Abstract
Iron is an essential nutrient that plays a role in bacterial differential gene expression and protein production. Accordingly, the comparative analysis of total lysate and outer membrane fractions isolated from A. baumannii ATCC 19606T cells cultured under iron-rich and -chelated conditions using 2-D gel electrophoresis-mass spectrometry resulted in the identification of 58 protein spots differentially produced. While 19 and 35 of them represent iron-repressed and iron-induced protein spots, respectively, four other spots represent a metal chelation response unrelated to iron. Most of the iron-repressed protein spots represent outer membrane siderophore receptors, some of which could be involved in the utilization of siderophores produced by other bacteria. The iron-induced protein spots represent a wide range of proteins including those involved in iron storage, such as Bfr, metabolic and energy processes, such as AcnA, AcnB, GlyA, SdhA, and SodB, as well as lipid biosynthesis. The detection of an iron-regulated Hfq ortholog indicates that iron regulation in this bacterium could be mediated by Fur and small RNAs as described in other bacteria. The iron-induced production of OmpA suggests this protein plays a role in iron metabolism as shown by the diminished ability of an OmpA isogenic deficient derivative to grow under iron-chelated conditions.
Keywords: A. baumannii, iron, iron regulation, total cell proteins, outer membrane proteins
1. Introduction
Acinetobacter baumannii is a Gram-negative aerobic coccobacillus recognized for its ability to cause severe nosocomial infections including pneumonia, urinary tract and burn infections, secondary meningitis and systemic infections [1, 2]. More recently, this pathogen has emerged as a threat to soldiers wounded during military operations in Iraq and Afghanistan [3, 4]. The high adaptability of this opportunistic pathogen coupled with its ability to resist a wide range of antibiotics and persist in medical environments underscores the clinical threat posed by this pathogen to critically ill hospitalized patients, including those suffering from heavy trauma such as military victims or victims of natural disasters [2, 5].
When colonizing a host, bacterial pathogens including A. baumannii, must compete with the host for essential nutrients. One of the most coveted nutrients in biological systems is iron, due to its essentiality to almost all living organisms and limited availability under physiological conditions [6]. This micronutrient plays an essential role in a diverse number of cellular processes including electron transport, nucleic acid biosynthesis, and protection from free radicals [7–10]. The iron concentration is tightly regulated within living systems because excess Fe can induce oxidative damage. Thus, in host tissues, the availability of free Fe is minimal, as most Fe is sequestered by high-affinity iron-binding proteins such as transferrin and lactoferrin [7–10]. Although low free-Fe concentration portends a non-specific host defense mechanism against infection, most successful pathogens use it as a stimulus to express not only active iron-acquisition systems, as it was described for A. baumannii [11], but also the expression of genes coding for virulence factors such as hemolysins, toxins, and proteases [12]. Iron is therefore considered to be not only an essential nutrient, but also an important signal for global regulation of gene expression in prokaryotes [13].
In Gram-negative bacteria, iron-regulated gene expression is generally under the control of the Fur (ferric uptake regulator) protein first described in Escherichia coli [14] and produced by a wide range of bacteria. This protein operates as a classical repressor when bound to Fe by inhibiting transcription from iron-regulated gene promoters in response to increase in Fe concentration [15]. Fur has also been shown to indirectly induce gene expression [16–18]. In E. coli, the positive regulatory effect of Fur on gene expression involves the Fur-mediated repression of the small RNA (sRNA) RhyB [19, 20]. A similar mechanism involving the PrrF1 and PrrF2 sRNA molecules was reported in Pseudomonas aeruginosa [21]. Although there is a large body of information related to iron acquisition and gene regulation in Gram-negative bacteria [13], little is known on the effect of this metal on differential gene expression in A. baumannii. This pathogen expresses active siderophore-mediated iron acquisition systems [11] and produces a Fur protein [22, 23], which is highly related to a large number of orthologs described in several other bacterial species. This iron repressor controls A. baumannii differential gene expression in response to changes in free-iron concentrations in the extracellular environment [23].
The field of proteomics is gaining recognition as a reliable and reproducible high-throughput approach to examine biological processes at the molecular level that in the case of A. baumannii has already provided important information on its metabolic versatility [24], the composition of outer membrane vesicles [25] and the effect of antibiotics and salts in differential protein production [26–28]. In this study, we employed a global proteomic approach based on 2-D gel electrophoresis (2-DE) and mass spectrometry to examine the differential production of proteins by the A. baumannii ATCC 19606T type strain when cultured under Fe-replete or Fe-chelated conditions. Our results show that free-iron availability affects the production of proteins involved in siderophore-mediated iron acquisition and storage functions, as well as proteins involved in metabolic processes. Our data also suggest that the expression of iron-regulated genes in this bacterium could be controlled by the iron repressor Fur and potential sRNA regulators.
2. Materials and Methods
2.1. Bacterial strains and culture conditions
The A. baumannii ATCC 19606T strain was routinely cultured in Luria-Bertani (LB) broth or agar [29] at 37 °C. Iron-rich and -chelated conditions were achieved by adding 100 μM FeCl3 dissolved in 0.1 M HCl and 100 μM 2,2′-dipyridyl (DIP), respectively. For proteomic experiments, cells were cultured with agitation (200 rpm) for 7 h at 37 °C in 1 ml of unsupplemented (−Fe/−DIP) LB broth or broth supplemented with 100 μM FeCl3 (+Fe/–DIP), 100 μM DIP (−Fe/+DIP), or 100 μM DIP plus 100 μM FeCl3 (+Fe/+DIP). The last three culture conditions were used to detect the production of iron-induced and iron-repressed proteins and determine whether protein changes observed in the presence of DIP were due to iron chelation, respectively. Each 1-ml culture was used to inoculate 100-ml LB broth samples of the corresponding composition that were incubated with shaking (200 rpm) at 37 °C for 14 h, at which time cultures reached stationary phase as determined by monitoring cell growth spectrophotometrically at OD600nm. Five culture replicates were used for each treatment condition and the entire culture procedure was repeated three times using fresh biological samples each time. Replicate samples cultured under the same conditions were pooled together and the cells were collected by centrifugation at 7,000g for 10 min at 4 °C. The cell pellets were suspended in 10 ml Tris-buffered saline (TBS) [30] and stored at −80 °C as the sample stock for each treatment.
2.2. Protein extraction
Total cell (TC) protein samples were prepared by collecting cells from 1 ml of each sample stock by centrifugation at 7,000g for 10 min at 4 °C. Cell pellets were suspended in 0.5 ml lysis buffer [5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM EDTA, 0.5% Tween-20, 25 mM Tris-HCl pH 7.0] and lysed by sonication while keeping samples on ice. Cell debris and unlysed cells were collected by centrifugation as described above and the supernatants, representing the TC protein samples, were carefully decanted into clean tubes and stored at −80 °C. Outer membrane (OM) enriched fractions were prepared by collecting cells from 9-ml sample stocks as described above. Cell pellets were rinsed with 25 ml ice-cold HEPES buffer (5 mM MgSO4, 1 mM PMSF, 2 mM EDTA, 50 mM HEPES potassium, pH 7.4) and then suspended in 10 ml ice-cold HEPES buffer and lysed by sonication on ice. After removing cell debris and unlysed cells as described above, the supernatants were centrifuged at 200,000g for 1 h at 4 °C. The pellets were washed unperturbed three times with 5 ml ice-cold HEPES buffer, suspended in 10 ml ice-cold HEPES buffer containing 2% Triton X-100 and incubated on ice for 1 h. The suspension was centrifuged again at 200,000g for 1 h at 4 °C and the pellet representing the OM fraction was suspended in 5 ml ice-cold HEPES buffer and stored at −80 °C until further analysis.
To precipitate proteins, one volume of ice-cold acetone was added to TC and OM fractions followed by a 30-min incubation on ice and centrifugation at 10,000g for 10 min at 4 °C. The pellet was solubilized in 0.5 ml of rehydration/isoelectric focusing (IEF) buffer [8M urea, 50 mM DTT, 4% (w/v) CHAPS, 0.2% (v/v) 3/10 ampholytes and 0.002% (w/v) bromophenol blue]. Insoluble material was removed by centrifugation at 10,000g for 10 min at RT and the protein content of the supernatant was determined with the bicinchoninic acid (BCA) protein quantification assay following the manufacturer’s instructions (Pierce, Rockford, IL, USA).
2.3. 2-DE fractionation and image analysis
For first dimension electrophoresis, 11-cm long pH 4-7 ReadyStrip IPG strips (Bio-Rad, Hercules, CA, USA) were passively rehydrated overnight at RT with 200 μg of protein extract suspended in IEF buffer. IEF was done using a PROTEAN IEF cell (Bio-Rad) at a current limit of 50 μA/strip at 10 °C with the following steps: active rehydration at 250 V for 11 h; 250 V (linear) for 15 min; 8 kV (linear) for 3 h; and 10 kV (rapid) until a total 80 kVh for a combined total of approximately 92 kVh. Each focused IPG strip was equilibrated by soaking, with mild stirring, in 4 ml of equilibration base buffer 1 (EBB1) [8M urea, 2% (w/v) sodium dodecyl sulfate (SDS), 50 mM Tris-HCl (pH 8.8), 20% (v/v) glycerol, 1% (w/v) DTT] for 10 min, followed by soaking in 4 ml of EBB2 [same content as EBB1 except DTT was replaced with 2.5% (w/v) iodoacetamide (IAA)]. Second dimension electrophoresis (SDS-PAGE) was performed in 8–16% gradient SDS-polyacrylamide Tris-HCl gels (Criterion precast gels, Bio-Rad) in a twelve-gel cell system (Criterion Dodeca Cell, Bio-Rad). Protein spots were visualized by staining with Coomassie Brilliant Blue using the method developed by Wang et al. [31]. Stained gels were scanned using the Bio-Rad VersaDoc Imaging System and gel images were analyzed using the PDQuest software package (version 7.3.0, Bio-Rad). Gels were preserved in 5% acetic acid at 4 °C until further analysis. All 2-DE experiments were carried out in triplicate.
Spots from TC or OM protein gels were matched so that a given spot had the same number across all gels. A master gel image containing matched spots across all gels was auto-generated for TC or OM gels. Spot volumes were normalized according to the total gel image density as suggested by the PDQuest software package. The gels from all four treatments (−Fe/−DIP, +Fe/−DIP, −Fe/+DIP and +Fe/+DIP) were compared by constructing four different replicate groups (each replicate group contained the gel images corresponding to a specific treatment). In each group, the average quantity for each spot was determined and pair-wise quantitative and statistical analysis sets were generated by comparing the same spot across each of the other groups. Pair-wise comparisons to determine significant differences in spot volumes between treatments were performed on standardized log10 values of protein spot volumes using the Student’s t-test analysis at a ≥ 95% confidence interval (P ≤ 0.05) as provided by the PDQuest software. Spots that showed at least two-fold change in volume (P ≤ 0.05) compared to at least one other treatment were further analyzed.
2.4. Protein digestion and identification via mass spectrometry
Protein identification was achieved by peptide mass fingerprinting (PMF) of trypsin digested protein spots. Briefly, protein spots were manually excised, reduced with DTT, alkylated with IAA and then digested with sequence grade trypsin in the presence of ProteaseMAX™ Surfactant according to the manufacturer’s protocol (Promega, Madison, WI). Tryptic-digests were then analyzed by Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (Bruker Ultraflex III MALDI-TOF/TOF Mass Spectrometer, Bruker, Billerica, MA, USA). The target plate was spotted with 0.02 ml of a 1:1 (v/v) mixture of tryptic-digest and matrix solution [10 mg/ml α-cyano-4-hydroxycinnamic acid (CHCA) in 50% ACN/0.1% TFA]. Spectra were acquired in positive reflectron mode with an accelerating voltage of 21 kV. Samples were analyzed within the 700–3,000 m/z range using as a standard a mixture of adrenocorticotropic hormone and angiotensin (Sigma, St. Louis, MO, USA). Monoisotopic peaks with S/N >3 were selected as the PMFs and common contaminants were eliminated using the PeakErazor software (v 2.01, Lighthouse data, Odense M, Denmark).
The MASCOT search engine (Matrix Science, London, UK) was used to match PMFs against all bacterial entries available at the NCBI nonreduntant database. Fixed and variable modifications (Cys carbamidomethylation and Met oxidization, respectively), one missed cleavage, and a mass tolerance of 50 ppm were considered during decoyed searches. Peptide mixtures matching Acinetobacter spp. products with the highest statistically significant search scores (P < 0.05) and accounting for the majority of the mass spectra peaks were assumed to be positively identified proteins.
2.5. Detection of OmpA by western blotting and testing of its role in iron acquisition
Proteins present in total cell membrane fractions were size fractionated by SDS-PAGE, transferred to nitrocellulose and probed with anti-OmpA polyclonal antibodies as described before [32]. The immunocomplexes were detected by chemiluminescence using horseradish peroxidase-labeled protein A. The intensity of the signals produced by each sample was determined by densitometry using an AlphaImager detection and quantification system (Alpha Innotech Corp., San Leandro, CA, USA) as described before [33]. The iron acquisition phenotype of the ATCC 19606T parental strain and the isogenic ompA insertion derivative 3:233 [32] was tested by culturing these strains in LB broth or agar containing increasing concentrations of DIP overnight at 37 °C as previously described [11]. Experiments were done twice in duplicate.
3. Results
3.1. Validation of the experimental approach
3.1.1. Culture conditions
To study the iron response in A. baumannii using a global proteomics approach, ATCC 19606T cells were grown in unsupplemented LB broth or in LB broth supplemented with 100 μM FeCl3 and/or 100 μM DIP. We used these conditions because preliminary work showed that growth of A. baumannii in LB broth supplemented with 100 μM DIP or 100 μM FeCl3 was sufficient to simulate free-iron-restricted and -rich conditions under which the production of BauA, the iron-regulated acinetobactin receptor [34], was induced and repressed, respectively, as determined by immunoblot analysis (data not shown). Accordingly, these conditions resulted in the identification of BauA (Table 1, spot 31 and 47) only in the TC and OM fractions isolated from cells cultured under iron-chelated conditions. Inspection of the gels displayed in Fig. 1 shows that there is an overall qualitative and quantitative similarity in the protein spot patterns generated for each type of subcellular fraction obtained from cells cultured under different iron conditions. This observation indicates that the iron-chelated and -rich conditions used in this study did not result in detectable cell toxicity, an outcome that could invalidate the data collected with the experimental approach used in this work. Additional variations in protein spots due to growth rate differences because of the iron content of the media were minimized by using stationary phase cells.
Table 1.
Acinetobacter baumannii ATCC 19606T Fe-repressed proteins.
| Spota | ASQb |
Protein function/namec Source is italicized |
Genec | NCBI Accession # | Theoreticald |
Se | Mf | Cg | Fh | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | Mr | pI | ||||||||
| Iron acquisition/Virulence | |||||||||||||
| 22 | 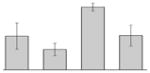 |
Outer membrane receptor for ferrienterochelin and colicins A. baumannii ACICU |
fepA | gi|184157262 | 82767 | 5.66 | 172 | 26 | 39 | TC | |||
| 27 | 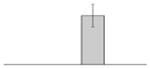 |
Ferric aerobactin receptor protein A. baumannii ATCC 19606T |
gi|260555123 | 85609 | 7.55 | 346 | 42 | 74 | TC | ||||
| 31 | 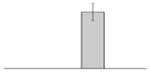 |
Ferric anguibactin receptor A. baumannii ATCC 19606T |
bauA | gi|260556700 | 82960 | 5.86 | 288 | 28 | 54 | TC | |||
| 33 |  |
Ferric aerobactin receptor protein A. baumannii ATCC 19606T |
gi|260555123 | 85609 | 7.55 | 138 | 25 | 38 | OM | ||||
| 37 |  |
Ferric enterobactin receptor precursor A. baumannii AYE |
fepA | gi|169796822 | 82746 | 5.66 | 146 | 23 | 42 | OM | |||
| 39 |  |
TonB-dependent vitamin B12 receptor A. baumannii ATCC 19606T |
btuB | gi|260557148 | 68121 | 5.22 | 277 | 28 | 57 | OM | |||
| 45 |  |
FhuE receptor A. baumannii ATCC 19606T |
fhuE | gi|260555105 | 80612 | 5.44 | 98 | 10 | 14 | OM | |||
| 46 |  |
FhuE receptor A. baumannii ATCC 19606T |
fhuE | gi|260555105 | 80612 | 5.44 | 226 | 25 | 42 | OM | |||
| 47 |  |
Ferric anguibactin receptor A. baumannii ATCC 19606T |
bauA | gi|260556700 | 82960 | 5.86 | 242 | 31 | 48 | OM | |||
| 48 | 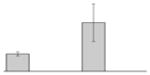 |
Outer membrane receptor protein, Fe transport A. baumannii ATCC 19606T |
gi|260557370 | 78171 | 5.47 | 339 | 33 | 58 | OM | ||||
| 49 |  |
Ferric enterobactin receptor precursor A. baumannii AYE |
fepA | gi|169796822 | 82746 | 5.66 | 113 | 19 | 28 | OM | |||
| 53 |  |
Ferric aerobactin receptor protein A. baumannii ATCC 19606T |
gi|260555123 | 85609 | 7.55 | 356 | 39 | 68 | OM | ||||
| 54 | 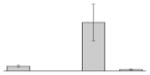 |
Outer membrane receptor for ferrienterochelin and colicins A. baumannii ACICU |
fepA | gi|184157262 | 82767 | 5.66 | 186 | 28 | 46 | OM | |||
| 55 | 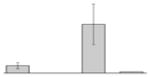 |
Ferric aerobactin receptor protein A. baumannii ATCC 19606T |
gi|260555123 | 85609 | 7.55 | 405 | 48 | 73 | OM | ||||
| 57 | 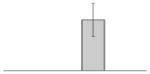 |
Ferric anguibactin receptor A. baumannii ATCC 19606T |
bauA | gi|260555123 | 85609 | 7.55 | 343 | 33 | 62 | OM | |||
| Porins | |||||||||||||
| 35 |  |
Carbapenem-associated resistance protein A. baumannii ATCC 19606T |
carO | gi|260556565 | 26855 | 4.66 | 133 | 12 | 76 | OM | |||
| Adaptation/Stress tolerance | |||||||||||||
| 26 | 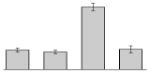 |
Fumarase C (fumarate hydratase class II) A. baumannii ATCC 19606T |
fumC | gi|260554730 | 50111 | 5.74 | 128 | 17 | 39 | TC | |||
| Energy/Metabolisms | |||||||||||||
| 4 | 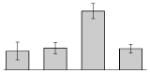 |
Serine hydroxymethyltransferase A. baumannii AYE |
glyA | gi|169795307 | 44967 | 5.44 | 107 | 11 | 36 | TC | |||
| 36 | 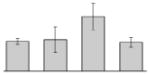 |
F0F1 ATP synthase α subunit A. baumannii ATCC 17978 |
atpA | gi|162286757 | 55363 | 5.29 | 207 | 25 | 45 | OM | |||
Average spot quantity per treatment group (n = 3); columns A, −Fe/−DIP; B, +Fe/−DIP; C, −Fe/+DIP; D, +Fe/+DIP.
Protein function, name and gene were determined by http:/www.ncbi.nlm.nih.gov/BLAST/.
Theoretical molecular mass (Mr) and isoelectric point (pI) were calculated by http:/www.expasy.org/.
Mascot score.
Number of matched peptide masses.
Percent sequence coverage.
Protein fraction, total cell (TC) or Outer membrane-enriched (OM) protein extracts.
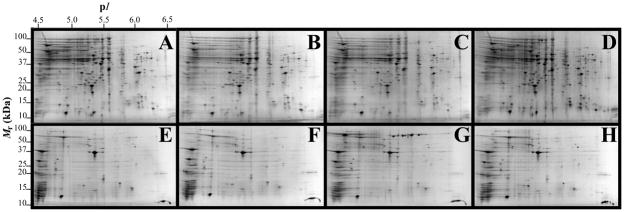
Fig. 1.
2-DE gel images of TC and OM-enriched protein fractions of A. baumannii ATCC 19606T. Panels A, B, C, and D represent TC fractions obtained from cells grown under −Fe/−DIP, +Fe/−DIP, −Fe/+DIP or +Fe/+DIP conditions, respectively. Panels E, F, G, and H represent OM fractions obtained from cells grown under −Fe/−DIP, +Fe/−DIP, −Fe/+DIP or +Fe/+DIP conditions, respectively. Mr, relative molecular weight; pI, isoelectric point.
Bacterial growth in the presence of 100 μM DIP plus 100 μM FeCl3 was used as a condition to determine whether changes in protein production were due to the chelation of iron or other metals present in the medium that could interact with DIP. The observation that the qualitative and quantitative spot patterns of cells cultured in LB broth supplemented with 100 μM FeCl3 and DIP were similar to those obtained with cells cultured in LB broth supplemented only with 100 μM FeCl3 indicate that the differential protein responses detected in this study are mostly due to DIP-mediated iron chelation. Proteins listed in Table 3 represent exceptions to this behavior as discussed below. It is important to mention that in certain instances more than one spot matched to a given protein, e.g., FepA (Table 1, spots 22, 37, 49, and 54). This could be due to several factors including multimerism, maturation state, and/or post-translational modifications.
Table 3.
Acinetobacter baumannii ATCC 19606T protein spots that were differentially expressed due to treatment with DIP but were non-responsive to supplementation with FeCl3.
| Spota | ASQb |
Protein function/namec Source is italicized |
Genec | NCBI Accession # | Theoreticald |
Se | Mf | Cg | Fh | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | Mr | pI | ||||||||
| Iron acquisition/Virulence | |||||||||||||
| 34 | 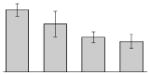 |
Putative outer membrane protein A. baumannii ATCC 17978 |
gi|126640934 | 22471 | 9.30 | 93 | 11 | 53 | OM | ||||
| Regulatory | |||||||||||||
| 50 | 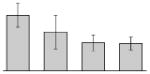 |
30S ribosomal protein S6 A. baumannii SDF |
rpsF | gi|169633238 | 14954 | 5.70 | 93 | 10 | 61 | OM | |||
| 52 | 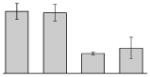 |
Peptidyl-prolyl cis-trans isomerase A. baumannii ATCC 17978 |
surA | gi|126641590 | 46526 | 5.75 | 197 | 19 | 47 | OM | |||
| Unknown | |||||||||||||
| 17 | 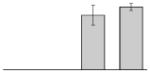 |
Short chain dehydrogenase A. baumannii SDF |
gi|169632063 | 26998 | 5.18 | 135 | 14 | 55 | TC | ||||
Average spot quantity per treatment group (n = 3); columns A, −Fe/−DIP; B, +Fe/−DIP; C, −Fe/+DIP; D, +Fe/+DIP.
Protein function, name and gene were determined by http:/www.ncbi.nlm.nih.gov/BLAST/.
Theoretical molecular mass (Mr) and isoelectric point (pI) were calculated by http:/www.expasy.org/.
Mascot score.
Number of matched peptide masses.
Percent sequence coverage.
Protein fraction, total cell (TC) or Outer membrane-enriched (OM) protein extracts.
3.1.2. Cell fractionation
The data reported in Tables 1 and 2 indicate that the selective detergent solubilization and ultracentrifugation method used to isolate outer membranes resulted in a significant and selective enrichment in this particular subcellular fraction. Most of the proteins detected in outer membrane samples are either bona fide or predicted proteins located in this component of the bacterial cell wall. Detection of the cytoplasmic Bfr and inner membrane AtpA proteins in the outer membrane fractions is because of the large complexes these proteins form as well as the ability of Bfr to aggregate and bind to cell membranes [35], which were recovered by ultracentrifugation.
Table 2.
Acinetobacter baumannii ATCC 19606T Fe-induced proteins.
| Spota | ASQb |
Protein function/namec Source is italicized |
Genec | NCBI Accession # | Theoreticald |
Se | Mf | Cg | Fh | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | Mr | pI | ||||||||
| Porins | |||||||||||||
| 32 | 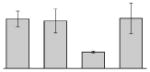 |
Putative outer membrane protein W A. baumannii ATCC 19606T |
ompW | gi|260556338 | 23137 | 6.29 | 93 | 8 | 57 | OM | |||
| 43 | 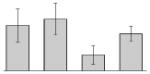 |
Putative outer membrane protein W A. baumannii ATCC 19606T |
ompW | gi|260556338 | 23137 | 6.29 | 83 | 8 | 57 | OM | |||
| 44 |  |
Outer membrane protein A A. baumannii |
ompA | gi|129307154 | 36939 | 5.42 | 109 | 12 | 37 | OM | |||
| 56 |  |
Outer membrane protein Omp38 A. baumannii ATCC 19606 |
omp38 | gi|260557183 | 38397 | 5.32 | 86 | 7 | 20 | OM | |||
| 58 |  |
Outer membrane protein Omp38 Acinetobacter sp. RUH2624 |
omp38 | gi|260548855 | 37972 | 5.72 | 139 | 15 | 39 | OM | |||
| Adaptation/Stress tolerance | |||||||||||||
| 5 | 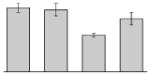 |
Chaperonin GroEL A. baumannii SDF |
groEL | gi|169632653 | 56886 | 4.92 | 273 | 34 | 55 | TC | |||
| 8 | 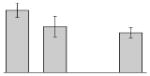 |
Superoxide dismutase (Fe) A. baumannii SDF |
sodB | gi|169632935 | 22905 | 5.56 | 123 | 6 | 41 | TC | |||
| 9 | 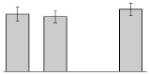 |
Bacterioferritin A. baumannii ATCC 17978 |
bfr | gi|126640856 | 18023 | 5.02 | 137 | 17 | 70 | TC | |||
| 10 |  |
Superoxide dismutase (Fe) A. baumannii SDF |
sodB | gi|169632935 | 22905 | 5.56 | 109 | 8 | 44 | TC | |||
| 24 | 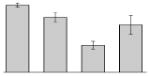 |
Superoxide dismutase (Fe) A. baumannii SDF |
sodB | gi|169632935 | 22905 | 5.56 | 83 | 8 | 44 | TC | |||
| 38 | 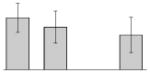 |
Bacterioferritin A. baumannii ATCC 19606T |
bfr | gi|260410611 | 18137 | 5.02 | 106 | 14 | 72 | OM | |||
| 41 | 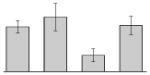 |
Bacterioferritin A. baumannii ATCC 19606T |
bfr | gi|260410611 | 18137 | 5.02 | 78 | 9 | 45 | OM | |||
| 42 | 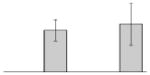 |
Bacterioferritin A. baumannii ATCC 19606T |
bfr | gi|260410611 | 18137 | 5.02 | 100 | 12 | 66 | OM | |||
| TCA cycle | |||||||||||||
| 14 |  |
Succinate dehydrogenase flavoprotein subunit A. baumannii ATCC 17978 |
sdh | gi|126642744 | 64049 | 6.07 | 92 | 6 | 18 | TC | |||
| 15 | 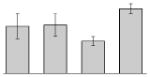 |
Aconitate hydratase 2 A. baumannii ATCC 19606T |
acnB | gi|260411150 | 95322 | 5.07 | 238 | 37 | 48 | TC | |||
| 19 | 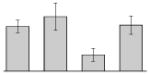 |
Fumarate hydratase A. baumannii ATCC 17978 |
fumA | gi|193076292 | 54948 | 5.44 | 129 | 18 | 49 | TC | |||
| 20 | 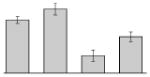 |
Aconitate hydratase 2 A. baumannii ATCC 19606T |
acnB | gi|260411150 | 95322 | 5.07 | 136 | 24 | 32 | TC | |||
| 21 | 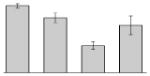 |
Aconitate hydratase 1 A. baumannii ATCC 19606T |
acnA | gi|260556094 | 100503 | 5.28 | 410 | 36 | 44 | TC | |||
| 30 | 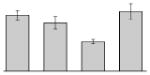 |
Succinate dehydrogenase flavoprotein subunit A. baumannii ATCC 17978 |
sdh | gi|126642744 | 64049 | 6.07 | 169 | 21 | 46 | TC | |||
| Lipid Metabolism | |||||||||||||
| 3 | 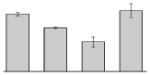 |
Beta-ketoadipyl CoA thiolase A. baumannii ATCC 19606 |
pcaF | gi|260555473 | 43107 | 5.96 | 138 | 20 | 54 | TC | |||
| 18 | 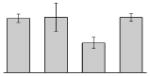 |
Acyl-CoA dehydrogenase A. baumannii ATCC 19606T |
gi|260554863 | 42780 | 5.30 | 185 | 26 | 46 | TC | ||||
| 25 | 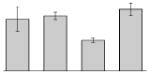 |
2-ketocyclohexanecarboxyl-CoA hydrolase A. baumannii ATCC 19606T |
gi|260555475 | 29342 | 5.75 | 231 | 27 | 76 | TC | ||||
| 29 | 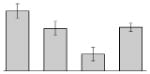 |
2-hydroxycyclohexanecarboxyl- CoA dehydrogenase A. baumannii ATCC 19606T |
fabG | gi|260555032 | 27802 | 5.96 | 100 | 10 | 50 | TC | |||
| Regulatory | |||||||||||||
| 2 | 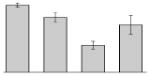 |
Host factor I for bacteriophage Q beta replication hfq A. baumannii AYE |
hfq | gi|169795564 | 16751 | 6.82 | 91 | 8 | 39 | TC | |||
| 51 |  |
50S ribosomal protein L3 A. baumannii AYE |
rplC | gi|169794594 | 22523 | 9.87 | 108 | 10 | 63 | OM | |||
| Energy/Metabolisms | |||||||||||||
| 1 | 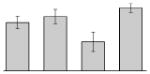 |
Phenylacetate-CoA oxygenase subunit PaaB A. baumannii ATCC 17978 |
paaB | gi|126641383 | 11317 | 6.04 | 176 | 15 | 86 | TC | |||
| 11 | 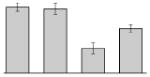 |
Putative flavohemoprotein (hemoglobin-like protein) A. baumannii ATCC 19606T |
gi|260409628 | 28814 | 4.91 | 188 | 15 | 60 | TC | ||||
| 12 | 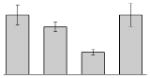 |
Phenylacetate-CoA oxygenase/ reductase PaaK subunit A. baumannii ATCC 17978 |
paaK | gi|193077015 | 39763 | 5.03 | 195 | 25 | 63 | TC | |||
| 16 |  |
Benzoate 1,2-dioxygenase, small subunit A. baumannii ATCC 19606T |
gi|260554867 | 18965 | 5.22 | 93 | 6 | 30 | TC | ||||
| 28 | 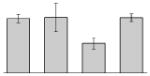 |
Phenylacetate-CoA oxygenase subunit PaaB A. baumannii ATCC 17978 |
paaB | gi|126641383 | 11317 | 6.04 | 196 | 16 | 91 | TC | |||
| Unknown | |||||||||||||
| 6 | 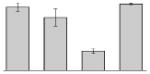 |
Putative 17-kDa Surface antigen A. baumannii ACICU |
gi|184157742 | 12424 | 4.70 | 87 | 8 | 83 | TC | ||||
| 7 | 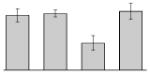 |
Signal peptide ydeN A. baumannii ATCC 19606T |
ydeN | gi|260556330 | 22987 | 4.75 | 92 | 7 | 34 | TC | |||
| 13 | 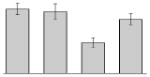 |
Hypothetical protein A1S_1295 A. baumannii ATCC 17978 |
gi|193076983 | 55265 | 5.07 | 129 | 17 | 39 | TC | ||||
| 23 |  |
Hemerythrin HHE cation binding domain protein A. baumannii AB900 |
gi|239501928 | 17516 | 5.68 | 88 | 9 | 53 | TC | ||||
| 40 |  |
Hypothetical protein A1S_1295 A. baumannii ATCC 17978 |
gi|193076983 | 55265 | 5.07 | 149 | 20 | 49 | OM | ||||
Average spot quantity per treatment group (n = 3); columns A, −Fe/−DIP; B, +Fe/−DIP; C, −Fe/+DIP; D, +Fe/+DIP.
Protein function, name and gene were determined by http:/www.ncbi.nlm.nih.gov/BLAST/.
Theoretical molecular mass (Mr) and isoelectric point (pI) were calculated by http:/www.expasy.org/.
Mascot score.
Number of matched peptide masses.
Percent sequence coverage.
Protein fraction, total cell (TC) or Outer membrane-enriched (OM) protein extracts.
3.2. 2-DE detection of A. baumannii differentially produced in response to free iron
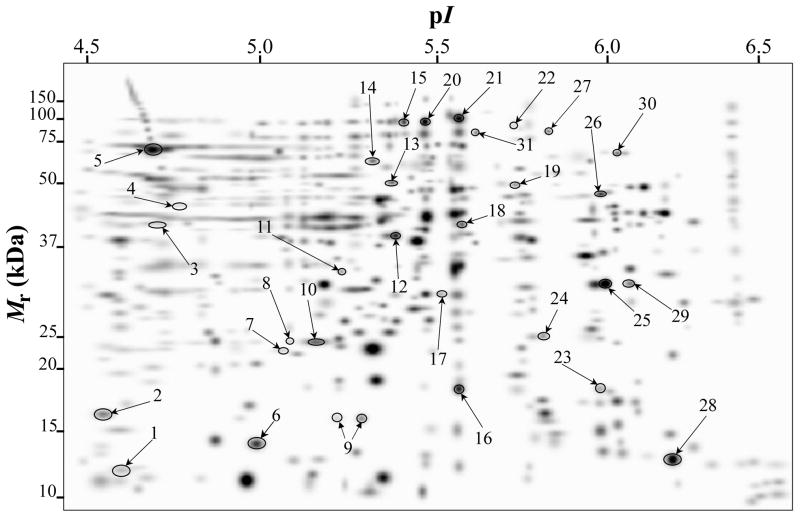
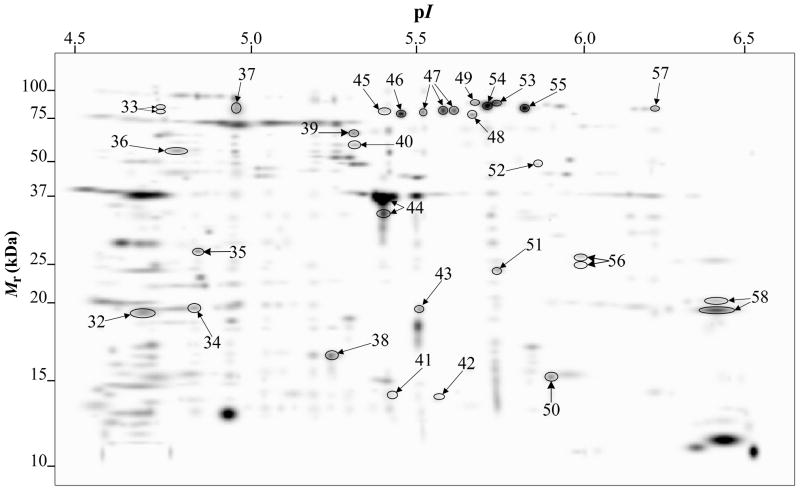
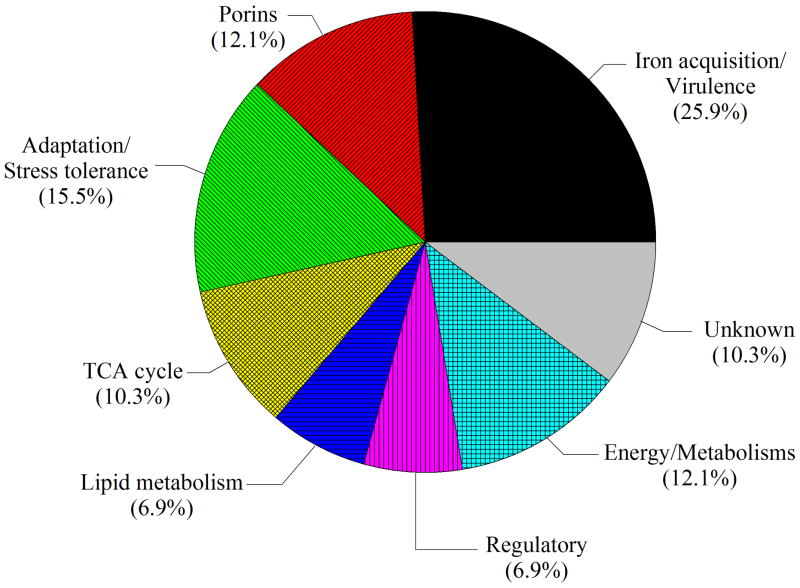
Fig. 1 shows representative 2-DE gel images of A. baumannii ATCC 19606T TC (upper panels) and OM (bottom panels) protein samples isolated from cells grown in LB broth without supplementation (panels A and E) or in LB broth supplemented with 100 μM FeCl3 (panels B and F), 100 μM DIP (panels C and G) or 100 μM FeCl3 plus 100 μM DIP (panels D and H). PDQuest analysis of these gels resulted in the reproducible identification of more than 600 and 400 spots in TC and OM protein gels, respectively. Quantitative analysis followed by PMF and database searching using BLASTp identified a total of 58 protein spots, 31 spots in TC protein extracts (Fig. 2) and 27 spots in OM protein fractions (Fig. 3), which were differentially produced in A. baumannii ATCC 19606T cells in response to inorganic Fe availability. A closeup view of the profiles of some selected spots from representative gels obtained with protein samples isolated from cells cultured under different conditions, which highlights variations in spot volumes, is shown in panels A-V of Fig. 4. A collated list of all proteins identified in this study is shown in supplemental Table S1. These proteins were mainly grouped into the following categories: (i) iron acquisition/virulence-related proteins, (ii) porins, (iii) adaptation/stress tolerance-related proteins, (iv) tri-carboxylic cycle (TCA)-related proteins, (v) proteins involved in lipid metabolism, (vi) regulatory proteins, (vii) proteins involved in energy production and general metabolisms and (viii) proteins with unknown functions (Fig. 5). The largest functional category was proteins involved in iron acquisition/virulence (25.9%) followed by adaptation/stress tolerance-related proteins (15.5%).
Fig. 2.
PDQuest-generated master gel image showing the general spot pattern of matched spots from TC fractions obtained from A. baumannii ATCC 19606T cells grown under −Fe/−DIP, +Fe/−DIP, −Fe/+DIP or +Fe/+DIP conditions. Differentially expressed spots are numbered. Mr, relative molecular weight; pI, isoelectric point.
Fig. 3.
PDQuest-generated master gel image showing the general spot pattern of matched spots from OM-enriched fractions obtained from A. baumannii ATCC 19606T cells grown under −Fe/−DIP, +Fe/−DIP, −Fe/+DIP or +Fe/+DIP conditions. Differentially expressed spots are numbered. Mr, relative molecular weight; pI, isoelectric point.
Fig. 4.
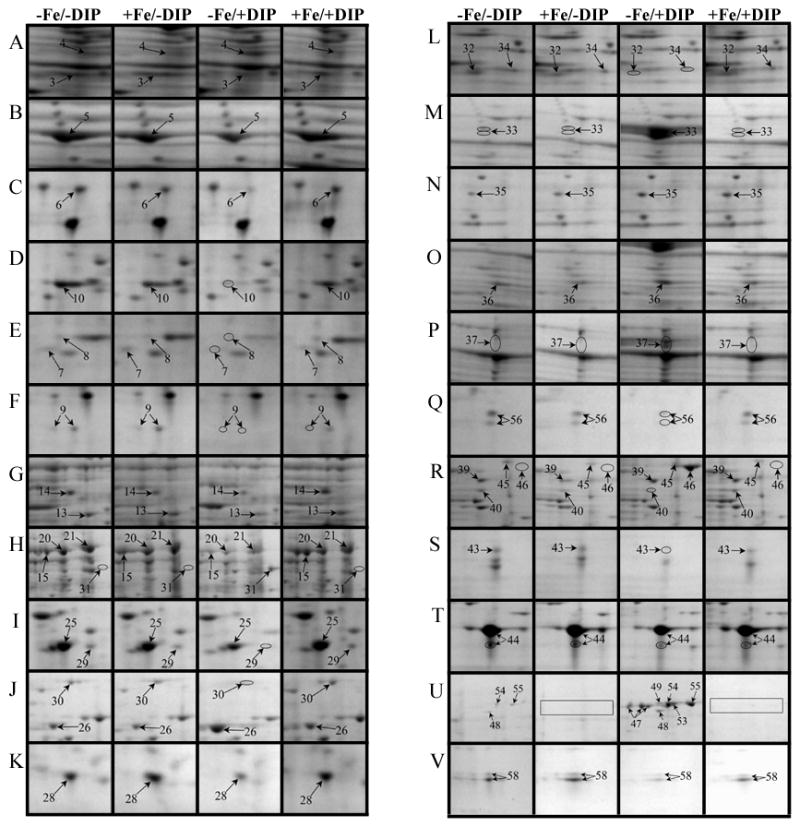
Close up view of some differentially expressed protein spots in representative 2-DE gels from TC (A-K) and OM (L-V) protein fractions obtained from A. baumannii ATCC 19606T cells grown under −Fe/−DIP, +Fe/−DIP, −Fe/+DIP or +Fe/+DIP conditions.
Fig. 5.
Functional category distribution of A. baumannii ATCC 19606T proteins differentially produced in response to free-iron changes in the culture medium.
3.3. Iron-repressed proteins
Our study identified 19 protein spots that were repressed by Fe (Table 1). Proteins belonging to the family of siderophore outer membrane receptors, some of which were detected in TC and OM protein fractions, were the most prevalently produced proteins in cells cultured in the presence of DIP. As expected from previous observations [34], the acinetobactin BauA receptor (Table 1, spots 31, 47 and 57) was in this group, which also includes proteins related to the enterobactin receptor protein FepA (Table 1, spots 22, 37, 49, and 54), a protein related to the ferric aerobactin receptor (Table 1, spots 27, 33, 53, and 55), a FhuE-like ferric rhodotorulic acid receptor (Table 1, spots 45 and 46), a BtuB-like vitamin B12 receptor (Table 1, spot 39), and a protein annotated as an outer membrane siderophore receptor (Table 1, spot 48). Iron chelation also induced the production of the outer membrane protein CarO (Table 1, spot 35), which plays a role in antibiotic resistance and ornithine transport [36], the predicted cytoplasmic proteins FumC (Table 1, spot 26) and the serine hydroxymethyltransferase (SHMT) GlyA (Table 1, spot 4), proteins that play a role in the TCA cycle and general metabolism, respectively. The increased production of the AtpA F0F1 ATP synthetase α subunit (Table 1, spot 36), a cytoplasmic membrane protein that was enriched with the OM fraction, could reflect the energy-intensive nature of the iron acquisition process [37, 38].
3.4. Iron-induced proteins
Our study identified 35 protein spots that were induced by Fe (Table 2), which represented the largest category of proteins differentially produced under the experimental conditions used in this study. As predicted from observations made previously [39], free-iron rich-conditions induced the overproduction of Bfr, an iron storage protein that was detected in the TC and OM-enriched fractions (Table 2, spots 9, 38, 41 and 42). Proteins associated with adaptation/stress responses, such as GroEL (Table 2, spot 5) and SodB (Table 2, spots 8, 10, and 24), were among those that were induced under iron-rich conditions. Proteins associated with the TCA cycle, such as Sdh (Table 2, spot 14 and 30), AcnB (Table 2, spot 15 and 20), AcnA (Table 2, spot 21) and FumA (Table 2, spot 19), as well as proteins associated with lipid metabolism, such as PcaF (Table 2, spot 3), an acyl-CoA dehydrogenase (Table 2, spot 18), a 2-ketocyclohexanecarboxyl-CoA hydrolase (Table 2, spot 25) and FabG (Table 2, spot 29), were also induced under Fe-repletion. General metabolism proteins, the production of which were enhanced in cells cultured in LB broth supplemented with free inorganic iron, include PaaB (Table 2, spots 1 and 28), PaaK (Table 2, spot 12), a putative flavohemoprotein (Table 2, spot 11) and a benzoate 1,2-dioxygenase small subunit (Table 2, spot 16). Synthesis of the RNA chaperone protein Hfq (Table 2, spot 2) and the ribosomal protein L3 RplC (Table 2, spot 51) is also increased when cells are cultured under iron-rich conditions. This category also includes five proteins with unknown cellular functions (Table 2, spot 6, 7, 13, 23 and 40). Table 2 also shows that the production of the outer membrane proteins OmpW (spot 32 and 43), OmpA (spot 44) and Omp38 (spot 56 and 58), an alternative name used to refer to the A. baumannii OmpA protein [40], were also induced under Fe-repletion.
While the differential production of OmpW in response to iron is in agreement with previous reports describing a similar behavior in other bacteria [41–45], the response displayed by OmpA seems to be novel. The proteomics data obtained with 2-DE was confirmed by western blotting using anti-OmpA. Fig. 6 shows that the production of OmpA decreases 2-fold in cells cultured in LB broth containing 100 μM DIP when compared with the amount of protein detected in the total membrane fraction isolated from cells grown in LB broth containing 100 μM FeCl3 either in the presence or absence of this synthetic iron chelator. The same trend was observed when this experiment was repeated with fresh biological samples using the same experimental conditions. These observations suggest that OmpA could play a role in iron acquisition in A. baumannii, a possibility that was tested using the ATCC 19606T 3:233 ompA isogenic insertion derivative we used to examine the interaction of this strain with eukaryotic cells [32]. Fig. 7 shows that, when compared with the response of the ATCC 19606T parental strain, the ability of the 3:233 ompA mutant to grow was significantly affected as the DIP concentration in the broth increased from 0 μM to 250 μM. A similar outcome was observed when cells were plated on LB agar containing increasing amounts of DIP. There were no noticeable differences in the growth of the ATCC 19606T parental strain on plates containing up to 200 μM DIP, while a noticeable growth reduction was observed on plates containing 250 μM DIP. The same analysis of the 3:233 ompA derivative showed that its growth was mildly and severely affected when the plates contained 150 μM and 200 μM DIP, respectively, and abolished by the presence of 250 μM DIP (data not shown).
Fig. 6.
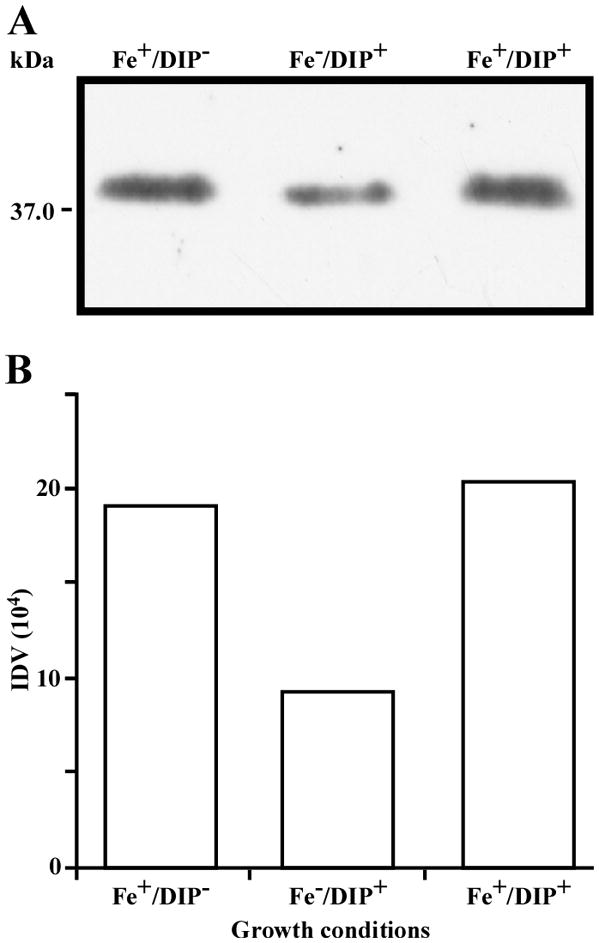
Differential production of OmpA. A. Proteins (2 μg/lane) from total cell membrane fractions isolated from A. baumannii ATCC 19606T cells grown under +Fe/−DIP, −Fe/+DIP or +Fe/+DIP conditions were sized fractionated by SDS-PAGE, electrotransferred to nitrocellulose and probed with polyclonal anti-OmpA antibodies. The immune complexes were detected with horseradish peroxidase-labeled protein A and chemiluminescence on an X-ray film. B. Densitometry of the signals shown in panel A. IDV, integrated density value = Σ (each pixel value - background).
Fig. 7.
Effects of iron chelation on bacterial growth. The A. baumannii ATCC 19606T parental strain (white bars) and the 19606T 3:233 OmpA mutant (black bars) were cultured in LB broth containing increasing amounts of DIP. Cell growth was recorded after overnight incubation at 37 °C. Error bars, 1 standard deviation (SD).
3.5. Protein spots that were differentially expressed due to treatment with DIP but were non-responsive to Fe supplementation
Our study identified four protein spots that were differentially expressed due to treatment with DIP but were non-responsive to supplementation with Fe (Table 3). These proteins include a putative outer membrane protein (spot 34) that could play a virulence role, the 30S ribosomal protein S6 RpsF (spot 50), the predicted periplasmic protein peptidyl-prolyl cis-trans isomerase SurA (spot 52), and a short chain dehydrogenase (spot 17) of unknown function.
4. Discussion
This study resulted in the identification of outer membrane proteins, particularly those involved in iron acquisition, as the largest group of proteins that are preferentially produced when A. baumannii ATCC 19606T cells are cultured under iron-chelated conditions. The fact that some of the iron-acquisition proteins are different from the BauA acinetobactin receptor protein suggests that this pathogen could acquire iron using different siderophore utilization pathways in addition to the acinetobactin-mediated system [34, 46] when co-existing as a component of the human microbiota. Similarly, the positive effect of DIP on the production of CarO, a protein involved in ornithine uptake [36], supports the possibility of the expression of alternative siderophore-mediated utilization systems, such as that mediated by pyoverdin in P. aeruginosa [47]. However, the modest reduction in CarO production observed when chelated LB broth was supplemented with FeCl3 suggests that other metals chelated by DIP could also play a role in its differential production.
The increased production of the serine hydroxymethyltransferase (SHMT) GlyA, which catalyses the formation of serine and tetrahydrofolate (THF), under iron-chelated conditions could be due to changes in glutamate abundance in response to changes in the free-iron content of the medium as proposed for B. pertussis [48]. Alternatively, the Fe-restricted induction of SHMT in A. baumannii could be part of a global cellular response based on the ability of this enzyme to bind mRNA and control gene expression as previously reported [49].
The coordinated differential production of FumA and FumC, enzymes that have similar fumarase activity but differ in structure and regulation [50–52], reflects the positive and negative regulatory roles iron plays in the A. baumannii physiology. FumA is an iron-dependent hydrolase that contains a 4Fe-4S active center that is highly produced under iron-rich conditions, while FumC is an iron-independent enzyme that is optimally produced under iron limitation. Based on these differential responses, it was proposed that FumC replaces FumA under iron-limiting conditions to ensure the operation of central metabolic pathways independently of changes in extracellular iron concentration [50]. Our data provide initial evidence in support of this hypothesis since the production of the A. baumannii FumA and FumC proteins was upregulated in cells cultured under iron-rich and iron-chelated conditions, respectively.
Iron-rich conditions promote the expression of genes coding for proteins involved in iron sequestration as well as iron-containing proteins that are part of the intracellular iron pool and play key metabolic functions [53]. Accordingly, our analysis shows that the production of proteins that sequester intracellular iron, such as Bfr, AcnA, AcnB, FumA, Sdh and SodB, is significantly enhanced when A. baumannii ATCC 19606T cells are cultured in Fe-rich medium. The transcriptional activation of the genes coding for these proteins is an iron-dependent process mediated by the Fur repressor via an indirect regulatory mechanism, which involves the production of RyhB in E. coli [19, 20, 53] and the PrrF1 and PrrF2 sRNAs in P. aeruginosa [21]. These observations together with the fact that A. baumannii also produces an iron-induced ortholog of Hfq, a protein that plays post-transcriptional functions particularly in bacterial iron metabolism [54, 55], is a strong indication for a similar transcriptional control of iron-regulated genes expressed by this pathogen.
The observation that OmpW, which could function as an iron user or exporter protein, belongs to the A. baumannii iron regulon is similar to that made in other bacteria such as E. coli [42, 43], Pasteurella multocida [44], Actinobacillus pleuropneumoniae [41] and Shewanella oneidensis [45]. In contrast, our experimental observation that the production of OmpA is also enhanced under iron-rich conditions is novel and suggest its potential role in iron metabolism. This possibility is supported by the finding that the A. baumannii ATCC 19606T 3:233 OmpA mutant, which we used to study the interaction of this strain with C. albicans filaments [32], has a defect in iron utilization under chelated conditions. Currently, we are investigating whether OmpA plays a role in the secretion of acinetobactin and/or the import of acinetobactin-iron complexes across the bacterial cell wall. The reduced production of OmpA and OmpW under iron-chelation could also be expected since this response would limit the influx of antimicrobial compounds mediated by these non-specialized porins [56] during the infection of the human host, which presents an iron-restricted environment to microbial pathogens.
5. Conclusion
A. baumannii responds to changes in extracellular iron concentrations by differentially producing proteins involved in a wide range of cellular functions. Iron-acquisition outer membrane proteins comprise the largest group of iron-repressed proteins while iron-induced proteins involved in a wide range of cellular and metabolic functions are the largest group of proteins identified in this study. While some of these observations agree with data obtained with other bacterial systems, the finding of several outer membrane siderophore receptors different from the BauA acinetobactin receptor reflects the unexplored possibility that members of the human microbiota interact with A. baumannii by sharing functionally compatible iron acquisition systems. Equally interesting is the observation that the A. baumannii iron stimulon includes proteins unknown to be iron regulated, the production of which could be controlled by a Fur regulon that could include sRNA regulators and the Hfq chaperone. Also significant is the novel observation of a potential role of OmpA in iron metabolism. Taken together, these findings open new avenues to explore and better understand the role of iron in the pathophysiology of A. baumannii. These avenues should combine proteomics with transcriptomic analysis, such as that recently described for the study of A. baumannii differential gene response induced by ethanol using RNA-sequencing [57]. This combined approach should provide information on the transcriptional and posttranscriptional processes controlling the expression of A. baumannii genes and gene products that play a role in the virulence of this human pathogen.
Supplementary Material
Acknowledgments
Funds from US Public Health AI070174 award and Miami University supported this work. We are grateful to Dr. J. Hawes, coordinator of the Miami University Center of Bioinformatics and Functional Genomics for his support and assistance with 2-D gel electrophoresis-mass spectrometry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–65. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. Acinetobacter baumannii infections among patients at military medical facilities treating U.S. service members, 2002–2004. MMWR. 2004;53:1063–6. [PubMed] [Google Scholar]
- 4.Davis KA, Moran KA, McAllister CK, Gray PJ. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg Infect Dis. 2005;11:1218–24. doi: 10.3201/1108.050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5:939–51. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 6.Crichton R. From molecular mechanisms to clinical consequences. 3. West Sussex, United Kingdom: John Wiley & Sons Ltd; 2009. Iron metabolism. [Google Scholar]
- 7.Crosa JH, Mey AR, Payne SM. Iron transport in bacteria. Washington, D.C: ASM Press; 2004. [Google Scholar]
- 8.Neilands J. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–31. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 9.Weinberg E. Acquisition of iron and other nutrients in vivo. In: Roth J, Bolin C, Brogden K, Minion F, Wannemuehler M, editors. Virulence Mechanisms of Bacterial Pathogens. Washington, DC: ASM Press; 1995. pp. 79–93. [Google Scholar]
- 10.Weinberg ED. Iron availability and infection. Biochim Biophys Acta. 2009;1790:600–5. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Zimbler DL, Penwell WF, Gaddy JA, Menke SM, Tomaras AP, Connerly PL, et al. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals. 2009;22:23–32. doi: 10.1007/s10534-008-9202-3. [DOI] [PubMed] [Google Scholar]
- 12.Litwin CM, Calderwood SB. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–49. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter BM, Whitmire JM, Merrell DS. This is not your mother’s repressor: the complex role of fur in pathogenesis. Infect Immun. 2009;77:2590–601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–92. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 15.de Lorenzo VL, Herrero M, Giovannini M, Neilands J. Fur (ferric uptake regulator) protein and CAP (catabolic-activator protein) modulate transcription of fur gene in Escherichia coli. Eu J Biochem. 1988;173:537–46. doi: 10.1111/j.1432-1033.1988.tb14032.x. [DOI] [PubMed] [Google Scholar]
- 16.Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol. 2004;52:1081–90. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- 17.Ernst FD, Bereswill S, Waidner B, Stoof J, Mader U, Kusters JG, et al. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology. 2005;151:533–46. doi: 10.1099/mic.0.27404-0. [DOI] [PubMed] [Google Scholar]
- 18.Litwin CM, Calderwood SB. Analysis of the complexity of gene regulation by Fur in Vibrio cholerae. J Bacteriol. 1994;176:240–8. doi: 10.1128/jb.176.1.240-248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–83. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–5. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, et al. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci USA. 2004;101:9792–7. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniel C, Haentjens S, Bissinger MC, Courcol RJ. Characterization of the Acinetobacter baumannii Fur regulator: cloning and sequencing of the fur homolog gene. FEMS Microbiol Lett. 1999;170:199–209. doi: 10.1111/j.1574-6968.1999.tb13375.x. [DOI] [PubMed] [Google Scholar]
- 23.Dorsey CW, Tolmasky ME, Crosa JH, Actis LA. Genetic organization of an Acinetobacter baumannii chromosomal region harbouring genes related to siderophore biosynthesis and transport. Microbiology. 2003;149:1227–38. doi: 10.1099/mic.0.26204-0. [DOI] [PubMed] [Google Scholar]
- 24.Soares NC, Cabral MP, Parreira JR, Gayoso C, Barba MJ, Bou G. 2-DE analysis indicates that Acinetobacter baumannii displays a robust and versatile metabolism. Proteome Sci. 2009;7:37. doi: 10.1186/1477-5956-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon SO, Gho YS, Lee JC, Kim SI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett. 2009;297:150–6. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- 26.Fernández-Reyes M, Rodriguez-Falcón M, Chiva C, Pachón J, Andreu D, Rivas L. The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics. 2009;9:1632–45. doi: 10.1002/pmic.200800434. [DOI] [PubMed] [Google Scholar]
- 27.Hood MI, Jacobs AC, Sayood K, Dunman PM, Skaar EP. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob Agents Chemother. 54:1029–41. doi: 10.1128/AAC.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun SH, Choi CW, Park SH, Lee JC, Leem SH, Choi JS, et al. Proteomic analysis of outer membrane proteins from Acinetobacter baumannii DU202 in tetracycline stress condition. J Microbiol. 2008;46:720–7. doi: 10.1007/s12275-008-0202-3. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Russell DW. A Laboratory Manual. 3. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning. [Google Scholar]
- 30.Bryan FT, Robinson CW, Jr, Gilbert CG, Langdell RD. N-ethylmaleimide inhibition of horseshoe crab hemocyte agglutination. Science. 1964;144:1147–8. doi: 10.1126/science.144.3622.1147. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Li X, Li Y. A modified Coomassie Brilliant Blue staining method at nanogram sensitivity compatible with proteomic analysis. Biotechnology Letters. 2007;29:1599–603. doi: 10.1007/s10529-007-9425-3. [DOI] [PubMed] [Google Scholar]
- 32.Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and the interaction of this pathogen with eukaryotic cells. Infect Immun. 2009;77:3150–60. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhodes ER, Tomaras AP, McGillivary G, Connerly PL, Actis LA. Genetic and functional analyses of the Actinobacillus actinomycetemcomitans AfeABCD siderophore-independent iron acquisition system. Infect Immun. 2005;73:3758–63. doi: 10.1128/IAI.73.6.3758-3763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorsey CW, Tomaras AP, Connerly PL, Tolmasky ME, Crosa JH, Actis LA. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology. 2004;150:3657–67. doi: 10.1099/mic.0.27371-0. [DOI] [PubMed] [Google Scholar]
- 35.Andrews SC, Harrison PM, Guest JR. Cloning, sequencing, and mapping of the bacterioferritin gene (bfr) of Escherichia coli K-12. J Bacteriol. 1989;171:3940–7. doi: 10.1128/jb.171.7.3940-3947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mussi MA, Relling VM, Limansky AS, Viale AM. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Lett. 2007;581:5573–8. doi: 10.1016/j.febslet.2007.10.063. [DOI] [PubMed] [Google Scholar]
- 37.Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 38.Postle K. TonB protein and energy transduction between membranes. J Bioenerg Biomemb. 1993;25:591–601. doi: 10.1007/BF00770246. [DOI] [PubMed] [Google Scholar]
- 39.Matzanke BF, Muller GI, Bill E, Trautwein AX. Iron metabolism of Escherichia coli studied by Mossbauer spectroscopy and biochemical methods. Eur J Biochem. 1989;183:371–9. doi: 10.1111/j.1432-1033.1989.tb14938.x. [DOI] [PubMed] [Google Scholar]
- 40.Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, et al. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol. 2008;10:309–19. doi: 10.1111/j.1462-5822.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 41.Deslandes V, Nash JH, Harel J, Coulton JW, Jacques M. Transcriptional profiling of Actinobacillus pleuropneumoniae under iron-restricted conditions. BMC Genomics. 2007;8:72. doi: 10.1186/1471-2164-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin XM, Wu LN, Li H, Wang SY, Peng XX. Downregulation of Tsx and OmpW and upregulation of OmpX are required for iron homeostasis in Escherichia coli. J Proteome Res. 2008;7:1235–43. doi: 10.1021/pr7005928. [DOI] [PubMed] [Google Scholar]
- 43.McHugh JP, Rodríguez-Quiñones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, et al. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem. 2003;278:29478–86. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 44.Paustian ML, May BJ, Kapur V. Pasteurella multocida gene expression in response to iron limitation. Infect Immun. 2001;69:4109–15. doi: 10.1128/IAI.69.6.4109-4115.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson DK, Beliaev AS, Giometti CS, Tollaksen SL, Khare T, Lies DP, et al. Transcriptional and proteomic analysis of a ferric uptake regulator (fur) mutant of Shewanella oneidensis: possible involvement of fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl Environ Microbiol. 2002;68:881–92. doi: 10.1128/AEM.68.2.881-892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mihara K, Tanabe T, Yamakawa Y, Funahashi T, Nakao H, Narimatsu S, et al. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology. 2004;150:2587–97. doi: 10.1099/mic.0.27141-0. [DOI] [PubMed] [Google Scholar]
- 47.Ge L, Seah SY. Heterologous expression, purification, and characterization of an l-ornithine N(5)-hydroxylase involved in pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7205–10. doi: 10.1128/JB.00949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vidakovics ML, Paba J, Lamberti Y, Ricart CA, de Sousa MV, Rodriguez ME. Profiling the Bordetella pertussis proteome during iron starvation. J Proteome Res. 2007;6:2518–28. doi: 10.1021/pr060681i. [DOI] [PubMed] [Google Scholar]
- 49.Liu X, Reig B, Nasrallah IM, Stover PJ. Human cytoplasmic serine hydroxymethyltransferase is an mRNA binding protein. Biochemistry. 2000;39:11523–31. doi: 10.1021/bi000665d. [DOI] [PubMed] [Google Scholar]
- 50.Park SJ, Gunsalus RP. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: role of the arcA, fnr, and soxR gene products. J Bacteriol. 1995;177:6255–62. doi: 10.1128/jb.177.21.6255-6262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tseng CP, Yu CC, Lin HH, Chang CY, Kuo JT. Oxygen- and growth rate-dependent regulation of Escherichia coli fumarase (FumA, FumB, and FumC) activity. J Bacteriol. 2001;183:461–7. doi: 10.1128/JB.183.2.461-467.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woods SA, Guest JR. Differential roles of the Escherichia coli fumarases and fnr-dependent expression of fumarase B and aspartase. FEMS Microbiol Lett. 1988;48:219–24. [Google Scholar]
- 53.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–71. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–33. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 55.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Blasi U. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 56.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 2010;6:e1000834. doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.