Abstract
Differential sensing is continuing to develop as an alternative to traditional, selective chemosensing techniques. This technique takes a cue from how the human senses of taste and smell operate in order to obtain qualitative and even quantitative data on single analytes and mixtures. Whereas classical chemosensing techniques inspired by the "lock-and-key" approach depend on the development of a selective receptor for a target analyte, pattern-based sensing depends on the development of an array of cross-reactive receptors, which produce a collection of responses upon the array’s interaction with a target analyte. This review focuses on an approach to differential sensing that diversifies synthetic receptors to be used in an array via appending combinatorial peptidic arms, metal ions, and indicators to a core binding unit.
Introduction
Differential sensing is now a prevalent technique that represents an alternative to the traditional "lock-and-key" approach to chemosensing [••1–3]. The technique surmounts the arduous process of developing highly selective receptor-indicator sensors because the necessity of selectivity is relaxed. Alternatively, the technique is based on the process of the human olfaction and gustation, which uses sensors biased toward classes of analytes [4]. The popularity of the technique is driven by the relative ease by which it can be adapted to many applications. Although numerous sensors are needed, these sensors need not be highly selective or specific for target analytes. Cross-reactive is the term commonly used to describe the sensor elements, which means that most all the individual sensors in the array respond to each analyte, but this response must have a level of variance that allows discrimination of the analytes. Evaluation of the collection of signals is done by chemometric analysis [3, ••5–8], such as principal component analysis (PCA), linear discriminant analysis (LDA), artificial neural networks (ANN), and hierarchical cluster analysis (HCA). The availability of these chemometric techniques greatly facilitates the development of differential sensing of various analytes, including peptides and proteins [9–15], sugars [16–19], ions [20–22], gases [23–25], terpenes [26], nitrated explosives [27,28], thiols [29], amines [30], and even cells [31,32]. Array sensing in combination with chemometric tools is also making possible the prediction of enantiomeric excess [33,34] of chiral compounds.
A major challenge normally besetting the development of an array sensing protocol is the production of sensing elements that have the ability to distinguish among structurally similar analytes, in simple, as well as complex solutions. As array sensing is developing, various approaches to array development are now being devised. Herein, we delineate one of our group’s approaches for the production of sensing elements in pattern-based sensing.
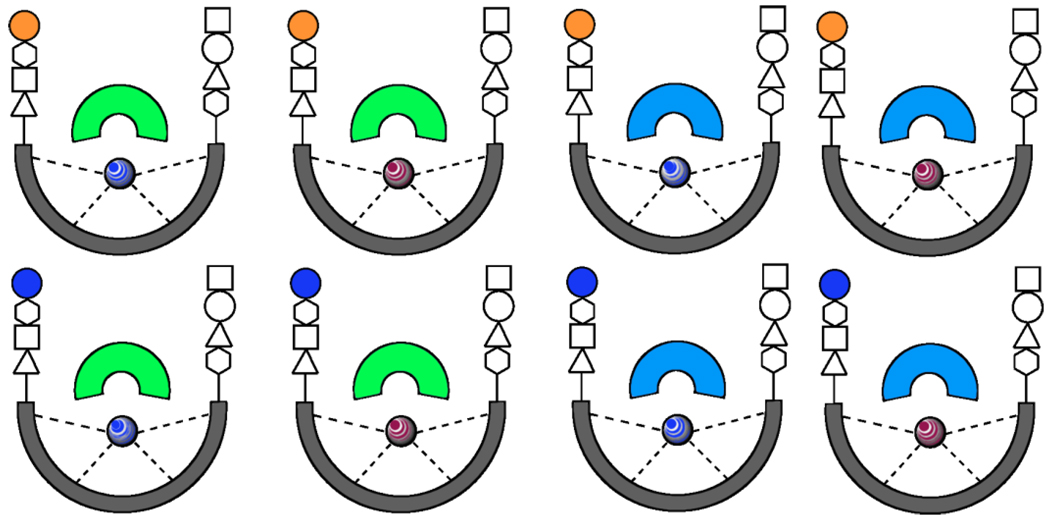
Although in pattern-based sensing it is generally viewed that synthetic receptors need not have a high degree of specificity or selectivity for target analytes, some degree of affinity and selectivity must be imparted to the synthetic receptors during their design [35,36]. Our group’s approach towards this goal involves employing a core unit, which binds the analyte class, while imparting differential binding by covalently or non-covalently appending variable moieties, such as peptidic chains, metal centers, as well as pH indicators, to the core unit (Figure 1). The inspiration for this approach derives from work by our group on the development of receptors designed through combinatorial chemistry [•37].
Figure 1.
Representation of the library of synthetic molecular receptors diversified for array sensing. The core unit (gray cup), which may be preorganized by means of a metal ion chelation (colored marbles), binds analytes of the same compound class. Diversity, and therefore cross-reactivity, in the array is made possible by the use of various indicators (green and blue inverted cup) and covalently appended moieties such as peptide chains (series of shapes). The array shown is a minimum array composed of a core binding unit × 2 metal ions × 2 indicators × 2 peptidic chains.
Nucleotide phosphate differentiation
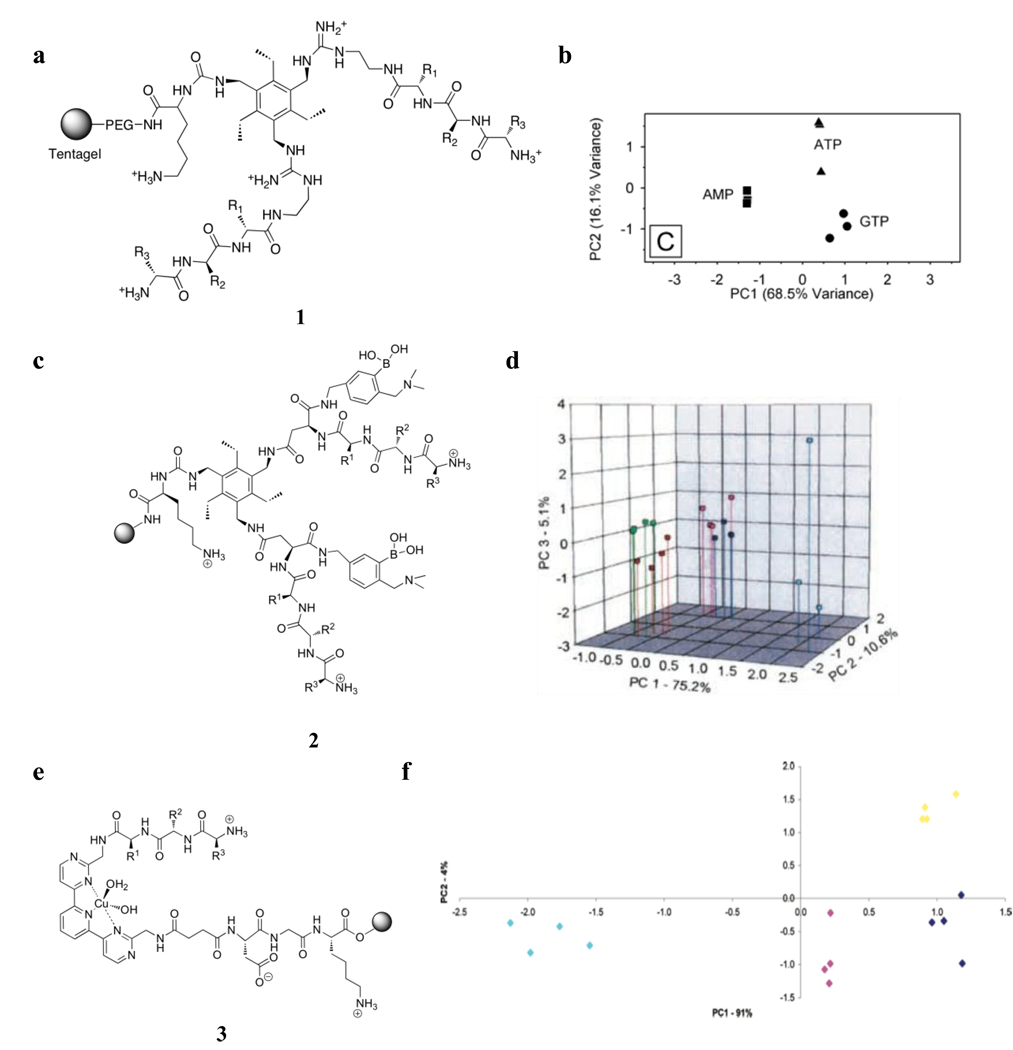
Our modular approach was first illustrated in the differentiation of nucleotide phosphates [38]. Here, the receptors were composed of a rationally designed, hexasubstituted aryl core covalently decorated with guanidinium groups that ensure binding of nucleotide triphosphates (1, Figure 2a). Differential binding by the array was achieved by appending combinatorially generated tripeptide chains. Randomly chosen members of the 4913-member receptor library were placed into a chip-based array platform, and were used in the differentiation of ATP, GTP, and AMP, via an indicator displacement assay [39] as a signaling protocol. The array of receptors showed excellent differentiation (Figure 2b) of the nucleotides by principal component analysis (PCA), and the receptors which contributed highly to this differentiation were found to contain amino acid residues that were consistent with those found in screening of the same library in a previous study [•37].
Figure 2.
(a) Receptor library, 1, used in the differentiation of ATP, GTP, and AMP. (b) Principal component analysis (PCA) score plot showing the separation of responses. (c) Receptor library, 2, used for the discrimination of some proteins. (d) PCA score plot of the response from the arrayed indicator uptake assay of the different proteins (red – lysozyme, green – elastin, blue – ovalbumin, fuschia – fetuin, teal – BSA). (e) Receptor library, 3, used for the discrimination of tripeptides and tripeptide mixtures. (f) PCA score plot of the response from the arrayed indicator uptake assay of the different peptides (light blue – HGT, pink – HKT, dark blue –HGT, yellow – GHT).
Differentiation of peptides, proteins, and phosphorylated peptides
We took advantage of the same approach to differentiate glycoproteins [40]. However, to increase the affinity of the receptor for glycopeptides and to ensure sugar binding, the peptidic arms of the receptor were functionalized with boronic acid groups (2, Figure 2c). The peptidic arms, once again, served to impart differential binding. The receptor library was found to differentiate ovalbumin, fetuin, lysozyme, and BSA. PCA of the responses from the different proteins in a microarray chip platform, using an indicator uptake assay as the signaling protocol, yielded good separation of the different proteins (Figure 2d).
A different core unit was employed in the discrimination of some simple tripeptides and tripeptide mixtures (3, Figure 2e) [•41]. A tridentate core that chelates Cu(II) was appended with two variable peptidic arms to increase the affinity for the peptide targets. Four tripeptides HKT, HET, HGT, and GHT as well as three mixtures containing two of the tripeptides, were used to test the discrimination by an array composed of 30 randomly chosen members of the receptor library. The resin-bound receptors were placed in a microarray chip and used in an indicator uptake assay using Orange G as dye. PCA of the responses, derived from CCD images of the array, showed clustering of the responses from the tripeptides (Figure 2f). However, the majority of variance is carried along only one axis, indicating little cross-reactivity of the receptors.
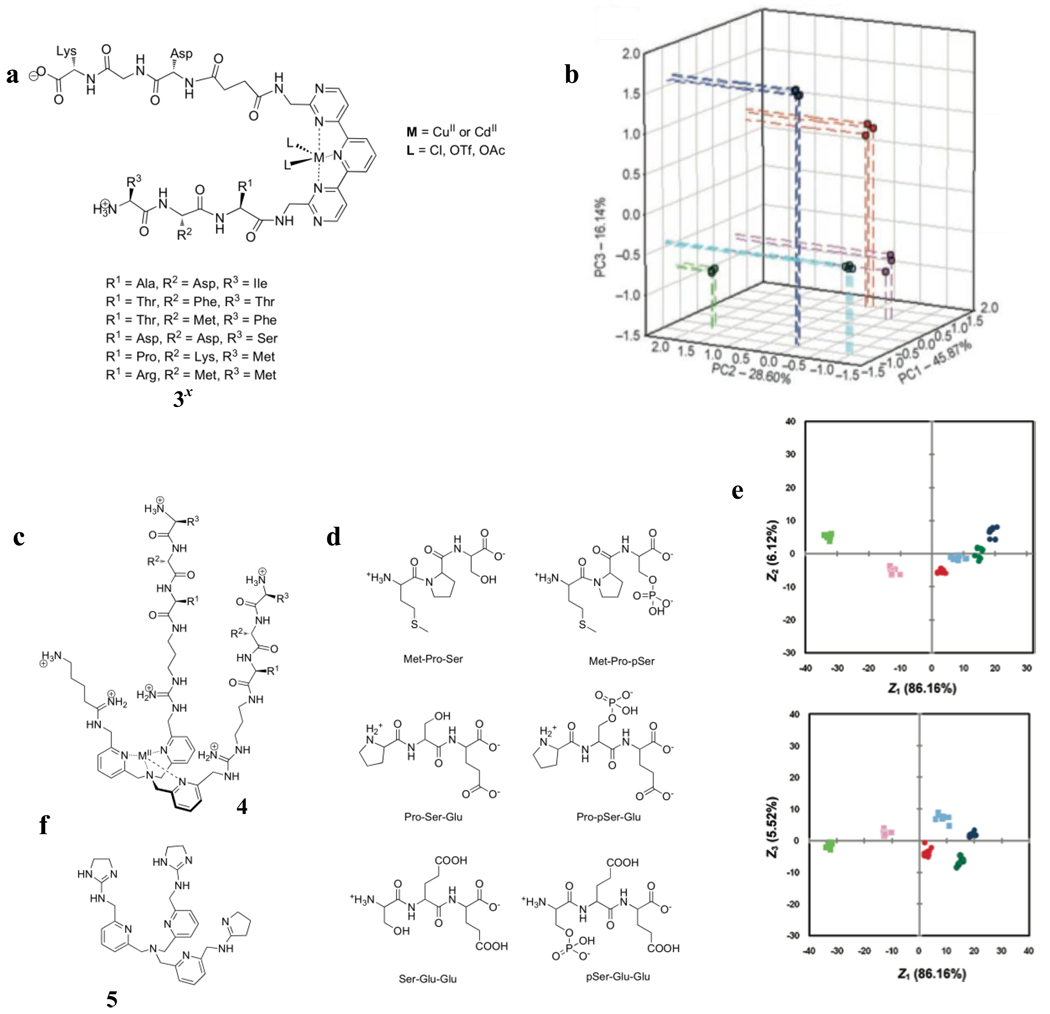
We turned to prescreening as a means to improve the cross-reactivity and our approach to differential sensing. This is illustrated in our group’s work on the differentiation of α-neurokinin (HKTDSFVGLM-NH2), substance P (RPKPQQFFGLM-NH2), and tachykinin analogues HKT, HKTD, and HET [42], which were chosen to challenge the discriminatory power of the same receptor library 3 (Figure 2e) in an array format. The combinatorially generated, resin-bound receptor library (Figure 3a) was prescreened for high binding receptors. These receptors were determined by visually identifying members of the library that bound to α-neurokinin that had been covalently modified with Disperse Red 1, a dye that was found to have little to no affinity for the receptor library. Six members of the library that bound to the α-neurokinin-Disperse Red 1 conjugate were sequenced, identified, and were used in the final array (3x, Figure 3a). Aside from appending variable peptidic arms to the tridentate core, the use of different metal ions and counterions contributed to the diversity in the array sensor. CuCl2, Cu(OTf)2, and Cd(OAc)2 were used, resulting in 18 receptor ensembles. This array gave an excellent classification of the peptides (Figure 3b), with good clustering and significant variance along three axes.
Figure 3.
(a) Receptor library, 3x, used in the discrimination of tachykinins and their analogues. (b) PCA score plot of response from α-neurokinin (HKTDSFVGLM-NH2) (blue), substance P (RPKPQQFFGLM-NH2) (light blue), and tachykinin analogues HKT (red), HKTD (green), and HET (violet). (c) Receptor library employed in the pattern based discriminaton of phosphorylated peptides. (d) Peptides and their phosphorylated versions analyzed. (e) LDA plots of the response of the analytes in b. (f) Structure of 5.
We recently studied phosphorylated peptides as analytes using our same general approach for pattern-based sensing. The receptor library (4, Figure 3c) in this case was derived from a core unit we previously had shown was selective for tetrahedral oxoanions [43]. Variation in the structures of the receptor library was brought about by random peptidic arms covalently appended to the amine-(tris)pyridine core, the use of different metal ions which were present to facilitate the preorganization of the binding site, as well as the use of various indicators. The signaling protocol used was an indicator displacement assay. Selection of metal ions and indicators proceeded by screening a model (5, Figure 3f) of the receptor library instead of the library itself. Indicator displacement assays with possible metals and indicators using the model receptor 5 revealed that three indicators and three metals gave the largest change in UV-vis absorbance upon exposure to phosphoserine. Phosphoserine was used as a model that represented the actual analytes (Figure 3d).
Prescreening of the combinatorial library 4 for binding the target analytes was done by incubating a Cu(II)-metallated on-bead receptor library with the dye Celestine Blue, manually picking the resulting intensely colored beads, and incubating these same beads with Pro-pSer-Glu (Figure 3d). Sequencing the beads which decolorized in the process gave five efficient receptors. These receptors were resynthesized and cleaved off the beads to make an array of 45 receptor-metal-indicator ensembles, which was then used in a solution phase assay to differentiate the tripeptides and their corresponding phosphorylated versions (Figure 3d). LDA of the spectroscopic data showed good classification of the analytes (Figure 3e).
Hamilton’s approach to differential sensing of proteins is complementary to our methodology. Recently, fluorescent DNA G-quadruplexes of protein-binding fragments were used in protein differentiation, [••9] in a dynamic combinatorial library array format [44,45]. Functionalization of the oligodeoxynucleotides with various fluorophores allowed formation of various combinations of the oligonucleotides, and hence, diversity in the array. Further, the monitoring of the individual quadruplexes enhances the pattern because each has fingerprint emission spectra by itself and in the presence of the analytes.
Differentiation of sugars
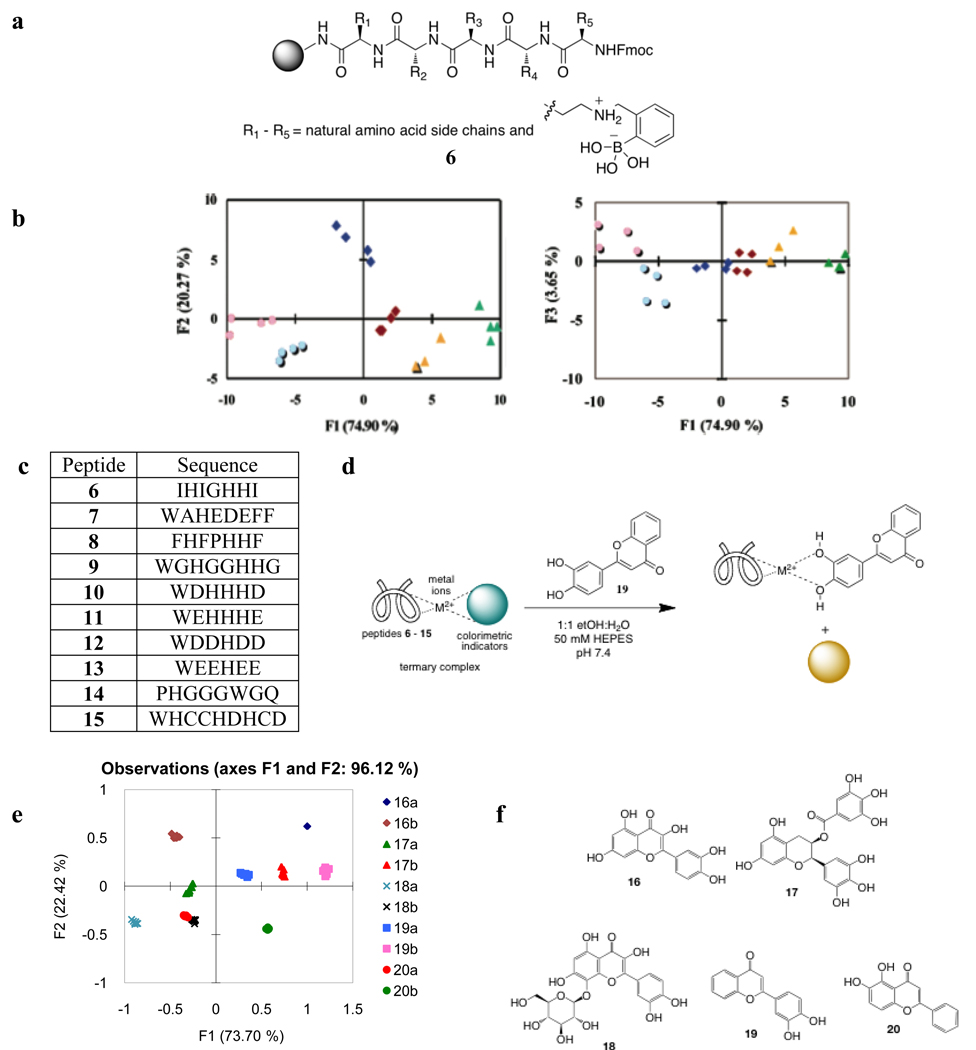
We have also used our strategy to target saccharides. Instead of using core-scaffolds that bias our libraries to the targets of interest, we took advantage of unnatural amino acids to ensure analyte binding. A bead-based, combinatorial pentapeptide library (6, Figure 4a) consisting of random amino acids, one of which is functionalized with a phenylboronic acid, was constructed for the array sensing of glucose, fructose, maltose, sucrose, maltilol, and sucralose [46]. The boronic acid side chain ensures sugar affinity to the library members. With an indicator uptake assay using bromopyrogallol red as a signaling protocol, the array was able to differentiate the sugars via LDA of the kinetic profiles of the indicator uptake by the beads (Figure 4b). These results were used as a training set for the identification of sucralose in complex mixtures, meaning actual beverages containing sucralose.
Figure 4.
(a) Boronic acid- functionalized receptor library used for the differentiation of sugars. (b) LDA plot of kinetic profiles of the indicator uptake of the array of receptors incubated with the sugars: fructose (orange triangle), glucose (green triangle), maltitol (light blue circle), maltose (blue diamond), sucralose (pink circle), sucrose (red diamond). (c) Peptide sequences used for the differentiation of tannins and wines. (d) Schematic representation of the indicator displacement assay of tannins using the peptidic ensembles in the array. (e) PCA score plot of responses of the different flavonoids (a - 0.060 mM, b - 0.12 mM) in (c). (f) Structures of flavonoids tested to determine the discriminatory properties of peptidic sensing ensembles.
Differentiation of flavonoids
Finally, the last example of pattern-based sensing that takes advantage of the use of core binding sites decorated with moieties that impart differential binding affinity for analytes, is our work on the differentiation of flavonoids found in wine. We explored random histidine-containing peptides that could potentially be preorganized by metal ions, and create binding sites for the analytes. The metal ions also ensure binding through the catechol functional group of the flavonoids. The sensing ensemble was then composed of a histidine-containing peptide, a metal ion, and a colorimetric indicator, all of which can be varied to give an array of cross reactive ensembles, and hence give differential response to analytes. An array composed of different peptides (Figure 4c), Cu(II), and pyrocatechol violet was able to differentiate different flavonoids (Figure 4f). The PCA score plot (Figure 4e) from this analysis showed that the flavonoids were classified according to identity as well as concentration.
Conclusions
In summary, our approach for differential sensing uses synthetic receptors biased to classes of analytes of interest. The receptors have a core unit decorated with moieties that impart cross-reactive binding. We have illustrated this approach in the differentiation of various analytes: nucleotide phosphates, peptides and their phosphorylated versions, proteins, sugars, and flavonoids in simple or complex mixtures. Variation in the members of the array, which gives differential affinity for analytes, is generated by appending combinatorially generated peptidic arms. Using indicator displacement or uptake assays as signaling protocols, diversity in the receptor array can also be generated using various indicators. In cases where a metal ion is needed to pre-organize the receptors, the use of various metal ions can further contribute to the diversity in the sensor array. Differentiation of the analytes can be achieved without prescreening of the receptor library, but prescreening improves the pattern recognition. We believe that our general protocol is broadly applicable to many classes of analytes, and we encourage others to adapt the approach to solve analytical problems where differential sensing is applicable.
Highlights
Combinatorial peptidic moieties, metal ions, and indicators appended to a core binding unit diversifies a receptor array
Such receptor arrays have effectively differentiated nucleotides, peptides, proteins, sugars, and flavonoids
Prescreening receptor libraries increases discriminatory power of receptor arrays
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
Papers of particular interest, published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1. Anslyn EV. Supramolecular analytical chemistry. J. Org. Chem. 2007;72:687–699. doi: 10.1021/jo0617971. Discusses principles behind array sensing using supramolecular sensors and applications of array sensing for quantitative analysis.
- 2.Wright AT, Anslyn EV. Differential receptor arrays and assays for solution-based molecular recognition. Chem. Soc. Rev. 2006;35:14–28. doi: 10.1039/b505518k. [DOI] [PubMed] [Google Scholar]
- 3.Collins BE, Wright AT, Anslyn EV. Combining molecular recognition, optical detection, and chemometric analysis. Top. Curr. Chem. 2007;277:181–218. [Google Scholar]
- 4.Albert KJ, Lewis NS, Schauer CL, Sotzing GA, Stitzel SE, Vaid TP, Walt DR. Cross-reactive chemical sensor arrays. Chem. Rev. 2000;100:2595–2696. doi: 10.1021/cr980102w. [DOI] [PubMed] [Google Scholar]
- 5. Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, Wold S. Multi- and Megavariate Data Analysis Part I: Basic Principles and Applications. edn 2nd. Umetrics AB: Umeå; 2006. Presents an easy to understand graphical explanation and applications of principal component analysis.
- 6.Jurs PC, Bakken GA, McClelland HE. Computational methods for the analysis of chemical sensor array data from volatile analytes. Chem. Rev. 2000;100:2649–2678. doi: 10.1021/cr9800964. [DOI] [PubMed] [Google Scholar]
- 7.Winquist F, Kratz-Rulcker C, Lundstrom I. Electronic tongues. MRS Bull. 2004:726–731. [Google Scholar]
- 8.Gouma P, Sberveglieri G. Novel materials and applications of electronic noses and tongues. MRS Bull. 2004:697–702. doi: 10.1557/mrs2004.205. [DOI] [PubMed] [Google Scholar]
- 9. Margulies D, Hamilton AD. Protein recognition by an ensemble of fluorescent DNA G-Quadruplexes. Angew. Chem. Int. Ed. 2009;48:1771–1774. doi: 10.1002/anie.200804887. Excellent example of receptors in a dynamic combinatorial library exquisitely incorporating diversifying elements to enable protein differentiation and detection in microliter quantities.
- 10.Zaubitzer F, Riss-Johannessen T, Severin K. Sensing of peptide hormones with dynamic combinatorial libraries of metal-dye complexes: the advantage of time-resolved measurements. Org. Biomol. Chem. 2009;7:4598–4603. doi: 10.1039/b912400d. [DOI] [PubMed] [Google Scholar]
- 11.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Sensing of proteins in human serum using conjugates of nanoparticles and green fluorescent protein. Nat. Chem. 2009;1:461–465. doi: 10.1038/nchem.334. S461/461-S461/426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda OR, Chen H-T, You C-C, Mortenson DE, Yang X-C, Bunz UHF, Rotello VM. Enzyme-Amplified Array Sensing of Proteins in Solution and in Biofluids. J. Am. Chem. Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rochat S, Gao J, Qian X, Zaubitzer F, Severin K. Cross-reactive sensor arrays for the detection of peptides in aqueous solution by fluorescence spectroscopy. Chem. Eur. J. 2010;16:104–113. doi: 10.1002/chem.200902202. [DOI] [PubMed] [Google Scholar]
- 14.Collins BE, Anslyn EV. Pattern-based peptide recognition. Chem. Eur. J. 2007;13:4700–4708. doi: 10.1002/chem.200700153. [DOI] [PubMed] [Google Scholar]
- 15.Viljanen J, Larsson J, Larsson A, Broo KS. A multipurpose receptor composed of promiscuous proteins. Analyte detection through pattern recognition. Bioconjugate Chem. 2007;18:1935–1945. doi: 10.1021/bc700247x. [DOI] [PubMed] [Google Scholar]
- 16.Schiller A, Vilozny B, Wessling RA, Singaram B. Recogntion of phospho sugars and nucleotides with an array of boronic acid appended bipyridinum salts. Anal. Chim. Acta. 2008;627:203–211. doi: 10.1016/j.aca.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Lim SH, Musto CJ, Park E, Zhong W, Suslick KS. A Colorimetric Sensor Array for Detection and Identification of Sugars. Org. Lett. 2008;10:4405–4408. doi: 10.1021/ol801459k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jagt RBC, Gomez-Biagi RF, Nitz M. Pattern-based recognition of heparin contaminants by an array of self-assembling fluorescent receptors. Angew. Chem. Int. Ed. 2009;48:1995–1997. doi: 10.1002/anie.200805238. [DOI] [PubMed] [Google Scholar]
- 19.Muller-Graff P-K, Szelke H, Severin K, Kramer R. Pattern-based sensing of sulfated glycosaminoglycans with a dynamic mixture of iron complexes. Org. Biomol. Chem. 2010;8:2269–2480. doi: 10.1039/c000420k. [DOI] [PubMed] [Google Scholar]
- 20.Palacios MA, Nishiyabu R, Marquez M, Anzenbacher PJ. Supramolecular chemistry approach to the design of a high-resolution sensor array for multianion detection in water. J. Am. Chem. Soc. 2007;129:7538–7544. doi: 10.1021/ja0704784. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Palacios MA, Anzenbacher PJ. Fluorescence sensor array for metal ion detection based on various coordination chemistries: general performance and potential application. Anal. Chem. 2008;80:7451–7459. doi: 10.1021/ac801165v. [DOI] [PubMed] [Google Scholar]
- 22.Palacios MA, Wang Z, Montes VA, Zyryanov GV, Anzenbacher PJ. Rational design of a minimal size sensor array for metal ion detection. J. Am. Chem. Soc. 2008;130:10307–10314. doi: 10.1021/ja802377k. [DOI] [PubMed] [Google Scholar]
- 23.Lim SH, Feng L, Kemling JW, Musto CJ, Suslick KS. An optoelectronic nose for the detection of toxic gases. Nat. Chem. 2009;1:562–567. doi: 10.1038/nchem.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng L, Musto CJ, Kemling JW, Lim SH, Suslick KS. A colorimetric sensor array for identification of toxic gases below permissible exposure limits. Chem. Comm. 2010;46:2037–2039. doi: 10.1039/b926848k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suslick BA, Feng L, Suslick KS. Discrimination of Complex Mixtures by a Colorimetric Sensor Array: Coffee Aromas. Anal. Chem. (Washington, DC, U. S.) 2010;82:2067–2073. doi: 10.1021/ac902823w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams MM, Anslyn EV. Differential sensing using proteins: exploiting the cross-reactivity of serum albumin to pattern individual terpenes and terpenes in perfume. J. Am. Chem. Soc. 2009;131:17068–17069. doi: 10.1021/ja908319m. [DOI] [PubMed] [Google Scholar]
- 27.Ponnu A, Edwards NY, Anslyn EV. Pattern recognition based identification of nitrated explosives. New J. Chem. 2008;32:848–855. [Google Scholar]
- 28.Hughes AD, Glenn IC, Patrick AD, Ellington A, Anslyn EV. A pattern recognition based fluorescence quenching assay for the detection and identification of nitrated explosive analytes. Chem. Eur. J. 2008;14:1822–1827. doi: 10.1002/chem.200701546. [DOI] [PubMed] [Google Scholar]
- 29.Hewage HS, Anslyn EV. Pattern-based recognition of thiols and metals using a single squarine indicator. J. Am. Chem. Soc. 2009;131:13099–13106. doi: 10.1021/ja904045n. [DOI] [PubMed] [Google Scholar]
- 30.Bang JH, Lim SH, Park E, Suslick KS. Chemically Responsive Nanoporous Pigments: Colorimetric Sensor Arrays and the Identification of Aliphatic Amines. Langmuir. 2008;24:13168–13172. doi: 10.1021/la802029m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bajaj A, Miranda OR, Kim I-B, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Detection and differentiation of normal, carcerous, and metastatic cells using nanoparticle-polymer sensor arrays. Proc. Nat. Acad. Sci. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bajaj A, Miranda OR, Phillips R, Kim I-B, Jerry DJ, Bunz UHF, Rotello VM. Array-Based Sensing of Normal, Cancerous, and Metastatic Cells using Conjugated Fluorescent Polymers. Journal of the American Chemical Society. 2010;132:1018–1022. doi: 10.1021/ja9061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shabbir SH, Joyce LA, da Cruz GM, Lynch VM, Sorey S, Anslyn EV. Pattern-based recogntion for the rapid determination of identity, concentration, and enantiomeric excess of subtly different threo diols. J. Am. Chem. Soc. 2009;131:13125–13131. doi: 10.1021/ja904545d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieto S, Lynch VM, Anslyn EV, Kim H, Chin J. High-throughput screening of identity, enantiomeric excess, and concentration using MLCT transitions in CD spectroscopy. J. Am. Chem. Soc. 2008;130:9232–9233. doi: 10.1021/ja803443j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavigne JJ, Anslyn EV. Sensing a paradigm shift in the field of molecular recognition: from selective to differential sensors. Angew. Chem. Int. Ed. 2001;40:3118–3130. doi: 10.1002/1521-3773(20010903)40:17<3118::AID-ANIE3118>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 36.Kitamura M, Shabbir SH, Anslyn EV. Guidelines for pattern recognition using differential receptors and indicator displacement assays. J. Org. Chem. 2009;74:4479–4489. doi: 10.1021/jo900433j. [DOI] [PubMed] [Google Scholar]
- 37. Schneider SE, O’Neil SN, Anslyn EV. Coupling rational design with libraries leads to the production of an ATP selective chemosensor. J. Am. Chem. Soc. 2000;122:542–543. Illustrates the use of combinatorial peptidic fragments in increasing the selectivity of ATP chemoreceptors.
- 38.McCleskey SC, Griffin MJ, Schneider SE, McDevitt JT, Anslyn EV. Differential receptors create patterns diagnostic for ATP and GTP. J. Am. Chem. Soc. 2003;125:1114–1115. doi: 10.1021/ja021230b. [DOI] [PubMed] [Google Scholar]
- 39.Wiskur SL, Ait-Haddou H, Lavigne JJ, Anslyn EV. Teaching old indicators new tricks. Acc. Chem. Res. 2001;34:963–972. doi: 10.1021/ar9600796. [DOI] [PubMed] [Google Scholar]
- 40.Wright AT, Griffin MJ, Zhenlin Z, McCleskey SC, Anslyn EV, McDevitt JT. Differential receptors create patterns that distinguish various proteins. Angew. Chem. Int. Ed. 2005;44:6375–6378. doi: 10.1002/anie.200501137. [DOI] [PubMed] [Google Scholar]
- 41. Wright AT, Anslyn EV, McDevitt JT. A differential array of metalated synthetic receptors for the analysis of tripeptide mixtures. J. Am. Chem. Soc. 2005;127:17405–17411. doi: 10.1021/ja055696g. The first paper to illustratre prescreening for increasing the discriminatory power of array sensors.
- 42.Wright AT, Edwards NY, Anslyn EV, McDevitt JT. The discriminatory power of differential receptor arrays is improved by prescreening-a demonstration in the analysis of tachykinins and similar peptides. Angew. Chem. Int. Ed. 2007;46:8212–8215. doi: 10.1002/anie.200701236. [DOI] [PubMed] [Google Scholar]
- 43.Zhang T, Edwards NY, Bonizzoni M, Anslyn EV. The use of differential receptors to pattern peptide phosphorylation. J. Am. Chem. Soc. 2009;131:11976–11984. doi: 10.1021/ja9041675. [DOI] [PubMed] [Google Scholar]
- 44.Buryak A, Pozdnoukhov A, Severin K. Pattern-based sensing of nucleotides in aqeuous solution with a multicomponent indicator displacement assay. Chem. Comm. 2007:2366–2368. doi: 10.1039/b705250b. [DOI] [PubMed] [Google Scholar]
- 45.Buryak A, Zaubitzer F, Pozdnoukhov A, Severin K. Indicator displacement assays as molecular timers. J. Am. Chem. Soc. 2008;130:11260–11261. doi: 10.1021/ja8037118. [DOI] [PubMed] [Google Scholar]
- 46.Edwards NY, Sager TW, McDevitt JT, Anslyn EV. Boronic acid based peptidic receptors for pattern-based saccharide sensing in neutral aqueous media, an application in real-life samples. J. Am. Chem. Soc. 2007;129:13575–13583. doi: 10.1021/ja073939u. [DOI] [PubMed] [Google Scholar]