Abstract
Nanoparticle-based arrays have been used to distinguish a wide range of biomolecular targets through pattern recognition. In this report, we highlight new “chemical nose” methodologies that use nanoparticle systems to provide high sensitivity sensing of biomolecular targets, including fluorescent polymer/gold nanoparticle complexes that can discriminate between different bioanalytes including proteins, bacteria, and mammalian cells as well as dye-based micellar systems for the detection of clinically important metallo- and non-metallo proteins.
1. Introduction
Most biological recognition processes occur via specific interactions. However, sensory processes such as taste and smell use “differential” binding where the receptors bind to their analytes through interactions that are selective rather then specific [1,2]. These array based sensing platforms can be trained to generate a response pattern analogous to olfaction, providing versatile detectors [3]. Recently, a variety of array based sensor platforms have been developed for biomacromolecule sensing, including porphyrins [4], oligopeptide functionalized resins [5], and polymers [6,7].
Nanoparticles (NPs) feature sizes commensurate with biomacromolecules, coupled with useful physical and optical properties [8,9]. Modulation of these physicochemical properties can be readily achieved by changing of core and/or ligand structure. In this report, we highlight the recent advances of array based/chemical nose sensors using materials such as gold, dendrimer, and magnetic nanoparticles for the detection and identification of analytes such as proteins, bacteria, and cells.
2. Nanoparticle arrays for sensing proteins
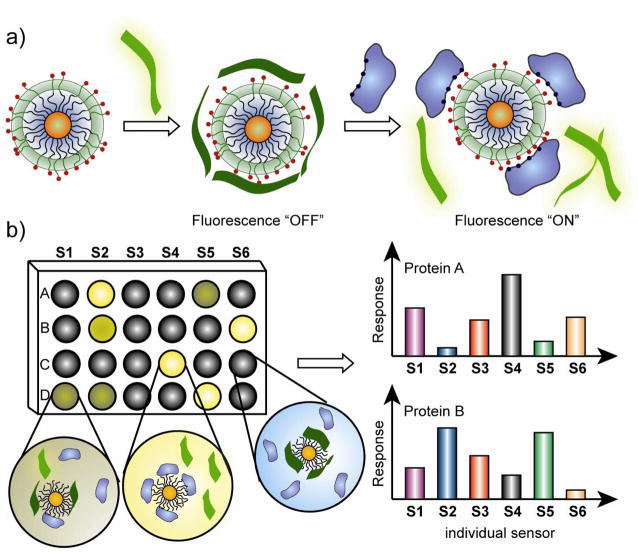
Irregular protein concentration levels in biofluids, e.g., serum, urine, and saliva, provide essential information for the early diagnosis of many pathological conditions [1,10,11,12,13,14]. Substantial efforts have been devoted to developing precise and efficient methods for protein sensing [15] including enzyme-labeled immunoassays [16], electrophoresis methods [17], and analytical techniques [18]. Detection and identification of imbalance through of an array-based sensing approach provides a promising alternative to these methods [5]. Array-based sensing approaches are complementary to more traditional immunosensing strategies (e.g. ELISA), providing versatile systems that can be “trained” to recognize analytes and potentially disease states. In 2007, Rotello et al. fabricated a sensor array composed of six cationic functionalized gold nanoparticles (AuNPs) and an anionic PPE polymer that can properly identify seven common proteins [19••]. The polymer fluorescence is quenched by gold nanoparticles; the presence of proteins disrupts the nanoparticle–polymer interaction (Figure 1a), producing distinct fluorescence response patterns (Figure 1b) based on particle-protein affinity. The effeciency of this system is attributed to both the quenching ability of AuNPs as well as the ‘molecular wire’ effect of PPE polymer [20]. Since the protein-nanoparticle interactions are determined by their respective structural features such as charged, hydrophobic, hydrophilic, and hydrogen-bonding sites [21], the differing affinities lead to a fluorescence response fingerprint pattern for individual proteins (Figure 1b). The raw data responses obtained were subjected to linear discriminant analysis (LDA) [22,23] to differentiate the fluorescence patterns of the nanoparticle–PPE systems against the different protein targets. This system showed a limit of detection of 4–215 nM depending on Mw protein and identifed correctly 52 out of 55 unknowns samples (94.5% accuracy) [19••].
Figure 1.
Schematic illustration of ‘chemical nose’ sensor array based on AuNP-fluorescent polymer conjugates. a) The competitive binding between protein and quenched polymer-AuNP complexes leads to the fluorescence light-up. b) The combination of an array of sensors generates fingerprint response patterns for individual proteins [19].
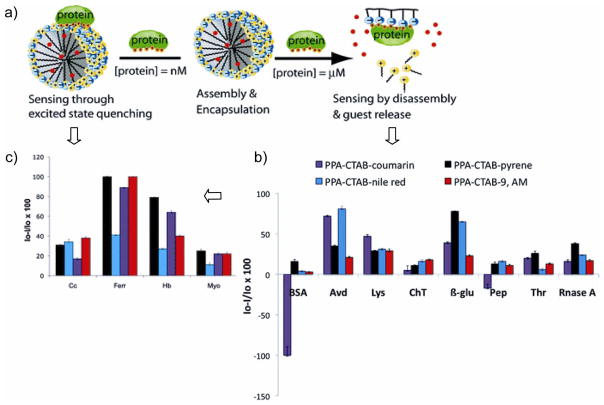
Polymeric nanoparticles provide a separate class of scaffolds for sensor design. Thayumanavan et al. developed a polymeric micellar nanosystem that responded to electronic complementarity, allowing the system to be selective for metalloproteins [24]. They used eight different fluorescence dye molecules non-covalently bound to the micellar interior of an amphiphilic homopolymer to generate a pattern that allowed the differentiation of four different metalloproteins with limits of detection of 1–200 μM. In another approach, Thayumanavan et al. reported a micellar disassembly process for transduction [25]. Five different noncovalently assembled receptors were generated, and the disassembly was studied by monitoring the encapsulated dye release in response to five different non-metalloproteins. The disassembly-induced fluorescence change of the guest molecule produces protein-specific patterns. The limit of detection in this approach was 8 μM. More recently, Thayumanavan et al. introduced a new method where the differential response was generated from a single polymer-surfactant complex with two approaches, i.e. the disassembly and guest release based pathways and photoinduced charge/energy transfer quenching (excited state quenching) (Figure 2a). By varying the transducer using non-metalloprotein and metalloproteins [26•] they were able to generate a limit of detection for non-metalloproteins of 8 μM (Figure 2b). In the case of the metalloproteins, the limit of detection was 80 nM (Figure 2c). Thayumanavan et. al also studied the use of a fluorescent anthracene-core dendrimer system that has carboxylic acid groups on the periphery that affords a differential response protein pattern though the binding energy transfer process at analyte concentrations between 1–5 μM [27]. Upon binding, an energy transfer process occurs with quenching of the fluorescent core. The interchange between quenching and binding lead to differential responses that allowed discrimination of 3 different metalloproteins.
Figure 2.
a) Schematic of the fluorescence response due to metalloproteins/nonmetalloprotein either due to the disassembly or due to the energy/electron transfer based quenching for metalloproteins. b) Analyte-dependent patterns from emission changes at 8 μM concentration of the nonmetalloprotein. c) Analyte-specific sensing patterns for metalloproteins with different dye molecules at 80 nM concentration of proteins (coumarin (10−6 M; λex = 400 nm); pyrene (10−6 M; λex = 339 nm); nile red (10−6 M; λex = 550 nm); 9-anthracene methanol (10−5 M; λex = 365 nm)). Figure reproduced with permission from reference [26].
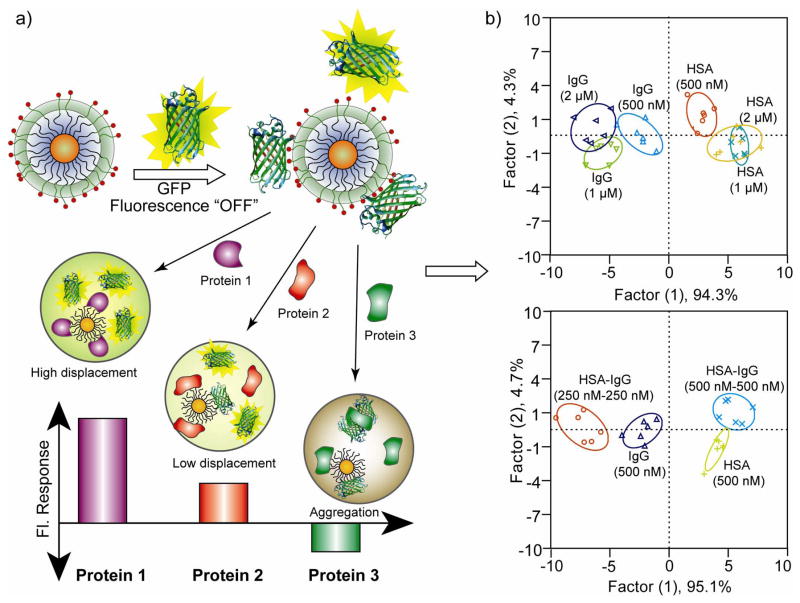
The above studies focused on protein sensing in buffer. The plasma/serum proteome, in contrast, contains more than 20,000 different proteins and an overall protein concentration of ~1 mM, (71 mg ml−1), providing a complex matrix for sensor design [28,29]. To construct a more effective protein sensing system, Rotello et al. selected green fluorescent protein (GFP) [30], a stable dimeric biomolecule used as transducer. The use of GFP minimizes aggregation, improving sensor efficiency as compared with the previous method using conjugated polymers. In this sensing process, the biocompatible AuNP/GFP conjugated array was able to identify five of the more abundant human serum proteins, i.e. human serum albumin, immunoglobulin G, transferrin, fibrinogen and α–antitrypsin, (Figure 3a) in an undiluted solution of human serum (overall protein concentration ~1 mM), obtaining a limit of detection of 500 nM [31•]. Further experiments indicated that mixtures of different proteins and the addition of one protein in different concentrations also led to a specific and reproducible change in the LDA-based patterns (Figure 3b).
Figure 3. GFP-NP sensor array.
a) Schematic illustration showing the competitive binding between proteins and quenched AuNP-GFP complexes whose aggregation leads to the fluorescence “turning on” or further quenching using a library of cationic nanoparticles. b) Discrimination of HAS and IgG at different concentrations and mixture of proteins. At the top, canonical score plot for the fluorescence patterns as obtained from LDA for HAS and IgG at different concentrations (500 nM, 1 μM, and 2 μM) with 95% confidence ellipses. At the bottom, HAS and IgG were mixed at 1:1 molar ratio with 250 nM each and 500 nM each, and added to five AuNP-GFP complexes. The canonical score plot obtained from LDA analysis were compared with those for the 500 nM individual proteins [31].
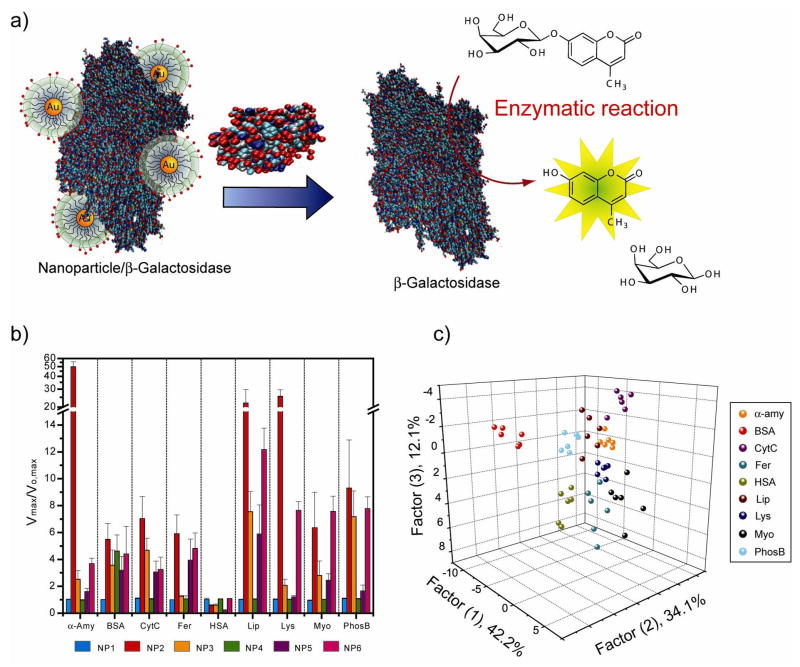
The sensitivity of fluorophore displacement strategies is limited by the inherent emissivity of the transducer. To overcome this limitation, Rotello et al. used enzyme-amplified array sensing (EAAS) to provide a platform with enhanced sensitivity [32••]. Functionalized cationic AuNPs electrostatically bind to the anionic β-Galactosidase (β-Gal), reversibly inhibiting the enzyme (Figure 4a) [33]. Displacement of the particle by analyte proteins restores β-Gal activity towards a fluorogenic substrate, generating a readout signal that is amplified through enzymatic catalysis. This EAAS system couples the signal amplification process of ELISA with the versatility of the “chemical nose” approach, as it is able to sense and identify a range of biomedically relevant proteins with a limit of detection of 1 nM in both buffer as well as in desalted urine solution (Figure 4b).
Figure 4.
A schematic representation of a sensor element in the sensor comprised of β-galactosidase (β-Gal) and cationic AuNPs and differentiation of proteins in 3-D. a) As shown, β-gal is displaced from the β-Gal/AuNP complex by protein analytes, restoring the catalytic activity of β-Gal towards the fluorogenic substrate 4-methylumbelliferyl-β-D-galactopyranoside, resulting in an amplified signal for detection. b) Differential protein pattern of the nine proteins at 1 nM. c) Canonical score plot of the first three factors of fluorescence response patterns obtained through β-Gal/AuNP sensor array against nine target proteins in 1 nM concentration [32].
3. Nanoparticle arrays for sensing bacteria and mammalian cells
The efficient detection of pathogenic microorganisms is of great importance in clinical, forensic, medical, and environmental sciences [34]. In clinical diagnostics, bacterial infections are identified by plating and culturing, a time-consuming methodology. While several newer methods including PCR, have been used to detect specific microorganisms [35], a facile wet-chemical method for the timely detection and identification of microorganisms would be of interest in both a clinical setting as well to test for food spoilage in industrial settings [36].
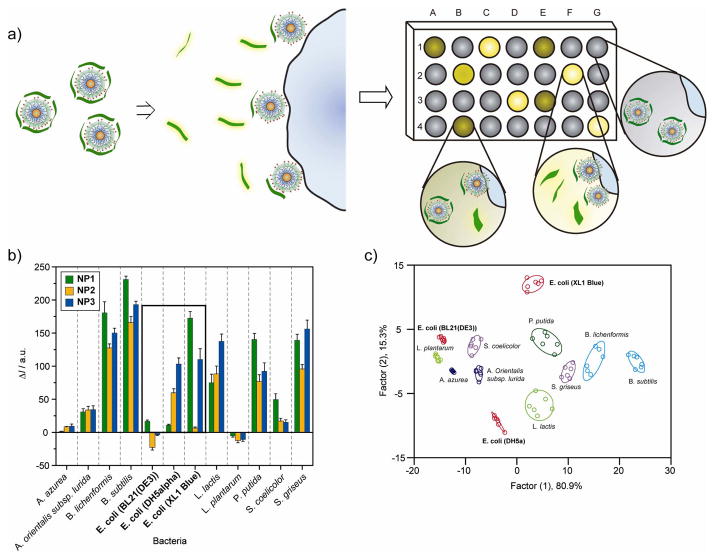
Rotello et al. created an array-based sensing system based on non-covalent conjugates of AuNPs and a modified PPE that allows the detection of bacteria within minutes [37•]. As shown in Figure 5, the anionic PPE polymer is bound initially to the functionalized cationic gold nanoparticle creating a fluorescence-quenched species. When these complexes are exposed to bacteria in solution, there is a competitive equilibrium for the positively charged AuNPs by both the polymer species and the surface of the bacteria. Depending on the interaction of bacteria with the functionalized head group, there is a differential polymer release from the surface of the AuNP whose fluorescence is restored providing a readable response. To test the efficacy of this methodology, they were able to differentiate between 12 different bacteria that contain both Gram-positive (e.g. A. azurea, B. subtilis) and Gram-negative (e.g. E. coli, P. putida) species. As shown in Fig. 5c, LDA analysis of the fluorescence responses discerns not only the species, but also betweens strain of E. coli at 2×105 cells/mL. In initial studies, they were able to successfully identify 61 out of 64 unknowns (95% accuracy) taken from the training set, showing the inherent reliability of this system.
Figure 5.
Array-based sensing of bacteria. a) Schematic diagram of the displacement assay between bacteria and the NP-PPE complex. b) Fluorescence response (ΔI) patterns of the NP-PPE sensory array against various classes of bacteria (2×105 bacteria/mL). As shown on the plot, the same strains of bacteria can also be identified. c) Canonical score plot for the first two factors of simplified fluorescence response patterns obtained with NP-PPE assembly arrays against bacteria (95% confidence ellipses shown) [37].
Recent research using nanoparticle arrays for cell surface identification has focused on the detection of cancer. Cancerous cells can be differentiated from non-cancerous ones on the basis of intracellular and extracellular (cell surface) biomarkers [38]. Cell detection based on cell surface protein biomarkers generally involves the development of specific antibodies [39,40,41]. Intracellular protein biomarkers [42] have been explored by emerging proteomic techniques, e.g. gel electrophoresis (2D-SDS-PAGE) [43] and mass spectrometry [44]. Although these lysate-based strategies provide a potential approach for cancer detection, they require both the presence and prior knowledge of intracellular biomarkers [45].
Rotello et al. have employed gold NP–fluorescent conjugated polymer constructs to differentiate normal cells from their cancerous and metastatic counterparts [46••]. Due to their cationic surfaces, the functionalized nanoparticles interact with cell surfaces through both electrostatic and hydrophobic/hydrophilic interactions. In their study, Three AuNP–PPE constructs were able to differentiate between cell types, but more significantly they are able to differentiate between isogenic cell lines are either normal, cancerous, or metastatic, e.g. CDBgeo, TD, and V14 using 20,000 cells. To lower the detection limit of these systems, they were able to develop a differential patterning array biosensor using a complex of functionalized AuNPs with green fluorescent protein (GFP) [47] that effectively identified and differentiated between several types of normal, cancerous, and metastatic isogenic mammalian cancer cells. Changing only the transducer, they have been able to achieve high sensitivity and full differentiation of the mammalian cells at concentrations of ~5000 cells, a four-fold enhancement of sensitivity relative to prior NP-polymer sensors.
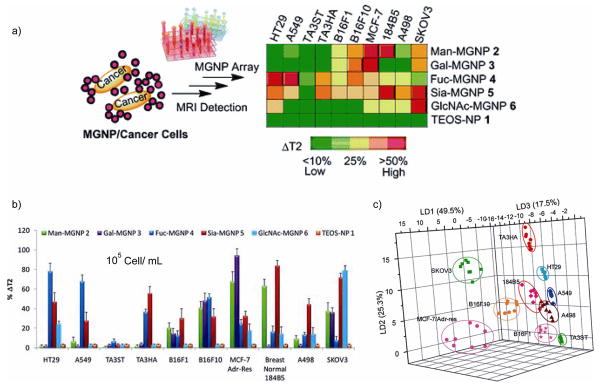
Using an alternate approach to transduction, Huang et al. reported a magnetic glyco-nanoparticle (MGNP) based nanosensor array system utilizing carbohydrates as the ligands to not only detect and differentiate cancer cells but also to quantitatively profile their carbohydrate binding abilities using magnetic resonance imaging (MRI) signatures (Figure 6) [48••]. These glyco-nanoparticles were incubated with the cell suspensions in media, and then analyzed by MRI for changes in transverse relaxation times (T2). Through this process, they were able to differentiate using LDA 10 different cell types at concentrations as low as 105 cells/mL (Figure 6c).
Figure 6.
a) Percentage changes of T2 relaxation time (%ΔT2) obtained upon incubating MGNPs 2–6 or the control NP 1 (20 μg/mL) with 10 cell lines (105 cells/mL). The ΔT2 was calculated by dividing the T2 differences between MGNP and MGNP/cancer cell by the corresponding highest ΔT2 from each MGNP category. b) LDA plots for the first three LDs of ΔT2 patterns obtained with the MGNP array upon binding with the 10 cell lines (105 cells/mL). Full differentiation of the 10 cell lines was achieved. Figure reproduced with permission from reference [48].
4. Conclusions
Based on the results presented in the review, it is apparent that nanoparticle-based sensors provide a powerful platform for analysis of proteins and cell surfaces. Through variation of sensor design it would appear that almost any system could be differentiated through appropriate sensor design. As we learn the strategies required for this differentiation, however, we will have to address the complexity of the target systems. Biofluids, such as undiluted human serum, contains a large amount of various proteins, salts, and cells that not only can inhibit or alter the sensing elements’ ability to detect target analytes, but also complicate pattern generation. Clearly, creation of clinically useful sensor will require co-evolution of chemical and data analysis strategies.
Acknowledgments
The research was supported by the National Science Foundation (NSF) Center for Hierarchical Manufacturing at the University of Massachusetts (NSEC, DMI-0531171), the NIH (GM077173 and AI073425) and the NSF IGERT (DGE-0504485) to B.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
[•] of special interest
[••] of outstanding interest
- 1.Bai VU, Kaseb A, Tejwani S, Divine GW, Barrack ER, Menon M, Pardee AB, Reddy GPV. Identification of prostate cancer mRNA markers by averaged differential expression and their detection in biopsies, blood, and urine. Proc Natl Acad Sci. 2007;104:2343–2348. doi: 10.1073/pnas.0610504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert KJ, Lewis NS, Schauer CL, Sotzing GA, Stitzel SE, Vaid TP, Walt DR. Cross-reactive chemical sensor arrays. Chem Rev. 2000;100:2595–2626. doi: 10.1021/cr980102w. [DOI] [PubMed] [Google Scholar]
- 3.Lavigne JJ, Anslyn EV. Sensing a paradigm shift in the field of molecular recognition: From selective to differential receptors. Angew Chem Int Ed. 2001;40:3119–3130. doi: 10.1002/1521-3773(20010903)40:17<3118::AID-ANIE3118>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Baldini L, Wilson AJ, Hong J, Hamilton AD. Pattern-based detection of different proteins using an array of fluorescent protein surface receptors. J Am Chem Soc. 2004;126:5656–5657. doi: 10.1021/ja039562j. [DOI] [PubMed] [Google Scholar]
- 5.Wright AT, Anslyn EV. Differential receptor arrays and assays for solution-based molecular recognition. Chem Soc Rev. 2006;35:14–28. doi: 10.1039/b505518k. [DOI] [PubMed] [Google Scholar]
- 6.Miranda OR, You CC, Phillips R, Kim IB, Ghosh PS, Bunz UHF, Rotello VM. Array-based sensing of proteins using conjugated polymers. J Am Chem Soc. 2007;129:9856–9857. doi: 10.1021/ja0737927. [DOI] [PubMed] [Google Scholar]
- 7.Bajaj A, Miranda OR, Phillips R, Kim IB, Jerry DJ, Bunz UHF, Rotello VM. Array-Based Sensing of Normal, Cancerous, and Metastatic Cells Using Conjugated Fluorescent Polymers. J Am Chem Soc. 2010;132:1018–1022. doi: 10.1021/ja9061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel MC, Astruc D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 9.You CC, Verma A, Rotello VM. Engineering the nanoparticle-biomacromolecule interface. Soft Matter. 2006;2:190–204. doi: 10.1039/b517354j. [DOI] [PubMed] [Google Scholar]
- 10.Masson JF, Battaglia TM, Khairallah P, Beaudoin S, Booksh KS. Quantitative measurement of cardiac markers in undiluted serum. Anal Chem. 2007;79:612–619. doi: 10.1021/ac061089f. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen SL, Andersen PL, Koch C, Jensenius JC, Thiel S. The level of the serum opsonin, mannan-binding protein in HIV-1 antibody-positive patients. Clin Exp Immunol. 1995;100:219–222. doi: 10.1111/j.1365-2249.1995.tb03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreasson U, Portelius E, Andersson ME, Blennow K, Zetterberg H. Aspects of beta-amyloid as a biomarker for Alzheimer’s disease. Biomarkers Med. 2007;1:59–78. doi: 10.2217/17520363.1.1.59. [DOI] [PubMed] [Google Scholar]
- 13.Daniels MJ, Wang YM, Lee MY, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 14.Abdelhafiz AH, Myint MP, Tayek JA, Wheeldon NM. Anemia, Hypoalbuminemia, and Renal Impairment as Predictors of Bleeding Complications in Patients Receiving Anticoagulation Therapy for Nonvalvular Atrial Fibrillation: A Secondary Analysis. Clin Ther. 2009;31:1534–1539. doi: 10.1016/j.clinthera.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Acharya G, Chang CL, Doorneweerd DD, Vlashi E, Henne WA, Hartmann LC, Low PS, Savran CA. Immunomagnetic diffractometry for detection of diagnostic serum markers. J Am Chem Soc. 2007;129:15824–15829. doi: 10.1021/ja073094m. [DOI] [PubMed] [Google Scholar]
- 16.Haab BB. Applications of antibody array platforms. Curr Opin Biotechnol. 2006;17:415–421. doi: 10.1016/j.copbio.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 17.McPherson RA, Pincus MR. Henry’s Clinical Diagnosis and Management by Laboratory Methods. Chapter 19 Saunders-Elsevier; 2007. [Google Scholar]
- 18.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–U710. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 19••.You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Detection and identification of proteins using nanoparticle-fluorescent polymer ‘chemical nose’ sensors. Nature Nanotech. 2007;2:318–323. doi: 10.1038/nnano.2007.99. The authors used different functional gold nanoparticles and a conjugated polymer to create a series of complex fluorescent “fingerprints” that were able to detect and identify proteins with high accuracy. This is the first time that this fashionable approach is used to detect biomolecules. [DOI] [PubMed] [Google Scholar]
- 20.Bunz UHF, Rotello VM. Gold Nanoparticle-Fluorophore Complexes: Sensitive and Discerning “Noses” for Biosystems Sensing. Angew Chem Int Ed. 2010;49:3268–3279. doi: 10.1002/anie.200906928. [DOI] [PubMed] [Google Scholar]
- 21.De M, Miranda OR, Rana S, Rotello VM. Size and geometry dependent protein-nanoparticle self-assembly. Chem Commun. 2009:2157–2159. doi: 10.1039/b900552h. [DOI] [PubMed] [Google Scholar]
- 22.SYSTAT 11.0. Systat Software; Richmond, CA 94804, USA: 2004. [Google Scholar]
- 23.Jurs PC, Bakken GA, McClelland HE. Computational methods for the analysis of chemical sensor array data from volatile analytes. Chem Rev. 2000;100:2649–2678. doi: 10.1021/cr9800964. [DOI] [PubMed] [Google Scholar]
- 24.Sandanaraj BS, Demont R, Aathimanikandan SV, Savariar EN, Thayumanavan S. Selective sensing of metalloproteins from nonselective binding using a fluorogenic amphiphilic polymer. J Am Chem Soc. 2006;128:10686–10687. doi: 10.1021/ja063544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savariar EN, Ghosh S, Gonzalez DC, Thayumanavan S. Disassembly of noncovalent amphiphilic polymers with proteins and utility in pattern sensing. J Am Chem Soc. 2008;130:5416–5417. doi: 10.1021/ja800164z. [DOI] [PubMed] [Google Scholar]
- 26•.Gonzalez DC, Savariar EN, Thayumanavan S. Fluorescence Patterns from Supramolecular Polymer Assembly and Disassembly for Sensing Metallo- and Nonmetalloproteins. J Am Chem Soc. 2009;131:7708–7716. doi: 10.1021/ja900579g. Using fluorescent transducers within an amphiphilic assembly, the authors demonstrate the ability to differentiate metallo and non-metallo proteins through pattern recognition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiwpanich S, Sandanaraj BS, Thayumanavan S. Fluorophore-cored dendrimers for patterns in metalloprotein sensing. Chem Commun. 2009:806–808. doi: 10.1039/b815263b. [DOI] [PubMed] [Google Scholar]
- 28.Anderson NL, Anderson NG. The human plasma proteome - History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 29.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects (vol 1, pg 845, 2002) Mol Cell Proteomics. 2003;2:50–50. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 30.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 31•.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Sensing of proteins in human serum using conjugates of nanoparticles and green fluorescent protein. Nature Chem. 2009;1:461–465. doi: 10.1038/nchem.334. This work not only shows sensing of proteins in a complex biofluid, but also by changing the fluorophore, one can obtain lower limits of detection for the system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Miranda OR, Chen HT, You CC, Mortenson DE, Yang XC, Bunz UHF, Rotello VM. Enzyme-Amplified Array Sensing of Proteins in Solution and in Biofluids. J Am Chem Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. The authors explored the use of an enzyme-nanoparticle conjugate as an amplifier to provide an array-based sensor with enhanced sensitivity. In addition, they demonstrated the robustness of their system using two different matrix systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De M, You CC, Srivastava S, Rotello VM. Biomimetic interactions of proteins with functionalized nanoparticies: A thermodynamic study. J Am Chem Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 34.Deisingh AK, Thompson M. Detection of infectious and toxigenic bacteria. Analyst. 2002;127:567–581. doi: 10.1039/b109895k. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maynor MS, Nelson TL, O’Sullivan C, Lavigne JJ. A food freshness sensor using the multistate response from analyte-induced aggregation of a cross-reactive poly(thiophene) Org Lett. 2007;9:3217–3220. doi: 10.1021/ol071065a. [DOI] [PubMed] [Google Scholar]
- 37•.Phillips RL, Miranda OR, You CC, Rotello VM, Bunz UHF. Rapid and efficient identification of bacteria using gold-nanoparticle - Poly(para-phenyleneethynylene) constructs. Angew Chem Int Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. Using a polymer-nanoparticle construct array, the authors were able to differentiate 12 bacteria strains including 3 bacteria of the same species. [DOI] [PubMed] [Google Scholar]
- 38.Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 39.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 40.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 41.Petty RD, Nicolson MC, Kerr KM, Collie-Duguid E, Murray GI. Gene expression profiling in non-small cell lung cancer: From molecular mechanisms to clinical application. Clin Cancer Res. 2004;10:3237–3248. doi: 10.1158/1078-0432.CCR-03-0503. [DOI] [PubMed] [Google Scholar]
- 42.Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 43.Shoshan SH, Admon A. Novel technologies for cancer biomarker discovery: humoral proteomics. Cancer Biomark. 2007;3:141–152. doi: 10.3233/cbm-2007-3305. [DOI] [PubMed] [Google Scholar]
- 44.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez-Carbayo M. Antibody arrays: Technical considerations and clinical applications in cancer. Clin Chem. 2006;52:1651–1659. doi: 10.1373/clinchem.2005.059592. [DOI] [PubMed] [Google Scholar]
- 46••.Bajaj A, Miranda OR, Kim IB, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Detection and differentiation of normal, cancerous, and metastatic cells using nanoparticle-polymer sensor arrays. Proc Natl Acad Sci. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. and reference therein. Using this strategy, the authors were able to distinguish different cell types based on the physicochemical nature of different cell surfaces. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Cell surface-based differentiation of cell types and cancer states using a gold nanoparticle-GFP based sensing array. Chem Sci. 2010;1:134–138. [Google Scholar]
- 48••.El-Boubbou K, Zhu DC, Vasileiou C, Borhan B, Prosperi D, Li W, Huang XF. Magnetic Glyco-Nanoparticles: A Tool To Detect, Differentiate, and Unlock the Glyco-Codes of Cancer via Magnetic Resonance Imaging. J Am Chem Soc. 2010;132:4490–4499. doi: 10.1021/ja100455c. Using magnetic nanoparticles bearing carbohydrate ligands, the authors detected and differentiated cancer cells through a magnetic resonance image (MRI) methodology. [DOI] [PubMed] [Google Scholar]