Abstract
Distinctin is a 47-residue antimicrobial peptide, which interacts with negatively charged membranes and is active against Gram-positive and Gram-negative bacteria. Its primary sequence comprises two linear chains of 22 (chain 1) and 25 (chain 2) residues, linked by a disulfide bridge between Cys19 of chain 1 and Cys23 of chain 2. Unlike other antimicrobial peptides, distinctin in the absence of the lipid membrane has a well-defined three-dimensional structure, which protects it from protease degradation. Here, we used static solid-state NMR spectroscopy to study the topology of distinctin in lipid bilayers. We found that In mechanically aligned lipid bilayers (charged or zwitterionic) this heterodimeric peptide adopts an ordered conformation absorbed on the surface of the membrane, with the long helix (chain 2), approximately parallel to the lipid bilayer (∼5° from the membrane plane) and the short helix (chain 1) forming a ∼24° angle. Since at lipid-to-protein molar ratio of 50:1 the peptide does not disrupt the macroscopic alignment of either charged or zwitterionic lipid bilayers, it is possible that higher concentrations might be needed for the hypothesized pore formation, or alternatively, distinctin elicits its cell disruption action by other mechanisms.
Introduction
Naturally occurring anti-microbial peptides (AMPs) are becoming a new weapon in the fight against bacterial drug resistance [1-7]. Traditional approaches for the treatment of bacterial infections rely on the use of compounds that become quickly ineffective due to the genetic plasticity of pathogenic microorganisms. AMPs represent a promising alternative to common antibiotics because of their ability to directly interact with the bacterial membrane causing cell lysis [1-3]. Resistance to AMPs is an unlikely event since it would imply severe modification of the lipid composition and architecture of the cell membrane.
Multicellular organisms produce AMPs as a defense against microbial pathogens. AMPs have been isolated from fungi, insects, amphibians, mammals and plants [4], with their expression triggered as a response to external stress or alternatively secreted from storage by superficial glands [8]. Due to their impact on pathogenic cells, they have also been shown to be effective against tumors, HIV infection, and pulmonary infections, as well as a role in modulating the immune system [4]. Over the past few decades, there have been many studies aimed at isolating and characterizing novel antimicrobial peptides with the potential to be used as new therapeutic agents against bacterial infections [5]. These studies have shown that AMPs do not possess any common amino acid motif, but do share some common structural features, including the presence of an amphipathic helix [9]. Several models have been proposed to explain how antimicrobial peptides interact with bacterial membranes to cause cell lysis, although molecular details on the mechanism of action have proved extremely challenging to elucidate [4, 10]. This is largely due to difficulty in studying these peptides in the presence of lipid membranes with conventional techniques such as X-ray crystallography.
Because of the potential therapeutic use of antimicrobial peptides, it is fundamental to understand the structural details of peptide/membrane interactions in order to design more specific and potent variants [11]. Solution NMR spectroscopy has been applied to a myriad of small and medium size antimicrobial peptides [12-17], giving important insights into the mechanism of insertion into micelles. However, micellar systems are only a coarse approximation of membrane bilayers; synthetic lipid bilayers (vesicles or planar bilayers) are the preferable system to test lipid/peptide interactions. The large size of the lipid/peptide complexes requires solid-state NMR (ssNMR) techniques. Specifically, ssNMR spectroscopy of peptide and proteins in mechanically and magnetically aligned lipid bilayers is able to provide structural and dynamic information [18-20]. First, it is possible to measure the degree of lipid bilayer alignment using 31P NMR spectroscopy [21]. Second, the orientation of peptide planes using selective or uniformly 15N or 13C labeled proteins can be determined [18, 22]. As a result, it is possible to interpret the changes in topology of these proteins following the perturbations to chemical shifts. Recent reports summarize the application of these methods to small membrane proteins as well as antimicrobial peptides [22-27].
In this work, we analyzed the topology of distinctin in both zwitterionic and charged oriented lipid bilayers by ssNMR spectroscopy. Distinctin is an antimicrobial peptide extracted from Phyllomedusa distincta, a tree frog that lives in the forests of Brazil [28]. It is active against Gram-positive and Gram-negative bacteria, but shows very little hemolytic activity against human erythrocytes [28, 29]. In a recent study [30, 31], distinctin was shown to be effective in the treatment of a severe staphylococcal infection (sepsis) in a murine model, underlying the potential role of this novel peptide for therapeutic purposes. These researchers found that distinctin can be associated with other antibiotic compounds, reducing the number of infections drastically [30].
Compared to other well-studied AMPs, distinctin presents some interesting peculiarities. It is a heterodimer composed of two polypeptide chains: chain 1 (22 residues) and chain 2 (25 residues) linked by a disulfide bridge between Cys19 of chain 1 and Cys23 of chain 2 (Figure 1A). Solution NMR studies in aqueous environment revealed that both chains are amphipathic α-helices (Figure 1B). It was also shown that distinctin, unlike other antimicrobial peptides, adopts a well-folded conformation in aqueous buffer, with a non-covalent parallel four-helical bundle (Figure 1C) [29, 31]. This conformation confers an increased stability and a marked resistance to proteolytic degradation [29].
Figure 1.
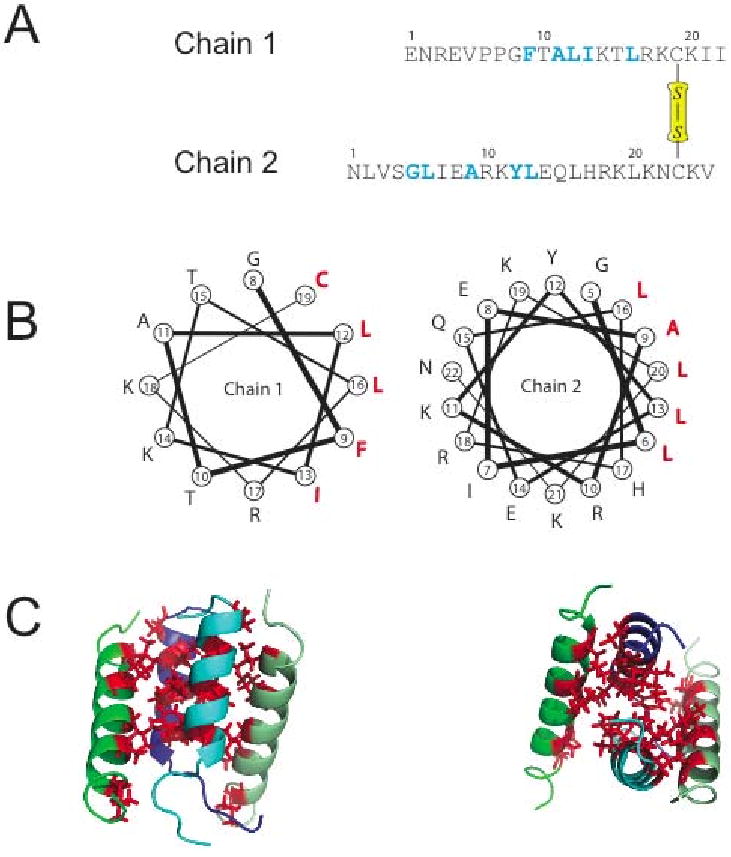
(A) Amino acid sequence of distinctin heterodimer. 15N labeled amino acids are highlighted in blue. Residue Cys-19 of chain 1 and Cys-23 of chain 2 are involved in a disulfide bridge. (B) Helical wheel plots showing the amphipathic nature of each chain. Hydrophobic residues are in red. (C) Three-dimensional structure of distinctin in aqueous buffer [29]. The four-helical bundle is stabilized by hydrophobic interactions of the residues, forming a hydrophobic core in the interior of the structure.
When incorporated into phosphatidylcholine/phosphatidylethanolamine planar bilayers, distinctin gave rise to voltage-dependent behavior typical of ion-channel formation [29]. Addition of negatively charged phosphatidylserine to the lipid preparations did not change this behavior, suggesting that the membrane composition does not play a major role in distinctin's function. Based on these data, different structural models were built to describe the pore formation in membranes. Molecular dynamics calculations based on structural and conductivity data suggested that distinctin could form pentameric pores in membranes with at least one of the helices crossing the lipid bilayer [31].
Recently, the topological preferences of distinctin in zwitterionic lipid bilayers were investigated by Bechinger and co-workers [32]. Using a combination of site specific 15N and 2H NMR spectroscopy, these researchers showed that distinctin in POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) lipid bilayers adopts an orientation with the two helical domains approximately parallel to the surface of the bilayers. This orientation is persistent both at lipid:protein ratios of 100:1 and 250:1. Although acquired in the absence of charged lipids and lipid:protein ratios higher than the limits observed for the formation of putative ion channels, the data from Bechinger and co-workers [32] do not support the mechanism of action proposed by Dalla Serra et al. [31].
In order to shed light on the topology of distinctin in charged and zwitterionic lipid membranes and its mechanism of action, we used 31P and 15N ssNMR spectroscopy in mechanically aligned lipid bilayers. We found that distinctin is absorbed on the surface of the lipid bilayers and that the charge of the lipid headgroups does not perturb its topology.
Materials and Methods
Peptide Synthesis
Distinctin peptide chains were independently synthesized by 9-flourenyl-methoxycarbonyl solid-phase chemistry as previously described [29]. Four different samples of distinctin labeled with 15N at the amide nitrogen were prepared: a) Phe-9 of chain 1 b) Ala-9 of chain 2 c) Ala-12-Leu-13-Ile-14-Leu-16 of chain 1 and d) Gly-5-Leu-6-Tyr-12-Leu-13 of chain 2. For the 15N isotopic enrichment, Fmoc-15N-Phe-OH, Fmoc-15N-Ala-OH, Fmoc-15N-Leu-OH, Fmoc-15N-Ile-OH, Fmoc-15N-Gly-OH, and Fmoc-15N-Tyr-OH (Sigma-Aldrich, St. Louis, MO) were used. To form the disulfide bond, the two peptide chains, dissolved in basic aqueous solution, were slowly mixed and dried under air flow [29]. All peptides were purified by reversed phase HPLC (98% purity) with the molecular masses confirmed by MALDI-TOF mass spectrometry (m/z 5482).
Solid-state NMR sample preparation
For zwitterionic lipid bilayers, we used 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (POPC/DOPE); while for charged bilayers we used 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1,2-dioleoyl-sn-glycero-3-phosphate (POPC/DOPA) lipid mixtures. Briefly, a stock solution of 1 mg/mL in doubly distilled H2O (ddH2O) of each distinctin sample was prepared and kept at -20 °C. For mechanically aligned samples, a mixture (4:1 mol:mol) of POPC:DOPE or a mixture (1:1) of POPC:DOPA from Avanti Polar Lipids (Alabaster, AL) was dried from chloroform in a glass vial to a thin film; then ddH2O (0.2% w/v) was added to the dried lipids and the resulting suspension was extruded ten times through a polycarbonate filter (Millipore, Billerica, MA) with 0.05 μm pore diameter using a Lipex extruder (Northern Lipids, Inc., Vancouver, BC, Canada). Ten passes were necessary to ensure that small unilamellar vesicles (SUVs) of homogeneous size were obtained. The suspension of SUVs was concentrated to 2 mL using an Amicon ultra-concentrator with a 10 kDa cutoff membrane. Distinctin was added to the lipids and the mixture was stirred for two hours at 25 °C. The mixture was then transferred onto 25 glass plates (5.7 mm × 12 mm × 0.030 mm; Marienfeld GmbH, Lauda-Königshofen, Germany) with water slowly evaporated by incubation for four hours at 40 °C. The plates were stacked, rehydrated for 96 hours in a humidity chamber to reach the liquid-crystalline phase and finally sealed in a glass cell. POPC:DOPE samples of Ala-9 and Phe-9 were prepared with a final lipid/protein (L:P) ration of 200:1, whereas distinctin labeled at chain 1 (A11-L12-I13-L16) and distinctiin labeled at chain 2 (G5-L6-Y12-L13) were reconstituted at a L:P ratio of 50:1. For the POPC/DOPA sample of Ala-9, the final L:P ratio was 50:1.
Solid state NMR spectroscopy
All spectra were acquired on a Varian spectrometer operating at field strength of 14.1 T (1H frequency of 600.1 MHz). 31P single pulse experiments were performed for all the samples in order to evaluate the macroscopic orientation of the lipid bilayers [21]. A doubly tuned flat coil probe (Varian Inc., Palo Alto, CA, BioStatic™) was used with the temperature for all experiments regulated at 5 °C. The following parameters were used: spectral width 100 kHz, acquisition time 5 ms, recycle delay time of 4 s and a decoupling field strength of 50 kHz, 90° pulse length of 6 μs. All spectra were processed with a 50 Hz line broadening window function. For each 31P spectrum, a total of 64 scans were collected and zero filled to 2048 points.
15N cross-polarization (CP) spectra of static aligned samples were acquired with the same low-E double resonance flat-coil probe (Varian Inc., Palo Alto, CA) using the following parameters: spectral width 100 kHz, acquisition time 5 ms, CP contact time 1 ms, a SPINAL64 heteronuclear decoupling [33] field strength of 50 kHz, and 5 μs 1H 90° pulse length. For each spectrum, a total of 24,000-56,000 scans were recorded and zero filled to 2048 points with a 100 Hz line broadening window function applied. The two-dimensional 15N chemical shift/1H-15N dipolar correlations were acquired using the SAMPI4 experiment [34] with 6,400 scans in the direct dimension and 10 increments used to evolve the dipolar coupling on the 15N Ala-9 selectively labeled sample. All of the spectra were referenced to the 15NH4Cl signal (41.5 ppm) as an external standard.
Results and Discussion
The first step in the present study was to assess the effect of different concentrations of distinctin peptide on the planarity of the lipid bilayers. Figure 2 shows 31P spectra of mechanically aligned POPC/DOPE lipid bilayers in the presence of distinctin at 200:1 L:P molar ratio. The presence of a single peak at ∼30 ppm is indicative of well aligned lipid bilayers with the normal parallel to the external magnetic filed. This is consistent with a recent report by Bechinger and co-workers [32] that shows no effect of distinctin peptide on the alignment of zwitterionic (POPC) lipid bilayers. Since distinctin interacts more strongly with membranes containing charged lipids (PA, PS, and PG) [29, 31], we investigated the effects of charged lipids by adding DOPA to the lipid preparations up to a molar ratio of 1:1, POPC:DOPA. Both the presence of the charged lipid and the low lipid:peptide ratio used (up to 50:1) did not cause a disruption of the lipid bilayers. Figure 3A shows the presence of a single resonance at ∼30 ppm, confirming the macroscopic orientation and integrity of the lipid bilayer. These data are consistent with the static 31P experiments carried out in lipid vesicles under the same lipid composition (POPC:DOPA) and lipid:peptide molar ratio (50:1) [29]. In those experiments, no changes in the 31P powder pattern were detected, ruling out detergent-like mechanism of action of distinctin or other major perturbations of the lipid membranes.
Figure 2.
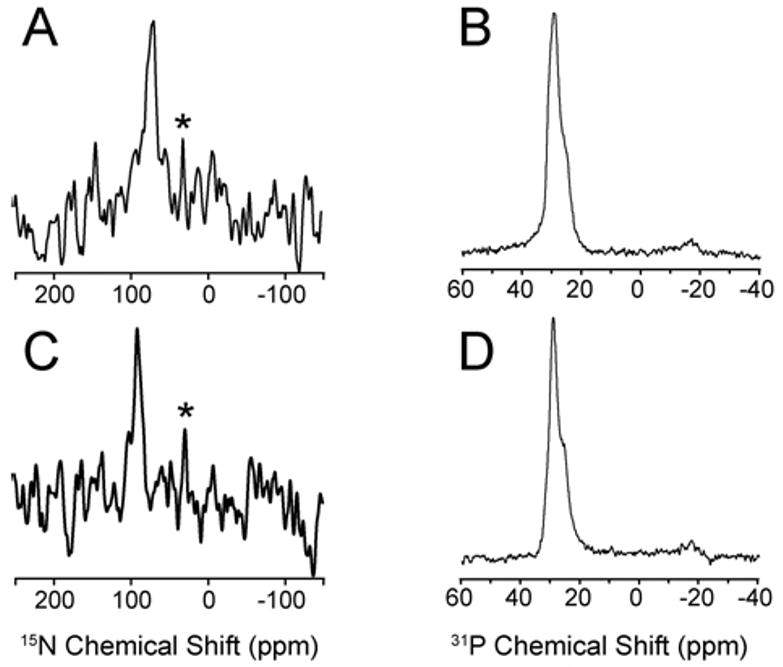
(A) and (C) One dimensional cross-polarization spectra of 15N Ala-9 (chain 2) and 15N Phe-9 (chain 1) distinctin, respectively in POPC/DOPE (4:1 mol:mol) at 200:1 lipid:protein. (B) and (D) One dimensional 31P spectra of aligned lipid bilayers for 15N Ala-9 (chain 2) and 15N Phe-9 (chain 1) distinctin, respectively. Asterisks indicate natural abundance 15N from lipid head groups.
Figure 3.
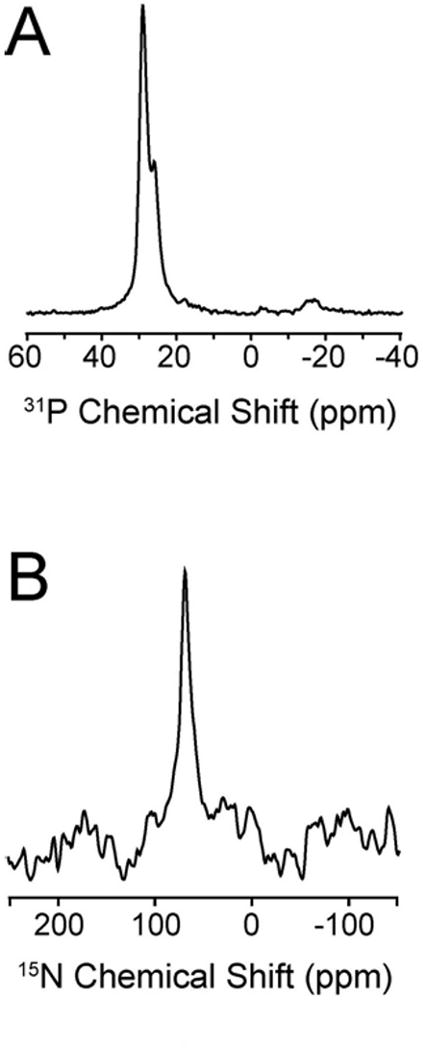
(A) 31P spectrum of POPC/DOPA (1:1 mol:mol) containing 15N Ala-9 (chain 2) distinctin at 50:1 lipid:peptide; (B) 15N proton decoupled cross-polarization spectrum of 15N Ala-9 (chain 2) distinctin at 50:1 lipid-to-peptide.
In order to ascertain the ability of each distinctin chain to penetrate the membrane, two samples were prepared: the first consisted of 15N labeled distinctin at Phe-9 of chain 1, and the second sample 15N at Ala-9 of chain 2 (Figure 1A). The 15N chemical shift of amide groups in helical segments is very sensitive to the orientation of the amide group with respect to the external magnetic field, which, in the case of aligned lipid bilayers, is collinear with the membrane normal. Qualitatively, chemical shifts greater than 200 ppm indicate that the helix is nearly perpendicular to the lipid bilayer plane (transmembrane). Conversely, chemical shifts smaller than 100 ppm entail that the helix is almost parallel to the membrane plane. Shifts around ∼110-120 ppm are usually indicative of isotropic or disordered regions of the protein.
Figure 2A and 2C show the proton decoupled cross-polarization spectra for the two singly labeled distinctin samples (15N Phe-9 or 15N Ala-9) in POPC/DOPE bilayers at a lipid:peptide molar ratio of 200:1. Assuming both distinctin chains are in a helical configuration, the Ala-9 amide nitrogen shift of ∼74 ppm corresponds to an almost parallel orientation of chain 2 with respect to the membrane plane, while Phe-9 resonates at 92 ppm, also indicating a parallel configuration for chain 1. The slight change in chemical shift can be attributed to a somewhat altered tilt angle of chain 1 with respect to chain 2, a different relative position of Phe-9 on the helical wheel, or alternatively, a difference in dynamics between chain 1 and chain 2, which would cause a scaling of the chemical shift toward the isotropic value (∼110-120 ppm). The extra peak at ∼30 ppm present in the Phe-9 spectrum can be assigned to the natural abundance 15N of the phospholipid head-groups [35]. The Phe-9 spectrum has a signal/noise ratio ∼4 times lower than Ala-9. This might be due to differences in the cross polarization efficiency or a slight change in peptide concentration. In the former case, both spectra were acquired using a contact time of 1 ms. It is well known that cross-polarization efficiency is linked to peptide or protein dynamics [36]. The differential behavior of the two sites is in agreement with the data obtained from solution NMR, circular dichroism and Fourier-transform Infrared spectroscopy [29], as well as the ssNMR work of Bechinger and co-workers [32], indicating that chain 1 is more dynamic than chain 2.
The experiments were repeated in POPC/DOPA oriented lipid bilayers. Figure 3B shows the proton decoupled cross-polarization spectrum of 15N Ala-9 (chain 2). In this case, we were able to reach a L:P molar ratio of up to 50:1, without disturbing the macroscopic orientation of the lipid bilayer (see Figure 3A). Predictably, the signal-to-noise ratio of this 15N spectrum is much higher than the previous spectra obtained at 200:1 molar ratio. However, the chemical shifts did not change substantially (∼74 ppm for distinctin in POPC/DOPE at L:P of 200:1 and ∼72 in POPC/POPA at L:P of 50:1). This demonstrates that the addition of the DOPA in the lipid preparations does not substantially modify the topological orientation of distinctin. The higher concentration of peptide reached under these conditions made it possible to carry out a two-dimensional separated local field experiment that resolves 15N chemical shifts with 1H-15N dipolar couplings. Given the predicted orientation based on 15N chemical shifts, we opted for the SAMPI4 experiment, which has a smaller 1H offset dependence on the measured dipolar coupling values [37]. Figure 4B shows the SAMPI4 spectrum of Ala-9 in mechanically oriented POPC/DOPA lipid bilayers. The measured dipolar coupling for Ala-9 is 4.0 kHz, a value typical of well-structured alpha helix absorbed on the lipid bilayer surface [38]. In order to ensure homogeneity of our sample preparations, we show the solution NMR [1H,15N]-HSQC spectrum of Ala-9 15N labeled distinctin in phosphate buffer at 37°C in Figure 4A, indicating homogenous linewidths and the isotropic chemical shift values for both 1H and 15N amide of Ala-9.
Figure 4.
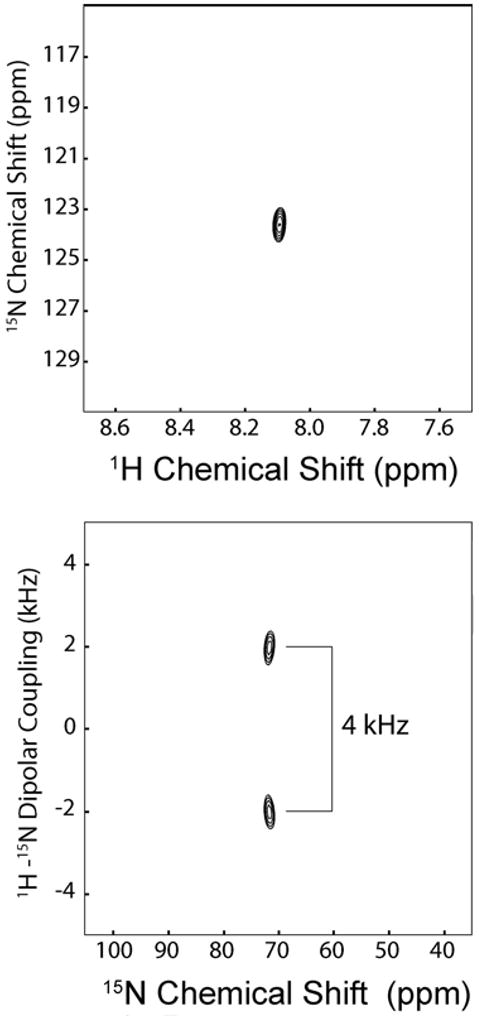
(A) Solution NMR [1H,15N]-HSQC spectrum acquired in phosphate buffer pH=6.5, showing the isotropic chemical shift of the 15N Ala-9 (chain 2) distinctin. (B) Solid-state NMR [1H,15N]-SAMPI4 spectrum of 15N Ala-9 (chain 2) distinctin reconstituted in oriented POPC/DOPA lipid bilayers.
To determine the topology of the distinctin heterodimer more accurately, two other samples were prepared: 1) 15N labels at Ala-11, Leu-12, Ile-13, Leu-16 of chain 1 and chain 2 unlabeled (Distinctin1) and 2) 15N labels at Gly-5, Leu-6, Tyr-12, Leu-13 of chain 2 and chain 1 unlabeled (Distinctin2).
Both samples were reconstituted in POPC/DOPE lipid bilayers at a L:P ratio of 50:1. Figures 5A and 5B show the proton decoupled 15N cross-polarization spectra of Distinctin1 and Distinctin2, respectively. Distinctin1 has four resolved resonances ranging from ∼75 to ∼110 ppm. The corresponding two-dimensional SAMPI4 spectrum is shown in Figure 5C. The four resonances are well dispersed in the nitrogen dimension, but have very similar dipolar coupling values, indicative of slightly tilted orientation of the helical axis.
Figure 5.
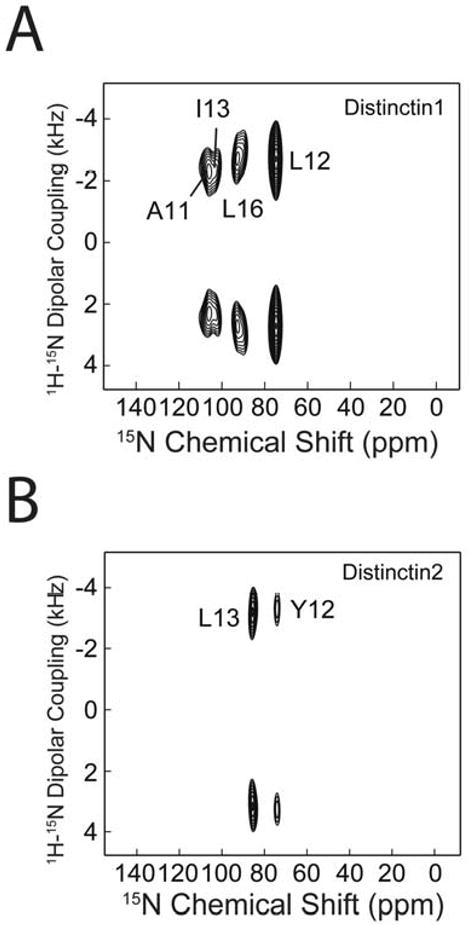
15N proton decoupled cross-polarization spectrum of distinctin 15N selectively labeled at (A) Ala-11, Leu-12, Ile-13, Leu-16 of chain 1 and at (B) Gly-5, Leu-6, Tyr-12, Leu-13 of chain2. (C) and (D) [1H,15N]-SAMPI4 spectra of the same samples as in (A) and (B).
Distinctin2, corresponding to the longer chain, presents only two clearly resolved resonances at ∼72 and ∼96 ppm, which were assigned to Leu-12 and Tyr-13, respectively. Although the sample had four labeled sites, two of them (Gly-5 and Leu-6) are located at the beginning of the α-helix [29], therefore highly dynamic resulting in inefficient cross-polarization. In contrast, the more structured residues Leu-12 and Tyr-13 are efficiently cross-polarized in our CP and SAMPI4 experiments.
In order to determine the tilt and rotation angles of the two distinctin chains in lipid bilayers more quantitatively, we implemented the chemical shift anisotropy values in the simulated annealing protocol according to Shi et al. [39] and refined the published solution structure of the distinctin heterodimer [29]. After refinement, the chemical shift anisotropy value of each residue was back-calculated from the energy minimized structure. Figure 6A and B show the anisotropic chemical shifts for each residue of the peptide calculated from the minimized structures (black traces) and the experimental values determined by oriented solid-state NMR (red points). Almost all of the experimental points are within the error of ± 5 ppm, following the periodic pattern typical of α-helices [40-44]. By fitting the CSA values we obtained tilt angles of ∼24° and 5° with respect to the bilayer plane, for chains 1 and 2, respectively.
Figure 6.
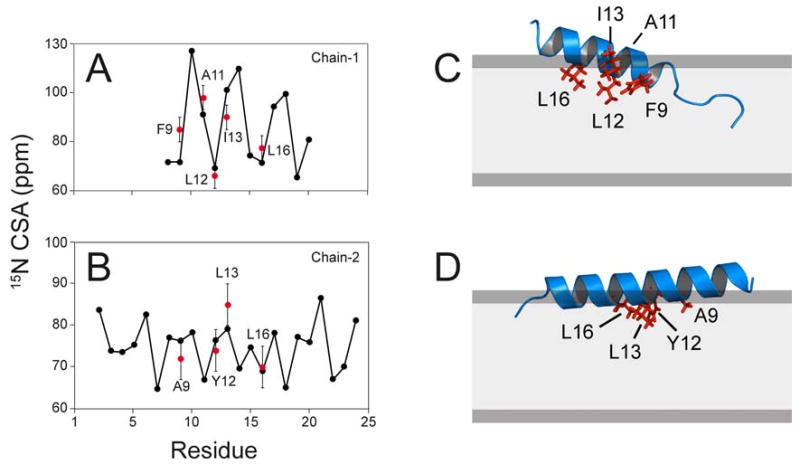
Calculated (black circles) and experimental (red circles) chemical shift anisotropy of distinctin chain 1 (A) and distinctin chain 2 (B). Error values in the experimental values are ± 5ppm. The chemical shift of Leu-16 in chain 2 is taken from ref [32]. Refined molecular structures of distinctin chain 1 (C) and chain 2 (D) obtained using the energy minimization protocol of Shi et al. [41].
To elicit antimicrobial activity (cell lysis), the two distinctin amphipathic helices need to interact strongly with bacterial membranes. This interaction is modulated by negatively charged lipids (DOPA1,2-dioleoylphosphatidylglycerol, phosphatidylinositol and phosphatidylserine) [29, 31]. However, our measurements show that lipid interactions occur even with the neutral lipids POPC and DOPE [31], without disrupting the lipid membranes. At a molar ratio of 50:1, the peptide is still absorbed on the surface of the membrane and there is no evidence of a transmembrane orientation of the longer distinctin chain (chain 2). This would support a “carpet-like” model of interaction with lipid bilayers [4].
A possible reconciliation of these observations is that distinctin might adopt a mixed mechanism of action between two extremes represented by the all-or-none [47, 48] and graded [9] antimicrobial mechanisms, an intermediate situation that Almeida and co-workers defined as the “grey zone” [9]. In other words, while distinctin may form transient pores even at lower concentrations, there is a substantial population of the peptide present on the surface of the lipid membrane, which is absorbed in a “carpet-like” fashion (see Figure 7). Increases in peptide concentration may drive the system to the formation of more organized structures similar to pores [31]. This ambivalence of distinctin may find a justification in its affinity for both zwitterionic and charged lipids. The latter does not fully justify the selectivity of distinctin for bacterial membranes versus erythrocytes and suggests that more experiments for the characterization of the thermodynamics and kinetics of membrane binding of distinctin need to go along with the structural studies in order to elucidate the mechanism of action of this important peptide.
Figure 7.
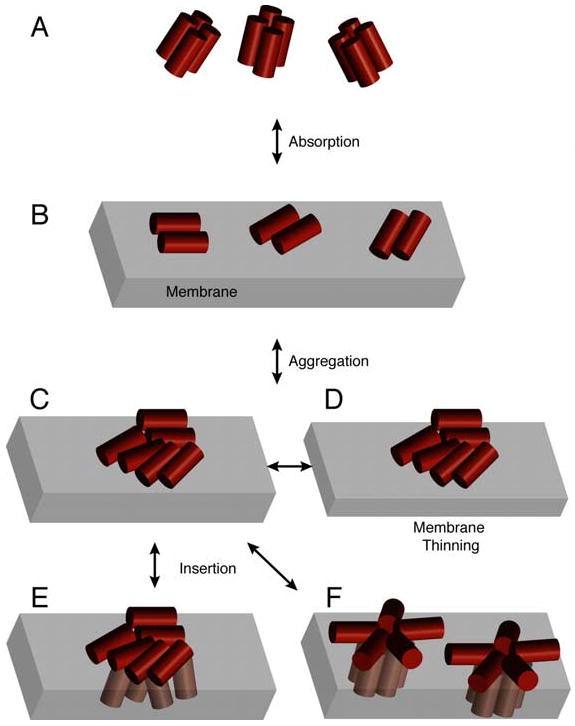
Proposed model of distinctin interacting with cell membranes: (A) in the soluble form, distinctin forms well-structured helical bundle (dimer of dimers) that are resistant to proteases, (B) The helical bundle deoligomerizes in the presence of the membrane to interact with the surface of the lipid bilayer; (C-F) aggregation of the peptide on the surface of the bilayer, and possible models to explain the channel-like behavior.
Conclusions
In conclusion, we report the solid-state NMR investigation of the distinctin peptide in three model membranes. We found that this peptide is absorbed on the surface of either zwitterionic or negatively charged lipid bilayers at a peptide to lipid ratios as high as 50:1. In agreement with previous studies [31], 31P spectra show that these high peptide concentrations do not disrupt model lipid bilayers (POPC/DOPE or POPC/DOPA). Two dimensional 15N chemical shift/1H,15N dipolar correlation spectra of several residues in chain 1 and chain 2 show a dipolar coupling of ∼4.0 kHz, confirming that distinctin, behaves as a typical amphipathic peptide, with the helical domains absorbed on the surface of the membrane and the helical axes approximately perpendicular to the normal of the membrane. In particular, the short chain 1 has a tilt angle of ∼24° with respect the plane of the bilayer, whereas chain 2 is almost parallel (∼5°). However, in lipid bicelles distinctin chain 2 can adopt both an approximately parallel or perpendicular (transmembrane) orientation. Taken with the recent electrochemical data on single-channel conductivity [29], this study suggests a mixed mechanism of action of distinctin, which is intermediate between the all-or-none and graded antimicrobial mechanism identified by Almeida et al. [9]. Even at low concentrations, distinctin can form stochastic pores that justify the ion conductivity; however, the peptide has high propensity to interact with the lipid membranes in a carpet-like topology.
Finally, our solid-state NMR investigation was carried out in mechanically aligned lipid bilayers spread onto glass plates. Although this system is widely employed in the structural biophysics of membrane proteins, further studies on distinctin with membrane models with higher degree of hydration are preferable. The latter will be part of our future endeavors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shai Y. Mechanism of the Binding, Insertion and Destabilization of Phospholipid Bilayer Membranes by a-Helical Antimicrobial and Cell Non-selective Membrane-lytic Peptides. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zasloff M. Antibiotic peptides as mediators of innate immunity. Curr Opin Immunol. 1992;4:3–7. doi: 10.1016/0952-7915(92)90115-u. [DOI] [PubMed] [Google Scholar]
- 4.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 5.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 6.Mookherjee N, Hancock RE. Cationic host defence peptides: innate immune regulatory peptides as a novel approach for treating infections. Cell Mol Life Sci. 2007;64:922–933. doi: 10.1007/s00018-007-6475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Easton DM, Nijnik A, Mayer ML, Hancock RE. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 2009;27:582–590. doi: 10.1016/j.tibtech.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soravia E, Martini G, Zasloff M. Antimicrobial properties of peptides from Xenopus granular gland secretions. FEBS Lett. 1988;228:337–340. doi: 10.1016/0014-5793(88)80027-9. [DOI] [PubMed] [Google Scholar]
- 9.Almeida PF, Pokorny A. Mechanisms of antimicrobial, cytolytic, and cell-penetrating peptides: from kinetics to thermodynamics. Biochemistry. 2009;48:8083–8093. doi: 10.1021/bi900914g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 11.Scott RW, DeGrado WF, Tew GN. De novo designed synthetic mimics of antimicrobial peptides. Curr Opin Biotechnol. 2008;19:620–627. doi: 10.1016/j.copbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porcelli F, Buck B, Lee DK, Hallock KJ, Ramamoorthy A, Veglia G. Structure and orientation of pardaxin determined by NMR experiments in model membranes. J Biol Chem. 2004;279:45815–45823. doi: 10.1074/jbc.M405454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porcelli F, Buck-Koehntop BA, Thennarasu S, Ramamoorthy A, Veglia G. Structures of the dimeric and monomeric variants of magainin antimicrobial peptides (MSI-78 and MSI-594) in micelles and bilayers, determined by NMR spectroscopy. Biochemistry. 2006;45:5793–5799. doi: 10.1021/bi0601813. [DOI] [PubMed] [Google Scholar]
- 14.Porcelli F, Verardi R, Shi L, Henzler-Wildman KA, Ramamoorthy A, Veglia G. NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry. 2008;47:5565–5572. doi: 10.1021/bi702036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zagorski MG, Norman DG, Barrow CJ, Iwashita T, Tachibana K, Patel DJ. Solution Structure of Pardaxin P-2. Biochem. 1991;30:8009–8017. doi: 10.1021/bi00246a019. [DOI] [PubMed] [Google Scholar]
- 16.Haney EF, Hunter HN, Matsuzaki K, Vogel HJ. Solution NMR studies of amphibian antimicrobial peptides: linking structure to function? Biochim Biophys Acta. 2009;1788:1639–1655. doi: 10.1016/j.bbamem.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Scott WR, Baek SB, Jung D, Hancock RE, Straus SK. NMR structural studies of the antibiotic lipopeptide daptomycin in DHPC micelles. Biochim Biophys Acta. 2007;1768:3116–3126. doi: 10.1016/j.bbamem.2007.08.034. [DOI] [PubMed] [Google Scholar]
- 18.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ketchem RR, Lee KC, Huo S, Cross TA. Macromolecular structural elucidation with solid-state NMR-derived orientational constraints. J Biomol NMR. 1996;8:1–14. doi: 10.1007/BF00198135. [DOI] [PubMed] [Google Scholar]
- 20.Ketchem RR, Hu W, Tian F, Cross TA. Structure and dynamics from solid state NMR spectroscopy. Structure. 1994;2:699–701. doi: 10.1016/s0969-2126(00)00070-8. [DOI] [PubMed] [Google Scholar]
- 21.Seelig J. Phosphorus-31 nuclear magnetic resonance and the head group structure of phospholipids in membranes. Biochim Biophys Acta. 1978;515:105–140. doi: 10.1016/0304-4157(78)90001-1. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez ID, Gehman DJ, Separovic F. Membrane interactions of antimicrobial peptides from Australian frogs. Biochim Biophys Acta. 2009;1788:1630–1638. doi: 10.1016/j.bbamem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Bechinger B, Kinder R, Helmle M, Vogt TCB, Harzer U, Schinzel S. Peptide structural analysis by solid-state NMR spectroscopy. Biopolymers (Peptide Science) 1999;51:174–190. doi: 10.1002/(SICI)1097-0282(1999)51:3<174::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 24.Hong M. Oligomeric structure, dynamics, and orientation of membrane proteins from solid-state NMR. Structure. 2006;14:1731–1740. doi: 10.1016/j.str.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Salgado J, Grage SL, Kondejewski LH, Hodges RS, McElhaney RN, Ulrich AS. Membrane-bound structure and alignment of the antimicrobial beta-sheet peptide gramicidin S derived from angular and distance constraints by solid state 19F-NMR. J Biomol NMR. 2001;21:191–208. doi: 10.1023/a:1012946026231. [DOI] [PubMed] [Google Scholar]
- 26.Lu JX, Damodaran K, Blazyk J, Lorigan GA. Solid-state nuclear magnetic resonance relaxation studies of the interaction mechanism of antimicrobial peptides with phospholipid bilayer membranes. Biochemistry. 2005;44:10208–10217. doi: 10.1021/bi050730p. [DOI] [PubMed] [Google Scholar]
- 27.Wang G. NMR of membrane-associated peptides and proteins. Curr Protein Pept Sci. 2008;9:50–69. doi: 10.2174/138920308783565714. [DOI] [PubMed] [Google Scholar]
- 28.Batista CV, Scaloni A, Rigden DJ, Silva LR, Rodrigues Romero A, Dukor R, Sebben A, Talamo F, Bloch C. A novel heterodimeric antimicrobial peptide from the tree-frog Phyllomedusa distincta. FEBS Lett. 2001;494:85–89. doi: 10.1016/s0014-5793(01)02324-9. [DOI] [PubMed] [Google Scholar]
- 29.Raimondo D, Andreotti G, Saint N, Amodeo P, Renzone G, Sanseverino M, Zocchi I, Molle G, Motta A, Scaloni A. A folding-dependent mechanism of antimicrobial peptide resistance to degradation unveiled by solution structure of distinctin. Proc Natl Acad Sci U S A. 2005;102:6309–6314. doi: 10.1073/pnas.0409004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cirioni O, Ghiselli R, Orlando F, Silvestri C, De Luca S, Salzano AM, Mocchegiani F, Saba V, Scalise G, Scaloni A, Giacometti A. Efficacy of the amphibian peptide distinctin in a neutropenic mouse model of staphylococcal sepsis. Crit Care Med. 2008;36:2629–2633. doi: 10.1097/CCM.0b013e318184430d. [DOI] [PubMed] [Google Scholar]
- 31.Dalla Serra M, Cirioni O, Vitale RM, Renzone G, Coraiola M, Giacometti A, Potrich C, Baroni E, Guella G, Sanseverino M, De Luca S, Scalise G, Amodeo P, Scaloni A. Structural features of distinctin affecting peptide biological and biochemical properties. Biochemistry. 2008;47:7888–7899. doi: 10.1021/bi800616k. [DOI] [PubMed] [Google Scholar]
- 32.Resende JM, Moraes CM, Munhoz VH, Aisenbrey C, Verly RM, Bertani P, Cesar A, Pilo-Veloso D, Bechinger B. Membrane structure and conformational changes of the antibiotic heterodimeric peptide distinctin by solid-state NMR spectroscopy. Proc Natl Acad Sci U S A. 2009;106:16639–16644. doi: 10.1073/pnas.0905069106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 34.Nevzorov AA, Opella SJ. Selective averaging for high-resolution solid-state NMR spectroscopy of aligned samples. J Magn Reson. 2007;185:59–70. doi: 10.1016/j.jmr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Mo Y, Cross TA, Nerdal W. Structural restraints and heterogeneous orientation of the gramicidin A channel closed state in lipid bilayers. Biophys J. 2004;86:2837–2845. doi: 10.1016/S0006-3495(04)74336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolodziejski W, Klinowski J. Kinetics of cross-polarization in solid-state NMR: a guide for chemists. Chem Rev. 2002;102:613–628. doi: 10.1021/cr000060n. [DOI] [PubMed] [Google Scholar]
- 37.Nevzorov AA, Park SH, Opella SJ. Three-dimensional experiment for solid-state NMR of aligned protein samples in high field magnets. J Biomol NMR. 2007;37:113–116. doi: 10.1007/s10858-006-9121-y. [DOI] [PubMed] [Google Scholar]
- 38.Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and Topology of Monomeric Phospholamban in Lipid Membranes Determined by a Hybrid Solution and Solid-State NMR Approach. Proc Natl Acad Sci U S A. 2009;106:10165–70. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi L, Traaseth NJ, Verardi R, Cembran A, Gao J, Veglia G. A refinement protocol to determine structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state NMR restraints. J Biomol NMR. 2009;44:195–205. doi: 10.1007/s10858-009-9328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denny JK, Wang J, Cross TA, Quine JR. PISEMA powder patterns and PISA wheels. J Magn Reson. 2001;152:217–226. doi: 10.1006/jmre.2001.2405. [DOI] [PubMed] [Google Scholar]
- 41.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J Magn Reson. 2000;144:150–5. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesleh MF, Lee S, Veglia G, Thiriot DS, Marassi FM, Opella SJ. Dipolar waves map the structure and topology of helices in membrane proteins. J Am Chem Soc. 2003;125:8928–8935. doi: 10.1021/ja034211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesleh MF, Veglia G, DeSilva TM, Marassi FM, Opella SJ. Dipolar waves as NMR maps of protein structure. J Am Chem Soc. 2002;124:4206–4207. doi: 10.1021/ja0178665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mascioni A, Veglia G. Theoretical analysis of residual dipolar coupling patterns in regular secondary structures of proteins. J Am Chem Soc. 2003;125:12520–12526. doi: 10.1021/ja0354824. [DOI] [PubMed] [Google Scholar]
- 45.Sanders CR, Oxenoid K. Customizing model membranes and samples for NMR spectroscopic studies of complex membrane proteins. Biochim Biophys Acta. 2000;1508:129–45. doi: 10.1016/s0005-2736(00)00308-4. [DOI] [PubMed] [Google Scholar]
- 46.Sanders CR, Hare BJ, Howard KP, Prestegard JH. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Progress in Nuclear Magnetic Resonance Spectroscopy. 1994;26:421–444. [Google Scholar]
- 47.Gregory SM, Cavenaugh A, Journigan V, Pokorny A, Almeida PF. A quantitative model for the all-or-none permeabilization of phospholipid vesicles by the antimicrobial peptide cecropin A. Biophys J. 2008;94:1667–1680. doi: 10.1529/biophysj.107.118760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yandek LE, Pokorny A, Floren A, Knoelke K, Langel U, Almeida PF. Mechanism of the cell-penetrating peptide transportan 10 permeation of lipid bilayers. Biophys J. 2007;92:2434–2444. doi: 10.1529/biophysj.106.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]


