element-dependent activities [10,31,25]. Group A comprised four related triphenyl compounds in addition to clotrimazole (Fig. 1A), all five of which had robust agonist ligand effects on PXR (Fig. 1D, Table 1). Three compounds had significant effects on CAR1; triphenylmethanol (A1) and DPPP (A2) were agonists, whereas TPMI (A3) acted as an inverse agonist (Fig. 1C). CAR3 responses were more similar to PXR than to CAR1 with four out of five compounds having agonist ligand effects, the exception being triphenylmethanol (Fig. 1B). The main structural difference between triphenylmethanol and the other four compounds is the absence of a bulky non-polar group in place of the hydroxyl substituent on the central carbon (Fig. 1A).
DPPP (A2) had the strongest agonist effect of this set on all three nuclear receptors. Differences between DPPP and TPMI (A3) were sufficient to cause a switch in effect from CAR1 agonist to inverse agonist. The addition of an ortho-chlorine to a phenyl group of TPMI (A3 → clotrimazole, A4) removed this CAR1 inverse agonist activity. TRAM-34 (A5), which differs from clotrimazole only in the substitution of a pyrazole for the imidazole ring, had no effect on CAR1 and lesser effects on CAR3 and PXR than A4, which implies that imidazoles could have greater nuclear receptor agonist activity than pyrazoles.
Interestingly, while triphenylmethanol (A1) was an agonist ligand of PXR and CAR1 with no significant effect on CAR3, closely related DPPP (A2) was an agonist ligand of all three nuclear receptors eliciting the greatest magnitude of response of this triphenyl subset. Para-OH groups are implicated as being important for the effects of artificial estrogens on the estrogen receptor (ERα) [32], and it appears these groups may be important for the activation of more general biosensors like CAR and PXR. DPPP is the only compound of this set with a para-OH group, which may partly explain the strength of response compared to the other compounds. While triphenylmethanol has a hydroxyl group it does not appear to be involved in agonist interactions with CAR3. For CAR3 specificity, the presence of a propyl group on DPPP in place of the hydroxyl group of triphenylmethanol may have greater importance. From the observations of group A, we would predict that hydrophobic interactions in the binding pocket of CAR3 are more important for agonist effects than potential hydrogen bond formation.
3.2 Group B - Phthaleins, Figure 2, Table 2
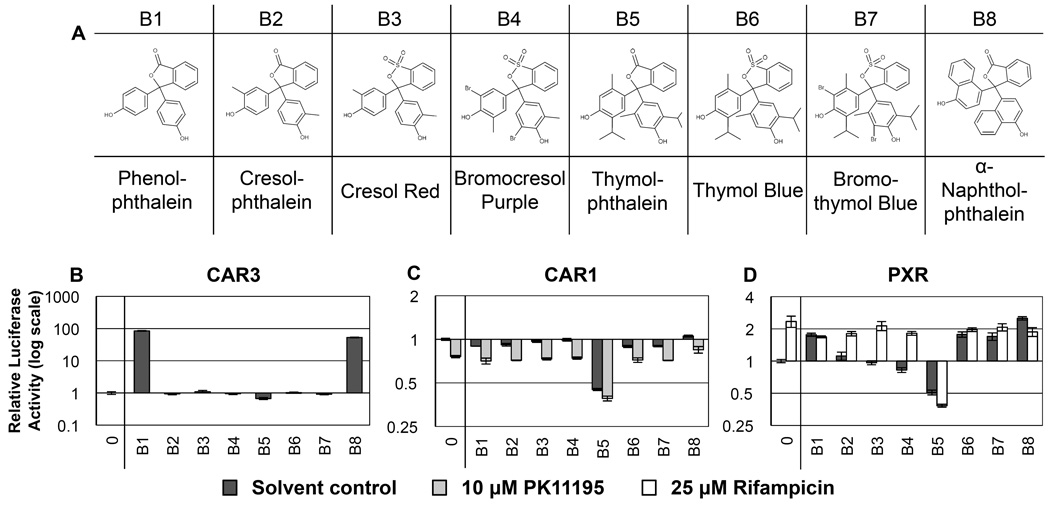
Figure 2. RQSAR for group B - Phthaleins.
(A) 2D structures of group B phthaleins. (B) CAR3-driven relative luciferase activity, (C) CAR1, and (D) PXR treated with 10 µM test compound, plotted using log scales. 0 = DMSO solvent control. Cotreatments were PK11195 (10 µM) for CAR1 and rifampicin (25 µM) for PXR. (n=4 except for B: B1 n=7, B2–8 n=8. C: B1 n=3).
Table 2.
Relative luciferase activity values for CAR1, CAR3 and PXR treated with group B compounds (Phthaleins).
| CAR3 | CAR1 | PXR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PK11195 | Control | Rifampicin | ||||||||
| ID | Chemical Name | Value | Siga | Value | Sig | Value | Sig | Value | Sig | Value | Sig |
| 0 | Solvent control | 1.00 | - | 1.00 | - | 0.76 | - | 1.00 | - | 2.35 | - |
| B1 | Phenolphthalein | 84.96 | *** | 0.90 | * | 0.71 | - | 1.76 | *** | 1.68 | - |
| B2 | Cresolphthalein | 0.94 | - | 0.92 | * | 0.72 | - | 1.11 | - | 1.80 | - |
| B3 | Cresol Red | 1.10 | - | 0.97 | - | 0.73 | - | 0.96 | - | 2.13 | - |
| B4 | Bromocresol Purple | 0.95 | - | 0.99 | - | 0.74 | - | 0.82 | * | 1.82 | - |
| B5 | Thymolphthalein | 0.67 | *** | 0.45 | *** | 0.39 | *** | 0.51 | *** | 0.38 | *** |
| B6 | Thymol Blue | 1.03 | - | 0.89 | ** | 0.72 | - | 1.77 | *** | 1.97 | - |
| B7 | Bromothymol Blue | 0.91 | - | 0.90 | ** | 0.71 | - | 1.70 | ** | 2.07 | - |
| B8 | α-Naphtholphthalein | 52.92 | *** | 1.05 | - | 0.85 | - | 2.51 | *** | 1.87 | - |
Sig = significance in unpaired t-test
 Significant agonist effect
Significant agonist effect
 Significant inverse agonist/antagonist effect
Significant inverse agonist/antagonist effect
0.01 < p < 0.05;
0.001 < p < 0.01;
p < 0.001
The phthaleins are a structural expansion of the triphenyl group A, which were chosen to further investigate the structure-activity relationship (SAR) of the triphenyl compounds. Phenolphthalein was used in certain laxatives until a 1997 FDA decision withdrew approval following findings that it caused tumors in rodents and could have estrogenic effects on human breast cancer cells in vitro [33]; however, no harmful effects in humans have yet been identified [34]. Given the known inter-species differences in NR ligands, compounds that cause toxic effects in rodents may activate NRs in humans leading to detoxification. Of the eight phthalein compounds tested (Table 2), six had ligand effects on PXR (B1 and B6–8 were agonists, B4 and B5 were antagonists (Fig. 2D)). Five phthaleins showed significant inverse agonist ligand effects on CAR1 (B1, B2, B5–7 (Fig. 2C)), and two had significant agonist ligand effects on CAR3 (B1 and B8 (Fig. 2B)). As with the group A compounds, CAR3 showed more similarity to PXR than to CAR1 in its responses to phthaleins.
Phenolphthalein (B1) and α-naphtholphthalein (B8) had robust agonist ligand effects on CAR3- and PXR-dependent transactivation of luciferase; conversely, phenolphthalein had inverse agonist activity on CAR1 and α-naphtholphthalein had no significant effect. A comparison of the ligand activity profiles of the phthalein set against their chemical structures indicated that the LBD of CAR3 might share more similarity with that of PXR, binding larger molecules with a greater surface area available for hydrophobic interactions within the pocket. The two naphthalene groups are the main difference between phenol- and α-naphtholphthalein, the presence of which may be sufficient to prevent effective interaction with the CAR1 pocket.
Cresol red (B3) had no effect on CAR1, CAR3 or PXR. Cresol red is closest in structure to cresolphthalein (B2), which was an inverse agonist of CAR1. Changing the sulfonyl group to a carbonyl group likely confers CAR1 inverse agonist activity. Thymolphthalein (B5) had antagonist/inverse agonist effects on all three nuclear receptors tested and further reduced the PK11195-repressed CAR1-dependent luciferase activity. Interestingly, thymolphthalein reduced the rifampicin-activated PXR signal to lower than that of thymolphthalein alone. No adverse effects on Renilla activity were seen indicating the repression was not due to toxicity, so it is possible that thymolphthalein may be a significant antagonist of a broad range of NRs.
Bromothymol blue (B7) had inverse agonist effects on CAR1 and tended towards antagonism of CAR3 yet showed agonist effects on PXR. While bromocresol purple (B4) is halogenated similarly to bromothymol blue, it had no significant ligand effect with either CAR isoform and had a small antagonist ligand effect on PXR in the absence of rifampicin. Bromothymol blue is brominated at identical positions to bromocresol purple but the methyl group is switched from the 6- position to the 3, and replaced by an additional propyl group on the 6- position of each hydroxyphenyl group (Fig 2A). These changes appear sufficient to convert a PXR antagonist to an agonist that also had inverse agonist effects on CAR1. Thymol blue lacks the bromine groups of bromothymol blue but retains the 2-methyl, 6-propyl groups on the hydroxyphenyl rings (using bromothymol blue numbering). Thymol blue caused effects of similar direction and magnitude to bromothymol blue on CAR1 and PXR indicating that the bromide groups are not necessary for the CAR1 inverse agonist effect. It is therefore likely that the bromide groups contribute less to the compound being a ligand than the methyl and propanyl groups that increase the hydrophobic surface area of the molecule.
3.3 Group C – Alkylphenols, Figure 3, Table 3
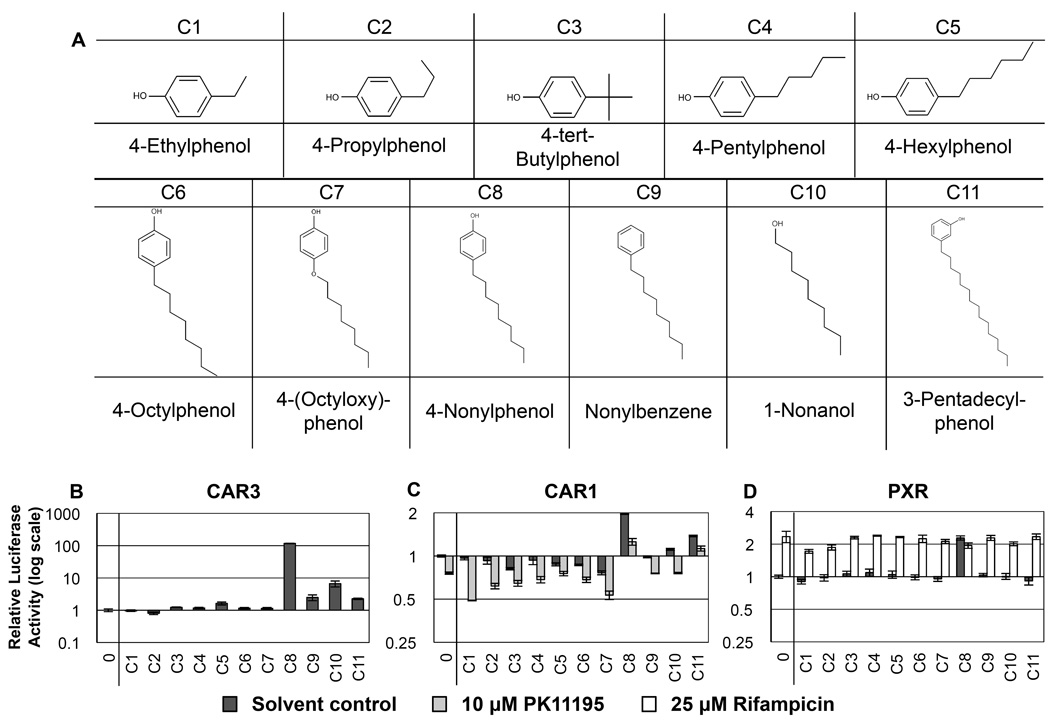
Figure 3. RQSAR for group C - Alkylphenols.
(A): 2D structures of group C alkylphenols. (B) CAR3-driven relative luciferase activity, (C) CAR1, and (D) PXR treated with 10 µM test compound, plotted using log scales. 0 = DMSO solvent control. Cotreatments were PK11195 (10 µM) for CAR1 and rifampicin (25 µM) for PXR. (n=4 except for B: C8 n=8).
Table 3.
Relative luciferase activity values for CAR1, CAR3 and PXR treated with group C compounds (Alkylphenols).
| CAR3 | CAR1 | PXR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PK11195 | Control | Rifampicin | ||||||||
| ID | Chemical Name | Value | Siga | Value | Sig | Value | Sig | Value | Sig | Value | Sig |
| 0 | Solvent control | 1.00 | - | 1.00 | - | 0.76 | - | 1.00 | - | 2.35 | - |
| C1 | 4-Ethylphenol | 0.97 | - | 0.97 | - | 0.49 | * | 0.90 | - | 1.72 | - |
| C2 | 4-Propylphenol | 0.81 | - | 0.93 | - | 0.62 | - | 0.98 | - | 1.86 | - |
| C3 | 4-tert-Butylphenol | 1.23 | - | 0.82 | ** | 0.65 | - | 1.06 | - | 2.30 | - |
| C4 | 4-Pentylphenol | 1.17 | - | 0.93 | - | 0.68 | - | 1.09 | - | 2.39 | - |
| C5 | 4-Hexylphenol | 1.62 | * | 0.88 | * | 0.75 | * | 1.05 | - | 2.32 | - |
| C6 | 4-Octylphenol | 1.15 | - | 0.87 | ** | 0.68 | - | 0.99 | - | 2.25 | - |
| C7 | 4-Octyloxyphenol | 1.14 | - | 0.76 | * | 0.53 | - | 0.96 | - | 2.12 | - |
| C8 | 4-Nonylphenol | 117.14 | *** | 1.97 | *** | 1.26 | *** | 2.28 | *** | 1.94 | - |
| C9 | Nonylbenzene | 2.48 | * | 0.99 | - | 0.76 | - | 1.04 | - | 2.29 | - |
| C10 | 1-Nonanol | 6.64 | ** | 1.11 | ** | 0.76 | - | 1.01 | - | 2.02 | - |
| C11 | 3-Pentadecylphenol | 2.26 | *** | 1.38 | *** | 1.13 | ** | 0.91 | - | 2.34 | - |
Sig = significance in unpaired t-test
 Significant agonist effect
Significant agonist effect
 Significant inverse agonist/antagonist effect
Significant inverse agonist/antagonist effect
0.01 < p < 0.05;
0.001 < p < 0.01;
p < 0.001
The xenoestrogen and persistent environmental contaminant, 4-nonylphenol, a breakdown product of alkylphenol ethoxylates widely used as surfactants and plasticizers, is a known agonist ligand of CAR1 and PXR [35,36]. Luciferase assay results here clearly demonstrated that 4-nonylphenol (C8) also has robust agonist effects on CAR3 (Fig 3B and Table 3). Routledge and Sumpter (1997) demonstrated that para hydroxyl alkylphenols are more estrogenic than ortho- and meta- forms [32]. Therefore, eight commercially available para-alkylphenols were tested in luciferase assays with plasmids for CAR1, CAR3 and PXR.
Only 4-nonylphenol (C8) showed significant agonist ligand effects with PXR, and gave the strongest CAR1 and CAR3 agonist responses of this group. Some of the shorter-chain alkylphenols had inverse agonist effects on CAR1 with 4-hexylphenol (C5) also having a small agonist ligand effect on CAR3. 4-Nonylphenol caused the greatest magnitude CAR1 response of any compound tested in this entire compound screen. Nonylbenzene (C9) and 1-nonanol (C10) were included as controls for the substituent components of nonylphenol. Neither compound had any significant effect with PXR; however, 1-nonanol had agonist ligand effects on CAR1 and CAR3 indicating the hydroxyl group on a 9-carbon chain is more important than the aryl group. Nonylbenzene had slight agonist activity with CAR3 only, despite its low solubility, which lends support to the idea that CAR1 has a greater requirement for polar substituents than CAR3. 3-pentadecylphenol (C11) was included in the screen to see if there might be an upper limit to chain length. Unfortunately, 4-pentadecylphenol was not commercially available to us at the time, nevertheless, 3-pentadecylphenol (C11) had small but significant agonist ligand effects on CAR1 and CAR3; it is possible that with increasing chain-length the para- position of the hydroxyl group may be less crucial to ligand effects. Studies are ongoing with 4-alkylphenols with alkyl chains greater than nine carbons.
Where alkyl chains contained fewer than nine carbons there was no effect on PXR. 4-tert-butylphenol (C3), 4-hexylphenol, 4-octylphenol and 4-(octyloxy)phenol (C5–7, respectively) had inverse agonist effects on CAR1. 4-hexylphenol (C5) had a small but significant agonist effect on CAR3. 4-Ethylphenol (C1) acted as a CAR1 inverse agonist only in the presence of PK11195. With increasing chain length, the alkylphenols comprise increasingly complex mixtures of branched isomers. More highly branched isomers have been shown to be more estrogenic [37] and it may be that the degree of branching also has an effect on CAR and PXR ligand activity. This is supported by the results for the phthalein and triphenyl groups of compounds (sections 3.1 and 3.2) where bulky hydrophobic substituents were associated with CAR and PXR ligand activity. The number of possible 3D conformations will vary considerably from the 2D representations shown here and it may be that it is a particular isomer or conformation that is responsible for the majority of the ligand effects. Further studies in this area are ongoing.
3.4 Group D – Stilbenes, Figure 4, Table 4
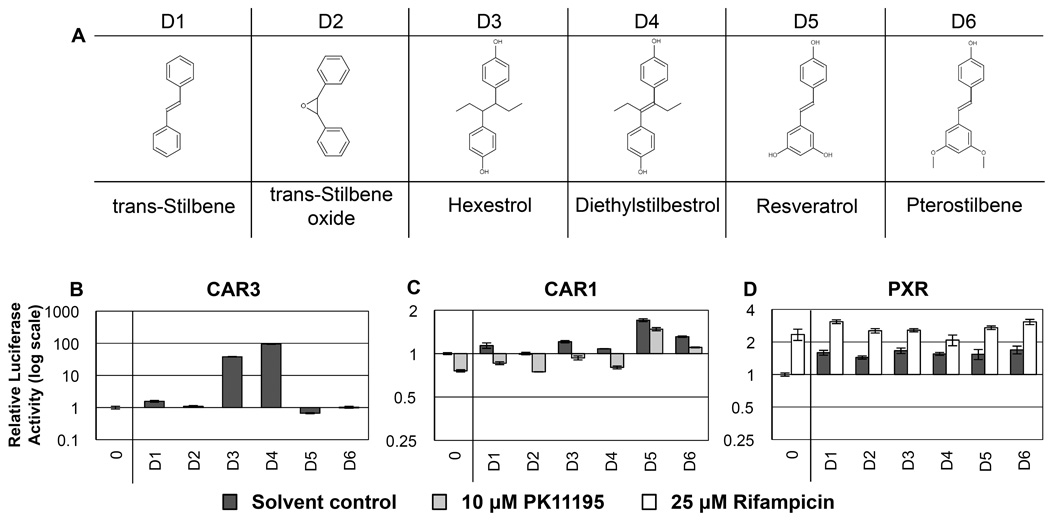
Figure 4. RQSAR for group D - Stilbenes.
(A) 2D structures of group D stilbenes. (B) CAR3-driven relative luciferase activity, (C) CAR1, and (D) PXR treated with 10 µM test compound, plotted using log scales. 0 = DMSO solvent control. Cotreatments were PK11195 (10 µM) for CAR1 and rifampicin (25 µM) for PXR. (n=4 except for B: D1–5 n=8. C: D4 n=3).
Table 4.
Relative luciferase activity values for CAR1, CAR3 and PXR treated with group D compounds (Stilbenes).
| CAR3 | CAR1 | PXR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PK11195 | Control | Rifampicin | ||||||||
| ID | Chemical Name | Value | Siga | Value | Sig | Value | Sig | Value | Sig | Value | Sig |
| 0 | Solvent control | 1.00 | - | 1.00 | - | 0.76 | - | 1.00 | - | 2.35 | - |
| D1 | trans-Stilbene | 1.56 | *** | 1.14 | * | 0.86 | ** | 1.59 | *** | 3.08 | - |
| D2 | trans-Stilbene oxide | 1.09 | - | 1.00 | - | 0.75 | - | 1.44 | *** | 2.54 | - |
| D3 | Hexestrol | 37.98 | *** | 1.21 | ** | 0.93 | - | 1.66 | *** | 2.57 | - |
| D4 | Diethylstilbestrol | 95.11 | *** | 1.08 | * | 0.80 | - | 1.55 | *** | 2.08 | - |
| D5 | Resveratrol | 0.67 | *** | 1.71 | *** | 1.48 | *** | 1.54 | * | 2.71 | - |
| D6 | Pterostilbene | 1.02 | - | 1.31 | *** | 1.11 | *** | 1.69 | ** | 3.07 | - |
Sig = significance in unpaired t-test
 Significant agonist effect
Significant agonist effect
 Significant inverse agonist/antagonist effect
Significant inverse agonist/antagonist effect
0.01 < p < 0.05;
0.001 < p < 0.01;
p < 0.001
Stilbene compounds are diarylethenes, some of which have estrogenic effects [38]. The artificial estrogen, diethylstilbestrol (DES; D4), in use as a metastatic prostate cancer therapy, was used to prevent miscarriage until teratogenic effects were observed [39]. The phytoalexins resveratrol and pterostilbene are hydroxylated stilbene derivatives, natural sources of which include the skins of red grapes and blueberries, respectively. There are reports of anti-cancer and anti-oxidant effects of these two compounds in mice, hamsters and prostate cancer cell lines and they are currently under intense investigation for their antioxidant, anti-inflammatory and anti-cancer properties [40,41]. This group of compounds also shares structural features with the bisphenols (group E, section 3.5).
That all six stilbene compounds tested (Table 4) had moderate agonist ligand effects on PXR (Fig. 4D) hints that encounters with plant secondary metabolites may have been a driving force behind the evolution of PXR activity seen today, and these features have been co-opted for the metabolism of structurally similar man-made compounds. CYP3A4, for example, is one of the main gene targets up-regulated by PXR and is involved in the metabolism a wide array of pharmaceutics and herbal supplements [42].
Certain members of this stilbene group had opposing ligand effects on CAR1 and CAR3 (Fig. 4C and 4B). Hexestrol (D3) and DES (D4) showed the greatest magnitude of agonist ligand effect on CAR3-dependent luciferase transactivation and moderate agonist activity on CAR1. Hexestrol (D3), a more flexible analog of DES, lacking a central double bond (Fig. 4A), had greater activity than DES on CAR1 and less on CAR3. While trans-stilbene (D1) had agonist ligand effects with CAR1 and CAR3, trans-stilbene oxide (TSO; D2) did not. The central epoxy group of TSO forces the molecule into a rigid structure similar to cis-stilbene. This structural change apparently disrupts ligand effects on both CAR isoforms but not PXR. Trans-resveratrol (D5) had opposite effects on CAR1 and CAR3, acting as a strong agonist ligand of CAR1 and antagonizing CAR3 compared to solvent control alone. Even though solvent control activation of CAR3 was very low, the repressive effect of resveratrol was clearly seen on multiple occasions (data not shown). Pterostilbene (D6) was also a CAR1 agonist having approximately half the agonist activity of resveratrol and no effect on CAR3. Cotreatment studies are ongoing to further understand this apparent antagonism of CAR3 by resveratrol.
Overall, the results from this group suggest that symmetrical para-hydroxyl groups on phenyl rings, and central, bulky hydrophobic groups make good candidate CAR3 agonists. Resveratrol and pterostilbene, both lacking the central ethyl side chains of DES and hexestrol, with meta-diol or meta– dimethoxy groups, on the second phenyl ring, respectively, provide clues to the key structural features of CAR1 agonists. Resveratrol (meta-diol) had a greater CAR1 agonist effect than pterostilbene, thus it is tempting to suggest meta-diol substituents may confer more potent CAR1 agonist feature than meta-methoxy groups.
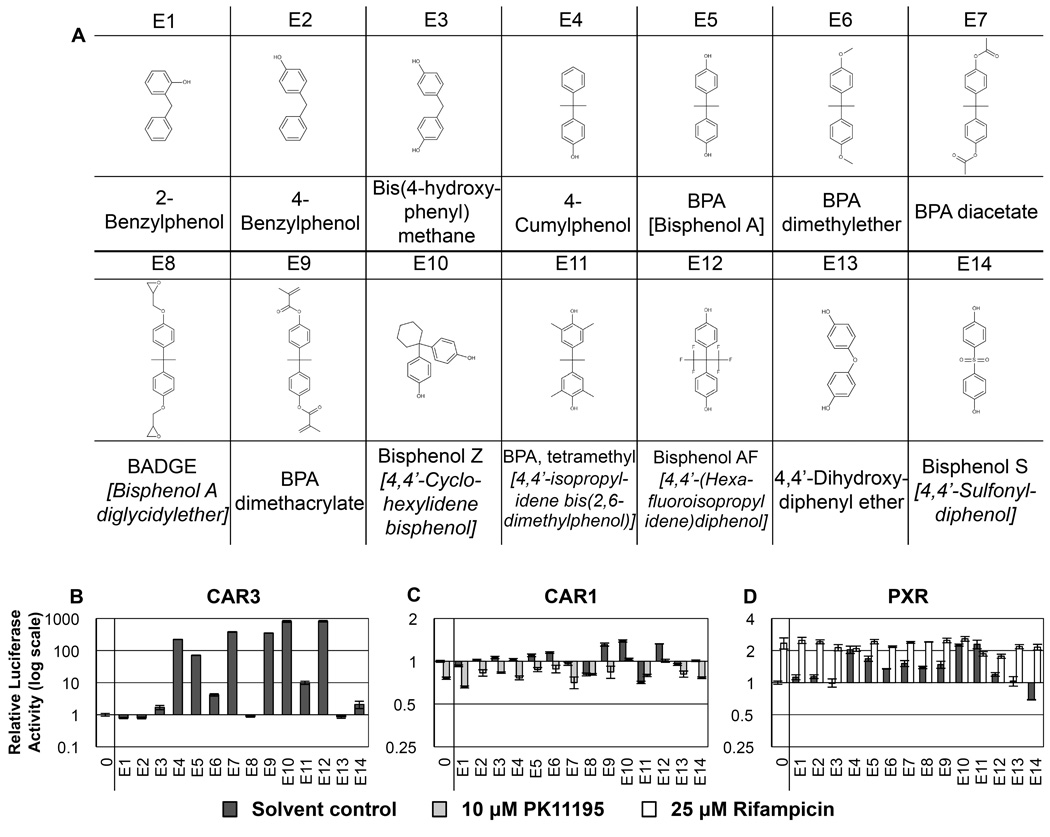
3.5 Group E - Diphenyls, Figure 5 and Table 5
Figure 5. RQSAR for group E - Diphenyls.
(A) 2D structures of group E diphenyl compounds. (B) CAR3-driven relative luciferase activity, (C) CAR1, and (D) PXR treated with 10 µM test compound, plotted using log scales. 0 = DMSO solvent control. Cotreatments were PK11195 (10 µM) for CAR1 and rifampicin (25 µM) for PXR. (n=4 except for B: E5 and E8 n=8. C: E5 and E12 n=3. D: E6 and E14 n=3).
Table 5.
Relative luciferase activity values for CAR1, CAR3 and PXR treated with group E compounds (Diphenyls).
| CAR3 | CAR1 | PXR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PK11195 | Control | Rifampicin | ||||||||
| ID | Chemical Name | Value | Siga | Value | Sig | Value | Sig | Value | Sig | Value | Sig |
| 0 | Solvent control | 1.00 | - | 1.00 | - | 0.76 | - | 1.00 | - | 2.35 | - |
| E1 | 2-Benzylphenol | 0.79 | - | 0.93 | - | 0.66 | - | 1.13 | - | 2.50 | - |
| E2 | 4-Benzylphenol | 0.80 | - | 1.02 | - | 0.83 | - | 1.14 | - | 2.42 | - |
| E3 | Bis(4-hydroxyphenyl)methane | 1.69 | - | 1.06 | * | 0.83 | ** | 1.00 | - | 2.13 | - |
| E4 | 4-Cumylphenol | 222.05 | *** | 1.03 | - | 0.76 | * | 2.04 | ** | 2.09 | - |
| E5 | BPA [Bisphenol A] | 71.17 | *** | 1.10 | - | 0.87 | - | 1.68 | *** | 2.43 | - |
| E6 | BPA dimethylether | 4.16 | *** | 1.15 | *** | 0.88 | - | 1.35 | *** | 2.17 | ** |
| E7 | BPA diacetate | 381.41 | *** | 0.96 | ** | 0.71 | * | 1.51 | ** | 2.39 | - |
| E8 | BADGE [BPA diglycidylether] | 0.88 | * | 0.81 | - | 0.81 | * | 1.39 | *** | 2.41 | - |
| E9 | BPA dimethacrylate | 351.40 | *** | 1.31 | *** | 0.84 | - | 1.48 | ** | 2.49 | - |
| E10 | Bisphenol Z [4,4′-Cyclohexylidenebisphenol] | 811.70 | *** | 1.39 | *** | 1.03 | *** | 2.25 | *** | 2.58 | - |
| E11 | BPA, tetramethyl [4,4′-isoproplyidene bis(2,6-dimethylphenol)] | 9.97 | *** | 0.71 | *** | 0.80 | - | 2.29 | ** | 1.86 | * |
| E12 | Bisphenol AF [4,4′-(Hexafluroisoproplyidene)diphenol] | 821.46 | *** | 1.33 | *** | 1.01 | *** | 1.20 | * | 1.77 | ** |
| E13 | 4,4′-Dihydroxydiphenyl ether | 0.86 | - | 0.95 | - | 0.81 | - | 1.03 | - | 2.18 | - |
| E14 | Bisphenol S [4,4′-Sulfonyldiphenol] | 2.09 | - | 1.01 | - | 0.77 | - | 0.69 | *** | 2.16 | - |
Sig = significance in unpaired t-test
 Significant agonist effect
Significant agonist effect
 Significant inverse agonist/antagonist effect
Significant inverse agonist/antagonist effect
0.01 < p < 0.05;
0.001 < p < 0.01;
p < 0.001
Out of 14 diphenyl compounds screened (Fig. 5A, Table 5), 11 had some ligand effect on at least one of the three nuclear receptors tested. Bisphenols are symmetrical molecules and, as a subset of the diphenyls, showed by far the greatest range of luciferase transactivation responses for CAR3 of all compound groups tested, from the barely apparent antagonist effects of BADGE (E8) to the strong agonist activities of bisphenol Z (E10) and bisphenol AF (E12). The variety within this group provides important clues as to which features are important for ligand activities and allow discrimination between these closely related compounds.
The lack of significant ligand effects of 2-benzylphenol (E1) and 4-benzylphenol (E2) on any NR tested implies that a single ortho- or para- hydroxyl group is not sufficient for a diphenyl compound to have ligand activity. Adding a second para-hydroxyl group to E2 makes bis(4-hydroxydiphenyl)methane (E3), a CAR1-specific agonist ligand. Adding two central methyl groups to E2, to make 4-cumylphenol (E4), gave E4 a strong CAR3 and PXR agonist activity with only slight agonist effects on CAR1 in the presence of PK11195 only. This would fit a hypothesis that the CAR3 LBD is more similar to PXR than CAR1 is to PXR. Also it appears that symmetrical para- polar groups are important features for CAR1 agonist activity, whereas central bulky hydrophobic groups are more specific for CAR3 responses.
BPA (E5) differs from 4-cumylphenol (E4) only by the addition of a second para-hydroxyl group (combining the structural features of E3 and E4). BPA had robust ligand effects on CAR3 and PXR but showed no significant effect on CAR1 (repeated observations, data not shown). The CAR3 response to BPA, although high, is approximately ⅔ its response to 4-cumylphenol, which implies that two polar groups at opposite ends of the molecule can diminish CAR3 ligand activity. BPA diacetate and BADGE (E7 and E8 respectively) had opposite effects on CAR1 and CAR3. BPA diacetate had agonist ligand effects on CAR3 and inverse agonist effects on CAR1 with solvent control and in addition to PK11195 cotreatment repression. BADGE caused significant repression of CAR3 luciferase transactivation (a significant effect even though basal luciferase activity was very low) and overcame PK11195 cotreated CAR1 repression without affecting solvent control treated cells. BPA dimethacrylate (E9) was an agonist ligand for CAR1, CAR3 and PXR. Structural comparison of E9 with BPA diacetate shows that changing the substituents from acetates to methacrylates makes no difference to CAR3 and PXR ligand effects (both strong) but changes the CAR1 effect from inverse agonist to agonist. BPA dimethylether (E6) had ligand effects on CAR3 but only to a low level of transactivation compared to other bisphenol compounds. E6 also showed low level PXR agonist activity in the presence of solvent control, but antagonized rifampicin transactivation. BPA dimethylether did act as an agonist ligand for CAR1, as did BADGE in the presence of PK11195, perhaps indicating that ether groups may contribute to more CAR1-specific ligand effects.
BPA dimethylether (E6), BPA tetramethyl (E11) and Bisphenol AF (E12) all had agonist ligand effects on PXR in the presence of solvent control but antagonized luciferase transactivation in cells cotreated with rifampicin (Fig. 5D). It is possible these compounds displace rifampicin or interact with rifampicin in some way to prevent its agonist effect. It is also possible that these three compounds are ligands of an endogenous accessory protein, only recruited to PXR when it is already in the active conformation (i.e. with rifampicin) in order to repress downstream targets. BPA tetramethyl (E11) shares the same core structure as BPA (E5) but had a lesser effect on CAR3 transactivation and acted as an inverse agonist of CAR1. Methyl groups at the 2- and 6- positions on both phenyl groups of E11 may be obscuring the para-hydroxyl groups, disrupting interactions with the CAR LBD necessary for agonist activity.
Bisphenol AF (E12) yielded the greatest magnitude of CAR3 response seen in the whole screen and also had robust agonist effects on CAR1 and PXR (in the absence of rifampicin). Building upon observations from group D, the six fluorine additions to the central isopropyl group, together with symmetrical, unobstructed para-hydroxyl groups, contributed to strong agonist effects with all three receptors. This ligand structure combines bulky side chains at the center of the molecule (features shared by CAR3 and PXR ligands) with the unobstructed para-hydroxyl groups seen to be common to many CAR1 agonists. Most diphenyl compounds in this screen were agonist ligands of PXR in the absence of rifampicin cotreatment (Table 5); E6, E11 and E12 antagonize luciferase transactivation by rifampicin-activated PXR.
4,4’-dihydroxydiphenyl ether (E13) and bisphenol S (E14) are two diphenyl compounds where the central atom is not a carbon. Both had no activity as CAR1 or CAR3 ligands and 4,4’-dihydroxydiphenyl ether had no ligand effect on PXR either. Bisphenol S, however, acted as a significant antagonist of PXR, an antagonism fully relieved by the addition of 25 µM rifampicin.
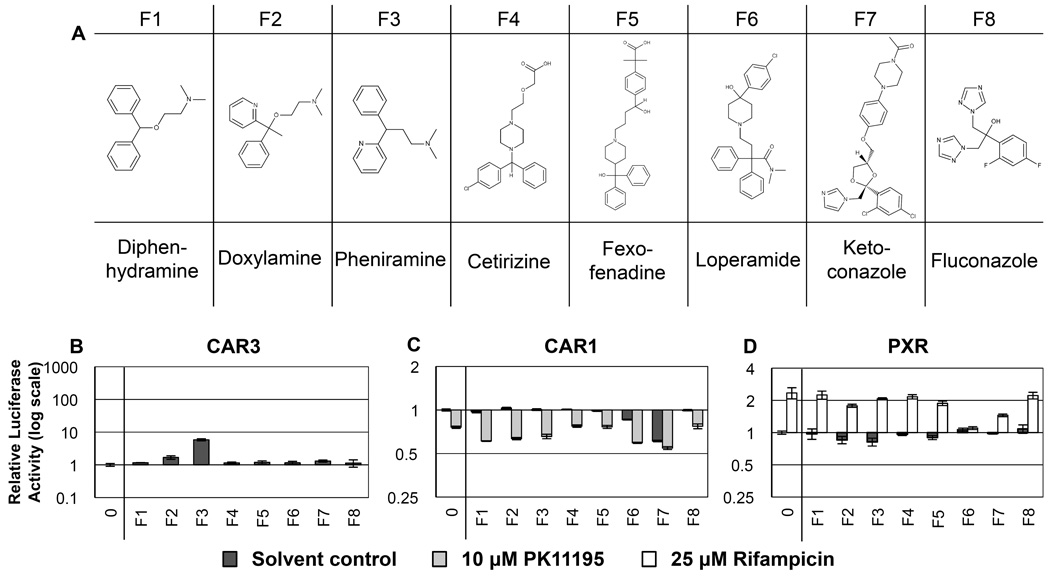
3.6 Group F - Antihistamines and antifungals, Figure 6, Table 6
Figure 6. RQSAR for group F - Antihistamines and Antifungals.
(A) 2D structures of group F antihistamine and antifungal compounds. (B) CAR3-driven relative luciferase activity, (C) CAR1, and (D) PXR treated with 10 µM test compound, plotted using log scales. 0 = DMSO solvent control. Cotreatments were PK11195 (10 µM) for CAR1 and rifampicin (25 µM) for PXR. (n=4 except for B: F1 and F8 n=3).
Table 6.
Relative luciferase activity values for CAR1, CAR3 and PXR treated with group F compounds (Antihistamines and Antifungals).
| CAR3 | CAR1 | PXR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PK11195 | Control | Rifampicin | ||||||||
| ID | Chemical Name | Value | Siga | Value | Sig | Value | Sig | Value | Sig | Value | Sig |
| 0 | Solvent control | 1.00 | - | 1.00 | - | 0.76 | - | 1.00 | - | 2.35 | - |
| F1 | Diphenhydramine | 1.16 | - | 0.97 | - | 0.61 | ** | 0.97 | - | 2.25 | - |
| F2 | Doxylamine | 1.68 | * | 1.03 | - | 0.63 | - | 0.85 | - | 1.78 | - |
| F3 | Pheniramine | 5.84 | *** | 1.01 | - | 0.66 | - | 0.82 | - | 2.07 | - |
| F4 | Cetirizine | 1.15 | - | 1.01 | - | 0.77 | - | 0.96 | - | 2.16 | - |
| F5 | Fexofenadine | 1.19 | - | 0.99 | - | 0.76 | - | 0.90 | - | 1.88 | * |
| F6 | Loperamide | 1.15 | - | 0.86 | ** | 0.59 | ** | 1.06 | - | 1.10 | ** |
| F7 | Ketoconazole | 1.30 | - | 0.61 | *** | 0.55 | *** | 0.98 | - | 1.45 | *** |
| F8 | Fluconazole | 1.13 | - | 1.00 | - | 0.77 | - | 1.08 | - | 2.22 | - |
Sig = significance in unpaired t-test
 Significant agonist effect
Significant agonist effect
 Significant inverse agonist/antagonist effect
Significant inverse agonist/antagonist effect
0.01 < p < 0.05;
0.001 < p < 0.01;
p < 0.001
Group F comprises currently licensed pharmaceutics that would be expected to have little or no activity on PXR. Diphenhydramine (F1), doxylamine (F2), pheniramine (F3), cetirizine (F4) and fexofenadine (F5) are all antihistamines. The antihistamine parent compound of fexofenadine, terfenadine (no longer licensed for medical use), was included in the screen but proved to be toxic to the HuH-7 cells at 10 µM (data not shown). Loperamide (F6) is an anti-diarrheal opioid modified so that it does not so readily cross the blood-brain barrier. Ketoconazole (F7) and Fluconazole (F8) are antifungal agents. Structural features within this set are more varied than in other groups, but members have in common two or more aromatic rings and five or more rotatable bonds.
None of the F compounds alone had any significant agonist ligand activity with PXR, however fexofenadine (F5), loperamide (F6) and ketoconazole (F7) all had antagonist effects on PXR in the presence of 25 µM rifampicin (Fig. 6D). Ketoconazole, and to a lesser extent loperamide, had inverse agonist effects on CAR1 in addition to the inverse agonism of PK11195. Ketoconazole antagonism of CAR1 and activated PXR has been reported previously [43,44,23]; we believe this is the first report of similar effects for loperamide and fexofenadine. It is possible these compounds could have antagonist effects on agonist activated CAR3; CAR3 agonist cotreatment studies may show antagonist effects similar to PXR.
Fluconazole (F8) differs from ketoconazole (F7) in chain-length and halogenation, having a less extensive structure and fluorines in place of chlorines. The C:F bond is one of the strongest in organic chemistry, having one of the shortest bond-lengths. Fluorine is smaller than chlorine in space-filling models and shares spatial similarities with C:OH groups; fluorine also has a greater capacity for hydrogen bond formation than chlorine [45]. Since fexofenadine and loperamide are not halogenated but share the overall extended shape of ketoconazole it is likely that the lack of competitive antagonism observed for fluconazole has more to do with the absence of an extended chain rather than the presence of fluorine over chlorine. Ketoconazole antagonism of PXR is caused by ligand binding outside of the LBD to the conserved, ligand-dependent, transactivation (AF2) domain of PXR where it disrupts interactions with coactivator protein steroid receptor coactivator 1 (SRC1,NCOA1) [23]. It is therefore possible that ketoconazole may interact with human CAR1 via its AF2 domain.
Diphenhydramine (F1), doxylamine (F2) and pheniramine (F3) are antihistamine compounds with two aryl substituents and a short aliphatic chain ending in a tertiary amine. While none of these three affected PXR, doxylamine and pheniramine both had significant CAR3 agonist ligand effects. Diphenhydramine had no effect on CAR3 but synergizes with PK11195 to further inverse agonize CAR1. The presence of at least one nitrogen in any of the phenyl groups seems to help confer CAR3 ligand status. The 3-ring antihistamine cetirizine (F4) had no significant effect on any of the three NRs.
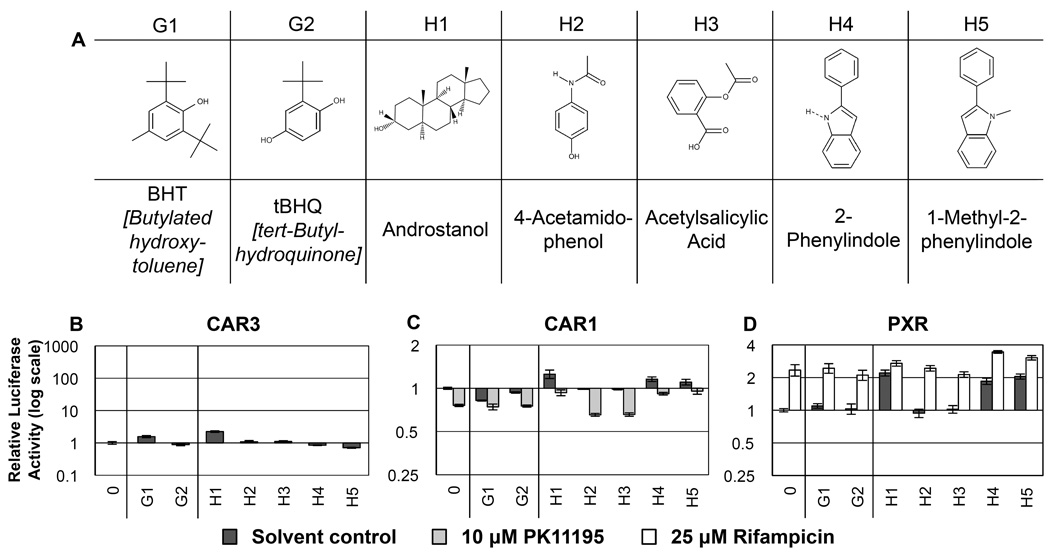
3.7 Group G – tert-Butyls, Figure 7, Table 7
Figure 7. RQSAR for group G and group H - tert-Butyl and miscellaneous.
(A) 2D structures of groups G and H, tert-butyl and miscellaneous compounds. (B) CAR3-driven relative luciferase activity, (C) CAR1, and (D) PXR treated with 10 µM test compound, plotted using log scales. 0 = DMSO solvent control. Cotreatments were PK11195 (10 µM) for CAR1 and rifampicin (25 µM) for PXR. (n=4 except for B: H1 and H4–5 n=8.
Table 7.
Relative luciferase activity values for CAR1, CAR3 and PXR treated with groups G and H compounds (tert-Butyls and miscellaneous).
| CAR3 | CAR1 | PXR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | PK11195 | Control | Rifampicin | ||||||||
| ID | Chemical Name | Value | Siga | Value | Sig | Value | Sig | Value | Sig | Value | Sig |
| 0 | Solvent control | 1.00 | - | 1.00 | - | 0.76 | - | 1.00 | - | 2.35 | - |
| G1 | BHT [Buytlated hydroxytoluene] | 1.57 | * | 0.82 | *** | 0.74 | - | 1.10 | - | 2.44 | - |
| G2 | tBHQ [tert-Butylhydroquinone] | 0.88 | - | 0.93 | ** | 0.75 | - | 1.03 | - | 2.11 | - |
| H1 | Androstanol | 2.24 | *** | 1.25 | * | 0.93 | - | 2.21 | *** | 2.71 | - |
| H2 | 4-Acetamidophenol | 1.09 | - | 0.98 | - | 0.65 | - | 0.94 | - | 2.44 | - |
| H3 | Acetylsalicylic acid | 1.11 | - | 0.99 | - | 0.66 | - | 1.02 | - | 2.13 | - |
| H4 | 2-Phenylindole | 0.85 | - | 1.16 | * | 0.92 | - | 1.85 | *** | 3.45 | * |
| H5 | 1-Methyl-2-phenylindole | 0.71 | ** | 1.10 | - | 0.96 | - | 2.05 | *** | 3.05 | - |
Sig = significance in unpaired t-test
 Significant agonist effect
Significant agonist effect
 Significant inverse agonist/antagonist effect
Significant inverse agonist/antagonist effect
0.01 < p < 0.05;
0.001 < p < 0.01;
p < 0.001
Three tert-Butyl compounds were included in this screen: butylated hydroxytoluene (BHT, G1), tert-butylhydroquinone (tBHQ, G2) and 4-tertbutylphenol (C3, Fig. 3A). While none had any ligand effect on PXR, all show significant inverse agonist effects on transactivation by CAR1. BHT (G1) had a small but significant ligand effect on CAR3. These three compounds are among the smallest to produce any CAR effects thus far when administered alone (4-ethylphenol (C1) had some activity in the presence of PK11195 only). It is tempting to postulate that these compounds represent the lower size limit of CAR ligands and that the activity of very small molecules on CAR1 will tend to be as inverse agonists.
3.8. Group H – other, Figure 7, Table 7
Androstanol was included as an example steroid that is usually an agonist ligand of PXR and inverse agonist of CAR1. In these assays, while having agonist ligand effects with PXR, androstanol actually had agonist ligand effects on CAR1 and small but significant ligand activity with CAR3. This response to androstanol may be cell line specific, as observed for clotrimazole [42]. Most other control compounds included in the screen behaved as previously reported. 4-Acetamidophenol (H2) and acetylsalicylic acid (H3) had no significant effect on any of the three receptors tested. H2 and H3 are similar to compounds G1 and G2 in terms of molecule size but had no CAR1 ligand effects. The indole compounds, 2-phenylindole (H4) and 1-methyl-2-phenylindole (H5), have an overall shape similar to the shorter diphenyl compounds; both showed agonist ligand effects with PXR despite their relatively small molecular size. H4 (2-phenylindole) had agonist ligand activity for CAR1, whereas H5 (1-methyl-2-phenylindole) did not; the methyl group could be obstructing the partially charged region favored by CAR1 for agonist ligand effects. Conversely, for CAR3, H4 had no effect and H5 had significant antagonist effects on luciferase transactivation.
3.9 Assay considerations
To test for general CAR ligand binding effects, the GAL4-UAS system of expression and reporter plasmids was used. Since this system is not limited to any one CAR consensus binding element it more accurately reflects CAR potential to bind to any target promoter, essentially controlling for any possible isomer-specific promoter preferences. Prior to performing the PXR screen a number of expression and reporter plasmids were tested, including pM-PXR (data not shown); the 3XFLAG-PXR expression plasmid, with additional 3XFLAG-RXRα, gave the greatest response with positive control treatment, 25 µM rifampicin, when using a CYP3A4 xenobiotic response element-driven luciferase reporter (p3A4-XREM-TK-luc).
The classic CAR ligand CITCO was not included as results with this compound were highly variable, pointing to a lack of stability as formerly noted [26]. Given that CITCO is wholly synthetic, and therefore not likely to have been a factor in the evolution of CAR and its responses, perhaps compounds that bind the receptor with lower affinity are of more biological relevance. It is possible that the reported high binding affinity of CITCO for CAR may not necessarily be representative of the normal physiological operation of CAR and it has been suggested that the xenosensor and metabolic status-sensing activities of CAR and PXR may in fact be regulated by low-affinity ligand interactions [46]. TR-FRET data published for CAR ligands shows that biologically active ligand pregnanedione has a much lower affinity for the CAR LBD than CITCO which lends support to this theory [47]. The groups of compounds represented and discussed here all come from chemical groups to which exposure, either through medication or environmental routes, is possible.
Different cell lines are routinely used for nuclear receptor ligand screening, and some cell-type specific effects have been noted. In our hands, the human hepatoma cell line HuH-7 has responded consistently to treatments over time and is easy to transfect with high efficiency. Future studies using human hepatocytes will look at subsets of compounds identified in this study to see if there is regulation of CAR and PXR target genes CYP2B6 and CYP3A4. Since primary hepatocytes express many nuclear receptors, we were interested firstly in using HuH-7 cells with transiently overexpressed receptors so that we could clearly delineate whether ligand effects were specific for CAR1, CAR3 or PXR.
4. Conclusions
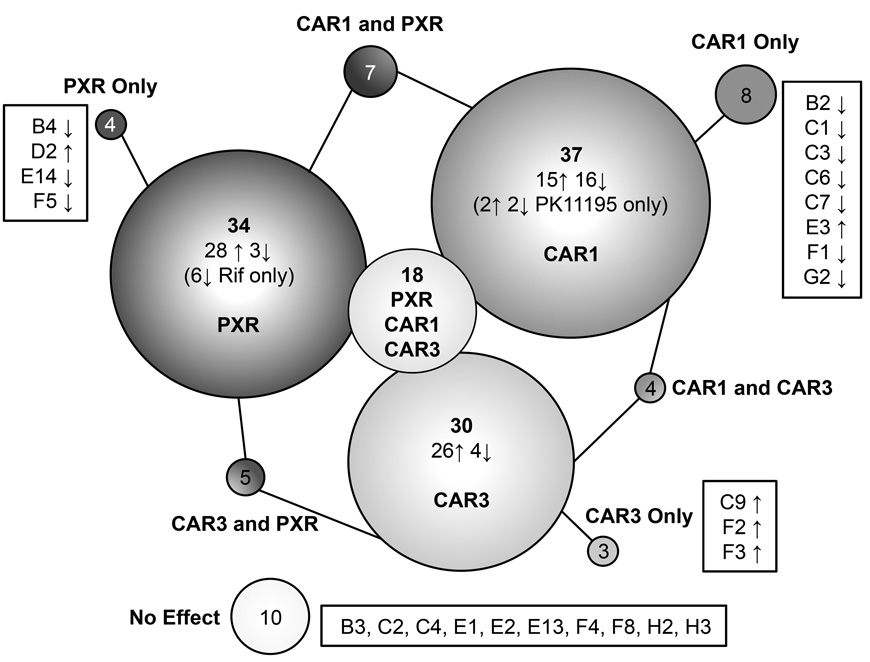
Out of 60 compounds screened, one (terfenadine) had to be excluded due to toxicity to HuH-7 cells at 10 µM. Luciferase assay screening of the remaining 59 using CAR1, CAR3 and PXR expression plasmids identified compounds with NR ligand activities from within structurally similar sets. Ten compounds had no significant ligand effects on CAR1, CAR3 or PXR. Of the 49 compounds with some significant effect on at least one receptor, 34 modulated PXR (four PXR only), 37 had activity with CAR1 (eight CAR1 only) and 30 had some ligand effect on CAR3 (three CAR3 only). A summary of these results is represented in Figure 8.
Figure 8. RQSAR summary Venn diagram.
Circle sizes are proportional to total number of ligand effects. Arrows indicate number of compounds with agonist (↑) and inverse agonist/antagonist (↓) activities, respectively.
The full screen shows that closely related structural variants of diverse types of chemical compounds have different effects on biologically relevant splice variants of CAR. Compounds with CAR3 ligand effects share structural features with ligands of CAR1 and PXR. It is possible the APYLT insertion which characterizes CAR3, although in a region outside of the binding pocket, causes a wider structural change to the protein making the pocket more permissive for larger molecules with bulkier groups, making it more similar to PXR than CAR1. Cotreatment with selective CAR1 antagonists identified in this screen, for example, 4-tert-butylphenol (C3), 4-octylphenol(C6) or 4-(octyloxy)phenol (C7), could be used to explore CAR3-specific effects on target gene expression.
RQSAR, using sets of compounds with different core structures, has yielded an interesting and valuable data set from which insights into structure-activity relationships may be inferred. Differences in ligand activity can be directly correlated with individual substituents or groups thereof. In addition to the identification of new ligands for the two most abundantly expressed hepatic CAR isoforms, the observations and analyses provided here are useful contributions to the field of structure-activity relationship studies, particularly as resources for the development of CAR and PXR ligand prediction algorithms.
Supplementary Material
Acknowledgement
Funding for this work was provided by a sub-award from the Rhode Island Institutional Development Award (IDeA) Network of Biomedical Research Excellence (RI-INBRE) Grant # P20RR016157 from the National Center for Research Resources (NCRR). AMD and LEA were support by RI-INBRE through a postdoctoral fellowship and a graduate research assistantship, respectively. SQ was supported through the RI-INBRE Summer Undergraduate Research Fellowship (SURF) program.
Abbreviations
- BPA
bisphenol A
- CAR
constitutive androstane receptor
- CITCO
6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime
- DMSO
dimethylsulfoxide
- LBD
ligand binding domain
- NR
nuclear receptor
- PK11195
1-(2-Chlorophenyl-N-methylpropyl)-3-isoquinolinecarboxamide
- PXR
pregnane X receptor
- QSAR
quantitative structure-activity relationship
- RQSAR
rational QSAR
Appendix
Supplementary Table S1: Table of chemicals used, suppliers and CAS numbers
Supplementary Table S2: Table of molecular properties calculated in Accelrys Discovery Studio Client
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.di MA, Marinis ED, Ascenzi P, Marino M. Nuclear receptors CAR and PXR: Molecular, functional, and biomedical aspects. Mol.Aspects Med. 2009 doi: 10.1016/j.mam.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Kodama S, Koike C, Negishi M, Yamamoto Y. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol.Cell Biol. 2004;24:7931–7940. doi: 10.1128/MCB.24.18.7931-7940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maglich JM, Watson J, McMillen PJ, Goodwin B, Willson TM, Moore JT. The nuclear receptor CAR is a regulator of thyroid hormone metabolism during caloric restriction. J.Biol.Chem. 2004;279:19832–19838. doi: 10.1074/jbc.M313601200. [DOI] [PubMed] [Google Scholar]
- 4.Roth A, Looser R, Kaufmann M, Meyer UA. Sterol regulatory element binding protein 1 interacts with pregnane X receptor and constitutive androstane receptor and represses their target genes. Pharmacogenet.Genomics. 2008;18:325–337. doi: 10.1097/FPC.0b013e3282f706e0. [DOI] [PubMed] [Google Scholar]
- 5.Moreau A, Vilarem MJ, Maurel P, Pascussi JM. Xenoreceptors CAR and PXR activation and consequences on lipid metabolism, glucose homeostasis, and inflammatory response. Mol.Pharm. 2008;5:35–41. doi: 10.1021/mp700103m. [DOI] [PubMed] [Google Scholar]
- 6.Konno Y, Negishi M, Kodama S. The roles of nuclear receptors CAR and PXR in hepatic energy metabolism. Drug Metab Pharmacokinet. 2008;23:8–13. doi: 10.2133/dmpk.23.8. [DOI] [PubMed] [Google Scholar]
- 7.Maglich JM, Lobe DC, Moore JT. The nuclear receptor CAR (NR1I3) regulates serum triglyceride levels under conditions of metabolic stress. J.Lipid Res. 2009;50:439–445. doi: 10.1194/jlr.M800226-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Gao J, He J, Zhai Y, Wada T, Xie W. CAR is an anti-obesity nuclear receptor that improves insulin sensitivity. J.Biol.Chem. 2009 doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinno H, Tanaka-Kagawa T, Hanioka N, Ishida S, Saeki M, Soyama A, Itoda M, Nishimura T, Saito Y, Ozawa S, Ando M, Sawada J. Identification of novel alternative splice variants of human constitutive androstane receptor and characterization of their expression in the liver. Mol.Pharmacol. 2004;65:496–502. doi: 10.1124/mol.65.3.496. [DOI] [PubMed] [Google Scholar]
- 10.Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J.Biol.Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- 11.Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J.Biol.Chem. 2003;278:17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol.Pharmacol. 2008;74:443–453. doi: 10.1124/mol.108.046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeKeyser JG, Stagliano MC, Auerbach SS, Prabhu KS, Jones AD, Omiecinski CJ. Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant CAR2. Mol.Pharmacol. 2009;75:1005–1013. doi: 10.1124/mol.108.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watkins RE, Maglich JM, Moore LB, Wisely GB, Noble SM, vis-Searles PR, Lambert MH, Kliewer SA, Redinbo MR. 2.1 A crystal structure of human PXR in complex with the St. John's wort compound hyperforin. Biochemistry. 2003;42:1430–1438. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- 15.Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, Moore JT. A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol.Cell. 2004;16:919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 16.Reschly EJ, Krasowski MD. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr.Drug Metab. 2006;7:349–365. doi: 10.2174/138920006776873526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoner MA, Auerbach SS, Zamule SM, Strom SC, Omiecinski CJ. Transactivation of a DR-1 PPRE by a human constitutive androstane receptor variant expressed from internal protein translation start sites. Nucleic Acids Res. 2007;35:2177–2190. doi: 10.1093/nar/gkm090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank C, Gonzalez MM, Oinonen C, Dunlop TW, Carlberg C. Characterization of DNA complexes formed by the nuclear receptor constitutive androstane receptor. J.Biol.Chem. 2003;278:43299–43310. doi: 10.1074/jbc.M305186200. [DOI] [PubMed] [Google Scholar]
- 19.Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol.Cell Biol. 1994;14:1544–1552. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J.Biol.Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Faucette S, Moore R, Sueyoshi T, Negishi M, LeCluyse E. Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J.Biol.Chem. 2004;279:29295–29301. doi: 10.1074/jbc.M400580200. [DOI] [PubMed] [Google Scholar]
- 22.Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc.Natl.Acad.Sci.U.S.A. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Huang H, Li H, Teotico DG, Sinz M, Baker SD, Staudinger J, Kalpana G, Redinbo MR, Mani S. Activated pregnenolone X-receptor is a target for ketoconazole and its analogs. Clin.Cancer Res. 2007;13:2488–2495. doi: 10.1158/1078-0432.CCR-06-1592. [DOI] [PubMed] [Google Scholar]
- 24.Huang W, Zhang J, Wei P, Schrader WT, Moore DD. Meclizine is an agonist ligand for mouse constitutive androstane receptor (CAR) and an inverse agonist for human CAR. Mol.Endocrinol. 2004;18:2402–2408. doi: 10.1210/me.2004-0046. [DOI] [PubMed] [Google Scholar]
- 25.Makinen J, Frank C, Jyrkkarinne J, Gynther J, Carlberg C, Honkakoski P. Modulation of mouse and human phenobarbital-responsive enhancer module by nuclear receptors. Mol.Pharmacol. 2002;62:366–378. doi: 10.1124/mol.62.2.366. [DOI] [PubMed] [Google Scholar]
- 26.Jyrkkarinne J, Windshugel B, Ronkko T, Tervo AJ, Kublbeck J, Lahtela-Kakkonen M, Sippl W, Poso A, Honkakoski P. Insights into ligand-elicited activation of human constitutive androstane receptor based on novel agonists and three-dimensional quantitative structure-activity relationship. J.Med.Chem. 2008;51:7181–7192. doi: 10.1021/jm800731b. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, LeCluyse EL. Role of orphan nuclear receptors in the regulation of drug-metabolising enzymes. Clin.Pharmacokinet. 2003;42:1331–1357. doi: 10.2165/00003088-200342150-00003. [DOI] [PubMed] [Google Scholar]
- 28.Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res. 2003;31:3194–3207. doi: 10.1093/nar/gkg419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auerbach SS, DeKeyser JG, Stoner MA, Omiecinski CJ. CAR2 displays unique ligand binding and RXRalpha heterodimerization characteristics. Drug Metab Dispos. 2007;35:428–439. doi: 10.1124/dmd.106.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J.Clin.Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toell A, Kroncke KD, Kleinert H, Carlberg C. Orphan nuclear receptor binding site in the human inducible nitric oxide synthase promoter mediates responsiveness to steroid and xenobiotic ligands. J.Cell Biochem. 2002;85:72–82. [PubMed] [Google Scholar]
- 32.Routledge EJ, Sumpter JP. Structural features of alkylphenolic chemicals associated with estrogenic activity. J.Biol.Chem. 1997;272:3280–3288. doi: 10.1074/jbc.272.6.3280. [DOI] [PubMed] [Google Scholar]
- 33.Ravdin PM, van BM, Jordan VC. Estrogenic effects of phenolphthalein on human breast cancer cells in vitro. Breast Cancer Res.Treat. 1987;9:151–154. doi: 10.1007/BF01807368. [DOI] [PubMed] [Google Scholar]
- 34.Cooper GS, Longnecker MP, Peters RK. Ovarian cancer risk and use of phenolphthalein-containing laxatives. Pharmacoepidemiol.Drug Saf. 2004;13:35–39. doi: 10.1002/pds.824. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez JP, Huang W, Chapman LM, Chua S, Moore DD, Baldwin WS. The environmental estrogen, nonylphenol, activates the constitutive androstane receptor. Toxicol.Sci. 2007;98:416–426. doi: 10.1093/toxsci/kfm107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuyama H, Hiramatsu Y, Kunitomi M, Kudo T, MacDonald PN. Endocrine disrupting chemicals, phthalic acid and nonylphenol, activate Pregnane X receptor-mediated transcription. Mol.Endocrinol. 2000;14:421–428. doi: 10.1210/mend.14.3.0424. [DOI] [PubMed] [Google Scholar]
- 37.Preuss TG, Gehrhardt J, Schirmer K, Coors A, Rubach M, Russ A, Jones PD, Giesy JP, Ratte HT. Nonylphenol isomers differ in estrogenic activity. Environ.Sci.Technol. 2006;40:5147–5153. doi: 10.1021/es060709r. [DOI] [PubMed] [Google Scholar]
- 38.Wu F, Khan S, Wu Q, Barhoumi R, Burghardt R, Safe S. Ligand structure-dependent activation of estrogen receptor alpha/Sp by estrogens and xenoestrogens. J.Steroid Biochem.Mol.Biol. 2008;110:104–115. doi: 10.1016/j.jsbmb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gouedard C, Barouki R, Morel Y. Induction of the paraoxonase-1 gene expression by resveratrol. Arterioscler.Thromb.Vasc.Biol. 2004;24:2378–2383. doi: 10.1161/01.ATV.0000146530.24736.ce. [DOI] [PubMed] [Google Scholar]
- 40.Perecko T, Jancinova V, Drabikova K, Nosal R, Harmatha J. Structure-efficiency relationship in derivatives of stilbene. Comparison of resveratrol, pinosylvin and pterostilbene. Neuro.Endocrinol.Lett. 2008;29:802–805. [PubMed] [Google Scholar]
- 41.Joseph JA, Fisher DR, Cheng V, Rimando AM, Shukitt-Hale B. Cellular and behavioral effects of stilbene resveratrol analogues: implications for reducing the deleterious effects of aging. J.Agric.Food Chem. 2008;56:10544–10551. doi: 10.1021/jf802279h. [DOI] [PubMed] [Google Scholar]
- 42.Chang TK, Waxman DJ. Synthetic drugs and natural products as modulators of constitutive androstane receptor (CAR) and pregnane X receptor (PXR) Drug Metab Rev. 2006;38:51–73. doi: 10.1080/03602530600569828. [DOI] [PubMed] [Google Scholar]
- 43.Huang H, Wang H, Sinz M, Zoeckler M, Staudinger J, Redinbo MR, Teotico DG, Locker J, Kalpana GV, Mani S. Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene. 2007;26:258–268. doi: 10.1038/sj.onc.1209788. [DOI] [PubMed] [Google Scholar]
- 44.Svecova L, Vrzal R, Burysek L, Anzenbacherova E, Cerveny L, Grim J, Trejtnar F, Kunes J, Pour M, Staud F, Anzenbacher P, Dvorak Z, Pavek P. Azole antimycotics differentially affect rifampicin-induced pregnane X receptor-mediated CYP3A4 gene expression. Drug Metab Dispos. 2008;36:339–348. doi: 10.1124/dmd.107.018341. [DOI] [PubMed] [Google Scholar]
- 45.Carosati E, Sciabola S, Cruciani G. Hydrogen bonding interactions of covalently bonded fluorine atoms: from crystallographic data to a new angular function in the GRID force field. J.Med.Chem. 2004;47:5114–5125. doi: 10.1021/jm0498349. [DOI] [PubMed] [Google Scholar]
- 46.Tzameli I, Moore DD. Role reversal: new insights from new ligands for the xenobiotic receptor CAR. Trends Endocrinol.Metab. 2001;12:7–10. doi: 10.1016/s1043-2760(00)00332-5. [DOI] [PubMed] [Google Scholar]
- 47.Invitrogen, Assay Pharmacology. LanthaScreen™ TR-FRET Constitutive Androstane Receptor Coactivator Assay Manual. 2007:11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.