Abstract
IL-26 is classified as a member of the IL-10 cytokine family because it has limited sequence homology to IL-10 and the IL-10-related cytokines. The human IL-26 gene, IL26, is located on chromosome 12q15 between the genes for two other important class-2 cytokines, IFNG (IFN-γ) and IL22 (IL-22). IL-26 is often co-expressed with IL-22 by activated T cells, especially Th17 cells. It signals through a heterodimeric receptor complex composed of the IL-20R1 and IL-10R2 chains. IL-26 receptors are primarily expressed on non-hematopoietic cell types, particularly epithelial cells. Signaling through IL-26 receptor complexes results in the activation of STAT1 and STAT3 with subsequent induction of IL-26-responsive genes. The biological functions of IL-26 have only begun to be defined.
Keywords: IL-26, Th17 cells, IL-20R1, STAT3, inflammation
1. Introduction
Interleukin-26 (IL-26) was originally discovered by Knappe et al. [1] as a novel cDNA clone, denoted AK155, that displays weak but significant sequence homology (approximately 25% identity) to IL-10. The human IL26 gene was first identified via its elevated expression in Herpesvirus saimiri (HVS)-transformed T cells. The encoded protein is a member of the IL-10 family of cytokines that also includes IL-10, IL-19, IL-20, IL-22, IL-24 [2, 3]. IL-26 is a secreted protein, and may be functional either as a monomer or homodimer, although definitive physicochemical data regarding the structure of IL-26 has not yet been published. However, it is clear that IL-26, like the other IL-10-related cytokines, is a member of the class-2 cytokine family. Although the IL-26 protein shares some sequence homology with IL-10, IL-26 binds to a distinct cell surface receptor consisting of the IL-20R1 and IL-10R2 chains, and induces functional activities that are different from those mediated by IL-10. The primary target cells for IL-10, including monocytes, neutrophils and lymphocytes, do not express IL-26 receptors.
The human IL-26 gene, IL26, maps to chromosome region 12q15 between two other important class-2 cytokine genes, IFNG (IFN-γ) and IL22 (IL-22) [4]. Several recent reports have shown that the IL-26 gene is often co-expressed with IL-22 at high levels by Th17 cells [5–7]. In fact, the phenotype of human Th17 cells is defined in part by the fact that these cells co-express IL-17, IL-22 and IL-26. Therefore, although IL-26 was originally discovered as an inducible gene in Herpesvirus saimiri (HVS)-transformed human T cells [1], it is now clear that the IL26 gene is more broadly expressed by Th17 cells following antigen-specific stimulation via clonotypic T cell antigen receptors.
2. Organization and evolutionary conservation of the IL-26 gene
IL-26 is classified as a member of the IL-10-related subfamily of cytokines that includes IL-10, IL-19, IL-20, IL-22, IL-24 [2,3]. With the exception of the IFN-λ genes, the IL-10-related genes are clustered on two loci located on two distinct chromosomes. The genes for IL-10, IL-19, IL-20 and IL-24 reside on a single locus on human chromosome 1q32. The genes for IL-22, IL-26 and IFN-γ are arranged in tandem on human chromosome 12q15, and are transcribed in the same orientation (Fig. 1). The human IL26 gene is composed of five exons that are separated by three introns. As is typical of all of the IL-10-related genes, the IL26 gene has a “phase 0” intron/exon boundary.
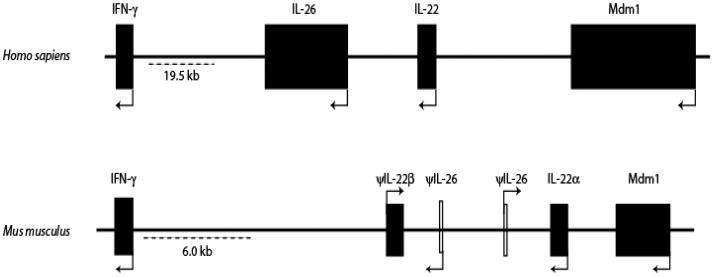
Figure 1.
A schematic comparison showing syntenic conservation of the IFN-γ, IL-26 and IL-22 loci in the human (Homo sapiens) and murine (Mus musculus) genomes. Information regarding the IFN-γ/IL-26/IL-22 locus was obtained from the NCBI map viewer for human chromosome-12 and murine chromosome-10. The closed boxes and arrowheads denote the positions and transcriptional orientations of the genes. The murine IL-26 gene fragments are shown as “open” boxes because they represent a bioinformatic prediction of an incomplete IL-26 gene. Abbreviations used: IFN-γ, interferon gamma; MDM, mouse double minute; IL, interleukin; ψ, pseudogene.
The IL-26 gene is conserved in most vertebrate species, but is curiously absent in mice. Paralogs of the IL-22, IL-26 and IFN-γ genes have been identified in several non-mammalian species, including fish, frogs and birds [8–10]. A recent analysis of the genomes of pufferfish (Takifugu rubripes), zebrafish (Danio rerio) and the western clawed frog (Xenopus tropicalis) showed syntenic conservation of this cytokine gene cluster [8–10]. Interestingly, there is an additional homolog of the IFN-γ gene in the fish genome, demonstrating that this is an active locus for lineage expansion of genes [8]. The genomic organization of both the fish and frog IL-26 genes are similar to the human IL-26 gene [9,10]. In mice, this cytokine gene cluster differs from most other species by the fact that the IL-22 gene is duplicated to generate two distinct IL-22 genes, denoted IL-22α and IL-22β [11]. However, in at least two strains, BALB/c and DBA/2, there is only a single copy of the IL-22 gene. The murine IL-22β gene is believed to be a pseudogene with a 658 nucleotide deletion in its non-coding exon at the 5’ end [11]. Moreover, in mice, the IL-26 gene is conspicuously absent from this cytokine gene cluster (Fig. 1) [12]. When this locus was examined more closely, remnant exons of the IL-26 gene that have substantial similarity to the human IL26 gene were identified (R.S. and H.A.Y., unpublished observations). Interestingly, these remnant exons of the IL-26 gene in mice indicate two copies of the gene in a tail-to-tail orientation. However, these exons do not form a full-length gene, and for unknown reasons, it appears that the IL-26 gene is disrupted only in rodents, but not in other vertebrates. So far, attempts to amplify the IL-26 gene sequence from various inbred and outbred strains of mice following stimulation have been unsuccessful. However, it has not been ruled out that a full length IL-26 gene paralog might be present in the genome of truly “wild” mice.
The amino acid sequence of IL-26 shares very low sequence homology (~15–25%) with other members of the IL-10 cytokine family or with IFN-γ [1,2,3]. In contrast, IL-26 shares very high amino acid sequence identity with its own mammalian paralogs (85–95%). However, as shown in Fig. 2, the amino acid sequence similarity of the human IL-26 protein to non-mammalian species such as zebrafish or frogs is much lower (17–30%). As also shown in Fig. 2, the human IL-26 protein contains a large number of positively charged amino acids, including 30 residues that are either lysine or arginine. As a consequence, the predicted isoelectric point for this protein is fairly alkaline: pI = 10.7. The overall net positive charge of the IL-26 protein provides an explanation for the reported heparin-binding activity of this molecule [1,4]. It is interesting to note that, like IL-26, IFN-γ also displays potent heparin-binding activity [13].
Figure 2.
A comparison of the deduced amino acid sequences for the IL-26 gene in several mammalian and lower vertebrate species. Residues conserved across the sequences of multiple species are shaded. Dashes (−) indicate gaps that were introduced for optimal alignment. Multiple sequence alignment was carried out using Bio-edit software which uses Clustal W [version 1.8] and BLOSUM series was used for Protein weight matrix. The accession numbers for sequences used in this analysis are as follows: human, Homo sapiens, GenBank accession no. EAW97181; bovine, Bos taurus, GenBank accession no. XP_001250652; chimpanzee, Pan troglodytes, XP_001152032; rhesus monkey, Macaca mulatta, GenBank accession no. XP_001117154; zebrafish, Danio rerio, GenBank accession no. AAI63119; western clawed frog, Xenopus tropicalis, GenBank accession no. ABU54058.
IL-26 signals through a heterodimeric receptor complex composed of two class II cytokine receptor proteins: IL-20R1 and IL-10R2. In contrast to the IL-26 gene, the genes encoding IL-20R1 (IL20RA) and IL-10R2 (IL10RB) have been highly conserved throughout evolution. Consequently, gene paralogs for both of these molecules have been identified in many mammalian species as well as lower vertebrates. Although the murine genome does not contain an intact IL-26 gene equivalent, it does contain the IL-26 receptor genes: Il20ra and Il10rb [14]. Therefore, although the IL-26 gene is not conserved in mice, the gene encoding the IL-26 receptor, IL-20R1, is conserved in rodents. This might be explained by the dual functions of the IL-20R1 chain because in addition to its function as the ligand-binding chain in the IL-26 receptor complex, IL-20R1 can also heterodimerize with the IL-20R2 chain to generate receptors for IL-19, IL-20 and IL-24. Therefore, in the absence of an IL-26 gene paralog in the murine genome, IL-20R1 must function predominantly as a receptor for IL-19, IL-20 and IL-24 in rodents.
3. Cellular Sources of IL-26
IL-26 can be produced by primary T cells, NK cells and T cell clones following stimulation with specific antigen or mitogenic lectins. IL-26 (AK155) was initially shown by several groups to be co-expressed together with another important IL-10-related cytokine gene, IL-22 [15,16]. IL-26 is co-expressed together with IFN-γ and IL-22 by human Th1 clones, but not by Th2 clones. It was subsequently found that IL-26 is co-expressed with IL-17 and IL-22 by Th17 cells, an important subset of CD4+ T-helper cells that is distinct from Th1 and Th2 cells [5–7]. More recently, a novel subset of CD56+ NKp44+ NK cells was identified that co-expresses IL-22 and IL-26, especially following treatment with IL-23 [17]. Furthermore, Hughes et al. described a different subset of immature NK cells that do not express CD56 or NKp44 but do express CD117 and CD161 and constitutively express IL-22 and IL-26 [18].
The mechanisms that regulate transcription of the human IL-26 gene are so far largely undefined. It is possible and perhaps likely that expression of the IL-26 gene is induced in an IL-23-dependent manner because IL-23 is known to induce differentiation of Th17 cells, and IL-23 amplifies expression of IL-17 and IL-22 by Th17 cells.
4. The IL-26 Receptor Complex
Although the IL-26 gene and its corresponding protein share some sequence homology with IL-10, IL-26 does not share IL-10’s primary functional activities. This is due in large part to the fact that the primary target cells for IL-10 such as monocytes and macrophages do not express IL-20R1, the ligand-binding chain of the IL-26 receptor complex. Instead, IL-26 is active on a variety of non-hematopoietic cell types, including many types of epithelial cells that do express the IL-20R1 chain.
In 2004, two groups independently identified the receptor for IL-26 [19, 20]. As illustrated in Figure 3A, IL-26 signals through a heterodimeric receptor complex composed of the IL-20R1 and IL-10R2 chains. IL-20R1 functions as the specific ligand-binding chain for IL-26, and IL-10R2 functions as an essential second chain to complete assembly of the active receptor complex. Neutralizing antibodies against either the IL-20R1 or IL-10R2 chains can block induction of IL-26 signaling [19,20]. IL-26 binds initially to IL-20R1 to form a binary complex: IL-26 + IL-20R1. The binding of IL-26 to the IL-20R1 chain rapidly induces conformational changes that facilitate recruitment of the IL-10R2 chain to complete assembly of the ternary complex. It is likely that the ternary IL-26 receptor complex is assembled in a sequential order as previously shown for the IL-10 receptor complex [21,22]. Once fully assembled, the receptor complex undergoes a conformational change(s) that induces activation of the receptor-associated Janus tyrosine kinases, Jak1 and Tyk2, and subsequent transient docking and phosphorylation of the STAT proteins, STAT1 and STAT3 [19,20].
Figure 3.
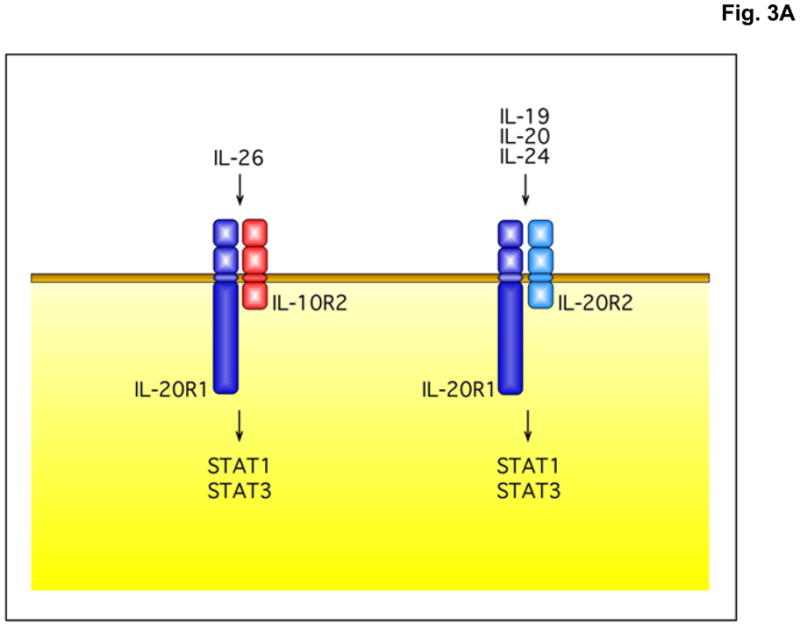
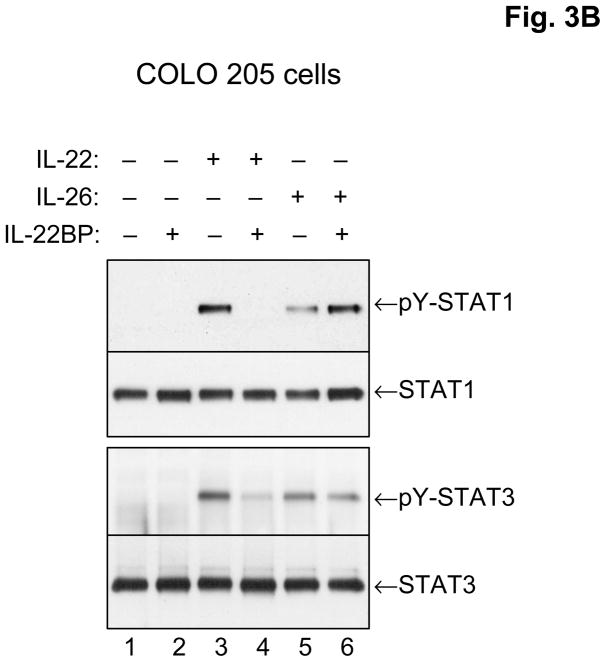
The IL-26 receptor complex. A. The IL-26 receptor complex is composed of two polypeptide chains: the ligand-binding chain, IL-20R1, and the accessory receptor chain, IL-10R2. IL-26 binds initially to IL-20R1, and then rapidly recruits the IL-10R2 chain to complete assembly of the active receptor complex. Ligand-induced heterodimerization of these receptor chains initiates a signal transduction cascade that results in the activation and nuclear translocation of STAT1 and STAT3. IL-20R1 can also dimerize with another class II cytokine receptor, IL-20R2, to generate receptors for IL-19, IL-20 and IL-24. B. IL-26 induces tyrosine phosphorylation of STAT1 and STAT3 in the human colorectal carcinoma cell line, COLO 205. COLO 205 cells were incubated with IL-22 or IL-26 (100 ng/mL) in the presence or absence of recombinant human IL-22 binding protein (IL-22BP) for 30 minutes at 37°C. At the end of this incubation period, whole cell lysates were prepared and analyzed by western blotting with antibodies specific for tyrosine-phosphorylated STAT1 or tyrosine-phosphorylated-STAT3 and total STAT1 and total STAT3.
Like IL-26, IL-10 also preferentially activates STAT3 in IL-10R1-positive cells such as macrophages and T cells. IL-10 is characterized in part by its ability to suppress production of proinflammatory cytokines such as TNF-α and IL-1 by activated macrophages [23]. However, IL-26 does not inhibit expression of genes such as TNF-α or IL-1 in monocytes or macrophages because these cells do not express the IL-26-binding chain, IL-20R1. Furthermore, studies have shown that IL-26 actually up-regulates expression of several proinflammatory cytokine genes such IL-6 and IL-8 in keratinocytes and intestinal epithelial cells [20,24]. Therefore, despite its structural relatedness to IL-10, IL-26 does not share IL-10’s ability to inhibit cytokine production by monocytes and macrophages.
4.1. IL-20R1: the ligand-binding chain for IL-26
The gene that encodes the human IL-20R1 chain, IL20RA, and its corresponding protein were originally identified as one of the two proteins that are required to form the receptor complex for IL-20, another IL-10-related cytokine [14]. In the case of IL-20, this cytokine binds initially to the IL-20R2 chain, and then this binary complex (IL-20 + IL-20R2) recruits the IL-20R1 chain to complete assembly of the active receptor complex [25]. The Type-I IL-20 receptor complex (IL-20R1 + IL-20R2) is structurally related to the IL-10 receptor complex (IL-10R1 + IL-10R2). The fact that the IL-20R1 chain also serves as an essential component of the IL-26 receptor complex illustrates the common theme that class II cytokine receptor complexes often share use of one or more polypeptide chains. For example, the class II cytokine receptor chain, IL-10R2, can heterodimerize with at least four different ligand-binding chains: IL-10R1, IL-20R1, IL-22R1 and IL-28R to generate the receptors for IL-10, IL-26, IL-22 and IL-28/IL-29 (IFN-λ), respectively [26].
IL-20 signals primarily but not exclusively through a heterodimeric receptor complex composed of the IL-20R1 and IL-20R2 chains [14]. These receptor proteins are also sometimes referred to as IL-20Rα and IL-20Rβ, respectively. This receptor complex was originally defined based on its ability to bind IL-20 [14]. However, subsequent studies showed that the IL-20R1:IL-20R2 complex is also used for signaling by two other IL-10-related cytokines, IL-19 and IL-24 [27,28]. Therefore, the IL-20R1-IL-20R2 receptor complex is shared by several cytokines, including IL-19, IL-20 and IL-24. The shared use of various receptor chains by several different cytokines is a common feature of signaling by many cytokines. For example, the common γ chain (γc) is an essential component of several cytokine receptors, including those for IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21. Similarly, the gp130 receptor is a shared component in the receptor complexes for IL-6, LIF and oncostatin M (OSM). Therefore, as illustrated in Figure 3A, the IL-20R1 chain can either heterodimerize with the IL-10R2 chain to form receptors for IL-26 or it can heterodimerize with the IL-20R2 chain to form receptors for IL-19, IL-20 and IL-24.
The IL-20R1 chain is expressed at moderate levels by many cell types [14–16]. Although IL-20R1 is expressed in skin and lung tissue, it is not expressed in lymphoid organs such as the thymus or spleen or by peripheral blood leukocytes [14,15,19]. IL-20R1 is also expressed at low levels in the brain, especially in the cerebellum, medulla and spinal cord [19]. The second chain of the IL-20 receptor complex, IL-20R2, has an even more limited tissue expression pattern, and is also not expressed at significant levels by leukocytes [14,15,28]. By comparison, the ligand-binding chain of the IL-10 receptor complex, IL-10R1 (IL10RA), is highly expressed by most if not all hematopoietic cells, including lymphocytes and macrophages, but not by non-hematopoietic cell types such as fibroblasts or endothelial cells [23,29]. In contrast, the second chain of the IL-10 receptor complex, IL-10R2 (IL10RB), is broadly expressed on most cell types and tissues [15,16,19,20,30].
Binding of IL-26 to its receptor complex (IL-20R1 + IL-10R2) induces rapid activation of STAT3 and, to a lesser degree, STAT1 [19,20]. Several groups have demonstrated that IL-26 can activate STAT1 and STAT3 using stably transfected tumor cell lines that overexpress the IL-20R1 and IL-20R2 genes. In addition, IL-26 can induce activation of STAT1 and STAT3 in certain human tumor cell lines that constitutively express the IL-20R1 and IL-10R2 chains. For example, as shown in Figure 3B, treatment of COLO 205 cells (human colorectal carcinoma) with recombinant human IL-26 induces activation (tyrosine phosphorylation) of STAT1 and STAT3. Like IL-26, IL-22 also induces tyrosine phosphorylation of STAT1 and STAT3 in COLO 205 cells, and as illustrated in Fig. 3B, the induction of STAT1 and STAT3 by IL-22 but not IL-26 can be markedly inhibited by the addition of recombinant soluble IL-22-binding protein (IL-22BP).
Little is known about the genes that are induced by signaling through the IL-26 receptor complex. However, the identification of human cell lines such as COLO-205 and HaCaT that constitutively express IL-26 receptors provides useful cellular targets for identifying genes that are inducible by IL-26. The repertoire of genes induced by IL-22 has already been examined in one published study [31]. It will be interesting to determine if the gene expression profiles induced by IL-26 are similar to those induced by IL-22 or if each of these cytokines activates its own unique repertoire of genes.
4.2. IL-10R2: the accessory receptor chain
The IL-10R2 chain (alias CRF2-4) is a short chain, R2-type, class-2 cytokine receptor. It functions primarily as an accessory chain to recruit the Janus tyrosine kinase, Tyk2, to the receptor complex. As mentioned earlier in this review, the IL-10R2 chain is an essential shared component in four distinct receptor complexes. The IL-10R2 chain can heterodimerize with either the IL-10R1, IL-22R1, IL-20R1 or IL-28R polypeptide chains to generate receptors for IL-10, IL-22, IL-26 and IL-28/-29 (alias IFN-λ), respectively [26]. In contrast to the respective ligand-binding R1 chains which have restricted tissue expression patterns, the IL-10R2 chain is broadly expressed on most cell types.
Both IL-22 and IL-26 are strong activators of STAT3 phosphorylation in target cells that express the relevant receptor complexes. The ligand-binding chains of the IL-22 and IL-26 receptor complexes (IL-22R1 and IL-20R1, respectively) are predominantly expressed by non-haematopoietic cell types, including epithelial cells of the skin and colon. The tyrosine residues present in so-called cytokine receptor box-3 motifs (YXXQ) at positions 446 and 496 on the intracellular domain of the human IL-10R1 chain are known to be critical for IL-10 function, and these motifs are highly conserved in the murine and human IL-10R1 polypeptides [32,33]. This motif has been described as being common to many STAT3-activating cytokine receptors, including IL-10R1, IL-20R1 and IL-22R1 [2,34,35]. Despite their common ability to induce activation of STAT3 in various target cells, IL-22 and IL-26 do not induce anti-inflammatory activities in their respective target cells. This suggests that the presence of a box-3 YXXQ motif and the ability to activate STAT3 are not sufficient to confer anti-inflammatory activity to the IL-22 or IL-26 receptor complexes as in the case for the IL-10 receptor complex. Other motifs within the IL-10 receptor complex must mediate the anti-inflammatory activities of IL-10.
IL-10 and the IL-10-related cytokines, IL-19, IL-20 and IL-24, are co-expressed by antigen-presenting cells such as monocytes, macrophages and dendritic cells. In contrast, IL-22 and IL-26 are produced primarily by activated T cells [5–7]. The genes encoding human IL-22, IL-26 and IFN-γ are located in close proximity to one another on chromosome 12q15. It is possible that these three genes (IFNG, IL26 and IL22) may be coordinately regulated under certain conditions. The close proximity of the IL-22 and IL-26 genes to the IFN-γ gene also suggests that IL-22 and/or IL-26 might induce certain functional activities in common with IFN-γ. Like IL-22 and IL-26, IFN-γ activates STAT1 and STAT3 in many cell types, however, unlike the receptors for IL-22 and IL-26 which are not present on leukocytes [15,16,36], IFN-γ receptors are expressed on leukocytes, including macrophages, neutrophils and lymphocytes.
Although T cells are the principal producers of IL-22 and IL-26, the primary target cells for these cytokines are non-hematopoietic cell types such as keratinocytes and hepatocytes. IL-22R1 is highly expressed in normal liver, kidney and pancreas. IL-20R1 is expressed at significant levels by epithelial cells in the skin, colon and small intestine. We and others have also found that many tumor cell lines derived from the liver or colon express high levels of IL-22R1, IL-20R1 or both [16,19,20,24]. As a result, these cell lines can respond to both IL-22 and IL-26.
5. Structure of IL-26 and the IL-26 Receptor Complex
Currently, high-resolution crystal structures of IL-26 or the IL-26/IL-20R1/IL-10R2 ternary complex have not yet been reported. Therefore, we developed an initial structural model of IL-26 and its interactions with IL-20R1 and IL-10R2. Blastp searches showed that IL-26 shares the greatest sequence identity (~24–27%) with IL-10, IL-22, and IL-19. Since crystal structures are available for each of these cytokines, we attempted to determine which cytokine provides the best scaffold for the IL-26 sequence [37–41]. To address this question, we first superimposed the IL-10, IL-22, and IL-19 crystal structures using STAMP [42]. The IL-26 amino acid sequence was then aligned to the IL-10/IL-22/IL-19 structural-sequence alignment using the program STACCATO [43]. The resulting alignment is shown in Figure 4A, and indicates that IL-19 provides the best structural scaffold to accommodate the IL-26 sequence. In particular, the IL-26 cysteine residues are properly aligned with the IL-19 cysteines with few alignment gaps. Our model is further supported by published receptor binding activity data that demonstrate that IL-20R1 can bind IL-19 as well as IL-26 [27,28].
Figure 4.
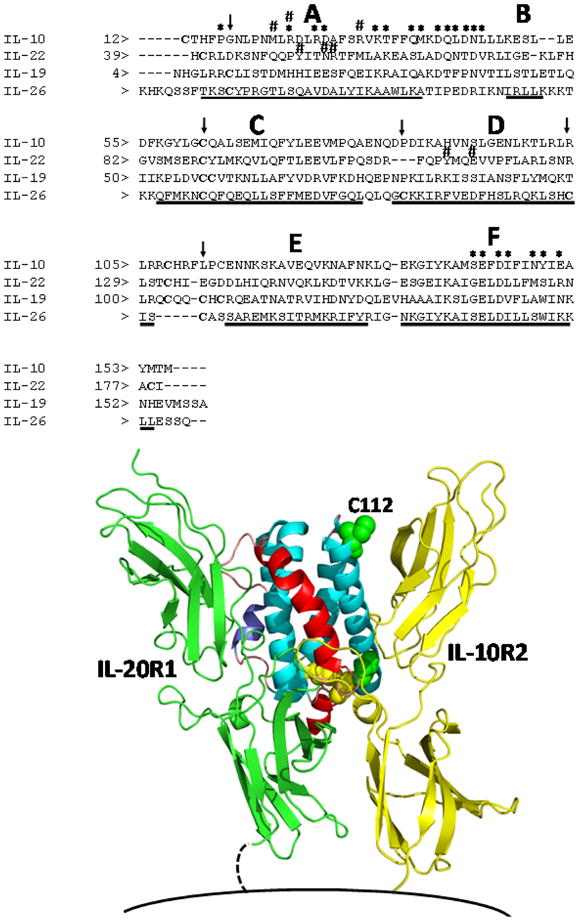
A structure-based sequence alignment of human IL-26 (A.) and a model of the human IL-26/IL-20R1/IL-10R2 ternary complex structure (B.). The sequence alignment was performed as described in the text. The six main helices (A–F) are denoted in Fig. 4A, and * and #s represent putative IL-20R1 and IL-10R2 binding residues, respectively. In the structure shown in Fig. 4B, helices A and B are colored red and purple, respectively. Cysteine residues are shown in yellow to denote residues that are conserved in the IL-19 sequence and green for residues that are not. The putative position of the cell membrane is shown at the bottom of the figure.
Our structural model of IL-26, based on the structure of IL-19, suggests that the cysteine pairs Cys-32/Cys-124 and Cys-79/Cys-121 form disulfide bonds, leaving Cys-112 at the N-terminal end of helix D accessible to form inter-molecular disulfides with other IL-26 molecules. In fact, IL-26 expressed in E. coli has been shown to form monomers and disulfide-linked homodimers [1,20]. Homodimers, as well as monomers, were also observed for Flag-tagged IL-26 expressed in COS-7 cells under non-denaturing conditions (presumably without β-mercaptoethanol). Unfortunately, dimer formation was not verified (only extracellular expression was verified) in the supernatants of virus-infected cells [1]. To date, all dimeric forms of IL-26 observed have been disulfide-linked homodimers, presumably via Cys-112. Our modeling efforts suggest that Cys-112 is near the IL-10R2 binding site, where disulfide formation could alter, or disrupt, IL-10R2 binding. One company (R&D Systems Inc., Minneapolis, MN) markets both monomeric and dimeric forms of recombinant human IL-26, and they claim that both forms are bioactive. The biological functions of IL-26 are not yet well defined, and further characterization has been hampered by difficulties in expressing significant quantities of this protein for biophysical and functional studies.
The same analytical method that we applied to the ligand, IL-26, was used to derive a model for the IL-26 receptor, IL-20R1. Structures of IL-10R1 [44] and IL-22R1 [45,46] were aligned using STAMP, followed by sequence/structural alignment using STACCATO. Based on conservation of disulfide bond pairings, the resulting sequence alignments indicate that the best model of IL-20R1 is derived using the D1 domain of IL-22R1 and the D2 domain of IL-10R1. A recent structural analysis of the IL-10/IL-10R1 and IL-22/IL-22R1 crystal structures demonstrated that cytokine recognition is predominantly a function of the D1 domain of the R1 receptor [46]. Based on this information, the alignment of IL-20R1 and IL-22R1 is consistent with similar receptor sharing properties of these receptors. In particular, IL-22R1 can bind to IL-20, which shares high sequence identity (~44%) and presumably structural similarity to IL-19, our molecular scaffold for the IL-26 structure. The D2 domains of IL-10R1 and IL-22R1 both form complexes with IL-10R2. We interpret the sequence alignment between IL-10R1 D2 and IL-20R1 D2 to suggest that these receptors make similar contacts with the IL-10R2 chain that are distinct from the IL-22R1/IL-10R2 interaction [46]. Based on these findings, we conclude that the IL-26/IL-20R1/IL-10R2 ternary complex is best approximated by the IL-10/IL-10R1/IL-10R2 ternary complex.
A ternary complex model showing the locations of IL-20R1 and IL-10R2 bound to IL-26 is shown in Figure 4B. As observed in the case of the IL-22/IL-22R1 and IL-10/IL-10R1 crystal structures, IL-20R1 contacts IL-26 helix A, the AB loop, helix B, and helix F. Based on the binary crystal structures, the IL-26 residues that are likely to interact directly with IL-20R1 are also shown in Figure 4B. Putative IL-20R1 binding residues on helix F are highly conserved among IL-10, IL-22, and IL-19, and conform to the IL-10 "fingerprint" sequence. In contrast to the conserved helix F residues, the helix A and AB loop residues, which make up part of the IL-20R1 as well as the IL-10R2 binding site, are highly variable. In addition, IL-10 and IL-22 residues important for interaction with IL-10R2 are found in different positions in the sequence alignment. This feature is consistent with our evolving understanding of IL-10R2 promiscuous binding, which appears to be based largely on the structural orientations of helices A and D [21,22]. A more thorough understanding of the IL-26/IL-20R1/IL-10R2 ternary complex will require more detailed structural studies complemented by functional testing.
6. IL-20R1 (IL20RA) variants generated by alternative RNA splicing
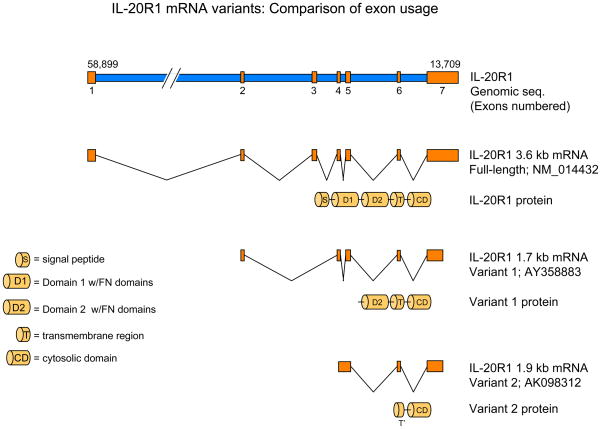
As discussed above, IL-20R1 is the ligand-binding chain for IL-26 and a functional component of several other cytokine receptor complexes, including the receptors for IL-19, IL-20 and IL-24 [27,28]. IL-20R1 can heterodimerize with IL-20R2 to generate receptor complexes for IL-19, IL-20 and IL-24, or it can dimerize with IL-10R2 to form IL-26 receptor complexes. While examining expression of the IL-20R1 gene, IL20RA, in various cell types, we found that this gene can generate three distinct mRNA transcripts [19]. One is the full-length 3.6 kb mRNA transcript that encodes the complete IL-20R1 polypeptide (GenBank accession no. NM_014432). The second is a 1.7 kb transcript that encodes a variant form of the receptor that lacks the first (membrane-distal) fibronectin type-III extracellular domain but retains the second (membrane-proximal) extracellular domain, transmembrane domain and the complete intracellular domain (GenBank accession no. AY358883). The third RNA variant contains a small portion of the transmembrane domain and the complete intracellular domain, but lacks the entire extracellular domain (GenBank accession no. AK098312). To evaluate the function of these IL-20R1 splice variants, we cloned and expressed their corresponding cDNAs in several cell lines that do not normally express these receptors. Only expression of the full-length 3.6 kb gene variant reconstituted IL-26 responsiveness in the human embryonic kidney epithelial cell line, HEK-293. The function(s) of the smaller 1.7 kb and 1.9 kb splice variants is unclear.
The differential expression of the variant forms of the IL-20R1 chain is analogous to the differential expression of variant forms of the IL-22 binding protein (IL-22BP, IL22RA2) [47–49]. Three splice variants of the human IL22RA2 gene have been identified: one encodes the full-length protein, a second encodes a variant that lacks the first fibronectin type-III domain, and a third encodes a variant that incorporates an additional peptide segment. Only the full-length protein appears to exhibit IL-22 binding activity. The two other variants are either inactive or mediate biologic functions that have not yet been defined.
7. The Potential Role of IL-26 in Human Autoimmune Diseases
Several groups have identified single nucleotide polymorphisms (SNPs) in or near the human IL26 gene that are associated with increased risk of developing certain autoimmune diseases, including multiple sclerosis (MS) and rheumatoid arthritis (RA) [50,51]. The region between the IFN-γ (IFNG) and IL-26 (IL26) genes was originally identified by Vandenbroeck and colleagues as a potentially important polymorphic region in patients with MS [50]. The same group subsequently identified two SNPs, denoted D12S2510 and D12S2511, that are strongly associated with susceptibility to autoimmune diseases such as RA and MS in a gender-specific manner [51]. In other words, these polymorphisms in the IL26 gene are associated with development of MS or RA in females but not in males. Related genome-wide association studies (GWAS) by Silverberg et al. identified a SNP in the human IL26 gene, rs2870946, that is strongly associated with development of inflammatory bowel disease (IBD) [52]. The mechanism by which these SNPs affect production or activity of IL-26 is currently unknown. However, it is noteworthy that a recent report from Stephan Brand’s group in Germany showed that inflamed colonic lesions from patients with IBD display markedly elevated expression of IL-26 mRNA [24]. The IL-26 gene expression in these patients correlated significantly with elevated expression of IL-22 and IL-8 mRNA. Together, these findings suggest that polymorphisms in the IL26 gene may provide useful genetic markers of increased risk for development of several human autoimmune diseases, including MS, RA and IBD. The levels of IL-26 gene expression might also be useful in monitoring the magnitude of inflammation in such patients.
8. Conclusions and Future Directions
IL-26 is frequently co-expressed together with IL-17 and IL-22 by activated Th17 cells. IL-26 might induce expression of a unique subset of genes in IL-26-responsive target cells. Alternatively, IL-26 might also amplify basal expression of genes that are induced by IL-17 and/or IL-22. For example, it has been shown that IL-22 synergizes with IL-17 to markedly up-regulate expression of many genes in primary keratinocytes [53]. Whether IL-26 can also synergize with IL-17 and/or IL-22 to amplify gene expression remains to be investigated.
Currently, there is very little information available regarding the biological functions of IL-26. In view of the demonstrated importance of Th17 cells in the pathogenesis of many inflammatory diseases, additional attention should be directed at defining the functions of IL-26. Are the biological functions induced by IL-26 similar to those induced by IL-22? Does IL-26 synergize with IL-17 and/or IL-22 to potentiate expression of pro-inflammatory genes in cells that express receptors for these cytokines? A recent report showed that IL-26 gene expression is elevated in inflamed colonic tissue of patients with inflammatory bowel disease [24]. The elevated expression of IL-26 mRNA correlated strongly with elevated levels of IL-22 mRNA. These findings provide an interesting clinical example of co-expression of the IL-22 and IL-26 genes in a relatively common inflammatory autoimmune disease. A fuller understanding of the biological functions of IL-26 might be derived from a comprehensive analysis of the genes that are expressed in IL-26-responsive target cells following treatment with this cytokine in vitro or in vivo. To date, there have been no reports of any microarray analyses of IL-26-responsive genes in any cell type or tissue. Such studies would clearly expand our knowledge of the biological activities of this cytokine.
Immortalization of primary human T cells by viral transformation with Herpesvirus saimiri induces production of IL-26 [1]. The secreted IL-26 protein can then bind and signal through IL-26 receptor complexes (IL-20R1 + IL-10R2) to induce expression of IL-26-responsive genes in various cell types, particularly epithelial cells. In view of the fact that viral transformation induces expression of IL-26, it is possible that IL-26 might induce antiviral activity in cells that express IL-26 receptors. To date, there are no published studies that directly address this question.
Although the murine genome does not appear to contain a paralog of the IL-26 gene, mice do express both receptor chains that are necessary to form IL-26 receptor complexes (i.e., IL-20R1 and IL-10R2). Consequently, it would be interesting to determine if human IL-26 can bind and signal in murine cell lines or primary mouse tissues that express the endogenous murine IL-20R1 chain. Recombinant human IL-10 can bind and signal through murine IL-10 receptor complexes, so it is certainly possible that the human IL-26 protein can bind and signal via murine IL-26 receptor complexes. If it does, it will be of interest to examine potential phenotypic changes induced by forced expression of the human IL-26 gene in a transgenic mouse model.
What are the primary biological functions of IL-26 in vivo? In view of the fact that IL-26 is highly expressed by Th17 cells, it is possible and perhaps likely that IL-26 contributes to pathologic conditions characterized by the presence of activated Th17 cells. If this is the case, then it might be desirable to block IL-26 activity using specific antagonists. Such antagonists could include neutralizing monoclonal antibodies against IL-26 or its receptor, IL-20R1. So far, IL-26 has received only limited attention from the global research community, however in light of the fact that this cytokine is frequently co-expressed with IL-17 and/or IL-22 by Th17 cells, more extensive functional studies are clearly warranted.
Figure 5.
The IL-20R1 gene, IL20RA, encodes several RNA splice variants. Three variants of the IL-20R1 gene can be expressed in cells that express this receptor chain. The first is the full-length 3.6 kb transcript (GenBank accession no. NM_014432) that encodes the complete receptor polypeptide, including both fibronectin type-III (FnIII) domains of the extracellular region. The second is a 1.7 kb variant (GenBank accession no. AY358883) that lacks the membrane-distal FnIII domain, but retains the membrane-proximal FnIII domain, the trans-membrane domain, and the complete intracellular region. This variant arises as a result of deletion of most of exon 1 and all of exon 3. The third transcript is a 1.9 kb variant (GenBank accession no. AK098312) that lacks the entire extracellular region, but retains a small part of the trans-membrane domain and the complete intracellular domain. This variant arises as a result of the complete deletion of exons 1, 2 and 3. Although the full-length (3.6 kb) transcript encodes a functional IL-26-binding protein, the functions of the shorter 1.7 kb and 1.9 kb variants are unknown.
Acknowledgments
This work was supported in part by U.S. Public Health Service Grants AI47300 and AI47300-08S from the National Institutes of Health (NIH) to M.R.W.
Biographies
 Raymond P. Donnelly, Ph.D. is a Senior Investigator in the Division of Therapeutic Proteins at the FDA Center for Drug Evaluation & Research (CDER) in Bethesda, Maryland. He received an M.S. in Microbiology from Oregon State University and a Ph.D. in Microbiology & Immunology from the Medical University of South Carolina in 1986. In 1987, he received a National Research Service Award to complete postdoctoral training at the Boston University School of Medicine. In 1989, he joined the FDA Division of Cytokine Biology as a Staff Fellow, and was tenured as a Principal Investigator at FDA in 1996. Dr. Donnelly serves as an expert regarding product manufacturing issues pertaining to a variety of therapeutic proteins, including cytokines and cytokine antagonists. He also manages a laboratory research program that is focused on defining the receptors for and the biological activities of various cytokines. Dr. Donnelly is a current or former member of several editorial boards, including the Journal of Immunology, Journal of Interferon & Cytokine Research, and Genes & Immunity. In 2005, he was awarded the FDA Scientific Achievement Award for Excellence in Laboratory Science.
Raymond P. Donnelly, Ph.D. is a Senior Investigator in the Division of Therapeutic Proteins at the FDA Center for Drug Evaluation & Research (CDER) in Bethesda, Maryland. He received an M.S. in Microbiology from Oregon State University and a Ph.D. in Microbiology & Immunology from the Medical University of South Carolina in 1986. In 1987, he received a National Research Service Award to complete postdoctoral training at the Boston University School of Medicine. In 1989, he joined the FDA Division of Cytokine Biology as a Staff Fellow, and was tenured as a Principal Investigator at FDA in 1996. Dr. Donnelly serves as an expert regarding product manufacturing issues pertaining to a variety of therapeutic proteins, including cytokines and cytokine antagonists. He also manages a laboratory research program that is focused on defining the receptors for and the biological activities of various cytokines. Dr. Donnelly is a current or former member of several editorial boards, including the Journal of Immunology, Journal of Interferon & Cytokine Research, and Genes & Immunity. In 2005, he was awarded the FDA Scientific Achievement Award for Excellence in Laboratory Science.
 Faruk Sheikh, Ph.D. is a Staff Fellow in the Division of Therapeutic Proteins at the FDA Center for Drug Evaluation & Research in Bethesda, Maryland. Dr. Sheikh received his Ph.D. in Biochemistry from Calcutta University, Calcutta, India in 1996. Dr. Sheikh was a recipient of a postdoctoral fellowship award from the Oak Ridge Institute for Science and Education (ORISE) to conduct studies at the FDA on the regulation of cytokine signaling and gene expression in the laboratory of Dr. Ray Donnelly. He continues to pursue this research interests while also serving as an FDA regulatory reviewer regarding the clinical application of recombinant cytokines and cytokine antagonists.
Faruk Sheikh, Ph.D. is a Staff Fellow in the Division of Therapeutic Proteins at the FDA Center for Drug Evaluation & Research in Bethesda, Maryland. Dr. Sheikh received his Ph.D. in Biochemistry from Calcutta University, Calcutta, India in 1996. Dr. Sheikh was a recipient of a postdoctoral fellowship award from the Oak Ridge Institute for Science and Education (ORISE) to conduct studies at the FDA on the regulation of cytokine signaling and gene expression in the laboratory of Dr. Ray Donnelly. He continues to pursue this research interests while also serving as an FDA regulatory reviewer regarding the clinical application of recombinant cytokines and cytokine antagonists.
 Harold Dickensheets, Ph.D., is a Staff Scientist in the Division of Therapeutic Proteins at the FDA Center for Drug Evaluation & Research in Bethesda, Maryland. He received his Ph.D. in Biochemistry in 1995 from the University of Kansas where he characterized auto-antigens involved in the pathogenesis of systemic lupus erythematosus. He then received a postdoctoral fellowship award from the Oak Ridge Institute for Science and Education (ORISE) to conduct postdoctoral research in Dr. Ray Donnelly’s laboratory at the FDA. Dr. Dickensheets conducts ongoing research regarding the cellular responses to various cytokines. He also serves as an expert reviewer regarding cytokine products that are regulated by the FDA.
Harold Dickensheets, Ph.D., is a Staff Scientist in the Division of Therapeutic Proteins at the FDA Center for Drug Evaluation & Research in Bethesda, Maryland. He received his Ph.D. in Biochemistry in 1995 from the University of Kansas where he characterized auto-antigens involved in the pathogenesis of systemic lupus erythematosus. He then received a postdoctoral fellowship award from the Oak Ridge Institute for Science and Education (ORISE) to conduct postdoctoral research in Dr. Ray Donnelly’s laboratory at the FDA. Dr. Dickensheets conducts ongoing research regarding the cellular responses to various cytokines. He also serves as an expert reviewer regarding cytokine products that are regulated by the FDA.
 Ram Savan, Ph.D. is currently a Visiting Fellow in the Laboratory of Experimental Immunology, Cancer and Inflammation Program, National Cancer Institute-Frederick. Maryland. He received his graduate training at United Graduate School of Agriculture Sciences, Kagoshima University, Japan. He was a recipient of the Japanese Society for Promotion of Science research fellowship. His current research interests are on regulation of cytokines through miRNA.
Ram Savan, Ph.D. is currently a Visiting Fellow in the Laboratory of Experimental Immunology, Cancer and Inflammation Program, National Cancer Institute-Frederick. Maryland. He received his graduate training at United Graduate School of Agriculture Sciences, Kagoshima University, Japan. He was a recipient of the Japanese Society for Promotion of Science research fellowship. His current research interests are on regulation of cytokines through miRNA.
 Howard A. Young, Ph.D. is a Principal Investigator in the Laboratory of Experimental Immunology, Cancer and Inflammation Program, National Cancer Institute-Frederick. The major work in his laboratory focuses on the regulation of interferon-gamma gene expression with a special emphasis on IFN-gamma expression in human and mouse Natural Killer cells. He is a Past President of the International Society for Interferon and Cytokine Research.
Howard A. Young, Ph.D. is a Principal Investigator in the Laboratory of Experimental Immunology, Cancer and Inflammation Program, National Cancer Institute-Frederick. The major work in his laboratory focuses on the regulation of interferon-gamma gene expression with a special emphasis on IFN-gamma expression in human and mouse Natural Killer cells. He is a Past President of the International Society for Interferon and Cytokine Research.
 Mark R. Walter, Ph.D. received a B.S. degree in Chemistry and Biology (Summa Cum Laude) from California Lutheran University. He received his Ph.D. and postdoctoral training at the University of Alabama at Birmingham (UAB) in the laboratories of Steve Ealick and Charlie Bugg, respectively. Dr. Walter is now a full Professor in the Department of Microbiology at UAB where his lab continues to elucidate structural mechanisms that lead to cytokine signaling. He also serves as a leader of the structural biology and drug development components of the UAB Comprehensive Cancer Center.
Mark R. Walter, Ph.D. received a B.S. degree in Chemistry and Biology (Summa Cum Laude) from California Lutheran University. He received his Ph.D. and postdoctoral training at the University of Alabama at Birmingham (UAB) in the laboratories of Steve Ealick and Charlie Bugg, respectively. Dr. Walter is now a full Professor in the Department of Microbiology at UAB where his lab continues to elucidate structural mechanisms that lead to cytokine signaling. He also serves as a leader of the structural biology and drug development components of the UAB Comprehensive Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knappe A, Hör S, Wittmann S, Fickenscher H. Induction of a novel cellular homolog of interleukin-10, AK155, by transformation of T lymphocytes with herpesvirus saimiri. J Virol. 2000;74:3881–3887. doi: 10.1128/jvi.74.8.3881-3887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotenko S. The family of IL-10-related cytokines and their receptors: related, but to what extent? Cytokine Growth Factor Rev. 2002;13:223–240. doi: 10.1016/s1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 3.Renauld JC. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat Rev Immunol. 2003;3:667–676. doi: 10.1038/nri1153. [DOI] [PubMed] [Google Scholar]
- 4.Fickenscher H, Pirzer H. Interleukin-26. Int Immunopharmacol. 2004;4:609–613. doi: 10.1016/j.intimp.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Wilson NJ, Boniface K, Chan JR, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immununol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 6.Manel N, Unutmaz D, Littman DR. The differentiation of human Th17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORγt. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pène J, Chevalier S, Preisser L, et al. Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol. 2008;180:7423–7430. doi: 10.4049/jimmunol.180.11.7423. [DOI] [PubMed] [Google Scholar]
- 8.Savan R, Ravichandran S, Collins JR, Sakai M, Young HA. Structural conservation of interferon-gamma among vertebrates. Cytokine Growth Factor Rev. 2009;20:115–124. doi: 10.1016/j.cytogfr.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi ZT, Nie P. Comparative study and expression analysis of the interferon-gamma gene locus cytokines in Xenopus tropicalis. Immunogenetics. 2008;60:699–710. doi: 10.1007/s00251-008-0326-y. [DOI] [PubMed] [Google Scholar]
- 10.Igawa D, Sakai M, Savan R. An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol Immunol. 2006;43:999–1009. doi: 10.1016/j.molimm.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Dumoutier L, Van RE, Ameye G, Michaux L, Renauld JC. IL-TIF/IL-22: genomic organization and mapping of the human and mouse genes. Genes Immun. 2000;1:488–494. doi: 10.1038/sj.gene.6363716. [DOI] [PubMed] [Google Scholar]
- 12.Schoenborn JR, Dorschner MO, Sekimata M, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadir R, Forest E, Lortat-Jacob H. The heparan sulfate-binding sequence of interferon-gamma increases the on-rate of interferon-gamma/interferon-gamma receptor complex formation. J Biol Chem. 1998;273:10919–25. doi: 10.1074/jbc.273.18.10919. [DOI] [PubMed] [Google Scholar]
- 14.Blumberg H, Conklin D, Xu WF, et al. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 2001;104:9–19. doi: 10.1016/s0092-8674(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 15.Wolk K, Kunz S, Asadullah K, Sabat R. Immune cells as sources and targets of the IL-10 family members. J Immunol. 2002;168:5397–5402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- 16.Nagalakshmi ML, Murphy E, McClanahan T, de Waal Malefyt R. Expression patterns of IL-10 ligand and receptor gene families provide leads for biological characterization. Int Immunopharmacol. 2004;4:577–592. doi: 10.1016/j.intimp.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Cella M, Fuchs A, Vermi W, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes T, Becknell B, McClory S, et al. Stage 3 immature human natural killer cells found in secondary lymphoid tissue constitutively and selectively express the TH 17 cytokine interleukin-22. Blood. 2009;113:4008–4010. doi: 10.1182/blood-2008-12-192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheikh F, Baurin VV, Lewis-Antes A, et al. Cutting Edge: IL-26 signals through a novel receptor complex composed of IL-20 receptor-1 and IL-10 receptor-2. J Immunol. 2004;172:2006–2010. doi: 10.4049/jimmunol.172.4.2006. [DOI] [PubMed] [Google Scholar]
- 20.Hör S, Pirzer H, Dumoutier L, et al. The T-cell lymphokine, interleukin-26, targets epithelial cells through the interleukin-20 receptor 1 and interleukin-10 receptor 2 chains. J Biol Chem. 2004;279:33343–33351. doi: 10.1074/jbc.M405000200. [DOI] [PubMed] [Google Scholar]
- 21.Yoon SI, Logsdon NJ, Sheikh F, Donnelly RP, Walter MR. Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex. J Biol Chem. 2006;281:35088–35096. doi: 10.1074/jbc.M606791200. [DOI] [PubMed] [Google Scholar]
- 22.Yoon SI, Jones BC, Logsdon NJ, Harris BD, Deshpande A, Radaeva S, Halloran BA, Gao B, Walter MR. Structure and mechanism of receptor sharing by the IL-10R2 common chain. Structure. 2010;18(5):638–648. doi: 10.1016/j.str.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnelly RP, Dickensheets H, Finbloom DS. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J Interferon Cytokine Res. 1999;19:563–573. doi: 10.1089/107999099313695. [DOI] [PubMed] [Google Scholar]
- 24.Dambacher J, Beigel F, Zitzmann K, De Toni EN, Göke B, Diepolder HM, Auernhammer CJ, Brand S. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut. 2009;58:1207–17. doi: 10.1136/gut.2007.130112. [DOI] [PubMed] [Google Scholar]
- 25.Pletnev S, Magracheva E, Kozlov S, et al. Characterization of the recombinant extracellular domains of human interleukin-20 receptors and their complexes with interleukin-19 and interleukin-20. Biochemistry. 2003;42:12617–12624. doi: 10.1021/bi0354583. [DOI] [PubMed] [Google Scholar]
- 26.Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukocyte Biol. 2004;76:314–321. doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- 27.Dumoutier L, Leemans C, Lejeune D, Kotenko SV, Renauld JC. Cutting edge: STAT activation by IL-19, IL-20 and mda-7 through IL-20 receptor complexes of two types. J Immunol. 2001;167:3545–3549. doi: 10.4049/jimmunol.167.7.3545. [DOI] [PubMed] [Google Scholar]
- 28.Parrish-Novak J, Xu W, Brender T, et al. Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J Biol Chem. 2002;277:47517–47523. doi: 10.1074/jbc.M205114200. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Wei SH, Ho AS, de Waal Malefyt R, Moore KW. Expression cloning and characterization of a human IL-10 receptor. J Immunol. 1994;152:1821–1829. [PubMed] [Google Scholar]
- 30.Gibbs VC, Pennica D. CRF2–4: isolation of cDNA clones encoding the human and mouse proteins. Gene. 1997;186:97–101. doi: 10.1016/s0378-1119(96)00690-7. [DOI] [PubMed] [Google Scholar]
- 31.Wolk K, Witte E, Wallace E, Döcke WD, Kunz S, Asadullah K, Volk HD, Sterry W, Sabat R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36:1309–23. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 32.O'Farrell AM, Liu Y, Moore KW, Mui AL. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and -independent pathways. EMBO J. 1998;17:1006–18. doi: 10.1093/emboj/17.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley JK, Takeda K, Akira S, Schreiber RD. Interleukin-10 receptor signaling through the JAK-STAT pathway: Requirement for two distinct receptor-derived signals for anti-inflammatory action. J Biol Chem. 1999;274:16513–21. doi: 10.1074/jbc.274.23.16513. [DOI] [PubMed] [Google Scholar]
- 34.Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10R-beta) is a common chain of both the IL-10 and IL-22 receptor complexes. J Biol Chem. 2001;276(4):2725–32. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 35.Lejeune D, Dumoutier L, Constantinescu S, Kruijer W, Schuringa JJ, Renauld JC. Interleukin-22 (IL-22) activates the JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma cell line. Pathways that are shared with and distinct from IL-10. J Biol Chem. 2002;277:33676–82. doi: 10.1074/jbc.M204204200. [DOI] [PubMed] [Google Scholar]
- 36.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–54. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Chang C, Magracheva E, Kozlov S, Fong S, Tobin G, Kotenko S, Wlodawer A, Zdanov A. Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J Biol Chem. 2003;278:3308–3313. doi: 10.1074/jbc.M208602200. [DOI] [PubMed] [Google Scholar]
- 38.Nagem RA, Colau D, Dumoutier L, Renauld JC, Ogata C, Polikarpov I. Crystal structure of recombinant human interleukin-22. Structure. 2002;10:1051–1062. doi: 10.1016/s0969-2126(02)00797-9. [DOI] [PubMed] [Google Scholar]
- 39.Walter MR, Nagabhushan TL. Crystal structure of interleukin 10 reveals an interferon gamma-like fold. Biochemistry. 1995;34:12118–12125. doi: 10.1021/bi00038a004. [DOI] [PubMed] [Google Scholar]
- 40.Xu T, Logsdon NJ, Walter MR. Structure of insect-cell-derived IL-22. Acta Crystallogr D Biol Crystallogr. 2005;61:942–950. doi: 10.1107/S0907444905009601. [DOI] [PubMed] [Google Scholar]
- 41.Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, Weatherbee J, Wlodawer A. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure. 1995;3:591–601. doi: 10.1016/s0969-2126(01)00193-9. [DOI] [PubMed] [Google Scholar]
- 42.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 43.Shatsky M, Nussinov R, Wolfson HJ. Optimization of multiple-sequence alignment based on multiple-structure alignment. Proteins. 2006;62:209–217. doi: 10.1002/prot.20665. [DOI] [PubMed] [Google Scholar]
- 44.Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity. 2001;15:35–46. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- 45.Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I. Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism. FEBS Lett. 2008;582:2985–2992. doi: 10.1016/j.febslet.2008.07.046. [DOI] [PubMed] [Google Scholar]
- 46.Jones BC, Logsdon NJ, Walter MR. Structure of IL-22 bound to its high-affinity IL-22R1 chain. Structure. 2008;16:1333–1344. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. Identification, cloning, and characterization of a novel soluble receptor that binds IL-22 and neutralizes its activity. J Immunol. 2001;166:7096–103. doi: 10.4049/jimmunol.166.12.7096. [DOI] [PubMed] [Google Scholar]
- 48.Gruenberg BH, Schoenemeyer A, Weiss B, Toschi L, Kunz S, Wolk K, Asadullah K, Sabat R. A novel, soluble homologue of the human IL-10 receptor with preferential expression in placenta. Genes Immun. 2001;2:329–34. doi: 10.1038/sj.gene.6363786. [DOI] [PubMed] [Google Scholar]
- 49.Weiss B, Wolk K, Grünberg BH, Volk HD, Sterry W, Asadullah K, Sabat R. Cloning of murine IL-22 receptor alpha 2 and comparison with its human counterpart. Genes Immun. 2004;5:330–6. doi: 10.1038/sj.gene.6364104. [DOI] [PubMed] [Google Scholar]
- 50.Goris A, Marrosu MG, Vandenbroeck K. Novel polymorphisms in the IL-10 related AK155 gene (chromosome 12q15) Genes Immun. 2001;2(5):284–6. doi: 10.1038/sj.gene.6363772. [DOI] [PubMed] [Google Scholar]
- 51.Vandenbroeck K, Cunningham S, Goris A, Alloza I, Heggarty S, Graham C, et al. Polymorphisms in the interferon-gamma/interleukin-26 gene region contribute to sex bias in susceptibility to rheumatoid arthritis. Arthritis Rheum. 2003;48(10):2773–8. doi: 10.1002/art.11236. [DOI] [PubMed] [Google Scholar]
- 52.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2010;42(4):332–7. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]