Abstract
Efficient mitochondrial function requires physical interactions between the proteins encoded by the mitochondrial and nuclear genomes. Co-evolution between these genomes may result in the accumulation of incompatibilities between divergent lineages. We test whether mitochondrial-nuclear incompatibilities have accumulated within the Drosophila melanogaster species subgroup by combining divergent mitochondrial and nuclear lineages and quantifying the effects on relative fitness. Precise placement of nine mtDNAs from D. melanogaster, D. simulans and D. mauritiana into two D. melanogaster nuclear genetic backgrounds reveals significant mitochondrial-nuclear epistasis affecting fitness in females. Combining the mitochondrial genomes with three different D. melanogaster X chromosomes reveals significant epistasis for male fitness between X-linked and mitochondrial variation. However, we find no evidence that the more than 500 fixed differences between the mitochondrial genomes of D. melanogaster and the D. simulans species complex are incompatible with the D. melanogaster nuclear genome. Rather, the interactions of largest effect occur between mitochondrial and nuclear polymorphisms that segregate within species of the D. melanogaster species subgroup. We propose that a low mitochondrial substitution rate, resulting from a low mutation rate and/or efficient purifying selection, precludes the accumulation of mitochondrial-nuclear incompatibilities among these Drosophila species.
Keywords: epistasis, mtDNA evolution, polymorphism, X chromosome
Introduction
In many animals and fungi the mitochondrial genome experiences significantly more mutations per base pair than the nuclear genome (Lynch et al. 2008; Montooth & Rand 2008). Combined with a potentially reduced effective population size due to the effects of selection on this non-recombining genome (Hill & Robertson 1966, Maynard-Smith & Haig 1974, Charlesworth et al. 1993, Muller 1964), the elevated mutation rate makes the mtDNA particularly prone to the fixation of deleterious mutations (Gabriel et al. 1993; Lynch 1996; Neiman & Taylor 2009). Additionally, cytoplasmic sweeps driven by selfish cytoplasmic elements can decrease diversity and increase the rate of deleterious substitution in the mtDNA (Shoemaker et al. 2004). The nuclear genome encodes many more genes necessary for mitochondrial function than does the mtDNA, making it a potentially large target size for mutations that rescue mitochondrial function. Stabilizing selection to preserve efficient mitochondrial function may result in compensatory evolution whereby fitness loss due to the fixation of slightly deleterious mutations in the mitochondrial genome is rescued by mutations in the nuclear genome (Rand et al. 2004; Dowling et al. 2008).
While the basic cellular function of the mitochondria has been conserved over long evolutionary timescales, this maintenance occurs across lineages that have diverged in their ecology, nutrient environment, behavior and life history over shorter timescales. Positive selection may therefore act on mitochondrial and nuclear genomes in concert to adapt cellular metabolism to new physiologies and ecologies (Ballard & Rand 2005; Ihmels et al. 2005; Dowling et al. 2008). Thus, rapid mitochondrial divergence and co-evolution between mitochondrial and nuclear genomes could in principle be driven by both positive and negative selection (Bazin et al. 2006; Meiklejohn et al. 2007; Oliveira et al. 2008). Either process could generate intergenomic incompatibilities between mitochondrial and nuclear genomes that have evolved in isolation from one another, resulting in decreased fitness in hybrids between isolated populations or closely related species (Burton et al. 2006).
Interpopulation hybrids of the marine copepod Tigriopus californicus have disrupted mitochondrial transcription and ATP synthesis, and lower fitness (Edmands & Burton 1999; Ellison & Burton 2006, 2008a,b). Maternal backcrosses that reconstitute the parental mitochondrial-nuclear combinations restore fitness, highlighting the potentially important role of mitochondrial-nuclear interactions in hybrid breakdown (Ellison & Burton 2008a). Co-evolution of mitochondrial and nuclear genomes results in sterile hybrids between the yeast species Saccharomyces cerevisiae and S. bayanus (Lee et al. 2008), and may underlie decreased hybrid fitness between species of the parasitic wasp Nasonia (Breeuwer & Werren 1995; Ellison et al. 2008; Niehuis et al. 2008). Nasonia and Tigriopus have high levels of mitochondrial divergence between species and among populations, respectively (Oliveira et al. 2008; Burton et al. 2006), and the regulatory regions of yeast mtDNAs evolve rapidly (Groth et al 2000), raising the possibility that the accumulation of mitochondrial-nuclear incompatibilities may scale with mtDNA divergence.
In Drosophila the predominant evolutionary force shaping mtDNA divergence is purifying selection (Rand & Kann 1996,1998; Ballard 2000; Montooth et al. 2009). However, evolutionary constraint varies across the mitochondrial proteome with particular OXPHOS complexes evolving at significantly different rates (Ballard 2000; Montooth et al. 2009). These complex-specific evolutionary rates also vary across Drosophila lineages (Montooth et al. 2009). While mtDNAs within D. simulans are associated with differences in fitness, life history traits and mitochondrial physiology (James & Ballard 2003; Ballard et al. 2007) and may decrease cytochrome C oxidase activity in hybrids with D. mauritiana (Sackton et al. 2003), the extent to which molecular evolutionary forces lead to an accumulation of mitochondrial-nuclear incompatibilities for fitness between Drosophila species is unknown.
To test for within- and between-species mitochondrial-nuclear interactions, we generated strains of D. melanogaster that carry nine mtDNAs of varying molecular divergence from within the D. melanogaster species subgroup in combination with two D. melanogaster nuclear genomes. To avoid retaining nuclear variants from the maternal parent that may accompany mitochondrial introgression during repeated backcrossing (James & Ballard 2003; Dowling et al. 2008), the nuclear genomes were precisely introduced using balancer chromosomes. Testing each mtDNA in two nuclear genetic backgrounds allows quantification of mitochondrial-nuclear epistasis for fitness. The phylogenetic context of this experiment allows a test of whether mitochondrial-nuclear incompatibilities accumulate as mitochondrial and nuclear genomes diverge together along species lineages. We find no evidence that the accumulation of fixed differences in the mtDNAs gives rise to mitochondrial-nuclear incompatibilities between these sibling species. Instead, we find that particular mitochondrial and nuclear variants that segregate within species, potentially as neutral, mildly deleterious or population-specific polymorphisms, interact to generate the strongest epistatic effects on fitness.
Materials and Methods
Generating mitochondrial-nuclear hybrids
We generated 35 D. melanogaster strains that combine nine mtDNAs from D. melanogaster and its sibling species (ore, aut, zim, siI, sm21, sm22, sm38, simw501, mau12) with two D. melanogaster nuclear backgrounds (Ore, Aut) and two additional D. melanogaster X chromosomes (P58, P89) (Table 1, Figure 1). Female offspring from crosses between D. simulans females and D. melanogaster males frequently die as embryos (Hadorn 1961), and those that survive are typically sterile. However, viable and weakly fertile F1 females can be obtained from crosses between D. simulans C167.4 females and D. melanogaster In(1)AB males (Davis et al. 1996), allowing the transfer of mtDNA from D. simulans into D. melanogaster. We backcrossed fertile F1 hybrid females to D. melanogaster males for multiple generations, after which we used balancer chromosomes to precisely replace the nuclear chromosomes with those from two D. melanogaster inbred lines. While this design avoids retention of nuclear variants from the maternal parent that may accompany mitochondrial introgression during backcrossing (James & Ballard 2003; Dowling et al. 2008), it does restrict tests for interspecific mitochondrial-nuclear interactions to D. melanogaster nuclear backgrounds.
Table 1.
Genotypes used to infer mitochondrial-nuclear and mitochondrial-X chromosome interactions for fitness
| Mitochondrial-X-nuclear genotype a | mtDNA haplotype | mtDNA source | X-chromosome genotype | Autosomal genotype |
|---|---|---|---|---|
| (oret); Ore; Ore; Ore | mel | D. melanogaster OreR | OreR | OreR |
| (zimt); Ore; Ore; Ore | zim | D. melanogaster Zim53 | OreR | OreR |
| (siIt); Ore; Ore; Ore | siI | D. simulans siI Hawaii | OreR | OreR |
| (sm21t); Ore; Ore; Ore | siII | D. simulans C167.4 | OreR | OreR |
| (sm22t); Ore; Ore; Ore | siII | D. simulans C167.4 | OreR | OreR |
| (sm38t); Ore; Ore; Ore | siII | D. simulans C167.4 | OreR | OreR |
| (simw501t); Ore; Ore; Ore | siII | D. simulans simw501 | OreR | OreR |
| (mau12t); Ore; Ore; Ore | maI | D. mauritiana mau12 | OreR | OreR |
| (auttp); Aut; Aut; Aut | mel | D. melanogaster AutW132 | AutW132 | AutW132 |
| (oretp); Aut; Aut; Aut | mel | D. melanogaster OreR | AutW132 | AutW132 |
| (zimtp); Aut; Aut; Aut | zim | D. melanogaster Zim53 | AutW132 | AutW132 |
| (siItp); Aut; Aut; Aut | siI | D. simulans siI Hawaii | AutW132 | AutW132 |
| (sm21tp); Aut; Aut; Aut | siII | D. simulans C167.4 | AutW132 | AutW132 |
| (sm22tp); Aut; Aut; Aut | siII | D. simulans C167.4 | AutW132 | AutW132 |
| (sm38tp); Aut; Aut; Aut | siII | D. simulans C167.4 | AutW132 | AutW132 |
| (simw501tp); Aut; Aut; Aut | siII | D. simulans simw501 | AutW132 | AutW132 |
| (mau12tp); Aut; Aut; Aut | maI | D. mauritiana mau12 | AutW132 | AutW132 |
| (auttp); P58; Aut; Aut | mel | D. melanogaster AutW132 | P58b | AutW132 |
| (oretp); P58; Aut; Aut | mel | D. melanogaster OreR | P58 | AutW132 |
| (zimtp); P58; Aut; Aut | zim | D. melanogaster Zim53 | P58 | AutW132 |
| (siItp); P58; Aut; Aut | siI | D. simulans siI Hawaii | P58 | AutW132 |
| (sm21tp); P58; Aut; Aut | siII | D. simulans C167.4 | P58 | AutW132 |
| (sm22tp); P58; Aut; Aut | siII | D. simulans C167.4 | P58 | AutW132 |
| (sm38tp); P58; Aut; Aut | siII | D. simulans C167.4 | P58 | AutW132 |
| (simw501tp); P58; Aut; Aut | siII | D. simulans simw501 | P58 | AutW132 |
| (mau12tp); P58; Aut; Aut | maI | D. mauritiana mau12 | P58 | AutW132 |
| (auttp); P89; Aut; Aut | mel | D. melanogaster AutW132 | P89b | AutW132 |
| (oretp); P89; Aut; Aut | mel | D. melanogaster OreR | P89 | AutW132 |
| (zimtp); P89; Aut; Aut | zim | D. melanogaster Zim53 | P89 | AutW132 |
| (siItp); P89; Aut; Aut | siI | D. simulans siI Hawaii | P89 | AutW132 |
| (sm21tp); P89; Aut; Aut | siII | D. simulans C167.4 | P89 | AutW132 |
| (sm22tp); P89; Aut; Aut | siII | D. simulans C167.4 | P89 | AutW132 |
| (sm38tp); P89; Aut; Aut | siII | D. simulans C167.4 | P89 | AutW132 |
| (simw501tp); P89; Aut; Aut | siII | D. simulans simw501 | P89 | AutW132 |
| (mau12tp); P89; Aut; Aut | maI | D. mauritiana mau12 | P89 | AutW132 |
The mitochondrial genotype is given in parentheses, followed by the X, 2nd and 3rd chromosome genotypes. Subscript “t” indicates that the stock has been cleared of Wolbachia using tetracycline and “p” indicates a P cytoplasm.
Two D. melanogaster X chromosomes (P58 and P89) from Davis, CA (S. Nuzhdin, USC) were also precisely substituted into the AutW132 panel of mitochondrial-nuclear genotypes.
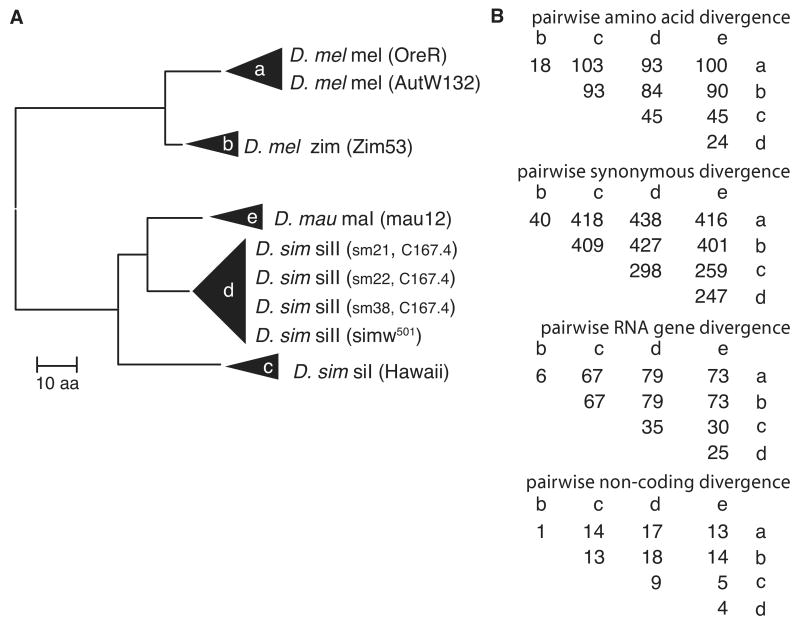
Figure 1.
Phylogenetic relationship between the five mitochondrial haplotype groups used to study mitochondrial-nuclear epistasis for fitness. A) Neighbor-joining tree using a concatenated proteome amino acid sequence (length = 3725 aa). Species names are followed by the mitochondrial haplotype and the line from which the mtDNA was isolated in parentheses. sm21, 22 and 38 are replicate mitochondrial introgressions from D. simulans strain C167.4. B) Pairwise divergence between the five mitochondrial haplotypes. The numbers of pairwise differences were estimated from 3724 codons for amino acid and synonymous site divergence, 3606 nucleotide sites for tRNA and rRNA gene divergence, and 147 non-coding sites. The difference in substitutions between branches d and e when compared to outgroups a, b or c is due to homoplastic substitution. Whole mtDNA sequence data from Ballard (2000) were aligned to the reference D. melanogaster and D. simulans siII genomes as in Montooth et al. (2009). NCBI accessions: a, NC_001709; b, AF200829; c, AF200835; d, AF200840; e, AF200831.
Females of D. simulans Hawaii (mitochondrial haplotype siI) and D. mauritiana mau12 (mitochondrial haplotype maI, which differs from the D. simulans siIII haplotype by one nucleotide substitution) (Ballard 2000) were repeatedly backcrossed to D. simulans C167.4 males. Subsequently, females carrying the siI or maI mtDNAs were crossed to D. melanogaster In(1)AB,w males. The In(1)AB chromosome carries a mutation that rescues F1 hybrid female viability and fertility (Hutter et al. 1990; Aruna et al. 2009). D. simulans simw501 (mitochondrial haplotype siII) females were crossed directly to In(1)AB,w males, as F1 female viability and fertility are also rescued in this cross. In addition, three strains of D. melanogaster (sm21, sm22, sm38) that carry the D. simulans C167.4 mtDNA (mitochondrial haplotype siII) (Sawamura et al. 2000), and a strain from the D. melanogaster Zimbabwe race (Zim53) were also used as a source of mtDNA. We backcrossed females carrying target mtDNAs to D. melanogaster Oregon-R (OreR) males for 3 generations, replacing much of the nuclear genome with D. melanogaster material.
To eliminate any D. simulans or D. mauritiana genomic regions that might have been retained during the introgression, we precisely replaced the nuclear genomes of all hybrid strains with chromosomes from either a D. melanogaster Ore-R (Ore) strain that was inbred for two generations via full-sib mating or from an isofemale line of D. melanogaster from Austria, AutW132 (Aut), obtained from Christian Schlötterer (Institut für Populations-genetik, Veterinärmedizinische Universität Wien, Vienna, Austria). To simultaneously replace the autosomes we used a strain of D. melanogaster carrying the second-chromosome balancer CyO and the third-chromosome balancer TM6B,Tb,Dr. To replace the X chromosome we used strains that carried the FM7c,B,sn balancer chromosome in an otherwise Ore or Aut genetic background. The mtDNA haplotypes of the constructed genotypes were confirmed by direct sequencing of mtDNA using the primer pairs, 3593F 5′-gaacagttcccgctttaggag/4528R 5′-gcagttaatcggacagctaatgtccc and 5314F 5′-gctccatttactattgcggactc/6195R 5′-cattaacagtgatacgcctc. Before the fitness assays, mitochondrial haplotypes were re-confirmed using PCR-RFLP analysis with the 3593/4528 primer pair and the Alu I, Dra I and Rsa I restriction enzymes.
Controlling the cytoplasm
The intracellular endosymbiont Wolbachia is co-transmitted with the mtDNA through the maternal cytoplasm of many D. melanogaster and D. simulans strains and can have myriad phenotypic effects, including cytoplasmic incompatibility (Merçot & Charlat 2004; Clark et al. 2005; Ikeya et al. 2009). To avoid confounding mtDNA and Wolbachia effects, we cured all cytoplasms of Wolbachia infection prior to the replacement of the nuclear chromosomes by rearing larvae on instant Carolina media mixed with a 0.03% tetracycline solution for two generations. Successful clearing of Wolbachia was confirmed by failure to amplify a PCR product using Wolbachia-specific primers (F 5′-tggtccaataagtgatgaagaaac; R 5′-aaaaattaaacgctactcca). All PCRs were run alongside a positive Wolbachia-infected control.
The ability to suppress mobilization of the P transposable element is also maternally inherited (Brennecke et al. 2008). When P element naïve (M cytotype) females mate with P element containing (P cytotype) males, mobilization of P elements can occur in offspring genomes (Kidwell et al. 1977, Brennecke et al. 2008). Failure to PCR amplify P element sequence (F 5′-taaaaggaggcgactcaacg; R 5′-ctcagctgctgctctaaacg) indicated that the balancer stocks to be used for replacing the nuclear genome and the Oregon-R stock that provided the nuclear genome were M cytotype. However, the wild lines that provided the Aut nuclear genome and the X chromosomes were P cytotype. To prevent mobilization of P elements during the substitution of Aut nuclear background chromosomes, the balancer strains and all mitochondrial-nuclear hybrid lines to be used for Aut chromosomal substitution were backcrossed to AutW132 males for multiple generations to establish a P cytotype.
Testing for mitochondrial-nuclear interactions affecting fitness
We used a chromosome segregation assay modified from Rand et al. (2001) to quantify fitness effects of mitochondrial-nuclear interactions (Figure 2). In this assay, individuals competed in the same vial with siblings carrying a visibly-marked X chromosome (FM6) that confers a Bar eye phenotype. Relative fitness was measured as the egg-to-adult viability of wild-type individuals relative to their FM6-bearing siblings across the 35 mitochondrial-nuclear genotypes (Table 1). Differences between nuclear genotypes in the relative competitive viability of wild-type flies compared to their FM6 siblings result from viability effects of the X chromosome directly competing with FM6, and genetic interactions between the wild-type or FM6 X chromosomes and the autosomes. Differences between mitochondrial-nuclear genotypes in relative competitive viability result from interactions between the mitochondrial genome and the wild-type or FM6 X chromosomes, or more complex interactions between the X chromosome, the autosomes, and the mtDNA. The advantages of this assay are that competing individuals share the same common rearing environment, and genotypes are easily inferred from the Bar phenotype. In males, differences in relative competitive viability arise from the hemizygous effects of X-linked variants in combination with the mitochondrial-nuclear genotype. In females, viability effects of wild-type X-linked variants that are completely dominant to FM6 when combined with particular mitochondrial-nuclear genomes will not be detected in this assay.
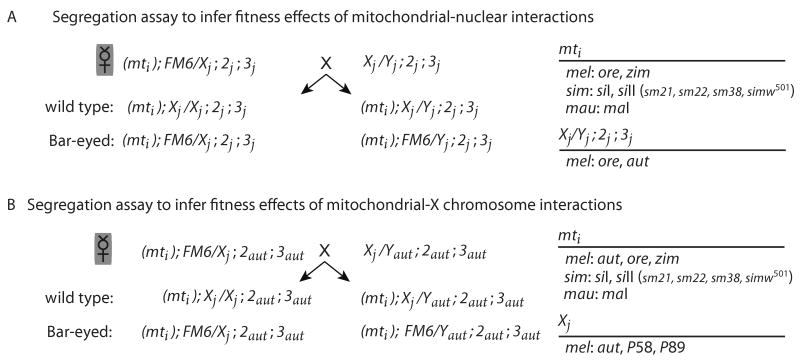
Figure 2.
The segregation assay used to measure relative fitness. A) The crossing design used to test for fitness effects of mitochondrial-nuclear interactions. B) The crossing design used to test for fitness effects of mitochondrial-X chromosome interactions. FM6 is a non-recombining X chromosome that results in a dominant, visible Bar-eyed phenotype. The mitochondrial genotype is given in parentheses followed by the X/Y, second and third chromosome genotypes.
We generated strains carrying an FM6 chromosome in both the Ore and Aut nuclear backgrounds. Females of each mitochondrial-nuclear genotype were mated to males carrying the FM6 chromosome in the same autosomal background. For the segregation assay, the resulting heterozygous females (Aut/FM6;Aut;Aut or Ore/FM6;Ore;Ore) were crossed to Aut or Ore males, respectively (Figure 2A). All offspring from these crosses inherit the mtDNA from the initial female. Female offspring were scored as either wild-type homozygotes or Bar heterozygotes, and male offspring were either wild-type or Bar hemizygotes. Relative competitive viability was calculated for each sex in each vial as the number of wild-type offspring divided by one plus the total progeny of that sex emerging from that vial (Haldane 1956), and is the measure of relative fitness used throughout.
Isolating mitochondrial-X chromosome interactions affecting fitness
In order to test specifically for fitness effects of interactions between the X chromosome and the mtDNA, we combined the nine mtDNAs with three X chromosomes (Aut, P58 and P89) in the Aut nuclear background (Table 1). Differences between mitochondrial-X chromosome genotypes in these strains result only from direct interactions between the mitochondrial genome and the wild-type or FM6 X chromosomes, as the autosomal genetic background is held constant. The P58 and P89 X chromosomes were derived from a D. melanogaster population in Davis, CA collected by Sergey Nuzhdin (University of Southern California). The P58 and P89 X chromosomes were substituted into the Aut autosomal background using balancer chromosomes. For the segregation assay, females heterozygous for FM6 and either the P58, P89 or Aut X chromosome in an Aut autosomal background were mated to males carrying the corresponding P58, P89 or Aut X chromosome in an Aut autosomal background (Figure 2B). All offspring from this cross inherit the mtDNA from the initial female. Female offspring were scored as either wild-type homozygotes or Bar heterozygotes for the Aut, P58, or P89 X chromosomes. Male offspring were either wild-type Aut, P58 or P89 hemizygotes or Bar hemizygotes. Relative fitness for each X-mitochondrial genotype was calculated as described above.
Experimental design and statistical analysis
Relative fitness of each genotype was measured in six replicate vials in which two males and two females were allowed to mate continuously and lay eggs for 3 days. Parents were transferred to a second vial to mate and lay a second brood of eggs for an additional 3 days. Wild-type and Bar-eyed progeny emerging from these vials were counted every other day for ten days. The entire experiment was conducted in two complete and independent blocks. The full design measured fitness for 35 mitochondrial-nuclear genotypes tested in six replicate assays for each of two replicate broods in two replicate blocks, with 118,932 individuals scored.
On average, 145 offspring emerged from a single vial. We eliminated observations where fewer than 10 flies of a given sex emerged from a vial. This removed less than 5% of the data, leaving 797 observations of male fitness and 807 observations of female fitness. The effects of mtDNA, nuclear genotype and the interaction between mtDNA and nuclear genotype were tested using mixed model analysis of variance (ANOVA) for each sex separately, due to the hemizygous versus heterozygous effects in the two sexes (see methods above). The complete ANOVA model was yijklm = μ + Mj + Nk + MNjk + Bl + bi + Pxijklm + εijklm, where yijklm is male or female relative fitness estimated from a single vial, Mj is the fixed effect of mtDNA, Nk is the fixed effect of either the nuclear or X-chromosome genotype, MNjk is the interaction between these factors, Bl is the fixed effect of brood, and bi is the random effect of block. The total number of offspring emerging from each vial (“vial productivity”) was used as a covariate in the model (Pxijklm) to control for any larval density effects on relative fitness. Separate slopes for the regression of fitness on vial productivity were fit for each nuclear or X-chromosome genotype. A significant mtDNA × nuclear interaction effect in the model was used to infer the presence of mitochondrial-nuclear epistasis for fitness. The biological interpretation of this interaction is that the fitness effect of substituting a particular mtDNA variant is conditional on the nuclear genetic background. Linear mixed models were performed using the nlme library in the software package R version 2.6.1 and verified using the mixed procedure in SAS version 9.1.3.
We employed two approaches to quantify the contributions of fixed interspecific mitochondrial differences and segregating intraspecific mitochondrial variation to mitochondrial-nuclear epistasis for fitness. Due to its similarity with the D. simulans siIII haplotype (Ballard 2000), we grouped the D. mauritiana maI haplotype with D. simulans for this analysis. First, we modified the linear mixed model above to test for a fixed effect of species mtDNA (D. melanogaster or D. simulans), within which we nested the random effect of within-species mitochondrial genotype. The absence of a species mtDNA effect in this model would indicate that, on average, there is no differential effect on fitness caused by substituting a D. simulans versus a D. melanogaster mtDNA into a D. melanogaster nuclear background. Additionally, we used the varcomp procedure in SAS to partition the variance in fitness into the between–species mtDNA effect and the within-species mitochondrial genotype effect using restricted maximum likelihood.
Three of the D. simulans siII mtDNAs (sm21, sm22, sm38) originated from replicate mtDNA introgressions from a single D. simulans strain. While these mtDNAs could potentially have diverged in sequence after the initial introgression, they are clearly not independent. When only lines containing these mtDNAs were analyzed, there was no significant difference in relative fitness between the three replicate siII mtDNAs (females: F = 1.08, P = 0.34, males: F = 1.74, P = 0.18) and no interaction with the nuclear genome (females: F = 0.58, P = 0.56, males: F = 1.09, P = 0.35). In the X chromosome experiment there was no main effect of the three siII mtDNAs on relative fitness in females (F = 1.01, P = 0.37), only a marginally significant effect in males (F = 3.28, P = 0.04), and no significant interaction with the X chromosome (females: F = 0.26, P = 0.90, males: F = 0.81, P = 0.52). Data for these three siII mtDNAs were therefore pooled in our analyses.
Results
Mitochondrial-nuclear fitness interactions
We assayed 12 genotypes that combined six mtDNAs (ore, zim, siI, siII, simw501, mau12) with two D. melanogaster nuclear backgrounds (Ore, Aut) to determine whether interactions between the mtDNA and the nuclear genome generate epistasis for fitness (Table 1). We used an X-chromosome segregation assay that competes individuals of each mitochondrial-nuclear genotype against competitors carrying a visibly marked FM6 X chromosome emerging from the same cross in replicate vials (see Methods, Figure 2A). The FM6 chromosome has low fitness, and the viability of wild-type individuals relative to FM6 competitors was, on average, greater than 0.5. Male relative fitness was greater than female relative fitness for all mitochondrial-nuclear genotypes (Figure 3), consistent with the presence of recessive deleterious mutations on the FM6 X chromosome that are masked in female heterozygous FM6 competitors.
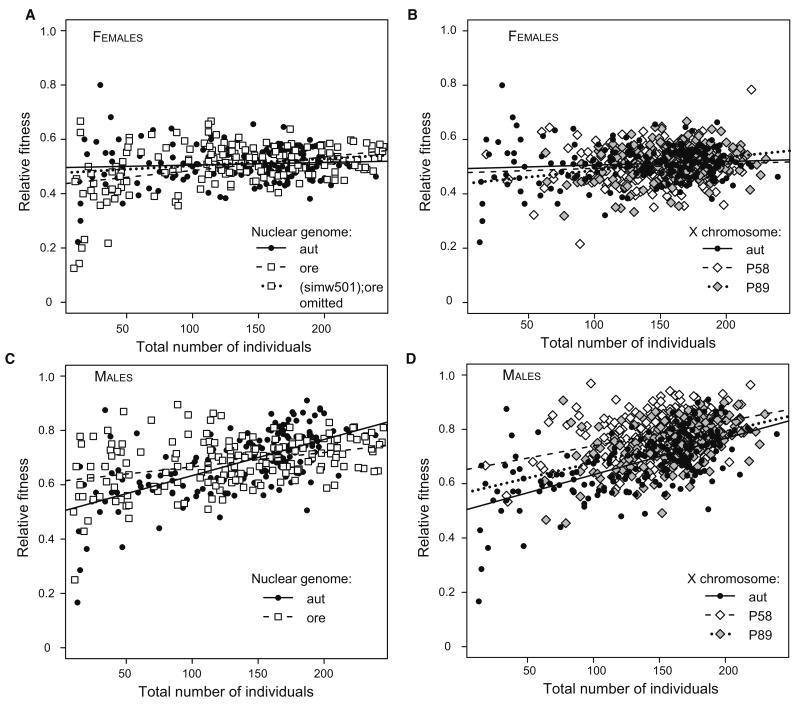
Figure 3.
Positive relationship between relative fitness estimates and the total number of individuals emerging from a vial for females and males in the mitochondrial-nuclear (A,C) and mitochondrial-X chromosome (B,D) experiments. Regression lines reveal a tendency for the slope to differ between autosomes and to differ between X chromosomes.
Relative fitness was weakly positively correlated with the total number of individuals emerging from a vial (Figures 3A, 3C), suggesting that FM6 flies are less competitive in crowded rearing conditions. This relationship was stronger in males than in females (Table 2), and the slope of the relationship differed between nuclear backgrounds, particularly in males (Figure 3C). We therefore included the number of individuals emerging from a vial (“vial productivity”) as a covariate in the mixed model ANOVA, allowing different slopes for the Ore and Aut nuclear backgrounds. Including vial productivity greatly increased the fit of the model for male relative fitness (LR = 56.78, P < 0.0001), but only mildly for female relative fitness (LR = 4.73, P = 0.03).
Table 2.
Mixed model ANOVA for effects of the mtDNA, nuclear genotype and the mitochondrial-nuclear interaction on relative fitness
| Dependent variable | Fixed effect | num df | den df | F-value | P-value | Random effect | σ2 |
|---|---|---|---|---|---|---|---|
| Female fitness | mtDNA | 5 | 346 | 6.727 | < 0.0001 | block | 8.17e-7 |
| nuclear | 1 | 346 | 0.779 | 0.3781 | residual | 4.82e-3 | |
| mtDNA × nuclear | 5 | 346 | 4.242 | 0.0009 | |||
| brood | 1 | 346 | 0.002 | 0.9646 | |||
| vial productivity × nuclear a | 2 | 346 | 5.138 | 0.0063 | |||
| Male fitness | mtDNA | 5 | 346 | 0.324 | 0.8982 | block | 5.02e-4 |
| nuclear | 1 | 346 | 0.028 | 0.8666 | residual | 6.45e-3 | |
| mtDNA × nuclear | 5 | 346 | 1.559 | 0.1710 | |||
| brood | 1 | 346 | 17.008 | < 0.0001 | |||
| vial productivity × nuclear | 2 | 346 | 39.995 | < 0.0001 |
The number of flies emerging from each vial was used as a linear covariate with different slopes estimated for the two nuclear genetic backgrounds.
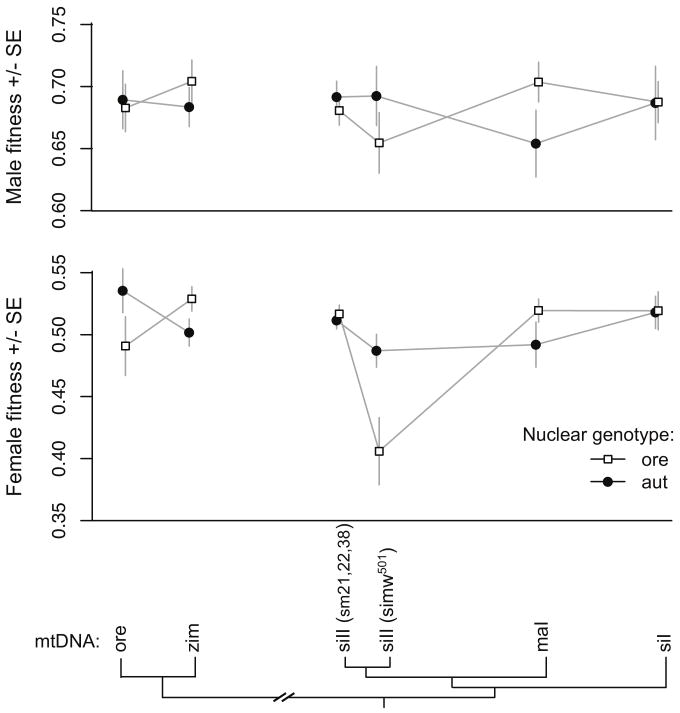
For male relative fitness, there was no main effect of mtDNA or interaction between mitochondrial and nuclear genotype (Figure 4, Table 2). However, for female relative fitness, there was a significant interaction effect between mitochondrial and nuclear genotypes for relative fitness (Figure 4, Table 2). Several mitochondrial-nuclear interactions contribute to female relative fitness. For example, while the D. melanogaster zim mtDNA has greater relative fitness than the D. melanogaster ore mtDNA in an Ore nuclear background, the fitness effects of these mtDNAs are reversed in an Aut nuclear background.
Figure 4.
Fitness values for males and females from twelve mitochondrial-nuclear genotypes relative to visibly marked sibling competitors reveal significant mitochondrial-nuclear epistasis for fitness in females. The mtDNAs are arranged along the x-axis by phylogenetic distance, highlighting the lack of relationship between mitochondrial molecular divergence and the fitness effect of mitochondrial substitution.
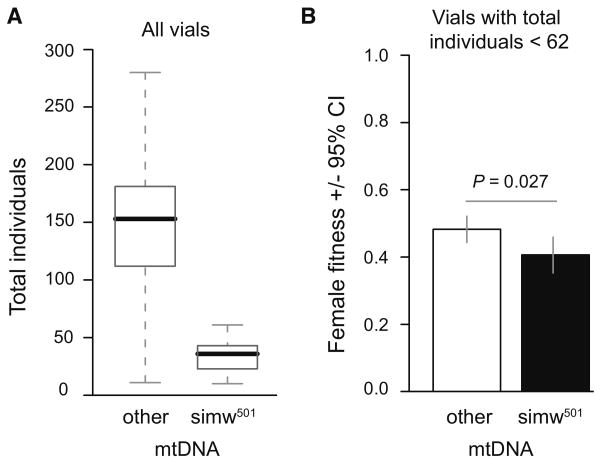
The strongest interaction results from the reduced relative fitness of females that have a D. simulans simw501 mtDNA paired with the D. melanogaster Ore nuclear genome, but not with the D. melanogaster Aut nuclear genome, providing strong evidence of mitochondrial-nuclear epistasis. The (simw501);Ore genotype also has lower mean vial productivity than other genotypes, although not outside the range observed for other genotypes (Figure 5A). To confirm that (simw501);Ore individuals have low relative fitness, even for their low level of productivity, we analyzed only vials producing fewer than 62 individuals, which was the maximum productivity of (simw501);Ore parents. Even with the reduced number of observations in this range, we found that (simw501);Ore females have significantly lower relative fitness than all other vials producing 62 or fewer offspring (Figure 5B).
Figure 5.
The (simw501);Ore mitochondrial-nuclear genotype confers low vial productivity, but also has low relative fitness given its productivity. A) Boxplots for (simw501);Ore compared to all other genotypes in the mitochondrial-nuclear interaction dataset demonstrate the low median number of total individuals emerging from vials of this genotype (bold horizontal bars), but also show that this level of productivity is within the range of productivity (dotted lines) of other genotypes. B) Relative fitness of (simw501);Ore females is significantly less than that observed for all other genotypes when only vials producing less than 62 individuals were analyzed.
Mitochondrial-X chromosome fitness interactions
To isolate two-way fitness interactions between mtDNA and X chromosomes, we combined seven mtDNAs (aut, ore, zim, siI, siII, simw501, mau12) with three D. melanogaster X chromosomes (Aut, P58, P89) in an Aut autosomal background (Table 1). As in the segregation assays above, there was a significant relationship between vial productivity and relative fitness that was stronger in males than in females and was dependent on the X-chromosome genotype (Figures 3B, 3D, Table 3). We treated vial productivity as a covariate in the analysis with separate slopes for the different X chromosomes. Including vial productivity increased the fit of the model for both male (LR = 156.9, P < 0.0001) and female viability (LR = 11.4, P = 0.0007).
Table 3.
Mixed model ANOVA for effects of the mtDNA, X-chromosome genotype and the mitochondrial-X chromosome interaction on relative fitness
| Dependent variable | Fixed effect | num df | den df | F-value | P-value | Random effect | σ2 |
|---|---|---|---|---|---|---|---|
| Female fitness | mtDNA | 6 | 613 | 1.11 | 0.3574 | block | 4.9e-13 |
| X chromosome | 2 | 613 | 2.73 | 0.0663 | residual | 3.80e-3 | |
| mtDNA × X chrom | 12 | 613 | 1.34 | 0.1922 | |||
| brood | 1 | 613 | 5.44 | 0.0200 | |||
| vial productivity × X chroma | 3 | 613 | 5.59 | 0.0009 | |||
| Male fitness | mtDNA | 6 | 613 | 3.383 | 0.0027 | block | 7.47e-5 |
| X chromosome | 2 | 613 | 98.191 | < 0.0001 | residual | 5.12e-3 | |
| mtDNA × X chrom | 12 | 613 | 2.534 | 0.0029 | |||
| brood | 1 | 613 | 88.847 | < 0.0001 | |||
| vial productivity × X chrom | 3 | 613 | 62.557 | < 0.0001 |
The number of flies emerging from each vial was used as a linear covariate with different slopes estimated for the three X-chromosome genetic backgrounds.
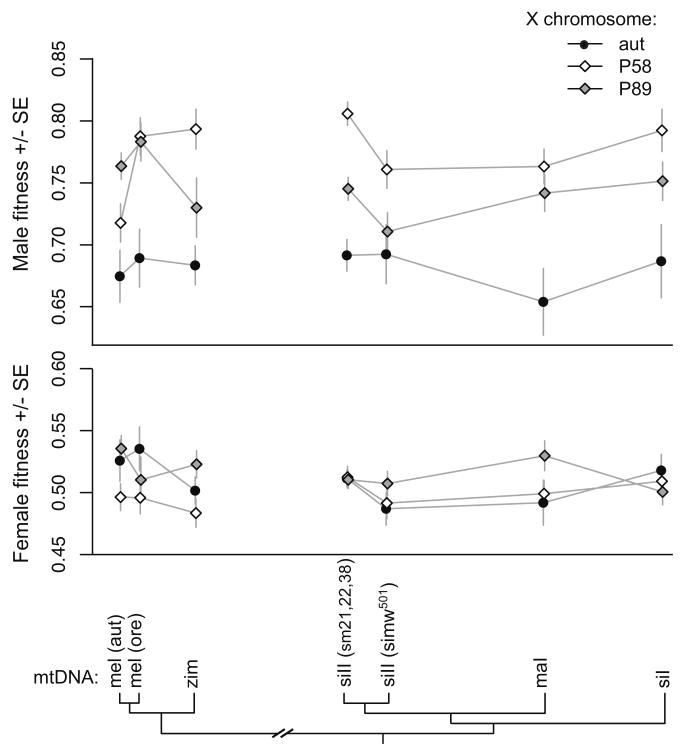
In contrast to the mitochondrial-nuclear segregation assay, male, but not female, relative fitness was significantly affected by interactions between the X chromosome and the mitochondrial genome (Figure 6, Table 3). The epistatic interactions are particularly evident in the changing rank order of male relative fitness values between X-chromosome genotypes among the D. melanogaster mtDNAs (Figure 6). Additionally, the fitness effects of D. simulans and D. mauritiana mtDNAs were dependent on the X chromosome. For example, there was no difference in relative fitness between males carrying the siII and simw501 mtDNAs in an Aut X-chromosomal background, but the siII mtDNA conferred higher relative fitness than the simw501 mtDNA in both the P58 and P89 X-chromosomal backgrounds (Figure 6).
Figure 6.
Fitness values for males and females from 21 mitochondrial-X chromosome genotypes relative to visibly marked sibling competitors reveal significant mitochondrial-X chromosome epistasis for fitness in males. The mtDNAs are arranged along the x-axis by phylogenetic distance, highlighting the lack of relationship between mitochondrial molecular divergence and the fitness effect of mitochondrial substitution.
Within- versus between-species effects on fitness
D. simulans and D. mauritiana mtDNAs have effects on relative fitness similar to those caused by D. melanogaster mtDNAs when combined with D. melanogaster nuclear genomes (Figures 4, 6). To quantify this observation, we compared the variance in relative fitness explained by variation among mitochondrial genotypes within D. melanogaster and within the D. simulans species complex to the variance in relative fitness explained by differences between these species mtDNA lineages.
Fitness consequences of mtDNA interactions with the Ore and Aut nuclear genomes were significant only in females (Table 2). Among females, 5.7% of the variance in relative fitness was explained by variation among mitochondrial genotypes within species, and 9.2% of the variance in relative fitness was attributed to within species mitochondrial genotypes interacting with the two nuclear genomes. However, 0% of the variance in relative fitness could be attributed to between-species differences in mtDNA. Mitochondrial-X chromosome interaction effects on relative fitness were significant only in males (Table 3), with 21.3% of the variance explained by variation among D. melanogaster X chromosomes. This X-linked variation interacted with within-species mitochondrial genotypes to explain 2.23% of the variance in relative fitness. However, none of the variance in male relative fitness could be explained by mtDNA differences between D. melanogaster and D. simulans.
Mixed models incorporating a random effect of mitochondrial genotype nested within a fixed effect of species mtDNA revealed no effect of species mtDNA on female fitness in the mitochondrial-nuclear experiment (F = 1.24, P = 0.47) and no effect of species mtDNA on male fitness in the mitochondrial-X chromosome experiment (F = 0.01, P = 0.94). On average, substituting a D. melanogaster mtDNA into a D. melanogaster nuclear background has as great of an effect on relative fitness as substituting a D. simulans mtDNA into a D. melanogaster nuclear background. Furthermore, there is no tendency for D. simulans or D. mauritiana mtDNA introgression to consistently increase or decrease fitness relative to D. melanogaster mtDNAs. These results suggest that mitochondrial-nuclear incompatibilities affecting egg-to-adult competitive viability do not scale with molecular divergence and have not accumulated between these species.
Male and female mitochondrial-nuclear epistasis
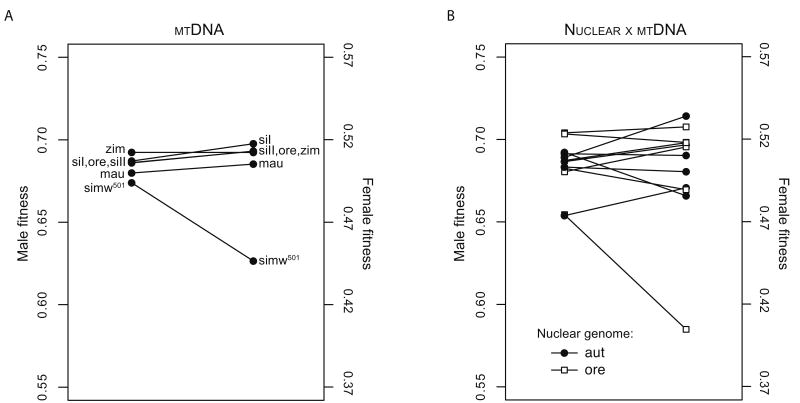
The segregation assay used here has greater power to detect relative fitness effects in males than in females, as males experience the full effects of all interactions between the mtDNA and the wild type or FM6 X chromosomes regardless of dominance. For example, a completely recessive synthetic lethal interaction between a D. simulans mtDNA and the FM6 chromosome would appear to be a male-specific mitochondrial-nuclear interaction in this assay. Despite this bias, we detected significant effects of mitochondrial-nuclear interactions on female fitness but not in males (Table 2). Although these interactions are stronger in females, Figure 4 reveals a consistent pattern of mitochondrial-nuclear genotype effects on male and female fitness. These effects are magnified in females, particularly the severe effect of the (simw501);Ore genotype. Comparing male and female fitness values reveals little sexual antagonism across mitochondrial-nuclear genotypes (Figure 7B). Mitochondrial-nuclear genotypic combinations that confer low fitness in females also confer low fitness in males, generating a significant correlation between male and female fitness values across the twelve mitochondrial-nuclear genotypes (Pearson's r = 0.71, P = 0.009).
Figure 7.
Comparisons of male and female relative fitness effects of mitochondrial (A) and mitochondrial-nuclear (B) genotypes. The y-axes are shifted relative to one another, but remain on the same scale. The nuclear genome has no main effect on relative fitness in either sex and is not shown.
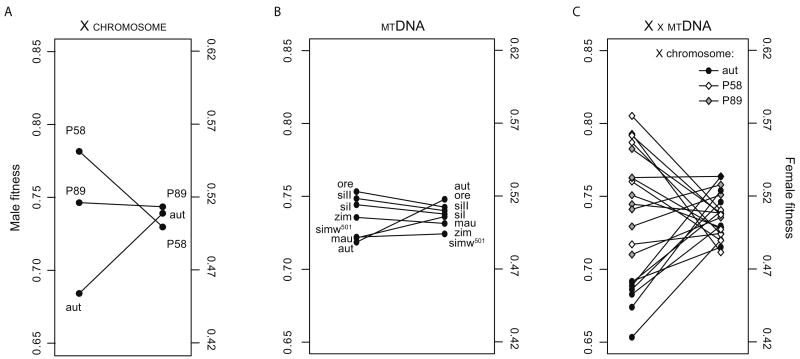
In contrast, the X-mitochondrial segregation assay indicated that some mitochondrial-X chromosome genotypes have different fitness effects in males and females. For example, the Aut mtDNA confers the lowest relative fitness in males but the highest relative fitness in females (Figure 8B). There is more male than female fitness variation among mitochondrial-X chromosome genotypes, and there is little congruence between the male and female fitness effects of any given genotype (Figure 8C). In contrast to the mitochondrial-nuclear data set, there is no correlation among male and female mean fitness values across mitochondrial-X chromosome genotypes (Pearson's r = -0.12, P = 0.60). However, this absence of a correlation may be driven by the strong sex-specific main effects of the X chromosomes (Figure 8A).
Figure 8.
Comparisons of male and female relative fitness effects of X chromosome (A), mitochondrial (B) and mitochondrial-X chromosome (C) genotypes. The y-axes are shifted relative to one another, but remain on the same scale.
Discussion
Mitochondrial-nuclear interactions for fitness result from segregating variation rather than fixed differences between species
Our results provide evidence for epistatic fitness interactions between mitochondrial polymorphisms segregating within D. melanogaster subgroup species and nuclear polymorphisms segregating in D. melanogaster. The strongest interactions in males were between D. melanogaster mtDNAs and D. melanogaster X chromosomes (Figure 6), two of which were sampled from the same population. In females there was a strong epistatic interaction between two D. simulans siII mtDNAs and the two D. melanogaster nuclear backgrounds (Figure 4). Unique among the D. simulans species complex mtDNAs, the simw501 mtDNA strongly impacts fitness when combined with the D. melanogaster Ore nuclear genetic backgound. The D. simulans simw501 mtDNA differs from the other D. simulans siII mtDNAs by only three nucleotide substitutions (Meiklejohn, Montooth and Rand, unpublished result). Thus, this case of intergenomic epistasis does not arise from the more than 500 substitutions that have fixed between the mtDNAs of D. melanogaster and the D. simulans species complex. Rather, a small number of polymorphic sites that distinguish the D. simulans simw501 mtDNA from other D. simulans siII mtDNAs interact epistatically with nuclear alleles that are segregating within D. melanogaster as potentially neutral, slightly deleterious or population-specific polymorphisms.
In both the mitochondrial-nuclear and mitochondrial-X chromosome experiments we observed no consistent increase or decrease in relative fitness associated with substituting the D. melanogaster versus the D. simulans species complex mtDNAs (Figures 4,6). As a result, 0% of the variance observed in relative fitness can be attributed to an effect of the species from which the mtDNA was derived. These results suggest that the fixed differences between the mtDNAs of these species are largely neutral and fully compatible with the D. melanogaster nuclear genome, although we cannot rule out strong effects of individual mtDNA substitutions that were subsequently compensated by other mitochondrial substitutions. This result is inconsistent with the accumulation of mitochondrial-nuclear incompatibilities that would arise from a process of divergent co-evolution between these mitochondrial and nuclear lineages. However, we tested for mitochondrial-nuclear effects on egg-to-adult competitive viability in a single environment. It remains possible that components of adult fitness are more sensitive to mitochondrial-nuclear incompatibilities and that these incompatibilities experience environment- or sex-specific effects (e.g. Chippindale et al. 2001).
D. melanogaster is strongly reproductively isolated from D. simulans, D. mauritiana, and D. sechellia, as interspecific crosses involving D. melanogaster normally produce sterile and/or inviable hybrid progeny (Sturtevant 1920; Lachaise et al 1986). Despite this, our experiments show that the approximately 100 non-synonymous and 400 synonymous fixed differences between D. melanogaster and its sibling species mtDNAs do not cause lethal or sterile incompatibilities in combination with the D. melanogaster nuclear genome, consistent with prior experimental introgressions of D. simulans mitochondria into D. melanogaster (Sawamura et al. 2000). Although we did not introgress mtDNAs in the reciprocal direction, the fact that viable hybrids of both sexes and fertile female hybrids can be recovered from D. melanogaster mothers and sibling species fathers (Hutter & Ashburner 1987; Barbash & Ashburner 2003) indicates that the D. melanogaster mtDNA is also largely compatible with the D. simulans nuclear genome. Mitochondrial-nuclear compatibility may be common among related Drosophilids, as mitochondrial introgression has occurred in nature between closely related species of Drosophila (Powell 1983; Machado & Hey 2003; Bachtrog et al. 2006).
Expression of X-linked variation and its interaction with the mtDNA was more pronounced in males than in females (Figure 8). However, due to the design of the segregation assay, any completely dominant alleles that differed between the wild-type X chromosomes would be detected in males but not in females. The male-specific effects of the X chromosome observed here should therefore be interpreted cautiously. In contrast, fitness effects in the mitochondrial-nuclear experiment were significant only in females. In particular, the (simw501);Ore genotype has a pronounced effect on female relative fitness, but little effect in males. This genotype exhibits a number of other deleterious phenotypes, including reduced female fecundity, and increased development time and compromised bristle development in both sexes (Meiklejohn, Montooth and Rand, unpublished results). These data suggest that nuclear and mitochondrial genomes in Drosophila can segregate variants, which alone may have little effect on fitness, but when combined result in strong epistatic effects that may be expressed differently in males and females.
Natural selection is not expected to maintain joint mitochondrial-autosomal polymorphism in the absence of frequency-dependent selection or differential selection in the sexes (Clark 1984; Gregorius & Ross 1984). However, the co-transmission of the mtDNA with the X chromosomes through females can maintain joint mitochondrial-X chromosome polymorphisms, particularly when these polymorphisms have sex-specific effects (Rand et al. 2001; Dowling et al. 2008). Thus, while the interactions we observed might result from segregating deleterious variants that have not yet been removed by purifying selection, it is also possible that they reflect variation maintained by sex-dependent fitness effects of mitochondrial-X chromosome interactions. The observed phenotypic effects of D. simulans mitochondrial haplotypes (James & Ballard 2003; Ballard et al. 2007), and the fact that interactions between nuclear and mitochondrial genomes are stronger between populations than within populations of D. melanogaster (Clark & Lyckegaard 1988; Rand et al. 2001; Dowling et al. 2007b) raises the possibility that local adaptation and population structure may also maintain mitochondrial variation within these species.
Population genetics of mitochondrial-nuclear interactions
Efficient purifying selection prevents the fixation of deleterious mutations, but, until removed, mildly deleterious polymorphisms will segregate within populations. D. melanogaster and D. simulans populations harbor an excess of non-synonymous mitochondrial polymorphism and these polymorphisms segregate at lower frequencies relative to the neutral expectation, consistent with a slightly deleterious model of molecular evolution (Ballard & Kreitman 1994; Rand & Kann 1996, 1998). The frequency and fate of such variants depends on their fitness effects and the effective population size (Ne). Complete linkage and uniparental inheritance reduces mtDNA Ne relative to nuclear loci and reduces the efficacy of natural selection, making the mtDNA particularly prone to the fixation of deleterious polymorphisms (Gabriel et al. 1993; Lynch 1996; Neiman & Taylor 2009).
Compensatory evolution at nuclear-encoded loci that recovers fitness loss due to high-frequency or fixed deleterious mtDNA variants has the potential to result in the accumulation of mitochondrial-nuclear incompatibilities between diverging lineages (Rand et al. 2004; Dowling et al. 2008). We find no evidence that these incompatibilities have accumulated within the D. melanogaster species group, consistent with strong purifying selection acting on the Drosophila mitochondrial genome (Ballard 2000; Montooth et al. 2009). In the D. melanogaster species subgroup, it appears that deleterious mitochondrial variants are efficiently removed from populations before compensatory mutations that might resolve any deleterious mitochondrial effects are fixed in the nuclear genome.
The compatibility between the mitochondrial and nuclear genomes of the D. melanogaster species group contrasts with the results from three opisthokont experimental systems. First, the inability of a S. bayanus nuclear-encoded protein to translate the S. cerevisiae mitochondrial OLI1 mRNA is responsible for a sporulation defect in hybrids (Lee et al 2008). This incompatibility appears to have been driven by rapid divergence between the S. cerevisiae and S. bayanus mitochondrial OLI1 5′ UTR (Lee et al 2008). Such rapid evolution is characteristic of yeast mitochondrial intergenic sequences (Groth et al 2000). Second, mitochondrial-nuclear incompatibilities have been identified among the parasitic wasps of the Nasonia species complex (Breeuwer & Werren 1995; Ellison et al. 2008; Niehuis et al. 2008). Nasonia has a mitochondrial substitution rate that is 30 times higher than the nuclear substitution rate (Oliveira et al. 2008). This substitution rate is hypothesized to have driven evolution of the nuclear-encoded genes of the oxidative phosphorylation pathway and the accumulation of mitochondrial-nuclear incompatibilities among Nasonia species that decrease hybrid fitness (Oliveira et al. 2008; Gibson et al. 2010; The Nasonia Genome Working Group 2010). Third, populations of the marine copepod T. californicus have mitochondrial substitution rates that are 25-fold higher than nuclear rates and experience mitochondrial-nuclear incompatibilities that decrease hybrid fitness (Burton et al. 2006; Ellison & Burton 2008a). In contrast, across Drosophilids, the mitochondrial synonymous substitution rate is only 2.75 times the nuclear substitution rate (Montooth et al. 2009).
These studies suggest that the rate of substitution in the mtDNA may be a critical parameter for the accumulation of incompatibilities driven by compensatory evolution between the mitochondrial and nuclear genomes. There are a number of possible reasons why D. melanogaster and D. simulans might have a lower ratio of mitochondrial to nuclear substitution rates than other taxa. First, mitochondrial mutation pressures could be reduced in Drosophila. In D. melanogaster mtDNA mutations occur almost exclusively at G:C base pairs and convert G:C to A:T (Haag-Liautard et. al 2008). This biased mutational process, combined with the low G+C content in the Drosophila mtDNA, may be why D. melanogaster has a lower ratio of mitochondrial to nuclear mutation rates than S. cerevisiae, C. elegans, and humans (Lynch et al. 2008; Montooth & Rand 2008). Second, Drosophila effective population sizes may simply be large enough that purifying selection can efficiently prevent the fixation of deleterious mtDNA mutations.
Mitochondrial evolutionary dynamics are also influenced by cytoplasmically inherited endosymbionts that sweep through populations via their effects on host reproduction (Hoffmann & Turelli 1997), and are posited to have driven mitochondrial-nuclear co-evolution in Nasonia (Oliveira et al. 2008, Raychoudhury et al. 2009, 2010). Cytoplasmic incompatibility from Wolbachia infection is currently weak and uncommon in D. melanogaster (Merçot & Charlat 2004; Fry et al 2004). However, there are at least five systems of Wolbachia-induced cytoplasmic incompatibility known from D. simulans (Clancy and Hoffmann 1996), suggesting that endosymbiont-driven cytoplasmic sweeps are likely to have occurred in Drosophila as well as Nasonia. Wolbachia has been implicated in mtDNA evolution in other Drosophila species groups (Shoemaker et al. 2004; Bachtrog et al. 2006), providing a comparative context in which to explore the relationship between cytoplasmic sweeps, mtDNA substitution rates and the accumulation of mitochondrial-nuclear incompatibilities.
Context dependence of intergenomic epistasis
If females and males are considered to constitute different cellular and physiological environments in which genes function, then the sex-dependence of mitochondrial-nuclear interactions reflect the complex interactions that exist between genetic and environmental factors (e.g. Bergland et al. 2008), which may be common for mitochondrial-nuclear interactions (Dowling et al. 2007a, Rand et al. 2001). The existence of this complexity highlights the need for fitness studies of mtDNA to include multiple nuclear backgrounds and rigorous genetic controls, as fitness differences in one background could be reversed in other genetic backgrounds. Resolving the relative contributions of mutation and substitution rates, genetic drift and genetic draft (Gillespie 2000; Bazin et al. 2006; Meiklejohn et al. 2007), and complex epistasis remains a significant challenge for the understanding of the evolution of the mtDNA and its interaction with nuclear loci. However, the mitochondrial-nuclear genotype as a unit of selection, particularly in species where this genotype can be manipulated, offers the potential to explore how complex genetic interactions influence the evolutionary process (Gillespie & Turelli 1989; Whitlock et al. 1995; Barton & Turelli 2004; Phillips 2008). The well-characterized physiological function encoded by the mitochondrial-nuclear genotype provides the opportunity to functionally dissect how genetic interactions are expressed in an environment- and sex-dependent fashion to influence metabolic fitness. This will be an important step in characterizing how genomic loci cooperate and conflict, as well as how metabolic physiologies evolve.
Acknowledgments
We thank Corbin Jones for sage advice on how to get D. mauritiana females to mate with D. simulans males. The manuscript was greatly improved by comments from Daven Presgraves and four anonymous reviewers. This work was funded by NIH grant GM067862 to D.M.R., and NIH NRSA fellowhips GM072399 to C.D.M. and GM076812 to K.L.M.
Literature Cited
- Aruna S, Flores HA, Barbash DA. Reduced fertility of Drosophila melanogaster Hybrid male rescue (Hmr) mutant females is partially complemented by Hmr orthologs from sibling species. Genetics. 2009;181:1437–1450. doi: 10.1534/genetics.108.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Thornton K, Clark A, Andolfatto P. Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution. 2006;60:292–302. [PubMed] [Google Scholar]
- Ballard JW. Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J Mol Evol. 2000;51:48–63. doi: 10.1007/s002390010066. [DOI] [PubMed] [Google Scholar]
- Ballard JW, Kreitman M. Unraveling selection in the mitochondrial genome of Drosophila. Genetics. 1994;138:757–72. doi: 10.1093/genetics/138.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JW, Melvin RG, Katewa SD, Maas K. Mitochondrial DNA variation is associated with measurable differences in life-history traits and mitochondrial metabolism in Drosophila simulans. Evolution. 2007;61:1735–1747. doi: 10.1111/j.1558-5646.2007.00133.x. [DOI] [PubMed] [Google Scholar]
- Ballard JW, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Evol S. 2005;36:621–642. [Google Scholar]
- Barbash DA, Ashburner M. A novel system of fertility rescue in Drosophila hybrids reveals a link between hybrid lethality and female sterility. Genetics. 2003;163:217–226. doi: 10.1093/genetics/163.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Turelli M. Effects of genetic drift on variance components under a general model of epistasis. Evolution. 2004;58:2111–2132. doi: 10.1111/j.0014-3820.2004.tb01591.x. [DOI] [PubMed] [Google Scholar]
- Bazin E, Glemin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- Bergland AO, Genissel A, Nuzhdin SV, Tatar M. Quantitative trait loci affecting phenotypic plasticity and the allometric relationship of ovariole number and thorax length in Drosophila melanogaster. Genetics. 2008;180:567–582. doi: 10.1534/genetics.108.088906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer JAJ, Werren JH. Hybrid breakdown between 2 haplodiploid species - the role of nuclear and cytoplasmic genes. Evolution. 1995;49:705–717. doi: 10.1111/j.1558-5646.1995.tb02307.x. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RS, Ellison CK, Harrison JS. The sorry state of F2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. The American Naturalist. 2006;168 6:S14–24. doi: 10.1086/509046. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Gibson JR, Rice WR. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc Natl Acad Sci USA. 2001;98:1671–5. doi: 10.1073/pnas.041378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Hoffmann AA. Cytoplasmic incompatibility in Drosophila simulans: evolving complexity. Trends Ecology Evolution. 1996;11:145–146. doi: 10.1016/0169-5347(96)30006-2. [DOI] [PubMed] [Google Scholar]
- Clark A. Natural selection with nuclear and cytoplasmic transmission. I. A deterministic model. Genetics. 1984;107:679–701. doi: 10.1093/genetics/107.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Lyckegaard EM. Natural selection with nuclear and cytoplasmic transmission. III. Joint analysis of segregation and mtDNA in Drosophila melanogaster. Genetics. 1988;118:471–481. doi: 10.1093/genetics/118.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark ME, Anderson CL, Cande J, Karr TL. Widespread prevalence of Wolbachia in laboratory stocks and the implications for Drosophila research. Genetics. 2005;170:1667–1675. doi: 10.1534/genetics.104.038901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AW, Roote J, Morley T, Sawamura K, Herrmann S, Ashburner M. Rescue of hybrid sterility in crosses between D. melanogaster and D. simulans. Nature. 1996;380:157–159. doi: 10.1038/380157a0. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Abiega KC, Arnqvist G. Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution. 2007a;61:194–201. doi: 10.1111/j.1558-5646.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Hailer F, Arnqvist G. Intergenomic epistasis for fitness: within-population interactions between cytoplasmic and nuclear genes in Drosophila melanogaster. Genetics. 2007b;175:235–244. doi: 10.1534/genetics.105.052050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Lindell J. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends in Ecology & Evolution. 2008;23:546–554. doi: 10.1016/j.tree.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Edmands S, Burton RS. Cytochrome C oxidase activity in interpopulation hybrids of a marine copepod: a test for nuclear-nuclear or nuclear-cytoplasmic coadaptation. Evolution. 1999;53:1972–1978. doi: 10.1111/j.1558-5646.1999.tb04578.x. [DOI] [PubMed] [Google Scholar]
- Ellison CK, Burton RS. Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution. 2006;60:1382–1391. [PubMed] [Google Scholar]
- Ellison CK, Burton RS. Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution. 2008a;62:631–638. doi: 10.1111/j.1558-5646.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- Ellison CK, Burton RS. Genotype-dependent variation of mitochondrial transcriptional profiles in interpopulation hybrids. Proc Natl Acad Sci USA. 2008b;105:15831–6. doi: 10.1073/pnas.0804253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Niehuis O, Gadau J. Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. Journal of Evolutionary Biology. 2008;21:1844–1851. doi: 10.1111/j.1420-9101.2008.01608.x. [DOI] [PubMed] [Google Scholar]
- Fry AJ, Palmer MR, Rand DM. Variable fitness effects of Wolbachia infection in Drosophila melanogaster. Heredity. 2004;93:379–89. doi: 10.1038/sj.hdy.6800514. [DOI] [PubMed] [Google Scholar]
- Gabriel W, Lynch M, Burger R. Muller's Ratchet and Mutational Meltdowns. Evolution. 1993;47:1744–1757. doi: 10.1111/j.1558-5646.1993.tb01266.x. [DOI] [PubMed] [Google Scholar]
- Gibson JD, Niehuis O, Verrelli BC, Gadau J. Contrasting patterns of selective constraints in nuclear-encoded genes of the oxidative phosphorylation pathway in holometabolous insects and their possible role in hybrid breakdown in Nasonia. Heredity. 2010;104:310–317. doi: 10.1038/hdy.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics. 2000;155:909–919. doi: 10.1093/genetics/155.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH, Turelli M. Genotype-Environment Interactions and the Maintenance of Polygenic Variation. Genetics. 1989;121:129–138. doi: 10.1093/genetics/121.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorius HR, Ross MD. Selection with Gene-Cytoplasm Interactions. I. Maintenance of Cytoplasm Polymorphisms. Genetics. 1984;107:165–178. doi: 10.1093/genetics/107.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth C, Petersen RF, Piskur J. Diversity in organization and the origin of gene orders in the mitochondrial DNA molecules of the genus Saccharomyces. Mol Biol Evol. 2000;17:1833–41. doi: 10.1093/oxfordjournals.molbev.a026284. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C, Coffey N, Houle D, Lynch M, Charlesworth B, Keightley PD. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 2008;6:e204. doi: 10.1371/journal.pbio.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn E. Zur Autonomie und Phasenspezifität der Latalität von Bastarden zwischen Drosophila melanogaster und Drosophila simulans. Rev Suisse Zool. 1961;68:197–207. [Google Scholar]
- Haldane J. The estimation of viabilities. Journal of Genetics. 1956;54:294–296. [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hoffmann AA, Turelli M. Cytoplasmic incompatibility in insects. In: O'Neill S, Hoffmann A, Werren J, editors. Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press; New York: 1997. pp. 42–80. [Google Scholar]
- Hutter P, Ashburner M. Genetic rescue of inviable hybrids between Drosophila melanogaster and its sibiling species. Nature. 1987;327:331–333. doi: 10.1038/327331a0. [DOI] [PubMed] [Google Scholar]
- Hutter P, Roote J, Ashburner M. A genetic basis for the inviability of hybrids between sibling species of Drosophila. Genetics. 1990;124:909–920. doi: 10.1093/genetics/124.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yania I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–40. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Broughton S, Alic N, Grandison R, Partridge L. The endosymbiont Wolbachia increases insulin/IGF-like signalling in Drosophila. Proc Biol Sci. 2009;276:3799–3807. doi: 10.1098/rspb.2009.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AC, Ballard JW. Mitochondrial genotype affects fitness in Drosophila simulans. Genetics. 2003;164:187–194. doi: 10.1093/genetics/164.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mtuation, sterility and male recombination. Genetics. 1977;86:813–833. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, David JR, Lemeunier F, Tsacas L. The reproductive relationships of Drosophila sechellia with D. mauritiana, D. simulans, and D. melanogaster from the Afrotropical region. Evolution. 1986;40(2):262–271. doi: 10.1111/j.1558-5646.1986.tb00468.x. [DOI] [PubMed] [Google Scholar]
- Lee HY, Chou JY, Cheong L, Chang NH, Yang SY, Leu JY. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135:1065–73. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Lynch M. Mutation accumulation in transfer RNAs: molecular evidence for Muller's ratchet in mitochondrial genomes. Mol Biol Evol. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, Thomas WK. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Hey J. The causes of phylogenetic conflict in a classic Drosophila species group. P Roy Soc Lond B Bio. 2003;270:1193–1202. doi: 10.1098/rspb.2003.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genetical Research. 1974;23:23–35. [PubMed] [Google Scholar]
- Meiklejohn CD, Montooth KL, Rand DM. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007;23:259–263. doi: 10.1016/j.tig.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Merçot H, Charlat S. Wolbachia infections in Drosophila melanogaster and D. simulans: polymorphism and levels of cytoplasmic incompatibility. Genetica. 2004;120:51–59. doi: 10.1023/b:gene.0000017629.31383.8f. [DOI] [PubMed] [Google Scholar]
- Montooth KL, Abt DN, Hofmann J, Rand DM. Comparative genomics of Drosophila mtDNA: Novel features of conservation and change across functional domains and lineages. J Mol Evol. 2009;69:94–114. doi: 10.1007/s00239-009-9255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth KL, Rand DM. The spectrum of mitochondrial mutation differs across species. PLoS Biol. 2008;6:e213. doi: 10.1371/journal.pbio.0060213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- The Nasonia Genome Working Group. Functional and Evolutionary Insights from the Genomes of Three Parasitoid Nasonia Species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman M, Taylor DR. The causes of mutation accumulation in mitochondrial genomes. Proc Biol Sci. 2009;276:1201–1209. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, Judson AK, Gadau J. Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics. 2008;178:413–426. doi: 10.1534/genetics.107.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, Raychoudhury R, Lavrov DV, Werren JH. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (hymenoptera: pteromalidae) Mol Biol Evol. 2008;25:2167–2180. doi: 10.1093/molbev/msn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. Epistasis - the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: evidence from Drosophila. Proc Natl Acad Sci U S A 80. 1983:492–5. doi: 10.1073/pnas.80.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Clark AG, Kann LM. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics. 2001;159:173–187. doi: 10.1093/genetics/159.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ. Cytonuclear cooperation: the genomics of cooperation. Trends in Ecology & Evolution. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–48. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Mutation and selection at silent and replacement sites in the evolution of animal mitochondrial DNA. Genetica. 1998;102-103:393–407. [PubMed] [Google Scholar]
- Raychoudhury R, Baldo L, Oliveira DC, Werren JH. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution. 2009;63:165–183. doi: 10.1111/j.1558-5646.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- Raychoudhury R, Grillenberger BK, Gadau J, Bijlsma R, van de Zande L, Werren JH, Beukeboom LW. Phylogeography of Nasonia vitripennis (Hymenoptera) indicates a mitochondrial-Wolbachia sweep in North America. Heredity. 2010;104:318–326. doi: 10.1038/hdy.2009.160. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Haney RA, Rand DM. Cytonuclear coadaptation in Drosophila: Disruption of cytochrome c oxidase activity in backcross genotypes. Evolution. 2003:2315–2325. doi: 10.1111/j.0014-3820.2003.tb00243.x. [DOI] [PubMed] [Google Scholar]
- Sawamura K, Davis AW, Wu CI. Genetic analysis of speciation by means of introgression into Drosophila melanogaster. PNAS 97. 2000:2652–2655. doi: 10.1073/pnas.050558597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker DD, Dyer KA, Ahrens M, McAbee K, Jaenike J. Decreased diversity but increased substitution rate in host mtDNA as a consequence of Wolbachia endosymbiont infection. Genetics. 2004;168:2049–2058. doi: 10.1534/genetics.104.030890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics. 1920;5:488–500. doi: 10.1093/genetics/5.5.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M, Phillips P, Moore F, Tonsor S. Multiple fitness peaks and epistasis. Annual Review of Ecology and Systematics. 1995;26:601–629. [Google Scholar]