Abstract
Proteomic analysis of cadaveric formalin-fixed, celloidin-embedded (FFCE) temporal bone tissue has the potential to provide new insights into inner ear disorders. We have developed a liquid chromatography-mass spectrometry (LC-MS) method for tissue sections embedded with celloidin. Q-TOF (Quadripole-time of flight mass spectrometry) MSE (mass spectrometry where E represents collision energy) and IdentityE™ were used in conjunction with nano-UPLC (capillary ultrahigh pressure liquid chromatography) for robust identification and quantification of a large number of proteins.
Formalin fixed paraffin-embedded (FFPE) mouse liver sections were used to evaluate formalin de-cross-linking by five different methods. Unfixed fresh mouse liver tissue was used as a control. Five different methods for preparation of FFPE tissue for MS analysis were compared, as well as four methods for celloidin removal with FFCE mouse liver tissue. The methods judged best were applied to FFCE 20 μm sections of mouse inner ear samples, and FFCE 20 μm human inner ear and human otic capsule bone sections.
Three of the five tissue extraction methods worked equally in detecting peptides and proteins from FFPE mouse liver tissue. The modified liquid tissue kit protocol was chosen for further studies. Four different celloidin removal methods were compared and the acetone removal method was chosen for further analysis. These two methods were applied to the analysis of FFCE inner ear and otic capsule sections. Proteins from all major cellular components were detected in the FFCE archival human temporal bone sections. This newly developed technique enables the use of FFCE tissues for proteomic studies.
Keywords: Proteomics, Inner ear, Celloidin, formalin-fixed, mass spectrometry
Introduction
Archival formalin-fixed tissue collections with attached clinical and outcome data represent a valuable and ready source for research studies. The use of formalin as a fixative induces protein cross-linking and has generally been assumed to render such tissues less suitable for proteomic studies. However, in recent years, several techniques have been developed for performing proteomic analyses on formalin fixed tissues (Crockett et al., 2005; Hood et al., 2005; Palmer-Toy et al., 2005; Becker et al., 2007; Hwang et al., 2007; Jiang et al., 2007). The tissues used in their studies were embedded in paraffin.
Celloidin embedding is widely used for cadaveric human temporal bone specimens (Schucknecht, 1993) because it provides far better preservation of the fine structures of inner ear, compared to paraffin (O'Malley et al., 2009). Proteomic analysis of existing cadaveric formalin fixed celloidin embedded (FFCE) temporal bone collections has the potential to provide new insights into cochlear physiology and pathophysiology of inner ear disorders (Palmer-Toy et al., 2005). No studies have been done that compare different celloidin removal or different tissue preparation methods for liquid chromatography-mass spectrometry (LC-MS) based proteomics of celloidin embedded tissues.
The present report concerns our attempts to develop a method for LC-MS analysis of FFCE tissue and optimize parameters for proteomic studies with these samples. In-order to evaluate and optimize methods for proteomic analysis of this valuable material, five different methods for preparation of formalin fixed tissue for MS analysis were compared, as well as four methods for celloidin removal. Our objective was to determine the best approach for accurate identification of the largest number of proteins. Q-TOF MSE (Quadripole-time of flight mass spectrometry where E represents collision energy) with the Waters IdentityE™ search engine for protein identification and absolute quantification were used in conjunction with nano UPLC (capillary ultrahigh pressure liquid chromatography). Our newly developed technique enables the use of FFCE tissues for proteomic studies. The initial experiments were performed on mouse sections that were processed in a manner similar to human temporal bone sections, so that we could minimize potential waste of precious human inner ear sections.
Materials and Methods
1.1. Mouse FFPE and FFCE tissue
Ten week old CBA/CAJ mice were used for these studies (Jackson Laboratory, Bar Harbor, ME). The mice were anesthetized with urethane (1000 mg/kg, Sigma, St. Louis, MO) and perfused intracardially first with 0.9% saline followed by 4% formaldehyde. The livers were removed and postfixed overnight in 4% formaldehyde at 4° C. To obtain fresh liver tissue, mice were anesthesized as above, exsanguinated and the liver removed. A portion of the liver tissue from the perfused animals was embedded in paraffin and the remainder embedded in celloidin (parlodion strips; Mallinckrodt Chemicals, Phillipsburg, NJ). The fresh tissue was kept frozen at −70° C until use.
The formalin fixed and paraffin embedded (FFPE) liver tissue was sectioned (10 μm) and the sections mounted on glass slides. The formalin fixed and celloidin embedded (FFCE) liver tissue was also sectioned (20 μm) and the sections were stored in 80% ethanol solution until use.
Mouse temporal bones were harvested at 0.5 hr post mortem. After 25 hours of additional fixation with 10 % formalin, the temporal bones were dissected out and decalcified in 0.12 M ethylenediaminetetraacetic acid (EDTA) for 7 days. The cochleae were then processed for celloidin embedding (Schuknecht, 1993). The celloidin blocks were serially sectioned (20 μm) and a few sections were stained with hematoxylin and eosin.
For the LC-MS studies, after celloidin removal, the sections were dissected under an operating microscope (Figure 1A). Two cochlear samples were studied. Eight sections were pooled for each sample. The cochlear partition of the temporal bone, containing all 2.5 turns was excised with a needle, was transferred to a collection tube and processed further.
Figure 1.
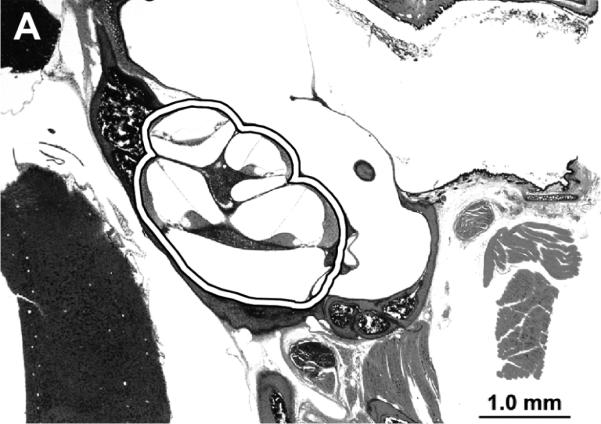
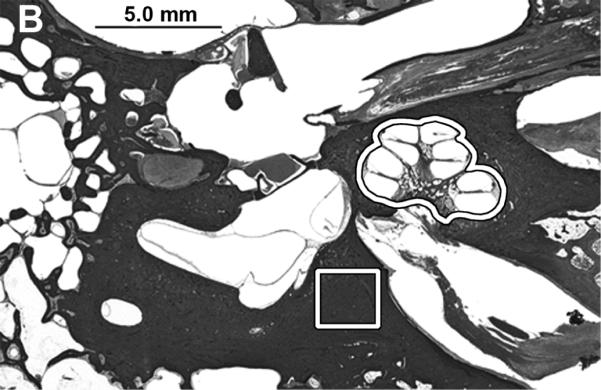
A) Photomicrograph of mouse celloidin embedded temporal bone section. The sections contained both cochleae and they were removed as outlined for further analysis. B) The cochleae and a sample of the otic capsule bone from cadaveric human celloidin embedded sections were removed (outlined areas). The scale bar is 1.0 mm in A and 5.0 mm in B.
1.2. Human FFCE inner ear tissue
The temporal bones of a 56 year old patient were removed in March 2006 after 9 hours postmortem time. The patient had no prior history of otologic diseases. The bones were fixed with 10% formalin and processed further according to Schucknecht et al. (1993).
The FFCE tissue blocks were cut into 20 μm sections and the sections were stored as described above. The sections for LC-MS analysis were treated as described above.
The cochlear partition was removed as described above. Samples of the bony otic capsule were also collected (Figure 1B). This study was approved by the MEEI institutional Review Board.
2. Tissue preparation for LC-MS
2.1. Fresh tissue
Fresh mouse liver tissue was homogenized mechanically for 10 minutes and sonicated with a water bath ultrasonicator (bath sonicator Cole-Parmer 8850, InnoCal, Vernon Hills, Illinois) for 30 minutes. The homogenization was done in water (proteomics grade) at room temperature and before the acetone precipitation step using a small hand held pellet pestle (Kimble/Kontes, Vineland, NJ). Six volumes of cold acetone was added, and after an incubation at −70° C for 3 h the preparation was centrifuged at 10,000 rcf for 15 min. Tissue (1.2 mg wet weight) was taken for further analysis. The acetone was removed and the pellet was lyophilized. In preparation for reduction, alkylation and trypsin digestion, the dried acetone powder was resuspended in 25 ul 0.2% Rapigest™ (Waters, Milford, MA), 50 mM ammonium bicarbonate. To start the reduction, 12.5 ul of 15 mM DTT (Dithiothreitol) was added for incubation at 60° C for 30 min. This was followed by addition of 12.5 μl of 60 mM iodoacetamide for incubation in the dark at room temperature for 30 min. Trypsin (2 μl, in 1 mM HCl at a concentration of 1 mg/ml, pH 3.0, proteomics grade Porcine Trypsin from Princeton Separation, Adelphia, NJ) was added for incubation overnight at 30° C. The sample was centrifuged at 10,000 rcf for 15 min, the supernatant was removed and stored at −20° C prior to LC-MS analysis. The different tissue extraction protocols used are described below.
2.2. SDS with heat
FFPE liver tissue was deparaffinized (10 min and 5 min in xylene, 5 min × 2 in 100% ethanol, 5 min × 2 in 95% ethanol, 5 min × 2 in 80% ethanol, 5 min × 2 in 50% ethanol and 5 min in proteomics grade water) and under a microscope, a 2 mm3 piece of tissue was transfered to a methanol rinsed polypropylene tube. The sample was sonicated for 45 min in 500 μl of 2% Sodium dodecyl sulfate (SDS) in 100 mM ammonium bicarbonate. DTT was added to a final concentration of 20 mM and the sample incubated at 70° C for one hour. Iodoacetamide was added to a concentration of 60 mM and the sample incubated in dark at room temperature for 30 min. Trypsin (2 µl, in 1 mM HCl at a concentration of 1 mg/ml, pH 3.0.) was added and the sample was incubated overnight at 30° C. After this the sample was centrifuged at 10000 rcf for 15 min, the supernatant was collected and stored at −20°C prior to LC-MS analysis.
2.3. 6M Guanidine HCl with heat
FFPE tissue was homogenized mechanically and sonicated in a water bath in 100 μl of a solution containing 40 mM TRIS, 6 M Guanine-HCl and 65 mM DTT, pH 8.2. The sample was incubated at 100° C for 30 min in a heating block followed by addition of 500 μl of 50 mM ammonium bicarbonate. Iodoacetamide was added to a concentration of 40 mM and the sample was incubated in the dark at room temperature for 30 min. Trypsin (2 μl, in 1 mM HCl at a concentration of 1 mg/ml, pH 3.0) was added, and incubated overnight at 30°C. The sample was centrifuged at 10,000 rcf for 15 min and the supernatant removed and stored at −20° C prior to LC-MS analysis.
2.4. QProteome kit (Qproteome FFPE Tissue Kit, Qiagen Inc., Valencia, CA)
The area of interest was excised with a needle and transferred to a collection tube. The manufacturer's instructions were followed. Extraction buffer (100 μl) was added to the tube containing the tissue sample. The sample was vortexed and incubated on ice for 5 min and in 80° C water bath for 2 h with agitation at 750 rpm. After incubation, the sample was cooled to 4° C for 10 min and centrifuged (15 min, 14000 g, 4° C). The supernatant was transferred to a new collection tube and stored at −20° C prior to LC-MS analysis.
2.5. Liquid Tissue kit® (MS Protein Prep Kit, Expression Pathology, Rockville, MD)
Extraction buffer (20 μl) was added to the tissue sample followed by incubation at 95° C for 90 min. Every 20 min the sample was microcentrifuged for 5 seconds. The sample was centrifuged at 10000 rcf for 1 min. The sample was cooled on ice for 2 min.
Trypsin (1 μg, in 1 mM HCl at a concentration of 1 mg/ml, pH 3.0) was added and the sample incubated for 18 hrs at 37° C. The sample was centrifuged at 10000 rcf for 1 min. DTT (2 μl of 100 mM) was added. The reaction tube was heated at 95° C for 5 min. The sample was centrifuged at 10,000 rcf for 1 min and stored until used at −20° C.
This kit was also used with a modified protocol. After cooling the sample on ice we added 2 μl of DTT (100 mM) and incubated the sample at 60° C for 30 min. Iodoacetamide (1 μl of 300 mM) was added and incubated in the dark at room temperature for 30 min. Trypsin (1 μg, in 1 mM HCl at a concentration of 1 mg/ml, pH 3.0) was added and the sample incubated for 18 hours in a 37° C water bath. The samples were centrifuged at 10,000 rcf for 1 min and stored at −20° C until used.
3. Celloidin removal methods
The sections were floated onto cigarette paper in a dish of 80% ethanol following which the paper was trimmed around each section. The sections were then placed face down onto a Superfrost® Plus (Thermo Fisher Scientific, Waltham, MA) slide. A piece of bibulous paper was placed over each section and a roller was used to flatten and smooth the section. After removing the bibulous paper, a second piece of bibulous paper (soaked in10% formalin) was placed over the cigarette paper, followed by a block of wood and a 500 g lead weight, and left to dry in air for 1 hour. The wood block and lead weights were removed, and the bibulous and cigarette papers were removed gently. Four different methods of removal of celloidin were used, as described below.
3.1 Celloidin removal with clove oil
The air-dried slides were dehydrated in 80%, 95%, and 100% (2 changes) ethanol for 3 min each and then transferred into clove oil overnight (Sone et al., 1998, 1999). The following morning, the slides were immersed in 100%, 100%, 95%, 80%, and 50% ethanol for 3 min each, followed by distilled water to remove the clove oil and rehydrate the tissue.
3.2. Celloidin removal with ether-alcohol
After the sections were adhered to the slides, the slides were transferred directly to ethyl ether-100% ethanol (Pawlowski et al., 1998) (1:1) for 30 min. The slides were repeatedly dipped in the solution to help remove the celloidin. The slides were transferred to a second change of ethyl ether-100% ethanol (1:1) for 30 min and agitated once again. The slides were then hydrated in 100%, 100%, 95%, 80%, 50% ethanol, (3 min each), followed by distilled water.
3.3. Celloidin removal with acetone
The celloidin was extracted with acetone for 30 min (Keithley et al., 1995). Sections were hydrated in 100%, 100%, 95%, 80%, 50% ethanol (3 min each) and transferred to distilled water prior to further analysis.
3.4. Celloidin removal with sodium hydroxide-methanol
Sodium hydroxide was dissolved in methanol (50g in 50 ml) and allowed to settle for 30 min at room temperature (Miguel-Hidalgo and Rajkowska, 1999). The solution on top of the undissolved pellets was diluted 1:2 with methanol and used immediately. Enough of the diluted sodium hydroxide-methanol solution to cover each section was applied by drops to the dried celloidin sections, allowed to stand for 5 min and then rinsed with 100% methanol (O'Malley et al., 2009). These two steps were repeated twice. Further rinsing with 100% and 70% methanol for 10 min followed. The slides were then transferred to distilled water.
4. LC-MSE
All samples were analyzed using the LC-MSE non-data dependant, absolutely quantitative proteomic platform described by Geromanos et al. (2009). Prior to analysis the samples were spiked with 100 fmol of pre-trypsin digested yeast alcohol dehydrogenase (Waters Inc., ALDH) as an internal standard. Each sample was dissolved in 24 μl of 0.1% formic acid and 2 μl injected using a NanoAquity auto sampler onto a Symmetry C18 trapping cartridge (180 μm i.d. × 2 cm length; Waters, Inc.) at a flow rate of 5 μL/min for 3 min. The peptides were then separated by in-line gradient elution onto a 75 μm i.d. × 10 cm column packed with 1.7 μm particle size BEH 130 (Waters Inc.), at a flow rate of 300 nL/min using a linear gradient from 2 to 40% acetonitrile in 0.1% formic acid over 60 min. The Q-Tof was operated in the LC/MSE mode of data acquisition, where alternating 2 s scans of low (4 V) or high (programmed from 15–40 V) collision energies are used to generate either intact peptide ions (low energy) or peptide product ions (high energy). Each raw data file was processed using ProteinLynx Global Server (PLGS) V2.3 software (Waters) to generate charge state reduced and deisotoped precursor mass lists as well as associated product ion mass lists for subsequent protein identification and quantification. Each processed file was searched against the Uniprot/Swissprot human databases using the IDENTITYE database search algorithm within PLGS V2.3. Parameters were set to: peptide tolerance, 30 ppm; fragment tolerance, 15 ppm; minimum fragment ion matches per peptide, 2; minimum fragment ion matches per protein, 6; minimum peptide ion matches per protein, 1; primary digest reagent, trypsin; reversed database false positive rate, 0; cysteine carbamidomethylation as fixed modification and methionine oxidation, asparagine and glutamine deamination as variable modifications. All samples were run in triplicate and only peptides identified in at least 2 of the replicates were selected. The proteins were quantified using the summed signal intensity of the three most intense tryptic peptides for each protein with reference to signal intensity of the three most intense ions for ALDH internal standard (100 fmol).
5. Data analysis
Formalin fixed paraffin embedded (FFPE) mouse liver sections were used to evaluate formalin de-cross-linking, trypsin digestion and recovery of tryptic peptides by five different methods. Unfixed fresh mouse liver tissue was used as a control. Four different celloidin removal protocols evaluated using FFCE mouse liver tissue. The method judged best was applied to FFCE 20 μm mounted sections of mouse inner ear samples and FFCE 20 μm human inner ear and human otic capsule bone. Tryptic digests of these were analyzed by nano-UPLC-MSE and IdentityE(™) for robust identification and quantification of proteins.
Results
Comparison of extraction methods with FFPE liver tissue
Of the five methods evaluated, three methods including Liquid Tissue Kit®, modified Liquid Tissue Kit, and Qiagen kit - proved to be clearly more proficient than the other methods, when the number of peptides and/or proteins identified were compared (data not shown). The number of peptides and proteins was lowest using the G-HCl protocol. The amount of tissue used was smallest with the Liquid Tissue kit protocols (Table 1). The number of proteins identified was highest using the Liquid tissue Kit protocol (Figure 2). Based on the sequence coverage of the proteins identified, the modified Liquid Tissue kit protocol was chosen for further studies of celloidin removal methods and application to FFCE otic samples.
Table 1.
A Table showing tissue consumption with all the protocols used
| Protocol | Volume of tissue digested (mm3) | Digest volume (μl) | mm3 tissue / μl | Injected | |
|---|---|---|---|---|---|
| μl | mm3 | ||||
| Liquid Tissue | 0.16 | 23 | 0.0070 | 2 | 0.014 |
| Mod Liquid Tissue | 0.16 | 23 | 0.0070 | 2 | 0.014 |
| QProteome | 1 | 52 | 0.0192 | 1 | 0.019 |
| SDS | 2 | 602 | 0.0033 | 4.8 | 0.016 |
| G-HCl | 2 | 607 | 0.0033 | 4.8 | 0.016 |
Figure 2.
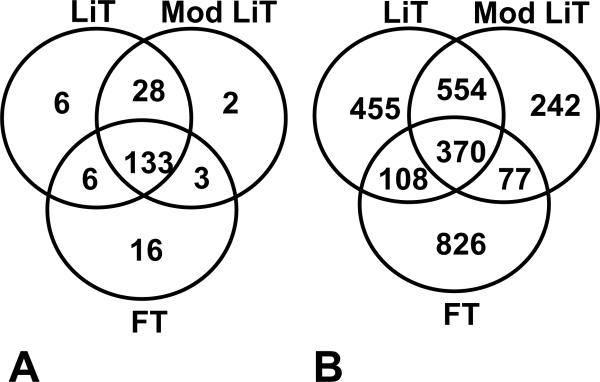
FFPE mouse liver data. Five different extraction methods were used. Results with two of the methods are shown here. The Liquid Tissue kit® and the modified liquid tissue kit proved to be the best methods for FFPE mouse liver tissue. Figure A) shows the number of proteins (unique and overlapping) and B) number of peptides (unique and overlapping). FT, fresh tissue.; LiT, Liquid Tissue extraction kit protocol; mod LiT, Liquid Tissue kit extraction kit with the added reduction and alkylation step before digestion.
Comparison of celloidin removal methods
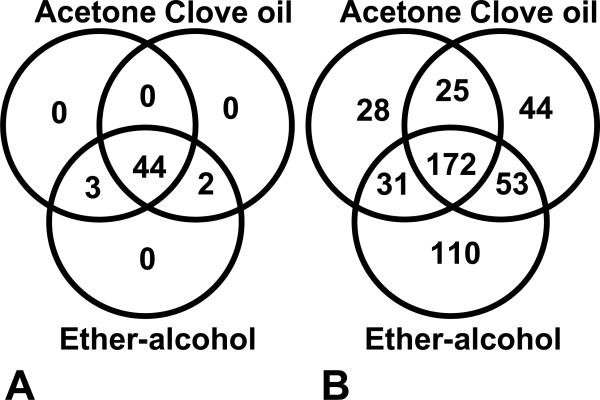
Celloidin removal was studied using four different protocols on FFCE mouse liver tissue. The modified Liquid Tissue kit extraction method was used for these analyses. Peptides and proteins could be detected using all 4 methods. Acetone, clove oil and ether-alcohol methods gave the best results. The results of these methods were nearly equal. The number of peptides and proteins identified using the three best removal methods are shown in figure 3. The acetone removal method was chosen for further analyses.
Figure 3.
A figure showing the celloidin removal data. The modified Liquid Tissue kit extraction method was used for these analyses. The number of proteins (A) (unique proteins and overlapping proteins) and number of peptides (B) (unique peptides and overlapping peptides) are shown. The samples were analyzed as duplicates.
FFCE temporal bone samples
Acetone celloidin removal with the Liquid Tissue kit, using the added reduction and alkylation step before digestion (the modified Liquid Tissue kit), were used for the analysis of FFCE cadaveric human inner ear and mouse inner ear sections and human otic capsule bone sections.
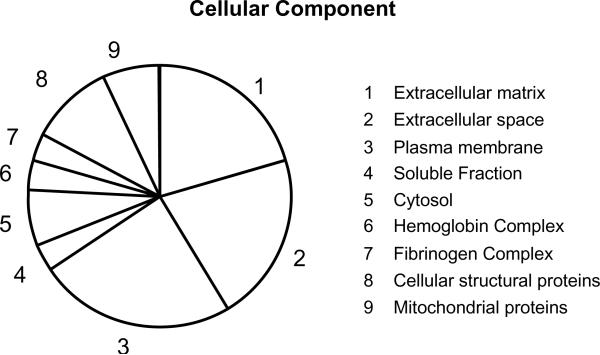
Mouse inner ear results are shown in table 2. The most abundant proteins identified and quantified were collagens. Again, with the human samples the most abundant proteins both in the inner ear and otic capsule bone samples were collagen alpha-1 and alpha-2 chain proteins (Table 3). 20 proteins were detected in inner ear samples and 8 proteins in otic bone samples. Cochlin was detected both in mouse and human inner ear samples. The dataset shows that proteins from all major cell compartments (i.e., cytoplasm, extracellular, membrane, and nucleus) were observed (Figure 4).
Table 2.
The mouse FFCE inner ear data. The number of unique peptides per protein is shown. The samples were analyzed as in triplicate (M2–3).
| Identified Proteins (17) | Accession Number | M2-1 | M2-2 | M2–3 | M3-1 | M3-2 | M3-3 |
|---|---|---|---|---|---|---|---|
| Histone H2A.x H2a/x. | P27661 | 2 | 2 | 2 | 4 | 3 | 4 |
| Cochlin precursor COCH-5B2. | Q62507 | 9 | 6 | 8 | 17 | 18 | 21 |
| Alpha-2-HS-glycoprotein precursor Fetuin-A Countertrypin. | P29699 | 7 | 10 | 11 | 11 | 13 | 13 |
| Collagen alpha-1I chain precursor Alpha-1 type I collagen. | P11087 | 4 | 6 | 4 | 4 | 4 | 3 |
| Collagen alpha-2I chain precursor Alpha-2 type I collagen. | Q01149 | 3 | 3 | 5 | 4 | 7 | 6 |
| Actin, cytoplasmic 1 Beta-actin. | P60710 (+1) | 1 | 1 | 1 | 3 | 4 | 2 |
| Hemoglobin subunit beta-1 | P02088 | 2 | 2 | 2 | 4 | 5 | 4 |
| Tubulin alpha-1C chain | P68373 | 1 | 2 | 1 | 2 | 2 | 2 |
| Histone H4. | P62806 | 0 | 2 | 2 | 3 | 5 | 4 |
| Serum albumin precursor. | P07724 | 0 | 0 | 0 | 3 | 2 | 4 |
| Histone H3.2. | P84228 | 0 | 0 | 0 | 3 | 2 | 3 |
| Hemoglobin subunit alpha | P01942 | 0 | 3 | 1 | 3 | 0 | 2 |
| Hemoglobin subunit beta-2 | P02089 | 1 | 1 | 0 | 1 | 2 | 1 |
| Thrombospondin-1 precursor. | P35441 | 1 | 0 | 0 | 2 | 2 | 2 |
| SPARC-like protein 1 | P70663 | 0 | 0 | 0 | 1 | 1 | 1 |
| Histone H1.2 H1 VAR.1 H1c. | P15864 (+2) | 0 | 0 | 0 | 0 | 1 | 2 |
| Myelin P0 protein precursor | P27573 | 0 | 2 | 0 | 0 | 0 | 0 |
Table 3.
A) FFCE human inner ear (HI1–5) and B) otic capsule bone (B1–5) data. The number of unique peptides identified for each protein in each run are shown. Five sections were used (HI1–5). B1–5 samples were taken from the same sections. All the samples were analyzed in triplicate.
| A) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identified Proteins (20) | Accession Number | HI1-1 | HI1–2 | HI1–3 | HI2-1 | HI2-2 | HI2–3 | HI3-1 | HI3-2 | HI3-3 | HI4-1 | HI4-2 | HI4-3 | HI5-1 | HI5-2 | HI5-3 |
| Collagen alpha 1 I chain | CO1A1_HUMAN | 4 | 6 | 4 | 4 | 3 | 6 | 12 | 14 | 14 | 14 | 12 | 17 | 9 | 11 | 9 |
| Collagen alpha 2 I chain | CO1A2_HUMAN | 2 | 3 | 2 | 1 | 1 | 1 | 7 | 8 | 10 | 2 | 5 | 7 | 2 | 7 | 6 |
| Vitronectin | VTNC_HUMAN | 4 | 5 | 5 | 4 | 6 | 7 | 9 | 10 | 8 | 9 | 8 | 8 | 5 | 8 | 7 |
| Cochlin | COCH_HUMAN | 1 | 1 | 1 | 4 | 4 | 2 | 5 | 3 | 8 | 9 | 6 | 4 | 2 | 4 | 3 |
| Tubulin alpha 1B chain | TBA1B_HUMAN | 2 | 0 | 1 | 1 | 3 | 2 | 4 | 5 | 3 | 3 | 3 | 5 | 3 | 3 | 3 |
| Prothrombin | THRB_HUMAN | 0 | 0 | 0 | 1 | 0 | 3 | 5 | 6 | 6 | 5 | 3 | 0 | 2 | 3 | 2 |
| Alpha 2 HS glycoprotein | FETUA_HUMAN | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 5 | 3 | 4 | 5 | 4 | 1 | 0 | 0 |
| Myelin P0 protein | MYP0_HUMAN | 0 | 0 | 1 | 3 | 1 | 5 | 4 | 3 | 4 | 2 | 0 | 3 | 3 | 2 | 0 |
| Thrombospondin 1 | TSP1_HUMAN | 0 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 3 | 2 | 2 | 3 | 1 | 1 | 1 |
| Vimentin | VIME_HUMAN | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 1 | 1 | 2 | 1 | 0 | 0 | 0 |
| Hemoglobin subunit gamma 2 | HBG2_HUMAN | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| Keratin type II cytoskeletal 1 | K2C1_HUMAN | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 4 |
| Histone H2A type 2 A | H2A2A_HUMAN (+3) | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 3 | 2 | 0 | 0 | 0 |
| Histone H3 3 | H33_HUMAN | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 2 | 2 | 1 | 0 | 0 | 0 |
| Actin cytoplasmic 1 | ACTB_HUMAN (+1) | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Histone deacetylase complex subunit SAP30L | SP30L_HUMAN | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Collagen alpha-1(II) chain | sp|P02458|CO2A1_HUMAN | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Tubulin beta 2C chain | FBB2C_HUMAN (+1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Histone H4 | H4_HUMAN | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ATP synthase subunit beta mitochondrial | ATPB_HUMAN | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| B) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identified Proteins (8) | Accession Number | B1-1 | B1–2 | B1–3 | B2-1 | B2-2 | B2–3 | B3-1 | B3-2 | B3-3 | B4-1 | B4-2 | B4-3 | B5-1 | B5-2 |
| Collagen alpha 1 I chain | CO1A1_HUMAN | 10 | 15 | 9 | 13 | 13 | 13 | 7 | 10 | 11 | 9 | 12 | 11 | 9 | 16 |
| Collagen alpha 2 I chain | CO1A2_HUMAN | 8 | 4 | 2 | 1 | 1 | 1 | 2 | 6 | 9 | 7 | 8 | 13 | 7 | 3 |
| Vitronectin | VTNC_HUMAN | 5 | 6 | 4 | 6 | 6 | 6 | 6 | 6 | 7 | 7 | 6 | 8 | 7 | 5 |
| Prothrombin | THRB_HUMAN | 3 | 4 | 2 | 6 | 6 | 6 | 4 | 6 | 7 | 10 | 8 | 10 | 4 | 3 |
| Alpha 2 HS glycoprotein | FETUA_HUMAN | 3 | 1 | 1 | 0 | 0 | 0 | 4 | 4 | 2 | 1 | 2 | 3 | 3 | 0 |
| Thrombospondin 1 | TSP1_HUMAN | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 2 | 0 |
| Histone deacetylase complex subunit SAP30L | SP30L_HUMAN | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Keratin type II cytoskeletal 1 | K2C1_HUMAN | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Biglycan | PGS1_HUMAN | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
Figure 4.
A chart showing the cellular components (%) of proteins found with FFCE sections of human cochlea based on gene ontology.
Discussion
The first reports in the literature demonstrating the ability to conduct an MS-based global proteomic analysis from FFPE tissues were by Hood et al. (2005), Crockett et al. (2005), and Palmer-Toy et al. (2005). Palmer-Toy et al. (2005) analyzed FFPE human temporal bone samples with the 2% SDS heating method. The spiral ligament was dissected from the sections and analyzed. They reported 125 proteins retrieved from this sample, 41 of which were identified by two or more peptides. Hood et al's report dealt with investigation of prostate tissue sections. Crockett et al. used a FFPE cell-block of human lymphoma cell line. Recently, Markaryan et al. (2010) used the Liquid Tissue Kit method to extract proteins from FFCE human cochlear tissue sections and spiral ganglion tissue isolated by laser microdissection. Protein identification was performed by bioinformatic analysis of high resolution tandem mass spectrometric data from fractionated tryptic peptide samples.
We have developed a technique for robust identification of proteins from FFCE human inner ear samples. To develop a MS analysis method for FFCE, we first selected an efficient method for removal of celloidin, trypsin digestion and extraction of peptides, which is the most critical step for proteomic analysis. A further advantage of celloidin removal using organic solvent extraction is that most tissue lipids that interfere both with trypsin digestion of proteins and LC-MS analysis of the resulting peptides will also be removed resulting in a larger number of protein identifications. Formalin fixation leads to formation of a net of covalent cross-links between side chains of proteins by methylene bridge formation. Proteins in the formalin-fixed tissue are difficult to extract by the conventional method because they are trapped in the network of cross-linked proteins (Jiang et al., 2007). Cross-linked protein networks may be de-cross-linked and further disrupted by trypsin digestion. Conventional protein analysis approaches are not applicable to formalin-fixed tissue because the intact proteins cannot be efficiently extracted. Several groups have reported methods to analyze formalin fixed archival tissue samples (Jiang et al., 2007; Hood et al., 2006; Nesvizhskii et al., 2005; Feng et al., 2006; Prieto et al., 2005; Hwang et al., 2007).
Hwang et al. (2007) developed a methodology for proteomics using a 2%SDS containing extraction buffer at high temperature. Presence of SDS in the sample is deleterious to reversed-phase HPLC separation and electrospray mass spectrometry. Therefore, it is preferable to develop protocols without utilizing SDS. Jiang et al.(2007) found that incubation of tissue in a lysis buffer containing 6M guanidine hydrochloride at high temperature led to the highest protein yield and the largest number of proteins identified. At least two commercial extraction kits for formalin-fixed tissue samples exist: QProteom (Becker et al., 2007) and Liquid Tissue (Hood et al., 2006). These methods have not been compared previously.
For this study, we choose five different methods: 2% SDS, 6M Guanidine-HCl, QProteom kit, Liquid Tissue kit and modified Liquid Tissue kit. Formalin fixed paraffin embedded (FFPE) mouse liver sections were used to evaluate formalin de-cross-linking by five different methods. Unfixed fresh mouse liver tissue was used as control. Five different methods for preparation of FFPE tissue for MS analysis were compared, as well as four methods for celloidin removal with the FFCE mouse liver tissue. The best method was applied to FFCE 20 μm mounted sections of mouse inner ear samples, and FFCE 20 μm human inner ear and human otic capsule bone sections.
Three of the five tissue extraction methods tested (the Liquid Tissue kit®, the modified Liquid Tissue kit, and the Qiagen kit) worked equally in detecting peptides and proteins from FFPE mouse liver tissue. The amount of tissue required for extraction was lowest with both Liquid Kit protocols. The modified liquid tissue kit protocol had the best coverage of proteins. The acetone removal method was applied to FFCE mouse liver sections. These two methods were used for FFCE inner ear and otic capsule sections. Proteins were identified from all major cellular compartments classified by gene ontology. Both the mouse and the human inner ear sample showed typical structural proteins for cochlea. The most abundant proteins in the human inner ear and otic capsule bone samples were collagens, as expected.
It appears that the number of proteins retrieved from FFCE human inner ears is less than the number from FFPE human inner ear tissue. In the present study we detected 20 proteins from the cochlea identified by two or more peptides. Similarly, Markaryan et al. (2010) identified 26 proteins with a minimum of two peptides. The present study and the study by Markaryan et al. were both done using FFCE tissue. In contrast, Palmer Toy et al's 2005 study using FFPE human cochlear tissue identified 41 proteins using two or more peptides. Future research is needed to determine if these differences are because of celloidin embedding or due to other factors such as postmortem time, cause of death etc.
Conclusions
Celloidin removal with acetone together with the modified Liquid Tissue kit protocol and the LC-MSE technique can be used to successfully study archival FFCE temporal bone samples. Proteins from all major cellular components were detected in the FFCE archival human temporal bone sections. This newly developed technique enables the use of FFCE tissues for proteomic studies
Acknowledgements
Supported in part by NIH grant U24 DC008559, Sigrid Juselius Foundation (A.A.A) and Academy of Finland (A.A.A.).
List of Abbreviations
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- FFPE
Formalin-fixed paraffin embedded
- FFCE
Formalin-fixed celloidin embedded
- LC-MS
Liquid chromatography-mass spectrometry
- MSE
Mass spectrometry, where E represents collision energy
- QTOF nano-UPLC
Quadripole-time of flight mass spectrometry with capillary ultrahigh pressure liquid chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Becker K-F, Schott C, Hipp S, Metzger V, Porschewski P, Beck R, Nahrig J, Becker I, Hofler H. Quantitative protein analysis from formalin-fixed tissues: implications for translational clinical research and nanoscale molecular diagnosis. J. Pathol. 2007;211:370–378. doi: 10.1002/path.2107. [DOI] [PubMed] [Google Scholar]
- Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KS. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab. Invest. 2005;85(11):1405–15. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- Feng S, Ye M, Jiang X, Jin W, Zou H. Coupling the immobilized trypsin micro-reactor of monolithic capillary with Urplc-ms/ms for shotgun proteome analysis. J. Proteome Res. 2006;5:422–428. doi: 10.1021/pr0502727. [DOI] [PubMed] [Google Scholar]
- Geromanos SJ, Vissers JPC, Silva JC, Dorschel CA, Li G-Z, Gorenstein MV, Bateman RH, Langridge JI. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9:1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- Hood BL, Conrads TP, Veenstra TD. Mass spectrometric analysis of formalin-fixed paraffin-embedded tissue: unlocking the proteome within. Proteomics. 2006;6(14):4106–14. doi: 10.1002/pmic.200600016. [DOI] [PubMed] [Google Scholar]
- Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Condrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-foxed prostate cancer tissue. Moll. Cell. Proteomics. 2005;4(11):1741–53. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- Hwang SI, Thumar J, Lundgren DH, Rezaul K, Mayya V, Wu L, Eng J, Wright ME, Han DK. Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene. 2007;26:65–76. doi: 10.1038/sj.onc.1209755. [DOI] [PubMed] [Google Scholar]
- Jiang X, Jiang X, Feng S, Tian R, Ye M, Zou H. Development of efficient protein extraction methods for shotgun proteome analysis of formalin-fixed tissues. J. Proteome Res. 2007;6(3):1038–47. doi: 10.1021/pr0605318. [DOI] [PubMed] [Google Scholar]
- Keithley EM, Horowitz S, Ruckenstein MJ. Na, K-ATPase in the cochlear lateral wall of human temporal bones with endolymphatic hydrops. Ann. Otol. Rhinol. Laryngol. 1995;104:858–863. doi: 10.1177/000348949510401106. [DOI] [PubMed] [Google Scholar]
- Li G-Z, Vissers JPC, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- Markaryan A, Nelson EG, Helseth LD, Hinojosa R. Proteomic analysis of formalin-fixed celloidin-embedded whole cochlear and laser microdissected spiral ganglion tissues. Acta otolaryngol. early online. 2010:1–6. doi: 10.3109/00016481003591749. [DOI] [PubMed] [Google Scholar]
- McGuire JF, Casado B. Proteomics: A Primer for otologists. Otology and Neurotology. 2004;25:842–849. doi: 10.1097/00129492-200409000-00032. [DOI] [PubMed] [Google Scholar]
- Miguel-Hidalgo JJ, Rajkowska G. Immunohistochemistry of neural markers for the study of the laminar architecture in celloidin sections from the human cerebral cortex. J. Neurosci. Methods. 1999;93(1):69–79. doi: 10.1016/s0165-0270(99)00114-4. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomics data: the protein inference problem. Mol. Cell. Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Mining the archival formalin-fixed paraffin-embedded tissue proteome: opportunities and challenges. Mol. Biosyst. 2008;4(7):712–20. doi: 10.1039/b800098k. [DOI] [PubMed] [Google Scholar]
- O'Malley JT, Merchant SN, Burgess BJ, Jones DD, Adams JC. Effects of Fixative and Embedding Medium on Morphology and Immunostaining of the Cochlea. Audiol. Neurotol. 2009;14:78–87. doi: 10.1159/000158536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Merchant SN. Efficient Method for the Proteomic Analysis of Fixed and Embedded Tissues. J. Proteome Res. 2005;4(6):2404–2411. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- Pawlowski KS, Wright CG, Meyerhoff WL. Histologic demonstration of glycosaminoglycans in inner ear fluids. Acta Otolaryngol. 1998;118:505–510. doi: 10.1080/00016489850154630. [DOI] [PubMed] [Google Scholar]
- Prieto DA, Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin fixed prostate cancer tissue. Mol. Cell. Proteomics. 2005;4:1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- Schuknecht HF. Pathology of the Ear. 2nd Edition Lea and Febiger; Philadelphia: 1993. [Google Scholar]
- Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77(7):2187–200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol. Cell. Proteomics. 2006;5(4):589–607. doi: 10.1074/mcp.M500321-MCP200. [DOI] [PubMed] [Google Scholar]
- Sone M, Paparella MM, Schachern PA, Morizono N, Le CT, Lin J. Expression of glycoconjugates in human Eustachian tubes with otitis media. Laryngoscope. 1998;108:1474–1479. doi: 10.1097/00005537-199810000-00010. [DOI] [PubMed] [Google Scholar]
- Sone M, Schachern PA, Paparella MM, Morizono N. Study of systemic lupus erythematosus in temporal bone. Ann. Otol. Rhinol. Laryngol. 1999;108:338–344. doi: 10.1177/000348949910800404. [DOI] [PubMed] [Google Scholar]
- Thalmann I. Proteomics and the inner ear. Brain Res. 2006;1091(1):103–12. doi: 10.1016/j.brainres.2006.01.099. [DOI] [PubMed] [Google Scholar]