Summary
Association studies of over 1 million SNPs capturing most of the human genome common variation became possible thanks to the information provided by the HapMap International project and the development of high-throughput genotyping technologies at accessible prices. Genome-wide scans analyzing thousands of individuals have now identified most if not all of the major genes involved in susceptibility for several systemic autoimmune diseases. In particular, results for rheumatoid arthritis (RA), systemic lupus erythematosus (SLE) and systemic sclerosis (SSc) are reviewed here. While most genes are shared between diseases, few seem to be unique reflecting that we still are long before knowing all genes, their interactions with other genes and the environment and their impact on biological functions.
Introduction
Previous studies on the genetics of complex diseases used small sample sizes leading to inconclusive results, with the exception of the strong genetic association between genes within the major histocompatibility complex (MHC) and the various autoimmune diseases. While GWAS have provided some surprises, they have also confirmed a few of the old results, but not all. The most important genes identified previously, IRF5 and STAT4 for SLE and PTPN22 and STAT4 for RA stand [1]. In this review we focus only on those genes identified in the last two to three years. As GWAS with extensive coverage have been used, except for a few examples (PDCD1 is a relevant one, as SNPs within this gene are lacking in the GWAS arrays), most of the genome has been screened with thousands of samples providing an overall picture of genetic susceptibility. We present only those genes that have been clearly replicated and whose role in genetic susceptibility is beyond doubt or the result of powerful GWAS with replication using large sample sizes.
Genetics of SLE
SLE is considered as a prototypic autoimmune disease, characterized by production of antinuclear autoantibodies, immune complex deposition and subsequent multiple organ damage.
The understanding of the genetic basis of SLE has expanded enormously over the past couple of years, driven principally by technological advances and the assessment of six genome-wide association studies (GWAS) in the last two years, in Caucasians and Asians [2–7].
The great majority of the identified genes are involved in innate and adaptive immune responses. The convincingly associated genes, as summarized in Table 1, are mainly implicated in immune-complex (IC) processing, T/B cell signaling and/or and Toll-like receptor (TLR) and type I interferon signaling. The strongest association in the GWAS era is undoubtedly the MHC region, but because it consists of strongly correlated polymorphisms in a ~6Mb region involving more than 400 genes it remains a challenge. Recent observations by Fernando, et al have shown at least two independent genetic effects within the MHC region in SLE: one signal is provided by the HLA-DRB1*0301 allele, and the second within the class III region specifically detecting the 6th intron of the SKIVL2 gene [8]. A more recent study genotyping over 1400 variants within the MHC region and over 10,000 individuals with various autoimmune diseases including SLE and RA showed the top disease-specific signal for SLE to be SNP [8,9] located between TNXB and CREBL1 and the HLA DRB1*0301 [9].
Table 1.
Susceptibility Genes for SLE
| Chromosome | Gene | SNPs | Population | References |
|---|---|---|---|---|
| 6p21 | HLA region | DRB1*0301 and several other alleles | European, several Asian, African-American, mixed European-Amerindian | [8,9] |
| 7q32 | IRF5 | 5bp promoter indel, rs2004640, rs2070197, rs10954213 | European, several Asian, mixed European-Amerindian, African-American | [3–5,7,20] |
| 2q32 | STAT4 | rs7574865, rs3821236, rs7601754 | European, mixed European-Amerindian, several Asian, African-American | [3–5,7,20] |
| 6q23 | TNFAIP3 | rs5029939, rs2230926 | European, Asian, African- American | [2–5,7,17,20] |
| 16p11 | ITGAM | rs9888739, rs1143679, rs4548893 | European, mixed European-Amerindian, Asian, African-American | [3–5,7,10,20] |
| 4q24 | BANK1 | rs10516487, rs17266594, rs3733197 | European, European-Amerindian, Asian | [3,6,7] |
| 1p13 | PTPN22 | rs2476601 | European | [4] |
| 8p23 | BLK | rs13277113, rs2736340 | European, several Asian | [3–5,7,20] |
| 2q37 | PDCD1 (CD279) | PD1.3A | European, European-Amerindian, Chinese | [49] |
| 1q25 | TNFSF4 | Risk haplotype; rs3850641 | European, Asian | [3,7,20] |
| 18q22.3 | CD226 | rs763361, rs727088 | European, European-Amerindian | [13] |
| 1q21-23 | FCGR2A | ARG131HIS | European, European-Amerindian, African American | [4,5,20] |
| 19p13.2 | TYK2 | rs280519 | European | [20] |
| 3p21.3 | TREX1 | rs72556554, R114H and other 11 nonsynonimous substitutions | European | [16] |
| Xq28 | MECP2-IRAK1 | rs2269368 | European, Chinese, Korean, European-Amerindian (Mexican) | [20] |
| 3p14.3 | PXK | rs6445975, rs2176082 | European | [4,20] |
| 2q24 | IFIH1 | rs1990760 | European | [20] |
| 11p15.5 | KIAA1542 (PHRF1) | rs4963128 | European | [4] |
| 8p23.1 | XKR6 | rs6985109 | European | [4] |
| 6q21 | ATG5-PRMD1 | rs6568431, rs2245214 | European, Chinese | [4,20] |
| 22q11.2 | UBE2L3 | rs5754217 | European, Chinese | [2,20] |
| 5q33.3 | PTTG1 | rs2431099 | European | [2,20] |
| 6p21 | UHRF1BP1 | rs11755393 | European | [20] |
| 5q32 | TNIP1 | rs7708392 | European, Chinese, Thai | [3,20] |
| 7p15.2 | JAZF1 | rs849142 | European | [20] |
| 7p21.3 | ICA1 | rs10156091 | European | [4,20] |
| 1q24 | IL10 | rs3024505 | European | [20] |
| 1q25.3 | NMNAT2 | rs2022013 | European, Chinese | [3,4] |
| 11q23.3 | ETS1 | rs6590330 | Chinese, Thai | [3,7] |
| 10q11.23 | WDFY4 | rs877819 | Chinese, Thai | [3,7] |
| 7p12.2 | IKZF1 | rs4917014 | Chinese | [3] |
| 12q24.32 | SLC15A4 | rs10847697, rs1385374 | Chinese | [3] |
| 2p22.3 | RASGRP3 | rs13385731 | Chinese | [3] |
Impaired IC clearance and deposition is an important pathological aspect in SLE. Susceptibility genes with important roles in IC processing known from previous studies are the FcGR family of genes and more recently ITGAM [10] coding for the surface antigen CD11b (or CR3). Signal transduction in immune cells, particularly T and B cells, is another pathway that has revealed to contain multiple lupus susceptibility genes, modulating T cell signaling such as TNFSF4 (OX40L) [11,12] and CD226 on NKT cells [13]. More recently BANK1 and BLK, thought to be involved in B cell activation and tolerance [5,6], respectively have been now clearly established [3,14,15].
One pathway that has been biologically and genetically strongly related to SLE pathogenesis is the type I interferon (IFN) pathway. Several genes for factors upstream and downstream of IFN production, such as IRF5, STAT4, and more recently TNFAIP3, TYK2, and TREX1, have been associated to susceptibility to SLE [2,16,17]. IRAK1 has been an interesting candidate but it is closely linked to MECP2, a gene that can regulate expression of IRAK1 also associated. It has proven as yet impossible to discern the genetic effects between these X chromosome genes [18–20]. TREX1 is mainly represented by rare but penetrant and mutations leading to high levels of type I interferon found in few patients with lupus, suggesting a potentially important role of rare variants that have remained undetected with the use of common variation mapping.
Other recently identified loci, such as PXK, XKR6 and KIAA1542 close to IRF7 [4], with no known function or correlation to SLE pathology, have the potential to lead to the discovery of novel pathways involved in SLE. It is unclear if the genetic association of KIAA1542 indeed represents an association with IRF7.
Finally, a very large replication study identified and replicated several genes among which are JAZF1, TNIP1, PRDM1 (or BLIMP1), UHRF1BP1, PTTG1 [2], UBE2L3 [2], IL10, IL21 and the IL21R [20]. Importantly, several other genes were also confirmed in this study such as ATG5, ICA1 and NMNAT2 found in a previous GWAS from the SLEGEN consortium [4]. ATG5 is an important component of the autophagy pathway. JAZF1 and UHRF1BP1 are transcription factors, while TNIP1 interacts with TNFAIP3, its function in regulating TNFAIP3 is not known. TNFAIP3 (or A20) regulates inflammation by turning off NFκB through polyubiquitination and degradation.
Studies in Asian populations have identified new susceptibility genes for lupus and replication of the hitherto identified genes in Europeans has revealed the presence of some genes but not others [3,7,14]. Two genes were clearly identified, ETS1 and WDFY4 [3,7]. ETS1 is involved in the development of TH17 cells while WDFY4 codes for a protein of unknown function. STAT4, IRF5, BANK1, BLK, TNFAIP3 and TNFSF4 have been confirmed in Asians [3,7]. Studies on African Americans and European-Amerindian admixed populations are ongoing.
Genetics of RA
Multiple GWAS have corroborated the MHC genes as major genetic contributors to the risk of developing RA [21–25]. Within the MHC, the strongest contribution to risk is given by the HLA-DRB1 gene, which codes for the third hypervariable region of the HLA-DR molecule β chain. Since the original report of Peter Stastny, diverse HLA-DRB1 alleles have been associated in European, Asian, African and European-Ameridian populations [26]. All the associated alleles were unified by Peter Gregersen under the hypothesis of the shared epitope (SE). SE alleles are associated with anti-citrulline antibody production, a major biomarker for RA and determine severity. A study analyzing over 1400 SNPs within the HLA regions from the IMAGEN Consortium found the peak association for RA between the gene BTNL2 and HLA-DRA (SNP rs2395175) and the allele for the DQA1 gene DQA1*0301 [9]. Dense typing of the MHC have revealed several DRB1-independent associations, including a signal at MICA, one in the border between class I and class III region and some in the class I region [27].
Although any other association outside the MHC is rather modest in RA, and the SE accounts for 18 to 37% of the genetic heritability [28], fine mapping of candidate non-MHC linkage regions successfully identified important susceptibility genes such as PTPN22 (1p13), PADI4 (1p36) and STAT4 (2q32). To date, seven GWAS conducted in collections of thousands of patients with RA and healthy controls reliably detect several new susceptibility genes [21–23,25,29–31] (Table 2). The loci supported by the best evidence are the TRAF1-C5 [22,24,30] and the 6q23 region [23–25,29], both with strongest effect in anti-CCP+ patients. GWAS studies have also confirmed the association of PTPN22 and STAT4, as well as previously reported candidate genes identified by studies that did not have enough power such as CTLA4 and CD40. The maximum power has been achieved by a recent meta-analysis of GWAS conducted in a total of 12,307 patients and 28,975 controls of European ancestry [24]. This allowed the identification and confirmation of IL6ST, SPRED2, RBPJ, CCR6 [25], IRF5 and PXK [24]. While SPRED2 has been found to be a negative regulator of the Ras-ERK cascade, IL6ST and CCR6 are inflammation regulators and RBPJ is a transcription factor important in dendritic cell function.
Table 2.
Susceptibility Genes for RA
| Chromosome | Gene | SNPs | Population | Reference |
|---|---|---|---|---|
| 6p21.32 | HLA-DRB1 | rs615672, rs660895, rs64576200, rs6910071, rs13192471 | European, Japanese | [21–25] |
| 1p13.2 | PTPN22 | rs6679677, rs2476601 | European | [21–24,30] |
| 2q32.3 | STAT4 | rs7574865 | European, Japanese | [24,25] |
| 9q34 | TRAF1-C5 | rs3761847, rs881375 | European | [22,24,30] |
| 6q23.3 | TNFAIP3, OLIG3 | rs10499194, rs6920220 | European, Japanese | [23–25,29] |
| 6p21.32 | HLA-DQA1, HLA-DQA2 | rs6457617 | European | [31] |
| 18q23 | SALL3 | rs2002842 | European | [31] |
| 20q13.12 | CD40 | rs4810485 | European | [23,24] |
| 9p13.3 | CCL21 | rs2812378 | European | [23] |
| 12q13.3 | KIF5A, PIP4K2C | rs1678542 | European | [23] |
| 1p36.32 | TNFRSF14 | rs3890745 | European | [23,24] |
| 10p15.1 | PRKCQ | rs4750316 | European | [23,24] |
| 7q21.2 | CDK6 | rs42041 | European | [23] |
| 2p16.1 | REL | rs13017599, rs13031237 | European | [24,30] |
| 2q33.2 | CTLA4 | rs231735, rs3087243 | European | [24,30] |
| 8p23.1 | BLK | rs2736340 | European | [30] |
| 2q11.2 | AFF3 | rs11676922, rs10865035 | European | [24] |
| 5q11.2 | ANKRD55, IL6ST | rs6859219 | European | [24] |
| 14q24.3 | BATF | rs7155603 | European | [24] |
| 5q21.1 | C5orf30 | rs26232 | European | [24] |
| 9p13.3 | CCL21 | rs951005 | European | [24] |
| 6q27 | CCR6 | rs3093023, rs3093024 | Japanese | [24,25] |
| 1q24.2 | CD247 | rs840016 | European | [24] |
| 17q12 | IKZF3 | rs2872507 | European | [24] |
| 4q27 | IL2, IL21 | rs13119723 | European | [24] |
| 10p15.1 | IL2RA | rs706778 | European | [24] |
| 7q32.1 | IRF5 | rs10488631 | European | [24] |
| 15q23 | KIF3 | rs17374222 | European | [24] |
| 1p34.3 | POU3F1 | rs12131057 | European | [24] |
| 3p14.3 | PXK | rs13315591 | European | [24] |
| 4p15.2 | RBPJ | rs874040 | European | [24] |
| 12q24.12 | SH2B3 | rs3184504 | European | [24] |
| 2p14 | SPRED2 | rs934734 | European | [24] |
| 21q22.3 | UBASH3A | rs11203203 | European | [24] |
It is important to note that major differences across racial groups have been noticed; for example, PTPN22 and CTLA4 were associated in Europeans whereas PADI4 and SLC22A4 are confirmed only in Asian population groups. In contrast, the STAT4 association is valid for European, Asian, and European-Amerindian but not African Americans [32]. All these findings add evidence of the complexity and the heterogeneity of the genetic basis of RA and justify the study of diverse populations. One GWAS has been performed in Japanese [25] but none in admixed populations of European-Amerindian or African American. The observed heterogeneity highlights the importance of conducting well-powered GWAS in non-European populations in order to dissect all the genetic contribution to the disease.
Genetics of Systemic Sclerosis
Systemic sclerosis (SSc) or scleroderma is an autoimmune disease characterized by an extensive fibrotic process that affects multiple organs and tissues. Until now genetic studies have not been particularly successful in the identification of risk factors for SSc. The controversial results found for the majority of genes, such as, PTPN22, CTGF or TGF-β, suggest that those studies were often limited by small sample size and clinical heterogeneity. Some were finally replicated when large sample sizes were used (Table 3). One of the first discoveries was the contribution, as in other autoimmune diseases, of HLA class II genes, which seem to be predominantly associated with the presence of specific autoantibodies rather than with SSc itself. These associations have been confirmed in a recent GWAS in SSc in the Korean population [33]. The first GWAS in a sample of European ancestry including 2,296 SSc patients and 5,171 controls has firmly established the role of the HLA genes in SSc [34]. STAT4 and IRF5 that had previously been identified as risk factors for SSc through candidate gene studies [35,36], were also identified in the GWAS of European patients with SSc [34]. The consistent association of these genes with SSc susceptibility provides compelling evidence that variation in genes with key functions in the innate immune system are involved in the pathogenesis of the disease.
Table 3.
Susceptibility Genes for SSc
| Chromosome | Gene | SNPs | Population | Reference |
|---|---|---|---|---|
| 6p21.32 |
HLA-DPB1, DPB2 HLA-DQB1 |
rs3128930, rs7764491, rs7763822, rs3128965, rs3117230, rs7763822, rs7764491, rs3117230, rs3128965 rs6457617 |
Korean, European European |
[33] [33] |
| 2q32.3 | STAT4 | rs7574865, rs11889341, rs8179673, rs10181656, rs3821236 | European, Japanese | [44] |
| 7q32 | TNPO3-IRF5 | rs2004640, rs2280714, rs10954213, rs10488631, rs12537284, rs4728142 | European, Japanese | [35,36] |
| 4q24 | BANK1 | rs10516487, rs17266594 | European | [41] |
| 8p23.1 | C8orf13-Blk | rs2736340, rs13277113 | European | [42,43] |
| 17q21.32 | TBX21 | rs11650354, rs17699436 | European | [[44] |
| 1q25.1 | TNFSF4 | rs1234314, rs2205960, rs844644, rs844648 | European | [47] |
| 10q24 | FAS | rs1800682 (G-670A) | European | [45,46] |
| 6q23.1 | CTGF | rs6918698 (G-945C), rs9399005 | European, Japanese | [37–40] |
| 1q22-23 | CD247 | rs2056626 | European | [34] |
The identification of the association of connective tissue growth factor (CTGF) gene with risk to SSc provides one of the most conflicting results to date. The potential functional −945 G allele of CTGF gene was primarily associated with susceptibility to SSc in Europeans and Japanese [37]. However, three additional studies in Europeans failed to replicate the association [38–40]. A recent study suggests that variant rs9399005 in the 3′UTR region of the CTGF gene is associated with both subtypes of SSc [39]. The lack of replication of the CTGF polymorphisms in three large cohorts of SSc suggests that CTGF may not be a strong genetic determinant for SSc susceptibility.
Genes involved in B cell receptor signaling contribute to SSc susceptibility. Association with diffuse SSc (dcSSc) has been identified with BANK1 [41]. The C8orf13-BLK region has been associated in European and Japanese patients with SSc [42,43]. These findings suggest an important role of B-cells in the pathogenesis of SSc.
A large multicenter study in Europeans found two polymorphisms (rs11650354 and rs17699436) in the TBX21 gene associated with SSc [44]. In addition, they also showed a gene-gene interaction between the TBX21 and STAT4.
Some others genes that appear to play a role in susceptibility to SSc are FAS, TNFSF4 and CD247. The promoter rs1800682 polymorphism in the FAS gene is a confirmed susceptibility variant for SSc in different populations [45,46]. In addition, the SLE susceptibility gene TNFSF4 [11,12,14], was associated with SSc in a large case-control study [47]. The CD247 gene, which encodes the T-cell receptor zeta (CD3ζ) subunit, was a new susceptibility gene for SSc in a GWAS and this association has been confirmed in an independent cohort [34].
Conclusions
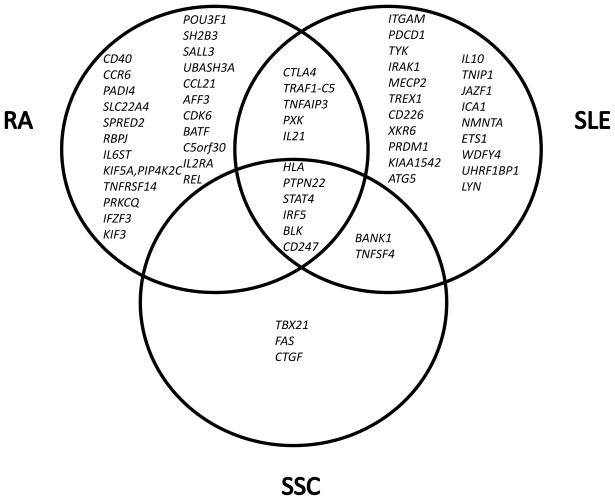
The genetics results deriving from GWAS are just the beginning of a new era of research, but new insight on disease genetics has been acquired. First, the risk alleles identified for these diseases explain only between 5–15% of the whole genetic contribution to disease and with odds ratios ranging from 1.01 to 2.4 at the most. Clearly, gene-gene interactions, gene-environment interactions and other genomic structural variation such as copy number variation, the role of rare variants and epigenetics need to be adressed. In this regard, the recent identification of rare, but highly penetrant mutations in the sialic acid acetylestarase gene (SIAE) involved in SLE and RA susceptibility and with an important functional impact in the gene [48] suggests that rare variants may have an important role not yet fully comprehended. Second, most autoimmune diseases, in particular systemic autoimmune diseases share several susceptibility genes. The differences seem to reside on the contribution of each gene in each disease. HLA alleles have been known to be different from disease to disease, but the risk alleles for non-MHC genes appear, until now to be the same. However while IRF5 and STAT4 are prominent genes in SLE, PTPN22 and TNFAIP3 are major genes in RA and CD247 in SSc. Very few genes are unique for each disease (Figure 1) and whether some genes may predominate in individuals with certain clinical manifestations is still a difficult nut to crack. Phenotyping of samples studied in genetics lack the detail required to define the correlation between clinical maifestations and genes. One possible exception is ITGAM in SLE, a unique gene for lupus importantly associated with kidney disease. Third, studies in different populations are important. Differences and similarities will lead to a comprehensive picture of genetic susceptibility, but may also pave the way to the very needed studies on gene-environmental interactions in autoimmune disease, a theme about which we know practically nothing.
Figure 1.
Unique and shared genes between SLE, RA and SSc
Genetics studies are a starting point to cell biology and immunology studies aimed at understanding disease pathogenesis and the influence of susceptibility genes on cell function. Indeed, we have challenging and exciting times ahead of us.
Acknowledgments
This work has been supported by the NIH grants AI083194, CA141700 and AR058621, the Instituto de Salud Carlos III (FIS) and the Consejería de Salud de Andalucía to MEAR. Funding from the Swedish Research Council, the Swedish Association Against Rheumatism, the King Gustaf Vth-80th Jubilee Fund and the Royal Swedish Academy of Sciences to MEAR for work supported in Sweden are greatly and sincerely acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wong M, Tsao BP. Current topics in human SLE genetics. Springer Semin Immunopathol. 2006;28:97–107. doi: 10.1007/s00281-006-0031-6. [DOI] [PubMed] [Google Scholar]
- *2.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *3.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- **4.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. This study is one of the two first GWAS fir SLE. Primarily ITGAM was identified as a major gene for lupus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **5.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. The other first GWAS in SLE. The risk polymorhisms close to the promoter of BLK and 3′ of C8orf13 (now FAM146A) correlate with expression levels of both genes. [DOI] [PubMed] [Google Scholar]
- **6.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jonsen A, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. This study identified BANK1 as a gene for lupus and also defined the functional variants involved in the risk for lupus. Two variants were identified, one leading to differences in isoform expression and a coding variant R61H substitution located in the second exon. The short isoform lacks the second exon and its expression correlated with protection. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, Hirankarn N, Ying D, Pan HF, Mok CC, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernando MM, Stevens CR, Sabeti PC, Walsh EC, McWhinnie AJ, Shah A, Green T, Rioux JD, Vyse TJ. Identification of Two Independent Risk Factors for Lupus within the MHC in United Kingdom Families. PLoS Genet. 2007;3:e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rioux JD, Goyette P, Vyse TJ, Hammarstrom L, Fernando MM, Green T, De Jager PL, Foisy S, Wang J, de Bakker PI, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A. 2009;106:18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **10.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, Chen W, Zhu C, McEver RP, Kimberly RP, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–154. doi: 10.1038/ng.71. This study identified a potential functional variant for ITGAM. In particular a coding R-H substitution is defined as the risk variants for SLE. The substitution is not conserved however and does not reside in a binding region for ITGAM (also known as CD11b or CR3) [DOI] [PubMed] [Google Scholar]
- 11.Delgado-Vega AM, Abelson AK, Sanchez E, Witte T, D’Alfonso S, Galeazzi M, Jimenez-Alonso J, Pons-Estel BA, Martin J, Alarcon-Riquelme ME. Replication of the TNFSF4 (OX40L) promoter region association with systemic lupus erythematosus. Genes Immun. 2009;10:248–253. doi: 10.1038/gene.2008.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunninghame Graham DS, Graham RR, Manku H, Wong AK, Whittaker JC, Gaffney PM, Moser KL, Rioux JD, Altshuler D, Behrens TW, et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat Genet. 2008;40:83–89. doi: 10.1038/ng.2007.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13.Lofgren SE, Delgado-Vega AM, Gallant CJ, Sanchez E, Frostegard J, Truedsson L, Garrido ED, Sabio JM, Gonzalez-Escribano MF, Pons-Estel BA, et al. A 3′UTR variant is associated with impaired expression of CD226 in T and NK T cells and susceptibility to systemic lupus erythematosus. Arthritis Rheum. doi: 10.1002/art.27677. [DOI] [PubMed] [Google Scholar]
- 14.Chang YK, Yang W, Zhao M, Mok CC, Chan TM, Wong RW, Lee KW, Mok MY, Wong SN, Ng IO, et al. Association of BANK1 and TNFSF4 with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2009;10:414–420. doi: 10.1038/gene.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito I, Kawasaki A, Ito S, Hayashi T, Goto D, Matsumoto I, Tsutsumi A, Hom G, Graham RR, Takasaki Y, et al. Replication of the association between the C8orf13-BLK region and systemic lupus erythematosus in a Japanese population. Arthritis Rheum. 2009;60:553–558. doi: 10.1002/art.24246. [DOI] [PubMed] [Google Scholar]
- 16.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- *17.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *18.Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, Alarcon-Riquelme ME, James JA, Vyse TJ, Gilkeson GS, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS ONE. 2008;3:e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19.Jacob CO, Zhu J, Armstrong DL, Yan M, Han J, Zhou XJ, Thomas JA, Reiff A, Myones BL, Ojwang JO, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2009;106:6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **20.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. A second GWAS and replication study identifying novel lupus susceptibility genes in Europeans with a large replication cohort. This study shows how large case control sets lead to identification of new susceptibility genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, Gianniny L, Korman BD, Padyukov L, Kurreeman FA, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **24.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. Study showing how the inclusion of large population sets leads to the identification of a large number of new disease loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *25.Kochi Y, Okada Y, Suzuki A, Ikari K, Terao C, Takahashi A, Yamazaki K, Hosono N, Myouzen K, Tsunoda T, et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet. 2010;42:515–519. doi: 10.1038/ng.583. [DOI] [PubMed] [Google Scholar]
- 26.Kochi Y, Suzuki A, Yamada R, Yamamoto K. Genetics of rheumatoid arthritis: underlying evidence of ethnic differences. J Autoimmun. 2009;32:158–162. doi: 10.1016/j.jaut.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Lee HS, Lee AT, Criswell LA, Seldin MF, Amos CI, Carulli JP, Navarrete C, Remmers EF, Kastner DL, Plenge RM, et al. Several regions in the major histocompatibility complex confer risk for anti-CCP-antibody positive rheumatoid arthritis, independent of the DRB1 locus. Mol Med. 2008;14:293–300. doi: 10.2119/2007-00123.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Woude D, Houwing-Duistermaat JJ, Toes RE, Huizinga TW, Thomson W, Worthington J, van der Helm-van Mil AH, de Vries RR. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009;60:916–923. doi: 10.1002/art.24385. [DOI] [PubMed] [Google Scholar]
- *29.Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, Pe’er I, Burtt NP, Blumenstiel B, DeFelice M, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, Seldin MF, Criswell LA, Plenge RM, Holers VM, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Julia A, Ballina J, Canete JD, Balsa A, Tornero-Molina J, Naranjo A, Alperi-Lopez M, Erra A, Pascual-Salcedo D, Barcelo P, et al. Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility. Arthritis Rheum. 2008;58:2275–2286. doi: 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- 32.Kelley JM, Hughes LB, Malik A, Danila MI, Edberg Y, Alarcon GS, Conn DL, Jonas BL, Callahan LF, Smith EA, et al. Genetic variants of STAT4 associated with rheumatoid arthritis in persons of Asian and European ancestry do not replicate in African Americans. Ann Rheum Dis. 69:625–626. doi: 10.1136/ard.2009.113183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Lee JE, Arnett FC, Xiong M, Park MY, Yoo YK, Shin ES, Reveille JD, Mayes MD, Kim JH, et al. HLA-DPB1 and DPB2 are genetic loci for systemic sclerosis: a genome-wide association study in Koreans with replication in North Americans. Arthritis Rheum. 2009;60:3807–3814. doi: 10.1002/art.24982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Radstake TR, Gorlova O, Rueda B, Martin JE, Alizadeh BZ, Palomino-Morales R, Coenen MJ, Vonk MC, Voskuyl AE, Schuerwegh AJ, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42:426–429. doi: 10.1038/ng.565. The first GWAS for SSc. Two large replication sets were used. Only one genes, CD247 (CD3z) was identified. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dieude P, Guedj M, Wipff J, Avouac J, Fajardy I, Diot E, Granel B, Sibilia J, Cabane J, Mouthon L, et al. Association Between the IRF5 rs2004640 Functional Polymorphism and Systemic Sclerosis. Arthritis and Rheumatism. 2009;60:225–233. doi: 10.1002/art.24183. [DOI] [PubMed] [Google Scholar]
- 36.Ito I, Kawaguchi Y, Kawasaki A, Hasegawa M, Ohashi J, Hikami K, Kawamoto M, Fujimoto M, Takehara K, Sato S, et al. Association of a Functional Polymorphism in the IRF5 Region With Systemic Sclerosis in a Japanese Population. Arthritis and Rheumatism. 2009;60:1845–1850. doi: 10.1002/art.24600. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi Y, Ota Y, Kawamoto M, Ito I, Tsuchiya N, Sugiura T, Katsumata Y, Soejima M, Sato S, Hasegawa M, et al. Association study of a polymorphism of the CTGF gene and susceptibility to systemic sclerosis in the Japanese population. Ann Rheum Dis. 2009;68:1921–1924. doi: 10.1136/ard.2008.100586. [DOI] [PubMed] [Google Scholar]
- 38.Rueda B, Simeon C, Hesselstrand R, Herrick A, Worthington J, Ortego-Centeno N, Riemekasten G, Fonollosa V, Vonk MC, van den Hoogen FH, et al. A large multicentre analysis of CTGF -945 promoter polymorphism does not confirm association with systemic sclerosis susceptibility or phenotype. Ann Rheum Dis. 2009;68:1618–1620. doi: 10.1136/ard.2008.100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granel B, Argiro L, Hachulla E, Fajardy I, Weiller PJ, Durand JM, Frances Y, Dombey AM, Marquet S, Lesavre N, et al. Association between a CTGF gene polymorphism and systemic sclerosis in a French population. J Rheumatol. 2010;37:351–358. doi: 10.3899/jrheum.090290. [DOI] [PubMed] [Google Scholar]
- 40.Gourh P, Mayes MD, Arnett FC. CTGF polymorphism associated with systemic sclerosis. N Engl J Med. 2008;358:308–309. author reply 309. [PubMed] [Google Scholar]
- 41.Rueda B, Gourh P, Broen J, Agarwal SK, Simeon C, Ortego-Centeno N, Vonk MC, Coenen M, Riemekasten G, Hunzelmann N, et al. BANK1 functional variants are associated with susceptibility to diffuse systemic sclerosis in Caucasians. Ann Rheum Dis. 2010;69:700–705. doi: 10.1136/ard.2009.118174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito I, Kawaguchi Y, Kawasaki A, Hasegawa M, Ohashi J, Kawamoto M, Fujimoto M, Takehara K, Sato S, Hara M, et al. Association of the FAM167A-BLK region with systemic sclerosis. Arthritis Rheum. 2010;62:890–895. doi: 10.1002/art.27303. [DOI] [PubMed] [Google Scholar]
- 43.Gourh P, Agarwal SK, Martin E, Divecha D, Rueda B, Bunting H, Assassi S, Paz G, Shete S, McNearney T, et al. Association of the C8orf13-BLK region with systemic sclerosis in North-American and European populations. J Autoimmun. 2010;34:155–162. doi: 10.1016/j.jaut.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gourh P, Agarwal SK, Divecha D, Assassi S, Paz G, Arora-Singh RK, Reveille JD, Shete S, Mayes MD, Arnett FC, et al. Polymorphisms in TBX21 and STAT4 increase the risk of systemic sclerosis: evidence of possible gene-gene interaction and alterations in Th1/Th2 cytokines. Arthritis Rheum. 2009;60:3794–3806. doi: 10.1002/art.24958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liakouli V, Manetti M, Pacini A, Tolusso B, Fatini C, Toscano A, Cipriani P, Guiducci S, Bazzichi L, Codullo V, et al. The −670G>A polymorphism in the FAS gene promoter region influences the susceptibility to systemic sclerosis. Ann Rheum Dis. 2009;68:584–590. doi: 10.1136/ard.2008.088989. [DOI] [PubMed] [Google Scholar]
- 46.Broen J, Gourh P, Rueda B, Coenen M, Mayes M, Martin J, Arnett FC, Radstake TR. The FAS −670A>G polymorphism influences susceptibility to systemic sclerosis phenotypes. Arthritis Rheum. 2009;60:3815–3820. doi: 10.1002/art.24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gourh P, Arnett FC, Tan FK, Assassi S, Divecha D, Paz G, McNearney T, Draeger H, Reveille JD, Mayes MD, et al. Association of TNFSF4 (OX40L) polymorphisms with susceptibility to systemic sclerosis. Ann Rheum Dis. 2010;69:550–555. doi: 10.1136/ard.2009.116434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Surolia I, Pirnie SP, Chellappa V, Taylor KN, Cariappa A, Moya J, Liu H, Bell DW, Driscoll DR, Diederichs S, et al. Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature. 466:243–247. doi: 10.1038/nature09115. The discovery that the gene SIAE, an enzyme that negatively controls B cell signaling, is related to autoimmunity. Eight homozygous mutations were foud in patients with rheumatoid arthritis and lupus and several heterozygous. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee YH, Woo JH, Choi SJ, Ji JD, Song GG. Association of programmed cell death 1 polymorphisms and systemic lupus erythematosus: a meta-analysis. Lupus. 2009;18:9–15. doi: 10.1177/0961203308093923. [DOI] [PubMed] [Google Scholar]