Abstract
The use of histone deacetylase (HDAC) inhibitors has revealed an essential role for deacetylation in transcription of IFN-responsive genes. The HDAC1 protein associates with both signal transducer and activator of transcription (STAT) 1 and STAT2, and IFN-α stimulation induces deacetylation of histone H4. Inhibition of HDAC1 by small interfering RNA (siRNA) decreases IFN-α responsiveness whereas expression of HDAC1 augments the IFN-α response, demonstrating that HDAC1 modulates IFN-α-induced transcription. Importantly, the innate antiviral response is inhibited in the absence of deacetylase activity. The requirement for deacetylase is shared by IFN-γ transcription response and may represent a general requirement for STAT-dependent gene expression.
Reversible posttranslational modifications and the enzymes that catalyze their attachment to and removal from protein targets are essential for the precise regulation of diverse cellular responses including signal transduction and gene regulation. Recent advances in the study of gene expression have revealed numerous posttranslational modifications to histone N termini. Acetylation, in particular, is regulated by the opposing actions of histone acetyl transferase (HAT) enzymes and histone deacetylase (HDAC) enzymes, and patterns of acetylated histone residues contribute to the chromatin code hypothesis for epigenetic regulation of gene expression (1–3). This hypothesis is supported by numerous reports of HAT or HDAC recruitment to promoters by interactions with promoter-specific transcriptional activators, and evidence indicates that the modified histone tails serve as docking sites for chromatin remodeling complexes and other transcriptional coactivators. A common view has emerged associating the recruitment of HAT activity with transcriptional activation, and HDAC activity with transcriptional repression (4). Several examples of elevated basal mRNA accumulation in the presence of HDAC inhibitors support this idea. However, a more global analysis of gene expression in leukemia cell lines estimates that at least 9% of the genome may be regulated by trichostatin A (TSA), with equal numbers of tested genes activated or suppressed (5). These data suggest that more specific patterns of acetylation and deacetylation may be required to comprise an interpretable epigenetic code for any individual gene or expression system.
For most cytokines, receptor binding triggers an intracellular signaling cascade involving one or more signal transducer and activator of transcription (STAT) proteins, producing active transcription factors that specify mRNA induction profiles (6). Prototypical STAT-signaling pathways in the IFN cytokine systems regulate both the cellular innate antiviral response and adaptive immune responses (7, 8). Binding of IFN-α/β to cell surface receptors leads to the tyrosine phosphorylation of cytoplasmic STAT1 and STAT2, which in combination with IFN regulatory factor 9 (IRF9), assemble into a heterotrimeric complex, the IFN-α/β-stimulated gene factor 3 (ISGF3; reviewed in refs. 9–13). ISGF3 rapidly enters the nucleus, binds to conserved IFN-stimulated response element (ISRE) sequences on the promoters of IFN-α/β-stimulated genes (ISG) and increases their transcription rates. The C-terminal STAT1 transcriptional activation domain (TAD) is dispensable for most ISGF3 transcriptional activity (14), and IRF9 contributes DNA-binding specificity but is transcriptionally inert in the absence of STAT proteins (15, 16). Instead, the STAT2 C terminus provides a potent and essential TAD for ISGF3, providing contact surfaces for coactivator recruitment (17). STAT2 interacts with the cAMP response element binding protein (CREB)-binding protein (CBP)/p300 HAT proteins (18–20) and a GCN5/TAFII130-containing (TAF = TATA-binding protein (TBP)-associated factor) HAT complex associated with localized transient histone H3 acetylation (21). For some but not all target promoters, STAT2 can also bind the Brahma-related gene (BRG) 1 subunit of the SWI-SNF chromatin remodeling complex (22). In addition, STAT2 recruits the metazoan Mediator complex through essential contacts with the vitamin D receptor-interacting protein (DRIP) 150/thyroid hormone receptor-associated protein (TRAP) 170 subunit (23).
Here, the requirement for deacetylase activity in IFN-α-inducible gene regulation was investigated. Inhibition of HDAC activity suppresses IFN-α transcriptional responses and prevents the IFN-α-induced innate antiviral response although no intrinsic defect in STAT protein tyrosine phosphorylation, ISGF3 oligomerization, nuclear transport, or DNA binding were observed. Results indicate that IFN-α stimulation induces local histone H4 deacetylation and that the deacetylase protein HDAC1 associates with both STAT1 and STAT2. Furthermore, specific reduction of HDAC1 by RNA interference inhibits IFN-α-induced transcription whereas HDAC1 expression enhances IFN-α-induced transcription. These findings indicate a fundamental role for deacetylase activity and HDAC1 in transcriptional activation in response to IFN-α/β, a requirement shared with IFN-γ signaling through STAT1 homodimers.
Experimental Procedures
Cell Culture, Cytokine and Drug Treatments, and Transfection. Human 2fTGH, 293T, and HeLa (S3) cells were maintained in DMEM supplemented with 10% cosmic calf serum (HyClone), except the RNA interference assay and the cytopathic effect assay, where DMEM containing 10% FBS or 2% cosmic calf serum was used, respectively. Transfection of 2fTGH cells was carried out by using SuperFect reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions. Treatment of cells with IFN was performed as indicated by using 1,000 units of IFN-α per ml or 5 ng of IFN-γ per ml. Trichostatin A (TSA, Upstate Biotechnology, Lake Placid, NY) was added at 400 ng/ml simultaneously with IFN as indicated, with the exception of Fig. 2D, where TSA was added to cells 1 h before harvesting, together with or followed by IFN-α for the indicated times. Sodium butyrate (NaB, Upstate Biotechnology) was added simultaneously with IFN at the final concentration of 5 mM.
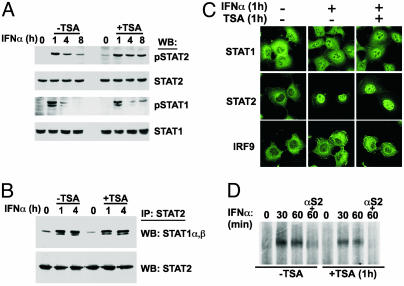
Fig. 2.
IFN-responsive STAT activation, dimerization, nuclear translocation, and DNA binding activity are intact in TSA-treated cells. (A) 2fTGH cells were treated with IFN-α alone or together with TSA for the indicated times. Protein extracts were analyzed by Western blot (WB) by using antibodies specific for tyrosine phosphorylated STATs (pSTAT1, pSTAT2), or total STATs (STAT1, STAT2). (B) 2fTGH cells were treated as in A, but protein extracts were subject to immunoprecipitation (IP) with STAT2 antiserum. Immune complexes were analyzed by Western blot for coprecipitated STAT1α and STAT1β. (C) 2fTGH cells were treated with IFN-α alone or together with TSA and processed for indirect immunofluorescence to detect STAT1, STAT2, and IRF9. (D) 2fTGH cells were treated with IFN-α and TSA for the indicated times, and lysates were subjected to EMSA by using a 32P-labeled ISG15-ISRE element probe. ISGF3 complex identity was confirmed by a STAT2 antibody supershift (αS2).
Plasmids and Reporter Gene Assays. Luciferase assays were carried out according to the manufacturer's instructions (Promega) by cotransfecting cDNA expression plasmids, reporter gene, and either pCMV-LacZ or Renilla luciferase (Dual-Luciferase Reporter Assay) to normalize for transfection efficiency. For IFN-γ responses, the reporter gene contained four copies of the M67-SIE linked to a TATA box and the firefly luciferase ORF (gift of J. Darnell and D. Besser, The Rockefeller University). The IFN-α/β responsive reporter gene contained five copies of the ISG54 ISRE element upstream of the TATA box and the firefly luciferase ORF. Cells were treated with IFN and TSA 24 h after transfection as indicated, except in Fig. 1D, where treatment began 18 h after transfection. FLAG-tagged HDAC1, -4, and -5 expression plasmids were a generous gift of S. L. Schreiber (Harvard University). IRF9 and IRF9-STAT2 plasmids have been described (15). Short treatment times for experiments in Fig. 3 (3 h) were chosen to maximize the effect of exogenously expressed HDAC1 before reporter gene accumulation reached its maximum. For all luciferase assays, data represent the mean ± SD of triplicate samples. For some figures, a student t test was used to determine significance.
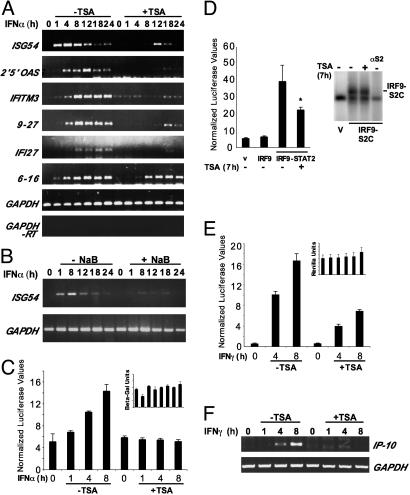
Fig. 1.
HDAC inhibition impairs IFN-α-stimulated gene activation. (A) 2fTGH cells were treated with IFN-α for the indicated amounts of time in the presence (+) or absence (–) of simultaneous TSA. RT-PCR was carried out with primers specific for the indicated genes. (B) Similar to A, but NaB was used. (C) 2fTGH cells were transfected with ISRE-luciferase reporter together with CMV-LacZ, treated as indicated, and analyzed by luciferase assay. Luciferase units were normalized to β-galactosidase. (Inset) β-Galactosidase levels, normalized to total protein concentration. (D Left) 2fTGH cells were transfected with empty vector or expression vectors for IRF9 or IRF9-S2C along with ISRE-luciferase reporter and Renilla luciferase control. *, P < 0.05. (Right) Lysates were tested for DNA binding by EMSA by using a 32P-labeled ISG15-ISRE oligonucleotide as probe. Supershift was performed with STAT2 antibody (αS2). (E) Similar to C, but cells were transfected with IFN-γ-responsive M67-luciferase reporter gene along with Renilla luciferase reporter plasmid for normalization, and treated as indicated. Luciferase values were normalized to Renilla luciferase values. (Inset) Renilla luciferase values normalized to total protein concentration. (F) 2fTGH cells were treated as indicated, and total RNA was analyzed by RT-PCR using primers specific for IP-10 and GAPDH.
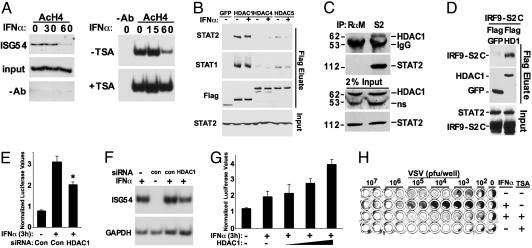
Fig. 3.
IFN deacetylates histone H4 and requires essential STAT–HDAC1 associations. (A Left) 2fTGH cells were treated with IFN-α for the times indicated and subjected to a ChIP assay using antibody specific for acetylated histone H4. Coprecipitated DNA was amplified by PCR by using primers specific for the ISG54 ISRE promoter element. (Right) HeLa cells were used in a similar assay in the presence or absence of TSA. (B) 2fTGH cells were transfected with expression vectors for FLAG epitope-tagged GFP, HDAC1, HDAC4, or HDAC5, stimulated with (+) or without (–) IFN-α as indicated, and lysates were precipitated with FLAG affinity resin. Eluted STAT1 and STAT2 were detected by Western blotting. Control FLAG immunoblot demonstrates equivalent precipitation of expressed proteins in the eluates; bottom, STAT2 immunoblot of unprecipitated extracts (Input) demonstrates equivalent loading. (C) Coimmunoprecipitation of STAT2 and HDAC1 from 2fTGH cell extracts. RαM, rabbit anti-mouse IgG; ns, nonspecific band. (D) Similar to B, but 2fTGH cells were transfected with IRF9-S2C and FLAG-HDAC1 or Flag-GFP expression plasmids before Flag affinity precipitation and analysis by Western blotting with STAT2 (Top) and Flag antibodies (Middle). Input panel demonstrates similar expression and loading levels. (E) 2fTGH cells were transfected with siRNA specific for HDAC1 or a scrambled siRNA control (Con) along with ISRE-luciferase reporter gene and Renilla luciferase control for normalization. *, P < 0.005. (F) Similar to E, except total RNA was isolated from cells and subjected to RT-PCR for ISG54 mRNA. IFN-α treatments were performed for 3 h. con = scrambled siRNA duplex. (G) 293T cells were transfected with ISRE-luciferase and Renilla luciferase in the absence (–) or presence of 1, 2, or 6 μg of HDAC1 expression plasmid, and treated with IFN-α as indicated. (H) 2fTGH cells were treated with (+) or without (–) IFN-α and/or TSA for 4 h followed by infection of duplicate wells with serially diluted vesicular stomatitis virus (VSV). Pfu, plaque-forming units.
Immunoprecipitations and Protein Assays. Polyclonal antisera to STAT1 (E23 and C-24), STAT2 (C-20), and HDAC1 (H-51) were obtained from Santa Cruz Biotechnology, and M2-FLAG antibodies were obtained from Sigma. These were all used according to the manufacturer's instructions. To immunoprecipitate FLAG epitope-tagged proteins, total extracts were prepared with whole-cell extract buffer [50 mM Tris (pH 8.0)/280 mM NaCl/0.5% IGEPAL/0.2 nM EDTA/2 mM EGTA/10% glycerol/1 mM DTT] supplemented with a protease inhibitor mixture (Complete; Roche). Lysates were treated with 1μg/ml DNase I (Sigma) in the presence of 1 mM DTT and 5 mM MgCl2 for 2 h followed by treatment with 50 μg/ml ethidium bromide for 1 h. Lysates were then incubated with M2-FLAG affinity agarose bead slurry overnight. After washing the beads with whole-cell extract buffer, immune complexes were eluted with 0.2 mg/ml FLAG tripeptide in protein elution buffer, PEB300 [20 mM Tris (pH 7.5)/300 mM NaCl/0.2 mM EDTA/0.1% Nonidet P-40/15% glycerol] for 2 h. Eluates were denatured in SDS loading buffer and separated by SDS/PAGE; Western blots were performed and chemiluminescent detection was carried out according to the manufacturer's protocol (NEN Renaissance). For endogenous coimmunoprecipitation assays, cellular lysates were treated with DNase I as above, and incubated with STAT2 antisera or rabbit anti-mouse IgG (DAKO) overnight. Western blots were performed by using HDAC1 antisera.
RNA Analysis. Total RNA was prepared by using TRIzol reagent (Invitrogen), digested with DNase I, and subjected to reverse transcriptase with SuperScript II RNase H-reverse transcriptase (Invitrogen) and PCR analysis. A mock reaction was carried out with no reverse transcriptase added (-RT). Onetenth of the resulting cDNA product was used as template for 25 cycles of PCR in the presence of [α-32P]dATP (Perkin–Elmer Life Sciences) using specific primers as indicated. As a control for genomic DNA contamination, PCR was carried out with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers by using the mock (-RT) reaction products as templates. After gel electrophoresis, products were detected by ethidium bromide staining or autoradiography, and quantified by phosphorimaging.
Primer sequences used were as follows: ISG54T, 5′-AATGCCATTTCACCTGGAACTTG-3′; ISG54B, 5′-GTGATAGTAGACCCAGGCATAGT-3; 2′5′OAST, 5′-AGGCAGAAAGGGATTTTATC-3′; 2′5′OASB, 5′-TAACCTAGGTCTCGACTCCA-3′; IFITM3T, 5′-ATGAGTCACACTGTCCAAAC-3′; IFITMB, 5′-AACAGGGACCAGACGACA-3′; 9–27T, 5′-CATCCTTCCAAGGTCCAC-3′; 9–27B, 5′-ATGTTCAGGCACTTGGCG-3′; IFI27T, 5′-TCACCTCATCAGCAGTGAC-3′; IFI27B, 5′-GACATCATCTTGGCTGCTA-3′; 6–16T, 5′-AACCTCTTCTTCCTTCTTGG-3′; 6–16B, 5′-TACTTGGGAGGTTGAGACAG-3′; GAPDHT, 5′-CCCTTCATTGACCTCAACT-3′; GAPDHB, 5′-GACGCCAGTGGACTCCA-3′.
Indirect Immunofluorescence. 2fTGH cells were grown on Permanox chamber slides (Nalgene Nunc) and treated with IFN-α and/or TSA for 1 h before fixation. Cells were fixed in 200 μl of 4% formaldehyde in PBS for 15 min and permeabilized in ice-cold 1:1 methanol-acetone for 10 min at –20°C. After five washes with PBS, samples were blocked with 1% BSA in PBS for 15 min at 37°C. After every subsequent antibody exposure, samples were washed and blocked. Antibody staining was performed for either STAT1 (0.02 μg/ml) or STAT2 (4 μg/ml) and detected with fluorescein isothiocyanate-conjugated rabbit IgG. STAT1 and STAT2 polyclonal antisera used for immunofluorescence were precleared on fixed and permeabilized STAT1-deficient U3A cells or STAT2-deficient U6A cells to reduce nonspecific background staining. Images were obtained by using a Leica (Deerfield, IL) TCSSP confocal microscope as described (24).
DNA Binding Analysis. Electrophoretic mobility-shift assays (EMSAs) were performed by using 32P-labeled double-stranded oligodeoxynucleotide-containing ISG15 promoter ISRE element as the probe. After treatment or transfection, whole-cell extracts were prepared as described and incubated with the probe in gel shift buffer [20 mM Hepes (pH 7.9)/4% Ficoll/1 mM MgCl2/0.1 mM EGTA/0.5 mM DTT/2 mg of poly(dI-dC)] for 20 min. The mixture was separated on a 5% polyacrylamide gel containing 0.25× Tris-borate EDTA and detected by autoradiography. For supershift, 0.1 μl of STAT2 antiserum was added to the reaction mixture.
RNA Interference. Individual wells of a 24-well culture dish containing 2fTGH cells were transfected with 100 pmol of small interfering RNA (siRNA) alone or together with ISRE-luciferase reporter gene and Renilla luciferase plasmid by using the TransIT-TKO reagent (Mirus, Madison, WI) following the manufacturer's recommendations. siRNAs (SMARTpool) specific for HDAC1 were obtained from Dharmacon (Lafayette, CO). For control, Scramble II Duplex siRNA was used. Reporter gene analysis or RT-PCR were performed 48 h after transfection.
Antiviral Assay. Antiviral responses measured by cytopathic effect assay were conducted by standard methodology using the Indiana strain of vesicular stomatitis virus as reported (25). Cells were fixed and stained with methylene blue (3% in 50% ethanol) at 18 h postinfection.
Chromatin Immunoprecipitation (ChIP) Assay. ChIP assays were performed by using the ChIP assay kit (Upstate Biotechnology). Protein–DNA complexes were crosslinked with formaldehyde, and DNA was either cleaved in purified nuclei with 100 units of micrococcal nuclease (see Fig. 3A Left), or sonicated to yield 500- to 1,000-bp fragments (see Fig. 3A Right). Acetylated histone H4 protein–DNA complexes were precipitated by using anti-acetyl-H4 antibody (Upstate Biotechnology). Precipitated DNA was amplified by radioactive PCR using primers encompassing 200 bp around the ISRE-binding site on the human ISG54 promoter as described (23). PCR products were separated on a 5% polyacrylamide gel and detected by autoradiography.
Results
IFN-Stimulated Gene Induction Is Blocked by HDAC Inhibitors. To examine the role for HDACs in IFN-stimulated transcription, human 2fTGH cells were stimulated with IFN-α in the presence or absence of pharmacological HDAC inhibitors, and total RNA was prepared over a 24-h time course. After reverse transcription with random primers, PCR was performed with primers specific for known ISGF3 targets. In all cases, the PCR profile indicated that the genes were induced rapidly and transiently in response to IFN stimulation (Fig. 1A). The presence of TSA during IFN-α stimulation dramatically inhibited induction of all of the genes tested, resulting in defective mRNA accumulation. Importantly, there was no inhibition of a control mRNA, GAPDH. To confirm that this inhibition was the result of the HDAC inhibitory activity of TSA and to rule out nonspecific effects of the compound, a second structurally distinct HDAC inhibitor, NaB, was used in a similar assay. Treatment of cells with NaB also blocked ISG induction (Fig. 1B), and similar ISG suppression was observed in several human cell lines tested, including HeLa, 293T, and 2fTGH (data not shown). These findings support the conclusion that HDAC activity is required for IFN-α-responsive transcription.
To test whether the requirement for HDAC activity was a general property of ISGF3-regulated transcription, an ISRE-luciferase reporter gene assay was carried out with similar methodology (Fig. 1C). Stimulation with IFN-α results in increased luciferase activity, but simultaneous treatment with TSA prevented reporter gene induction. This finding indicates that the TSA inhibition of IFN-α-responsive transcription is independent of specific promoter context. To determine whether the inhibitory effects of TSA on IFN signaling are specifically due to transcriptional responses and not another aspect of IFN-α/β signal transduction, an IFN-independent, ISRE-dependent minimal model system was tested for TSA sensitivity. A hybrid transcription factor, IRF9-S2C, is an IFN-independent ISGF3 mimic with the DNA binding subunit of ISGF3 linked to the TAD of STAT2 (26). Expression of IRF9-S2C potently induced the transcription of the ISRE-luciferase reporter gene, and treatment with TSA for 8 h reduced reporter gene activity by ≈44% (Fig. 1D). Previous studies (27–29) have suggested that acetylation of IRF proteins might interfere with their DNA-binding activity, and therefore, it was important to test the DNA-binding ability of the IRF9-S2C protein. EMSAs indicate that TSA treatment has no effect on the ISRE DNA-binding activity of IRF9-S2C. Together, these findings indicate that the TSA treatment specifically targets IFN transcriptional regulation rather than some other aspect of ISGF3 function and furthermore implicate the STAT2 TAD as a potential target of HDAC inhibition.
These results clearly signify that HDAC activity is required for ISGF3-dependent transcriptional responses. To test whether this deacetylase requirement is shared by different STAT-dependent transcription responses, similar assays were conducted for IFN-γ-dependent transcription. In a similar signaling pathway, IFN-γ activates a gamma-IFN-activated factor, GAF, that is a homodimer of STAT1. Although similar in principle to ISGF3, the STAT1 homodimer binds to a distinct response element (the gamma IFN-activated sequence, GAS) to regulate a subset of IFN target genes (30). Treatment of cells with IFN-γ resulted in reporter gene activation, which was significantly reduced in the presence of TSA (Fig. 1E). In addition to reporter gene inhibition, RT-PCR assays indicate that transcription of IP-10, an endogenous IFN-γ target gene, was also prevented by HDAC inhibition (Fig. 1F). Together, these results demonstrate that deacetylation is required for both STAT1- and STAT2-dependent transcriptional responses.
TSA Does Not Target the IFN-Janus Kinase (JAK)-STAT-ISGF3 Pathway. Although important for the regulation of histones, several examples of direct transcription factor modification by acetylation have been described (27–29, 31). Because negative regulation of IFN signaling to the nucleus could provide one mechanistic explanation for the observed defect in ISG activation, the effects of HDAC inhibitors on IFN-α signal transduction were tested directly by examining ISGF3 phosphorylation, dimerization, nuclear translocation, and DNA binding (Fig. 2). Immunoblotting with phosphotyrosine-specific antibodies indicates that IFN-α-dependent activation of STAT1 and STAT2 is not inhibited by TSA treatment, and coimmunoprecipitation assays indicate that STAT heterodimerization remains intact. Indirect immunofluorescence was used to test the subcellular localization of ISGF3 components after IFN-α stimulation. Regardless of TSA treatment, IFN-α induced nuclear translocation of both STAT1 and STAT2, and no differences in IRF9 localization were observed. Finally, EMSAs indicate that TSA treatment had no effect on ISGF3 DNA-binding activity. These results indicate that IFN-α-responsive ISGF3 signaling from the cytoplasm to the nucleus remains intact in the presence of TSA, suggesting that ISGF3 or upstream pathway components are not targets of deacetylase activity.
ISGF3 Recruits HDAC1. Because ISGF3 activation and nuclear translocation are not affected by deacetylase inhibitors, evidence for IFN-α-induced histone deacetylation was sought. It was previously reported (21) that histone H3 becomes hyperacetylated in ISGF3 target promoters in response to IFN-α, so the effects of IFN-α on histone H4 acetylation were tested. Crosslinking ChIP assays were carried out to examine the acetylation status of histone H4 at the IFN-α-responsive ISG54 promoter. Basal H4 acetylation was readily detected in this assay, but IFN-α stimulation resulted in reduction of acetylated H4 levels at the ISG54 locus (Fig. 3A Left). Importantly, this H4 deacetylation was prevented by TSA treatment (Fig. 3A Right), consistent with the notion that H4 may be a target of IFN-α-responsive deacetylation. Although the direct enzyme-substrate relationship is not clearly established, we postulate that that TSA mediates its inhibitory effect on ISGF3-regulated transcription by interfering with a requisite step between ISGF3 and histone H4 deacetylation.
One simple explanation for these results invokes the ISGF3-dependent recruitment of an HDAC enzyme to the target promoter(s). To test for STAT–HDAC interactions, FLAG epitope-tagged HDAC proteins representing two classes of HDAC enzymes (HDAC1, -4, and -5) were expressed in cells and collected on FLAG affinity resin (Fig. 3B). Immunoblotting the eluates with specific antisera indicates that both STAT1 and STAT2 are coprecipitated with HDAC1 whereas little or no interactions were observed between STATs and HDAC4 or HDAC5. No obvious differences in STAT–HDAC1 interaction patterns were observed in these experiments after IFN-α treatment, suggesting that IFN-induced posttranslational modifications are not required for STAT–HDAC1 association. Instead, their interaction is regulated on the basis of subcellular distribution because HDAC1 is an exclusively nuclear protein and can interact with the nascent nuclear ISGF3 only after IFN stimulation. The STAT2–HDAC1 interaction was further confirmed by coimmunoprecipitation of endogenous proteins from wholecell extracts. HDAC1 was readily detected in the STAT2 immunoprecipitation, but not in the control precipitation (Fig. 3C). Because HDAC1 has been previously implicated as a TSA-sensitive H4 deacetylase (32), it is a reasonable candidate for both the observed effects of IFN-α stimulation on H4 deacetylation and the inhibition of ISG transcription by TSA. Nonetheless, it remains possible that redundancy with closely related deacetylase proteins may be involved in the regulating of full IFN responses. Interactions between IRF9-S2C and HDAC1 were also observed (Fig. 3D), and the function of this IFN-independent transcription factor was inhibited by TSA (Fig. 1D). Both ISGF3 and IRF9-S2C require deacetylase activity for transcriptional stimulation and bind to HDAC1, consistent with the model that the STAT2 TAD is at least one determinant for the recruitment of HDAC1 to the ISG promoter.
To verify a role for HDAC1 in ISGF3 transcriptional activity, IFN-α responses were evaluated in cells where HDAC1 expression levels were either reduced or enhanced. RNA interference was used to reduce HDAC1 expression in the context of an ISRE-dependent transcription assay. IFN-α-dependent reporter gene activity was unaffected by transfection of a control non-specific “scrambled” siRNA duplex. In contrast, siRNA duplexes specific for HDAC1 significantly reduced IFN-responsive reporter gene activity (Fig. 3E). The role of HDAC1 in IFN activation of an endogenous ISGF3 target gene, ISG54, was tested by using a similar RNA interference assay and RT-PCR analysis. Reduced levels of ISG54 accumulation were observed in the presence of HDAC1 siRNA duplexes compared with “scrambled” control. Quantification of ISG54 signals by phosphorimaging and normalization to GAPDH indicates a 40% reduction in ISG54 mRNA in the presence of HDAC1 siRNA compared with control samples transfected with a scrambled siRNA (Fig. 3F). In a complementary assay designed to test the HDAC1-positive regulatory role in ISGF3-mediated transcription, an ISRE-luciferase reporter gene assay was carried out in 293T cells where HDAC1 expression was increased by transfection with HDAC1 expression vector. Treatment with IFN-α resulted in relatively modest activation of the reporter after 3 h, which was enhanced by HDAC1 expression in a dose-dependent manner (Fig. 3E). These results strongly implicate HDAC1 as a critical positive coactivator for ISGF3-dependent transcriptional responses.
IFN Antiviral Response Requires HDAC Activity. The biological activity of IFN-α/β has been well characterized as the establishment of a cellular antiviral state capable of restricting virus replication. Fully functional STAT proteins and ISGF3 transcriptional activity are required for this innate immune response (25, 33). To validate the requirement for HDAC activity in the IFN-α biological response, the effects of TSA on antiviral activity were tested in a standard assay for virus-induced cytopathic effects (25, 34) (Fig. 3F). Treatment of cells with IFN-α for 4 h results in an antiviral state that is capable of reducing the cytopathic effects of vesicular stomatitis virus by two orders of magnitude. In contrast, simultaneous treatment with TSA completely prevents the IFN-α-induced antiviral state. This biological response assay provides a powerful demonstration of the importance of HDAC activity in IFN-induced innate antiviral immunity.
Discussion
The results of this study indicate that deacetylase activity is required for positive transcriptional regulation of IFN-α/β responsive genes. Specifically, results demonstrate that the deacetylase protein HDAC1 can interact with both the STAT1 and STAT2 subunits of ISGF3. The inhibition of deacetylase activity has no effect on IFN-α signaling leading to STAT activation, the assembly and nuclear import of the ISGF3 transcription factor complex, or ISGF3 DNA binding, but instead targets a downstream event required for activated gene expression. Inhibition of HDAC1 by RNA interference results in a reduction in both the ISRE reporter and endogenous ISG54 mRNA accumulation whereas exogenous expression of HDAC1 augmented the response. Furthermore, it was observed that histone H4 is deacetylated at a target promoter in response to IFN-α stimulation. Irrespective of whether the H4 deacetylation is an indirect prerequisite for transcriptional activation or a direct consequence of HDAC1 recruitment, TSA prevents the observed IFN-α-induced H4 deacetylation, in agreement with an active role of HDAC activity in positive ISG regulation. The effects of HDAC inhibition are sufficient to obliterate IFN function in the innate immune response because TSA treatment prevents the primary biological outcome, induction of a cellular state refractory to virus infection. Moreover, we have demonstrated that the IFN-γ-STAT1 system similarly requires deacetylase activity, and that STAT1 can independently interact with HDAC1. Consideration of these data along with a preliminary finding that STAT3-dependent transcription is similarly inhibited by TSA (I.N. and C.M.H., unpublished observation), and prior reports of TSA inhibition of IL-2 responses and HDAC recruitment to STAT5 target genes (35, 36), lead to a hypothesis that recruitment of HDAC activity might be a general requirement for STAT-dependent transcriptional regulation. Because HDAC inhibitors are effective in the treatment of cancer, their ability to modify cytokine-dependent transcriptional regulation vital for innate and adaptive immunity is an important clinical consideration that might guide the development of second generation deacetylase inhibitors (37, 38).
Although seeming to undermine the general correlation of HDAC activity with transcriptional repression, transient and reversible modifications to specific histone tail lysine residues are a central tenet of the chromatin code hypothesis. This idea is supported not only by the current data set and other examples of STAT-dependent transcription that require deacetylase activity, but also by the direct observation of several transiently acetylated histone lysine residues during temporal induction of the IFN-β promoter enhanceosome (39). Therefore, it is tempting to speculate that the requirement for HDAC activities and other modification removal systems are likely to be the rule rather than the exception in the regulation of activated transcription. The role for HDAC1 and deacetylase activity in positive IFN-dependent transcription demonstrated here raises several additional basic mechanistic questions for future consideration. For example, it remains possible that HDAC1 is only one of the deacetylase enzymes responsible for full IFN responses. Requirements for other class I HDACs, such as the close homolog HDAC3, may account for the partial effects of HDAC1 siRNA relative to the more complete inhibition by TSA. Our FLAG coprecipitation data suggest a weak capacity for STAT interactions with class II HDACs (Fig. 3B). The identification of the specific pattern(s) of individual lysine residue modifications on histone H4 that are altered by IFN stimulation, and whether additional targets for deacetylation exist in the IFN pathway will surely illuminate the various roles for deacetylation in cytokine signaling. The role of HDAC1 in IFN responses might reflect an independent activity of the deacetylase or perhaps an alternate function of one of the multisubunit HDAC-containing complexes (e.g., NuRD, Sin3) that are typically associated with transcriptional repression. Complete understanding of IFN-dependent gene regulation will require more information on the dynamic relationships between recruited deacetylases and acetylases, as well as the precise coupling of these regulatory events to establish a chromatin code platform that, in concert with additional coactivating partners, signals transcription activation. The research described here clearly implicates HDAC1 and HDAC activity as an essential element of the coactivation system for IFN-induced gene regulation and the IFN-induced innate antiviral response.
Acknowledgments
We thank Dr. Stuart Schreiber for providing FLAG-HDAC cDNAs, Jason Rodriguez for assistance with immunofluorescence, and Christina Ulane for assistance with FLAG affinity chromatography. This work was supported by National Institutes of Health Grant GM063872 and American Cancer Society Grant RSG-02-049-01 (to C.M.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: STAT, signal transducer and activator of transcription; ISRE, IFN-stimulated response element; ISG, IFN-stimulated gene; ISGF, ISG factor; HDAC, histone deacetylase; HAT, histone acetyltransferase; TSA, trichostatin A; NaB, sodium butyrate; siRNA, small interfering RNA; IRF, IFN regulatory factor; TAD, transcriptional activation domain; EMSA, electrophoretic mobility-shift assay; ChIP, chromatin immunoprecipitation.
References
- 1.Cheung, W. L., Briggs, S. D. & Allis, C. D. (2000) Curr. Opin. Cell Biol. 12, 326–333. [DOI] [PubMed] [Google Scholar]
- 2.Roth, S. Y., Denu, J. M. & Allis, C. D. (2001) Annu. Rev. Biochem. 70, 81–120. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 4.Struhl, K. (1998) Genes Dev. 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 5.Chambers, A. E., Banerjee, S., Chaplin, T., Dunne, J., Debernardi, S., Joel, S. P. & Young, B. D. (2003) Eur. J. Cancer 39, 1165–1175. [DOI] [PubMed] [Google Scholar]
- 6.Levy, D. E. & Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662. [DOI] [PubMed] [Google Scholar]
- 7.Samuel, C. E. (2001) Clin. Microbiol. Rev. 14, 778–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biron, C. A. (2001) Immunity 14, 661–664. [DOI] [PubMed] [Google Scholar]
- 9.Horvath, C. M. (2000) Trends Biochem. Sci. 25, 496–502. [DOI] [PubMed] [Google Scholar]
- 10.Darnell, J. E., Jr., Kerr, I. M. & Stark, G. M. (1994) Science 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- 11.Kessler, D. S., Veals, S. A., Fu, X.-Y. & Levy, D. E. (1990) Genes Dev. 4, 1753–1765. [DOI] [PubMed] [Google Scholar]
- 12.Aaronson, D. S. & Horvath, C. M. (2002) Science 296, 1653–1655. [DOI] [PubMed] [Google Scholar]
- 13.Lau, J. F. & Horvath, C. M. (2002) Mt. Sinai J. Med. 69, 156–168. [PubMed] [Google Scholar]
- 14.Muller, M., Laxton, C., Briscoe, J., Schindler, C., Improta, T., Darnell, J. E., Jr., Stark, G. R. & Kerr, I. M. (1993) EMBO J. 12, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraus, T. A., Lau, J. F., Parisien, J. P. & Horvath, C. M. (2003) J. Biol. Chem. 278, 13033–13038. [DOI] [PubMed] [Google Scholar]
- 16.Veals, S. A., Schindler, C., Leonard, D., Fu, X.-Y., Aebersold, R., Darnell, J. E., Jr., & Levy, D. E. (1992) Mol. Cell. Biol. 12, 3315–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qureshi, S. A., Leung, S., Kerr, I. M., Stark, G. R. & Darnell, J. E., Jr. (1996) Mol. Cell. Biol. 16, 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulson, M., Pisharody, S., Pan, L., Guadagno, S., Mui, A. L. & Levy, D. E. (1999) J. Biol. Chem. 274, 25343–25349. [DOI] [PubMed] [Google Scholar]
- 19.Park, C., Lecomte, M. J. & Schindler, C. (1999) Nucleic Acids Res. 27, 4191–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya, S., Eckner, R., Grossman, S., Oldread, E., Arany, Z., D'Andrea, A. & Livingston, D. M. (1996) Nature 383, 344–347. [DOI] [PubMed] [Google Scholar]
- 21.Paulson, M., Press, C., Smith, E., Tanese, N. & Levy, D. E. (2002) Nat. Cell Biol. 4, 140–147. [DOI] [PubMed] [Google Scholar]
- 22.Huang, M., Qian, F., Hu, Y., Ang, C., Li, Z. & Wen, Z. (2002) Nat. Cell Biol. 4, 774–781. [DOI] [PubMed] [Google Scholar]
- 23.Lau, J. F., Nusinzon, I., Burakov, D., Freedman, L. P. & Horvath, C. M. (2003) Mol. Cell. Biol. 23, 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez, J. J., Parisien, J. P. & Horvath, C. M. (2002) J. Virol. 76, 11476–11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath, C. M. & Darnell, J. E., Jr. (1996) J. Virol. 70, 647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus, T. A., Lau, J. F., Parisien, J.-P. & Horvath, C. M. (2003) J. Biol. Chem. 278, 13033–13038. [DOI] [PubMed] [Google Scholar]
- 27.Caillaud, A., Prakash, A., Smith, E., Masumi, A., Hovanessian, A. G., Levy, D. E. & Marie, I. (2002) J. Biol. Chem. 277, 49417–49421. [DOI] [PubMed] [Google Scholar]
- 28.Masumi, A., Yamakawa, Y., Fukazawa, H., Ozato, K. & Komuro, K. (2003) J. Biol. Chem. 278, 25401–25407. [DOI] [PubMed] [Google Scholar]
- 29.Masumi, A. & Ozato, K. (2001) J. Biol. Chem. 276, 20973–20980. [DOI] [PubMed] [Google Scholar]
- 30.Decker, T., Lew, D. J., Mirkovitch, J. & Darnell, J. E., Jr. (1991) EMBO J. 10, 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu, W. & Roeder, R. G. (1997) Cell 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 32.Ferreira, R., Naguibneva, I., Mathieu, M., Ait-Si-Ali, S., Robin, P., Pritchard, L. L. & Harel-Bellan, A. (2001) EMBO Rep. 2, 794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durbin, J. E., Hackenmiller, R., Simon, M. C. & Levy, D. E. (1996) Cell 84, 443–450. [DOI] [PubMed] [Google Scholar]
- 34.Friedman, R. M. (1981) Interferons: A Primer (Academic, New York).
- 35.Xu, M., Nie, L., Kim, S. H. & Sun, X. H. (2003) EMBO J. 22, 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyama, Y., Adachi, M., Sekiya, M., Takekawa, M. & Imai, K. (2000) Blood 96, 1490–1495. [PubMed] [Google Scholar]
- 37.Yoshida, M., Furumai, R., Nishiyama, M., Komatsu, Y., Nishino, N. & Horinouchi, S. (2001) Cancer Chemother. Pharmacol. 48, Suppl. 1, S20–S26. [DOI] [PubMed] [Google Scholar]
- 38.Grozinger, C. M. & Schreiber, S. L. (2002) Chem. Biol. 9, 3–16. [DOI] [PubMed] [Google Scholar]
- 39.Agalioti, T., Chen, G. & Thanos, D. (2002) Cell 111, 381–392. [DOI] [PubMed] [Google Scholar]