Abstract
Chemical reactions that facilitate the attachment of synthetic groups to proteins are useful tools for the field of chemical biology and enable the incorporation of proteins into new materials. We have previously reported a pyridoxal 5′-phosphate (PLP) mediated reaction that site-specifically oxidizes the N-terminal amine of a protein to afford a ketone. This unique functional group can then be used to attach a reagent of choice through oxime formation. Since its initial report, we have found that the N-terminal sequence of the protein can significantly influence the overall success of this strategy. To obtain short sequences that lead to optimal conversion levels, an efficient method for the evaluation of all possible N-terminal amino acid combinations was needed. This was achieved by developing a generalizable combinatorial peptide library screening platform suitable for the identification of sequences that display high levels of reactivity toward a desired bioconjugation reaction. In the context of N-terminal transamination, a highly reactive alanine-lysine motif emerged, which was confirmed to promote the modification of peptide substrates with PLP. This sequence was also tested on two protein substrates, leading to substantial increases in reactivity relative to their wild type termini. This readily-encodable tripeptide thus appears to provide a significant improvement in the reliability with which the PLP-mediated bioconjugation reaction can be used. This study also provides an important first example of how synthetic peptide libraries can accelerate the discovery and optimization of protein bioconjugation strategies.
Introduction
Protein bioconjugates are increasing being used as functional and structural components of new materials.1 In a typical approach, a synthetic functional group of interest is attached to a specific amino acid side chain using one of a relatively limited set of chemical reactions that can modify unprotected biomolecules in aqueous solution.2,3 However, most strategies (such as lysine targeting) lead to complex product mixtures due to the many instances of this functional group on protein surfaces. Although this is acceptable for some applications, improved site selectivity is required to access architecturally precise biomolecular materials. This is often achieved by targeting cysteine residues, but there are many instances in which it is inconvenient or impossible to introduce a single copy of this residue without sacrificing protein function. Also, a growing number of applications (such as FRET-based sensors,4 pollutant responsive hydrogels,5 and therapeutic delivery systems6) require the modification of proteins in two distinct locations. While cysteine modification may be used to install the first group, compatible modification methods that can be used to install the second functional group are in short supply.
As one solution to this challenge, we have previously reported7,8,9 a protein bioconjugation reaction that achieves site-specificity by targeting the N-terminus, a unique position in the sequence. When incubated with pyridoxal 5′-phosphate (PLP), the N-terminal amine undergoes a transamination reaction that installs a ketone or an aldehyde in that position without modifying lysine side chain amines.7 The unique reactivity of the new carbonyl group relative to native side chain functionalities allows for further conjugation to alkoxyamine-containing probes via oxime formation10,11 (Figure 1). This technique provides a convenient and readily scalable way to install a single functional group in a single location, and has been shown to tolerate free cysteine residues.4,5,12
Figure 1.
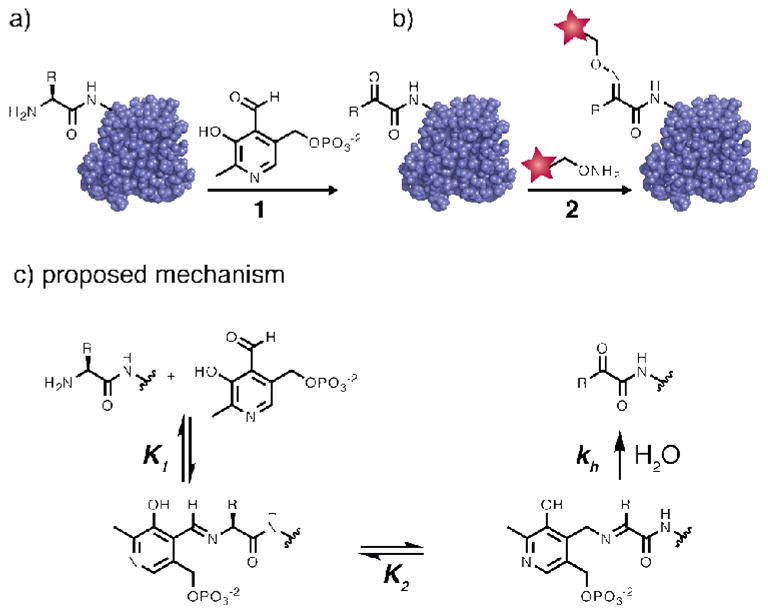
The general scheme of PLP-mediated bioconjugation. (a) In the first step, a protein is incubated with PLP (1) under mild, aqueous conditions. This oxidizes the N-terminus of the protein to a ketone or an aldehyde, providing a unique functional group for further modification. (b) In the second step the ketone is conjugated to an alkoxyamine-bearing reagent (2) through oxime formation. (c) The proposed mechanism begins with Schiff base formation between the N-terminal amine and the PLP aldehyde. Tautomerization, followed by hydrolysis, affords the keto-protein product. Based on this mechanism, the reaction rate is expected to depend on both the concentration of protein and PLP.
There are other methods of N-terminal modification,13,14,15,16,17 each of which may be suitable for different applications and proteins. N-terminal tryptophan residues16 can ligated to aldehyde reagents via a Pictet-Spengler reaction, but it can be difficult to obtain proteins with a tryptophan in the N-terminal position.18 N-terminal serines and threonines can be cleaved using sodium periodate to yield aldehyde functionalities for futher derivitization.17 However sodium periodate may not be compatible with some applications, such as glycoproteins and proteins with large numbers of cysteine or methioine residues.5 PLP-mediated transamination is complementary to these other methods of N-terminal modification and is especially useful because of its compatibility with other protein modification strategies. For instance it can be used in combination with cysteine alkylation12 as well as C-terminal modification using native chemical ligation (NCL).19,20
PLP-mediated bioconjugation has been used to modify a number of different proteins in applications ranging from surface and polymer attachment21 to the dual modification of viral coat proteins with different chromophores for light harvesting systems.12 Since the initial report of this method for protein functionalization, our group has sought to explore the scope of this reaction and to improve its reliability for new protein targets. Our previous work has shown that the transamination reactivity of a protein is dependent on the identity of the N-terminal residue, as well as those in the second and third positions.9 The terminal and internal residues were found to have a complex synergistic effect on the overall reactivity, quickly outpacing our ability to screen sequence candidates individually.
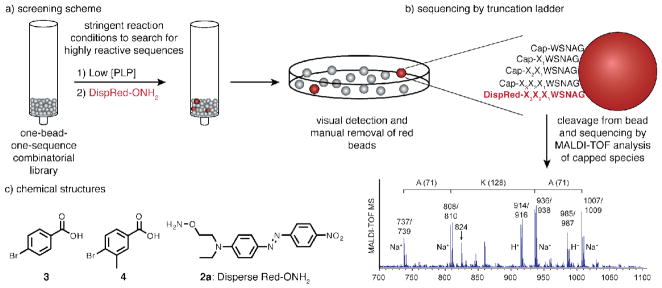
Based on the observation that the penultimate and antepenultimate residues are significant in determining the reactivity of peptides and proteins towards PLP-mediated N-terminal transamination, we sought to identify the most highly reactive sequences of amino acids from the chemical space spanning all combinations of three N-terminal residues. In order to identify such sequences, we developed a combinatorial peptide library screening platform based on a colorimetric detection of desired reactivity, as outlined in Figure 2. Using stringent reaction conditions to screen for the most active sequences, a highly conserved alanine-lysine motif was identified. The improved reactivity of this motif was verified on peptide substrates, and the scope was explored using positional scanning experiments. Additionally, we introduced this sequence on two proteins using site directed mutagenesis. In both cases, the alanine-lysine mutants exhibited improved yields relative to the wild type termini. It is anticipated that this new, highly reactive motif will serve to increase the reactivity of many other protein targets.
Figure 2.
Library screening scheme. (a) The 8,000-member combinatorial peptide library of the form XXXWSNAG was subjected to PLP-mediated transamination and subsequent oxime formation with Disperse Red alkoxyamine 2a. Stringent reaction conditions were used during the PLP reaction such that only sequences with high reactivity transaminated to form a keto group. These active sequences were colorimetrically distinguished from the others during the next step, where the Disperse Red dye formed a covalent oxime linkage with the transaminated sequences. The library was then examined under a microscope, and the red beads were manually removed for sequencing. (b) Each bead contained a truncation ladder in addition to the full-length peptide that had participated in the reaction. Sequencing was performed by cleaving the peptide species from selected beads and identifying the ladder peptides using MALDI-TOF MS. The mass differences between adjacent capped species corresponded to the amino acid in that position. Capped species were easily identified by the unique isotope pattern of the bromine atoms. The mass of the uncapped peptide can be identified at 824 m/z, confirming the assignment. The stuctures of the bromobenzoic acid caps 3 and 4, and the Disperse Red alkoxyamine used in these experiments are shown in (c).
Results/Discussion
Design of a Peptide Library Screening Platform
Although the reactivity of full-sized proteins will undoubtedly be influenced by their tertiary structure, previous work9 had demonstrated that general trends in N-terminal reactivity were consistent between peptide and protein substrates. This observation suggested that it would be possible to screen peptide libraries to identify sequences that would retain their reactivity in the protein context. To do this, we developed a screening platform by adapting a number of traditional techniques used for the construction of solid phase combinatorial peptide libraries.22 As our objective was to find a short genetically encodable sequence that could be conveniently introduced through mutagenesis, we limited our randomization to the terminal three residues, which were followed by an invariant region that could include purification handles or other sequences of interest in future studies. The resulting library included all 20 natural amino acids in each of the three N-terminal positions, corresponding to 8,000 members.
The library was synthesized on resin beads using a split-and-pool technique.22 Additional steps were performed after the coupling of each variable position to cap a portion of the growing chain to facilitate sequencing by mass spectrometry, as described below. A five-amino acid base sequence, WSNAG, was synthesized before the variable positions to provide the library members with enough mass to separate their signals from matrix noise peaks during MALDI-TOF MS analysis. Standard Fmoc solid phase peptide chemistry was used during the synthesis, which was based on Tentagel resin to allow aqueous screening conditions to be performed while the peptides were still attached to the beads.
A key step of the library screening scheme was the development of a simple and rapid method of determining the sequence on active beads. To this end, we created a sequencing technique based on a built-in truncation ladder.23 Compared to de novo sequencing based on tandem MS/MS fragmentation data, this approach allowed for facile identification of the peptide sequence using only intact ions. To create a truncation ladder, a small portion (<10%) of the growing peptide on each bead was capped with bromobenzoic acid (3) during synthesis of the variable positions. Once cleaved, the unique isotope patterns of these truncated species enabled their unambiguous identification, allowing the sequence on each bead to be determined by simple calculation of the mass differences between the molecular ions (Figure 2b). Additionally, we were able to differentiate between isobaric amino acids (isoleucine and leucine) and nearly isobaric amino acids (glutamine and lysine) by capping one of each pair with methylbromobenzoic acid (4) during the synthesis. Supporting Information Figure S1 shows the analysis of a representative library member, in which the relative amounts of peptide and capped species on an individual bead can be seen.
In order to identify beads bearing sequences with high levels of transamination activity, we developed a colorimetric detection method for the ketone and aldehyde products (Figure 2a). Peptides that yielded these functional groups were easily distinguished from those that did not upon subsequent treatment with a Disperse Red alkoxyamine (2a) to form a bright red oxime product (the synthesis of the Disperse Red alkoxyamine is described in the Supporting Information). The oxime formation was performed over 3 h using 10 mM Disperse Red alkoxyamine in DMF. Washing to remove unbound DAFPerse Red led to a clear contrast between beads with and without covalent modification (Supporting Information Figure S2). Spatial separation of the library components on a Petri dish allowed visual detection and manual removal of the red beads for determination of the corresponding sequences.
Screening for Highly Reactive N-Terminal Sequences
The standard conditions reported for PLP-mediated transamination of peptide substrates are 10 mM PLP in 50 mM pH 6.5 phosphate buffer for 18 h at room temperature. When the library was subjected to these reaction conditions (Figure 3a), the majority of the library members were modified by the alkoxyamine dye. As expected, varying degrees of conversion for different sequences could be observed in the various shades of red ranging from nearly colorless to light orange to deep red. This visual representation of reactivity corresponded well with our previous experience of PLP-mediated bioconjugation on protein substrates. To access sequences with greater than average reactivity, we next applied more stringent reaction conditions by lowering the concentration of PLP. Subjecting the library to 1 mM PLP resulted in fewer and lighter red beads (Figure 3b). When the library was treated with 100 μM PLP, a stark contrast in relative reactivity was seen: almost all of the beads were colorless except for a very small number (~1 in 300) that were bright red (Figure 3c). The library was screened multiple times with these conditions using approximately 24 mg of resin, which corresponded to 24,000 beads (three times the library size, a ratio that has been calculated to represent 95% of the members24).
Figure 3.
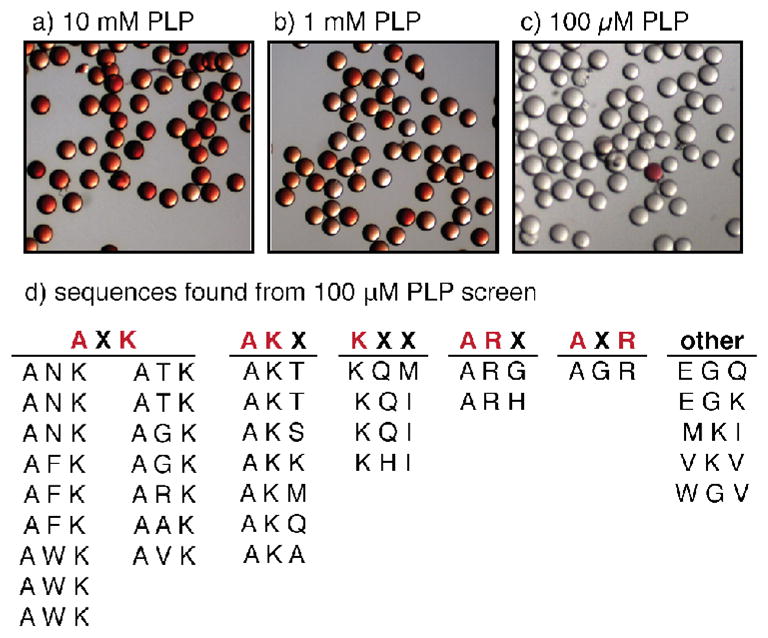
Visual evaluation of the library at different screening conditions, including sequences identified from a 100 μM PLP reaction. (a) When incubated with 10 mM PLP, the majority of the library members turned red, confirming that at standard reaction conditions many sequences exhibit at least some degree of conversion. (b) Using 1 mM PLP, fewer library members turned red, and most showed a lighter color. (c) Using only 100 μM PLP, most of the sequences showed no conversion (as evidenced by the colorless beads) but a small number of red beads were still identified. (d) The sequences corresponding to the red beads at 100 μM PLP revealed AXK and AKX as the predominant N-terminal motifs.
The sequences that were identified after selection and sequencing of the red beads are listed in Figure 3d. These sequences showed a high degree of consensus and were categorized by a few convergent patterns. The preferred N-terminal amino acid showed a striking preference for alanine, which was typically followed by a positively charged amino acid in the second or third position, with lysine being more common than arginine. There were also several lysine terminal sequences, as well as other sequences that did not match this motif. The prevalence of N-terminal alanine fit our previous observations of this reaction,9 but the benefits of the adjacent positive charge had not been formally observed. Interestingly, our previous reactivity screens also contained a lysine residue in position 2. This was included for the purpose of increased solubility and to check for lysine modification in addition to N-terminal transamination. Although it was not modified directly, it now seems that this residue was more than an innocent bystander in these reactions.
Verification of Identified Consensus Sequences
Although the red color of the beads identified during library screening was indicative of the oxime product, the MALDI-TOF mass spectra of the peptide mixtures cleaved from individual beads were not sufficient for quantification of the reaction yield. To get a more detailed understanding of the product distribution of the alanine-positive charge motif, we resynthesized the AKTWSNAG sequence. Upon subjecting beads (10 mg) bearing this peptide to identical transamination conditions, they were visually found to react with the Disperse Red alkoxyamine much as they had during the library screen. Quantification of the reaction conversion was achieved by reacting the N-terminal ketones with benzylalkoxyamine (250 mM in H2O, 3 h), as we found that the Disperse Red oxime tag suppressed ionization. After analysis by LC-QTOF MS, integration of the extracted ion chromatograms of the reaction products was used to obtain the relative yields of each species.9 As confirmation of these values, the tryptophan fluorescence ratios of the products were also compared in some cases. Similar results were obtained.
Under conditions identical to those that were used to screen the library (100 μM PLP, 18 h, rt), the AKT peptide provided a 30% yield of oxime product (Figure 4). Peptides containing only one of these components (i.e. AET- and LKT-WSNAG) were found to be unreactive under these conditions, confirming the synergistic effect of the Ala-Lys combination. At higher PLP concentrations the yield of oxime product increased for all three peptides, with the AKT sequence consistently producing higher reactivity. The mechanism of the transamination reaction almost certainly involves reversible imine formation as the first step, followed by tautomerization. It is likely that the positive charge of the lysine residue serves to recruit the PLP molecule through interactions with the phosphate group, thus increasing the value of K1 in Figure 1b. The origin of the beneficial alanine effect is less clear, as this residue could influence the equilibrium position of the tautomerization step (K2) as well as encourage hydrolysis and/or oxime formation.
Figure 4.
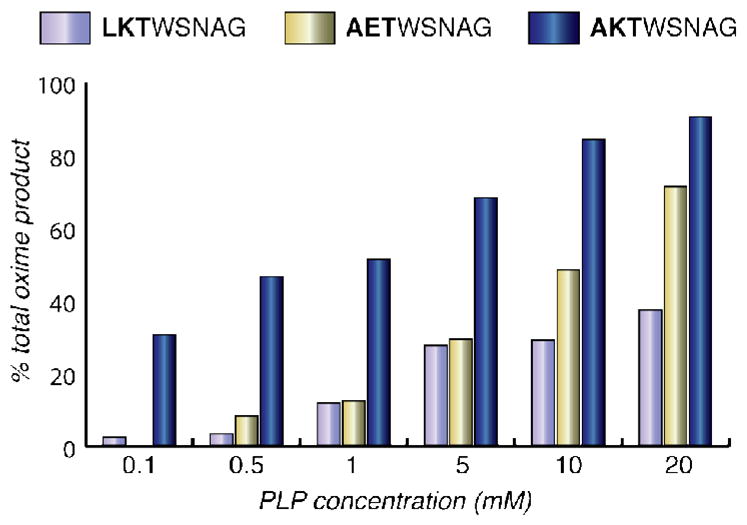
Analogs of the reactive alanine-lysine motif identified during library screening were synthesized to verify the reactivity of this sequence. AKT was compared to LKT to confirm the importance of alanine as the first residue and AET was used to probe the role of a positive charge in the second position. At 0.1 mM PLP, the concentration of library screening, AKT showed a significantly higher yield than the other sequences. AKT also resulted in notably improved yields at higher concentrations of PLP.
Positional Scanning
A positional scanning experiment was performed to determine the ideal position of lysine (in either the second or third position), and to identify any residues that might suppress the reactivity of the motif, which would be important to avoid when using this motif as a tag sequence on proteins. We synthesized 38 octapeptides, in which the three N-terminal residues were of the form AXK or AKX. The identity of X was varied to include each of the 20 natural amino acids, with the exception of cysteine. After subjecting these peptides to standard PLP transamination conditions (10 mM, 18 h), we found that all of the sequences with alanine in the first position and lysine in the second or third position gave relatively high yields of oxime product (Figure 5). Despite some variation in yield, the lowest observed was still above 50% and no residues were found to quench the reactivity of the motif substantially. The most apparent trend was a slight preference for lysine in the second position over the third position. Because alanine-lysine-threonine had one of the highest yields of the synthesized peptides and it occurred multiple times in the library screen, we chose this specific sequence to pursue as an optimized protein tag.
Figure 5.
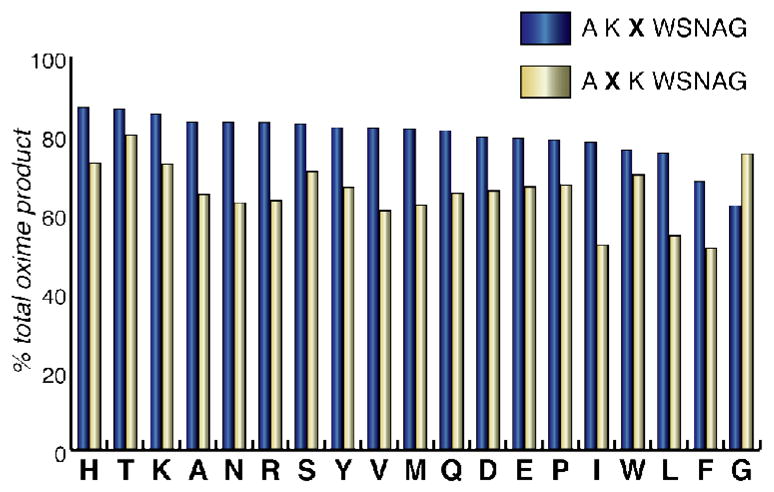
Positional scanning was performed to explore the scope of the AKX and AXK motifs. The total oxime yield after reaction with standard PLP conditions (10 mM, 18 h) was determined for 38 peptides with varied amino acids in the X positions (all natural amino acids except cysteine were included). Although high yields were seen in all cases, peptides with lysine in the second position consistently provided higher conversion.
Application to Protein Substrates
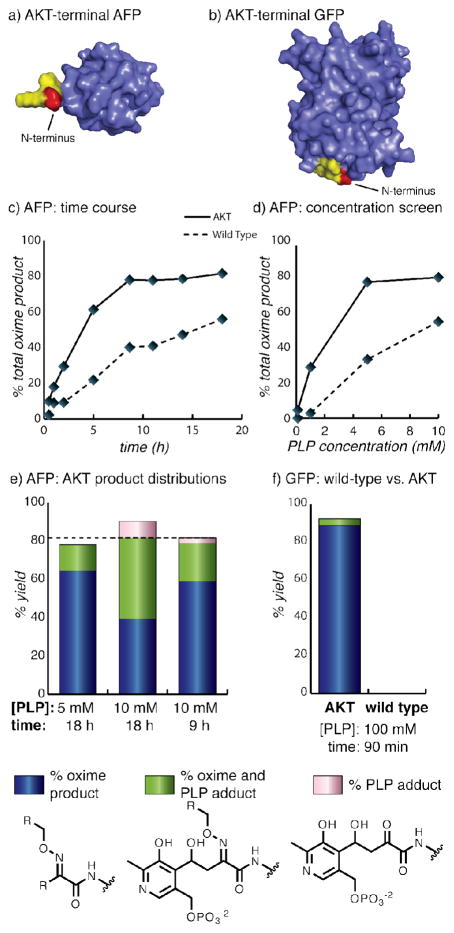
Protein modification reactions are typically performed at substrate concentrations that are significantly lower than those used for peptide modification. Although we have not yet determined the rate law for the transamination reaction, it is reasonable to expect it to depend on the concentration of both PLP and the N-terminal group. Correspondingly, the low concentration of a protein should require a higher PLP concentration to compensate, and will be particularly sensitive to the magnitude of K1 in Figure 1b. To determine if the alanine-lysine-threoine motif would improve the reactivity of proteins through this effect, we installed it on two different substrates: Green Fluorescent Protein (GFP) and the Type III “Antifreeze” Protein (AFP).25 Figure 6a,b indicates the locations of the new sequences on the protein surfaces. On AFP, the AKT motif was a two-residue extension of the wild type N-terminus and on GFP the AKT residues replaced the wild type sequence with no extension of the N-terminus. The wild type N-terminal sequence of GFP (MVS) has minimal reactivity towards PLP at RT to 37 °C,20 and the wild type AFP has fair-to-good reactivity. After expression and purification, the wild-type and AKT variants of these proteins were incubated with PLP at 37 °C, concentrated to remove excess PLP after the reaction, and then subjected to benzylalkoxyamine (25 mM in 25 mM phosphate buffer pH 6.5, 18 h) to allow quantification of the product distribution using mass spectrometry.
Figure 6.
The performance of wild-type and AKT-terminal sequences were compared for the transamination of two protein substrates. The positions of the new termini are shown for (a) an “Antifreeze” Protein (AFP) and (b) GFP. The Ala residue is in red and the Lys-Thr portion is in yellow. (c) The transamination of the AFP mutant showed that the AKT sequence outperformed the wild-type terminus (GNQ) at every time point analyzed. (d) The AKT-terminal AFP also outperformed the wild-type sequence at different concentrations of PLP. (e) The product distribution of the AKT-terminal AFP mutant shows that maximum oxime yield (blue plus green) and minimum PLP adduct (green plus pink) could be achieved by optimizing the reaction conditions. The dotted line marks the highest total oxime yield acheived, demonstrating that the strategies that decreased byproduct formation did not sacrifice overall yield. (f) Using the strategy of a high concentration of PLP for a short reaction time, conditions were found that gave a high oxime yield with minimal byproducts for AKT-terminal GFP in only 90 minutes. The wild type GFP mutant achieved no modification under the same conditions, highlighting the enhanced reactivity conferred by the AKT sequence.
Wild type AFP showed intermediate levels of oxime product formation, which followed the expectations of a GNQ N-terminal sequence. However, the AKT mutant outperformed the wild type under every set of reaction conditions screened (including variation of the concentration of PLP and the reaction time), as shown in Figure 6c and 6d. Upon inspection of the product distribution for the AKT-terminal mutant (Figure 6e), we observed a significant amount of covalent PLP addition to the N-terminus, which corresponded by mass to an aldol addition of PLP to the introduced ketone group (Supporting Information Figure S3). However, the aldol addition of PLP did not preclude oxime formation, as the green portions of the bars correspond to the reaction of the PLP adduct with the alkoxyamine agents. Thus, in many applications where the goal is simply to ligate the target protein to another material through oxime formation, the PLP addition byproduct may be innocuous.
As would be expected for a bimolecular reaction, we observed that the yield of PLP adduct increased with the same parameters as transamination (i.e. higher concentrations of PLP and increased reaction times). Since the PLP adduct likely forms after the transamination has taken place (Supporting Information Figure S4), we hypothesized that a balance of these two reaction conditions might minimize PLP adduct formation without sacrificing the total oxime yield. For AFP, using a lower concentration of PLP for the full reaction time (5 mM for 18 h) or using the standard concentration for a shorter time (10 mM for 9 h) resulted in decreased amounts of PLP adduct (green bar, Figure 6e) while preserving the total amount of oxime yield, shown as a dotted line. Therefore to minimize the PLP adduct byproduct, it is recommended that lower concentrations of PLP be used for longer reaction times, or that higher concentrations of PLP be used for shorter incubation periods. In practice, the optimal balance varies somewhat for each protein substrate, and, like most other organic reactions, a preliminary screen of the reaction variables is recommended.
Applying these strategies for maximizing oxime yield with minimal PLP addition byproduct led to striking results for AKT-terminal GFP. Using the high concentration of PLP/short reaction time strategy, we observed over 90% yield of oxime product with negligible amounts of byproducts by using 100 mM PLP for 90 min (Figure 6f). Under the same conditions, wild-type GFP (N-terminal sequence: MVS) provided no conversion, highlighting the importance of the AKT sequence for achieving increased reactivity. Screening of the full peptide library with these reaction conditions resulted in high reactivity for a number of beads, with too many being observed to allow meaningful sequencing. The product distributions of AKT-terminal GFP under other reaction conditions are shown in Supporting Information Figure S5. As GFP is commonly fused to protein domains to allow visualization using fluorescence, we anticipate that the availability of a fast and reliable method for the introduction of a second functional group will benefit many applications.
Conclusion
The proteins AFP and GFP demonstrate that mutating the N-terminal sequence of a protein to AKT can increase reactivity toward PLP-mediated bioconjugation and thus these results serve to validate our combinatorial peptide library screening process. We have also identified reaction conditions for AKT-terminal proteins that maximize yield and purity. We anticipate that the development of this encodable motif will be of interest to those in the chemical biology and biomolecular materials communities who wish to apply this mild, site-specific transamination reaction to their own protein substrates. Based on the success of these studies, we are continuing to use this combinatorial library screening platform to accelerate the discovery and optimization of new bioconjuation reactions. In particular, we are using this platform to identify new transamination agents that have a reduced tendancy to form aldol addition products.
Supplementary Material
Acknowledgments
This work was generously supported by the NIH (R01 GM072700). LSW was supported by a predoctoral fellowship from the NSF. LSW, BWT, and APEK were supported by the Berkeley Chemical Biology Graduate Program (NRSA Training Grant 1 T32 GMO66698).
Footnotes
SUPPORTING INFORMATION AVAILABLE. Full experimental details and additional characterization spectra. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.For lead examples, see: Douglas T, Young M. Nature. 1998;393:152–155.Abedin MJ, Liepold L, Suci P, Young M, Douglas T. J Am Chem Soc. 2009;131:4346–4354. doi: 10.1021/ja8079862.Lee S, Mao C, Flynn CE, Belcher AM. Science. 2002;296:892–895. doi: 10.1126/science.1068054.Nam YS, Shin T, Park H, Magyar AP, Choi K, Fantner G, Nelson KA, Belcher AM. J Am Chem Soc. 2010;132:1462–1463. doi: 10.1021/ja908812b.Wang Q, Lin T, Tang L, Johnson JE, Finn MG. Angew Chem, Int Ed. 2002;41:459–462. doi: 10.1002/1521-3773(20020201)41:3<459::aid-anie459>3.0.co;2-o.Niemeyer CM. Angew Chem Int Ed. 2001;40:4128–4158. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S.McMillan RA, Paavola CD, Howard J, Chan SL, Zaluzec NJ, Trent JD. Nature Materials. 2002;1:247–252. doi: 10.1038/nmat775.Wang P, Sergeeva MV, Lim L, Dordick JS. Nat Biotechnol. 1997;15:789–93. doi: 10.1038/nbt0897-789.Heredia KL, Maynard HD. Org Biomol Chem. 2007;5:45–53. doi: 10.1039/b612355d.
- 2.Hermanson GT. Bioconjugate Techniques. 1. Academic Press; San Diego: 1996. [Google Scholar]
- 3.Tilley SD, Joshi NS, Francis MB. The Chemistry and Chemical Reactivity of Proteins. In: Begley T, editor. The Wiley Encyclopedia of Chemical Biology. Wiley-VCH; Weinheim: 2008. [Google Scholar]
- 4.Crochet AP, Kabir M, Francis MB, Paavola CD. Biosensors and Bioelectronics. 2010 doi: 10.1016/j.bios.2010.05.012. in press. [DOI] [PubMed] [Google Scholar]
- 5.Esser-Kahn AP, Iavarone AT, Francis MB. J Am Chem Soc. 2008;130:15820–15822. doi: 10.1021/ja807095r. [DOI] [PubMed] [Google Scholar]
- 6.(a) Kovacs EW, Hooker JM, Romanini DW, Holder PG, Berry KE, Francis MB. Bioconjugate Chemistry. 2007;18:1140–1147. doi: 10.1021/bc070006e. [DOI] [PubMed] [Google Scholar]; (b) Tong GJ, Hsiao SC, Carrico ZM, Francis MB. J Am Chem Soc. 2009;131:11174–11178. doi: 10.1021/ja903857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. Angew Chem, Int Ed. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 8.Scheck RA, Francis MB. ACS Chem Biol. 2007;2:247–251. doi: 10.1021/cb6003959. [DOI] [PubMed] [Google Scholar]
- 9.Scheck RA, Dedeo MT, Iavarone AT, Francis MB. J Am Chem Soc. 2008;130:11762–11770. doi: 10.1021/ja802495w. [DOI] [PubMed] [Google Scholar]
- 10.Jencks WP. J Am Chem Soc. 1959;81:475–481. [Google Scholar]
- 11.Rose K. J Am Chem Soc. 1994;116:30–33. [Google Scholar]
- 12.Dedeo MT, Duderstadt KE, Berger JM, Francis MB. Nano Lett. 2010;10:181–186. doi: 10.1021/nl9032395. [DOI] [PubMed] [Google Scholar]
- 13.Tam JP, Yu Q, Miao Z. Biopolymers. 1999;51:311–332. doi: 10.1002/(SICI)1097-0282(1999)51:5<311::AID-BIP2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 14.Dixon HBF. J Protein Chem. 1984;3:99–108. [Google Scholar]
- 15.Dixon HBF, Fields R. Methods Enzymol. 1972;25:409–419. doi: 10.1016/S0076-6879(72)25036-4. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Zhang L, Hall SE, Tam JP. Tetrahedron Lett. 2000;41:4069–4073. [Google Scholar]
- 17.Geoghegan KF, Stroh JG. Bioconjugate Chem. 1992;3:138–146. doi: 10.1021/bc00014a008. [DOI] [PubMed] [Google Scholar]
- 18.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Proc Natl Acad Sci. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Dawson PE, Muir TW, Clarklewis I, Kent SBH. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]; (b) Hofmann RM, Muir TW. Curr Opin Biotech. 2002;13:297–303. doi: 10.1016/s0958-1669(02)00326-9. [DOI] [PubMed] [Google Scholar]; (c) Tolbert TJ, Wong CH. J Am Chem Soc. 2000;122:5421–5428. [Google Scholar]
- 20.Esser-Kahn AP, Francis M. Angew Chem, Int Ed. 2008;120:3811–3814. [Google Scholar]
- 21.(a) Christman KL, Broyer RM, Tolstyka ZP, Maynard HD. J Mater Chem. 2007;17:2021–2027. [Google Scholar]; (b) Lempens EHM, Helms BA, Merkx M, Meijer EW. Chem Bio Chem. 2009;10:658–662. doi: 10.1002/cbic.200900028. [DOI] [PubMed] [Google Scholar]; (c) Gao W, Liu W, Mackay JA, Zalutsky MR, Toone EJ, Chilkoti A. Proc Natl Acad Sci. 2009;106:15231–15236. doi: 10.1073/pnas.0904378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Lam K, Lebl M, Krchnak V. Chem Rev. 1997;97:411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]; (b) Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 23.Youngquist RS, Fuentes GR, Lacey MP, Keough T. J Am Chem Soc. 1995;117:3900–3906. [Google Scholar]
- 24.Burgess K, Liaw AI, Wang N. J Med Chem. 1994;37:2985–2987. doi: 10.1021/jm00045a001. [DOI] [PubMed] [Google Scholar]
- 25.Esser-Kahn AP, Trang V, Francis MB. J Am Chem Soc. 2010 doi: 10.1021/ja103038p. available as ASAP article. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.