Abstract
Interleukin-10 (IL-10) family of cytokines includes a number of its viral homologs, and eight cellular cytokines (IL-19, IL-20, IL-22, IL-24, IL-26, IL-28A, IL-28B, and IL29. The latter three proteins are also known as IFN-λ2, IFN-λ3, and IFN-λ1 and are recognized as type III (or λ) interferons. Most of the cellular homologs of IL-10 are monomeric in solution, whereas IL-10 and its viral homologs are intercalated dimers consisting of two helical bundle domains topologically similar to the monomeric members of the family. A classical four-helix bundle, a signature element of all helical cytokines, is always found as part of the domain of each member of the IL-10 family. The only crystal structures of the cytokine receptors that have been determined to date are for their extracellular domains (ECDs). Each ECD consists of two β-sandwich domains connected in the middle by a linkage. Signal transduction occurs when a cytokine binds to its two appropriate receptor chains. IL-10 and its viral homologs use the same IL-10 receptor system, whereas the cellular homologs of IL-10 use their own receptors, which in some cases may overlap and be used in different pairwise combinations. The known structures of binary complexes allowed marking of the receptor binding site, which always includes helix A, loop AB and helix F (IL-10 notations) on the side of a ligand, and loops of the N-terminal and C-terminal domains directed toward the ligand, and the interdomain linkage of the ECD. An analysis of the published structures of both the binary and ternary complexes of all helical cytokines allowed generation of a model of the signaling complex of IL-10. The receptor binding site I of the high affinity receptor IL-10R1 is exactly the same as in the crystal structure of the binary IL-10/sIL-10R1 complex, whereas the receptor binding site II is located on the surface of the first and the third helices of the four-helix bundle. The receptor/receptor interface, or site III, is formed between the C-terminal domains of IL-10R1 and IL-10R2.
Keywords: Class II cytokines, interleukin-10, ligand/receptor interactions, helix bundle, signal transduction
Introduction
The structure-function relationships are the key factors to our understanding of how cytokines play their biological role in communication between cells on molecular level. This review is exclusively devoted to discussing the known structures of cellular cytokines belonging to the IL-10 family. Since the other chapters have described the biological activity of each protein in great detail, it is unnecessary to repeat such information here. It is important to understand that the IL-10 family is only a subfamily of a large group of proteins that are collectively named cytokines, unifying interleukins, interferons, and some growth factors. The modern classification of cytokines is far from perfect, even though a number of different approaches, functional and structural, have been attempted [1–5]. It is now accepted that helical cytokines can be divided on two classes by the structure of their receptors. The whole IL-10 family belongs to class II cytokines, identified in early nineties [1, 2, 6] (reviewed in ref. [7–12]). This classification is mainly based on the primary structural features of the ECDs of the receptors, characterized by the presence of a single, mostly conserved disulfide bridge in each (N- and C-terminal) subdomain of the ECD, and the lack of the intact “WSXWS” motif in the proximity of the cell membrane [1]. The family of Class II cytokines consists of type I IFNs, including IFN-α, IFN-β, IFN-ω, IFN-ε, IFN-κ, IFN-τ, IFN-ς/lumitin, IFN-δ and IFN-ν [13–19]; IFN-γ, the single type II IFN (reviewed in [19, 20]); and the IL-10 family of cytokines [21] that includes four viral homologs (Epstein-Barr virus [22–24], equine herpersvirus type 2 [25], Orf parapoxvirus [26, 27], simian cytomegalovirus [28, 29]), and eight cellular homologs (IL-19 [30], IL-20 [31], IL-22 [32, 33], IL-24 [34, 35], IL-26 [36]) (reviewed in [5, 8, 37–42]), and also including type III IFNs [43, 44]: IFN-λ1 (IL-29), IFN-λ2 (IL-28A), IFN-λ3 (IL-28B) (reviewed in [45]). While the sequence similarity between IL-10 and some of its viral homologs may be very high, the cellular homologs are much less homologous and differ from both IL-10 and from each other in their biological functions.
Signal initiation
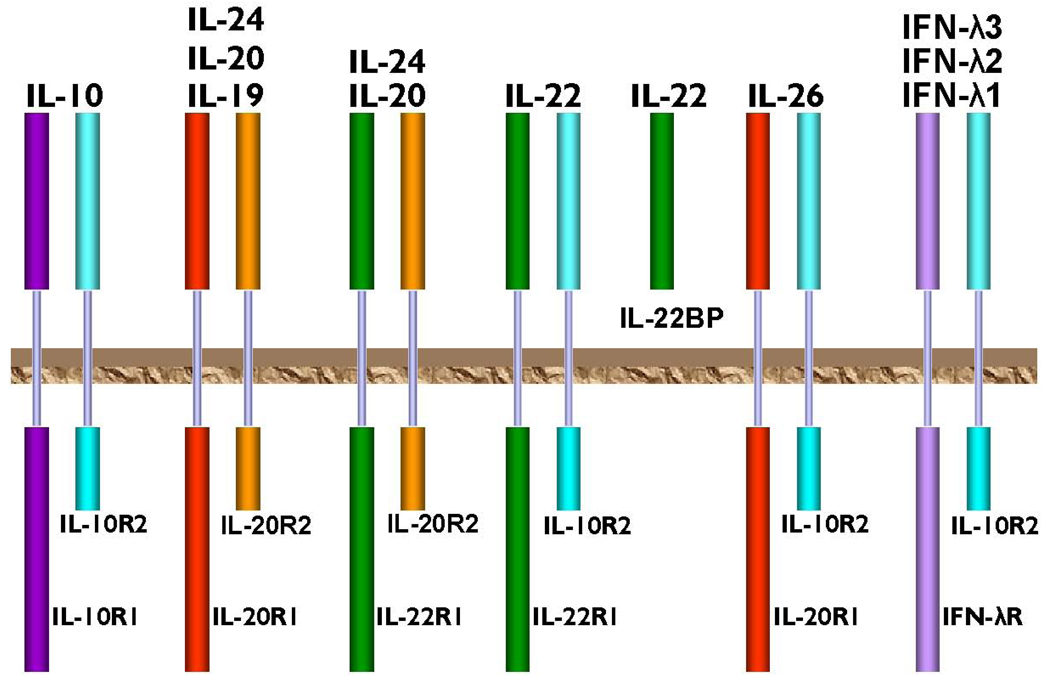
All class II cytokines, including members of the IL-10 family [46], initiate their biological signals by association with two appropriate membrane receptors. Both receptors have topologically similar ECDs, composed of two fibronectin type III domains [47, 48] folded in the conformation of a seven stranded β-sandwich, a short transmembrane domain, and intracellular/cytoplasmic domains of different lengths (Fig. 1). The chain with a long cytoplasmic domain is called receptor chain one, and the one with a short cytoplasmic domain is receptor chain two (Fig.1). The dimerization of receptor chains upon ligand binding leads to engagement of the intracellular Jak/STAT pathways [49, 50] that eventually initiates transcription of appropriate genes. Besides their sizes, the receptor chains also differ by their affinity toward cytokines. Chain one exhibits usually, although not always [51], higher affinity, and, with the exception of IL-26 which signals through two low affinity receptor chains [52], all other signaling complexes always include one high and one low affinity chain. In addition, the receptor chains can also be used by different cytokines in different combinations. IL-10 and its viral homologs use high affinity IL-10R1 [53] and low affinity IL-10R2 [54], whereas the cellular homologs of IL-10 use combinations of different receptor chains: IL-20R1, IL-20R2 [31, 55, 56], IL-22R1 [33, 57, 58], IFN-λR [43, 44] and IL-10R2 (Fig.1).
Fig.1.
Schematic representation of the IL-10 family of cytokines. Different combinations of receptor chains are shown. The brown plate represents a hypothetical cell membrane.
Some of these cytokines have the ability to bind to a few different combinations of the receptor chains. For example, IL-20 and IL-24 form stable complexes with either a pair IL-20R1/IL-20R2, or with IL-22R1/IL-20R2 [55, 56, 59]. There are altogether six known different membrane-bound receptor chains and one naturally occurring soluble receptor called IL-22 binding protein (IL-22BP) [60–62] that are utilized by the IL-10 family of cytokines.
Structures of the cytokines and receptors deposited in the PDB
Table I lists all structural information about members of IL-10 family deposited to date in the Protein Bank. Most of the crystal structures represent free ligands determined at medium-to-high resolution, along with a few structures of binary complexes. Unfortunately, there is as yet no available experimentally-determined structure of a ternary or signaling complex. Whereas such complexes have been prepared in a soluble form necessary for crystallization [51], some remaining technical problems must be overcome before crystals will be obtained.
Table I.
Structures of the proteins that belong to the Class II cytokines family deposited in the PDB.
| Protein | Biol. relev. aggregation | Species | Expression system | PDB code/Reference | Resolution/Remarks |
|---|---|---|---|---|---|
| IL-10 | Dimer | Human | E. coli | 1ILK/[66] | 1.8 Å, room temp |
| IL-10 | Dimer | Human | E. coli | 2ILK/[67] | 1.6 Å, 100 K. |
| IL-10 | Dimer | Human | E. coli | 1INR/[94] | 2.0 Å |
| IL-10 | Dimer | Human | E. coli | 2H24/[88] | 2.0 Å |
| Epstein-Barr IL-10 | Dimer | EBV | E. coli | 1VLK/[95] | 1.9 Å |
| IL-19 | Monomer | Human | DES2 | 1N1F/[64] | 1.95 Å |
| IL-22 | Monomer | Human | E. coli | 1MR4/[65] | 2.0 Å |
| IL-22 | Monomer | Human | DES2 | 1YKB/[96] | 2.6 Å |
| IFN-λ3 | Monomer | Human | E.Coli | 3HHC[74] | 2.8 Å |
| IL-10/sIL-10R1 | Tetramer | Human | E. coli, DES2 | 1J7V/[78] | 2.9 Å |
| IL-10/sIL-10R1mut | Tetramer | Human | E. coli, DES2 | 1Y6K/not publ. | 2.52 Å |
| EBV IL-10/sIL-10R1, and mut. | Tetramer | EBV | E. coli, DES2 | 1Y6M,1Y6N/[84] | 2.8 Å, 2.7 Å |
| CMV IL-10/sIL-10R1 | Tetramer | CMV | E. coli, DES2 | 1LQS/[83] | 2.7 Å |
| IL-10 monomer/9D7 Fab | Trimer | Human Rat | E. coli | 1LK3/[97] | 1.91 Å |
| IL-22/IL-22R1 | Dimer | Human | E. coli | 3DLQ[79] | 1.9 Å |
| IL-22/IL-22R1 | Dimer | Human | DES2 | 3DGC[80] | 2.5 Å |
| IL-22/ILBP | Dimer | Human | E. coli | 3G9V[81] | 2.76 Å |
Chinese hamster ovary cells;
Drosophila expression system, Sf9 cells.
Structures of the ligands
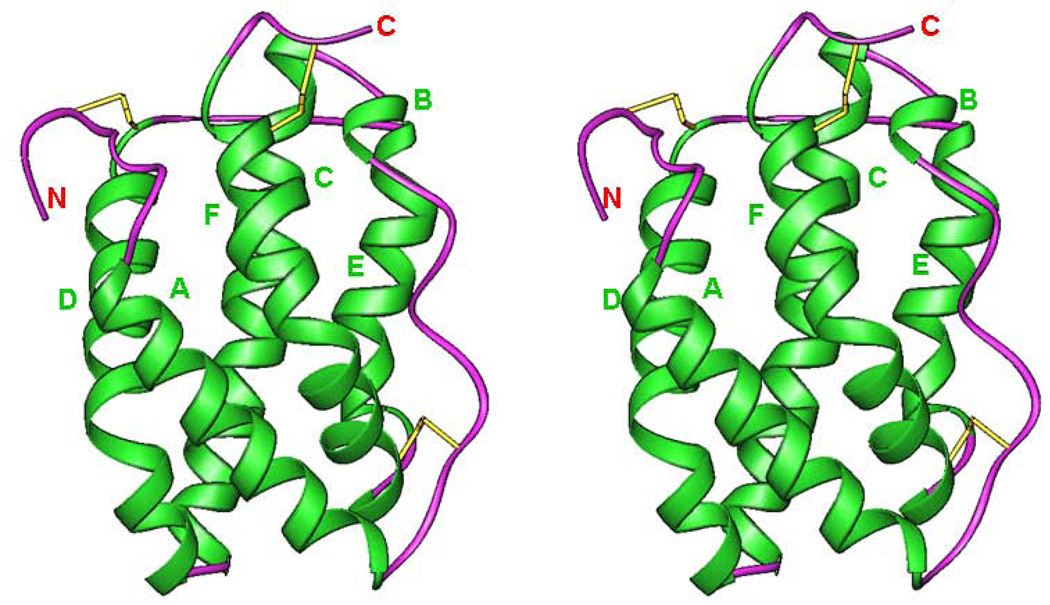
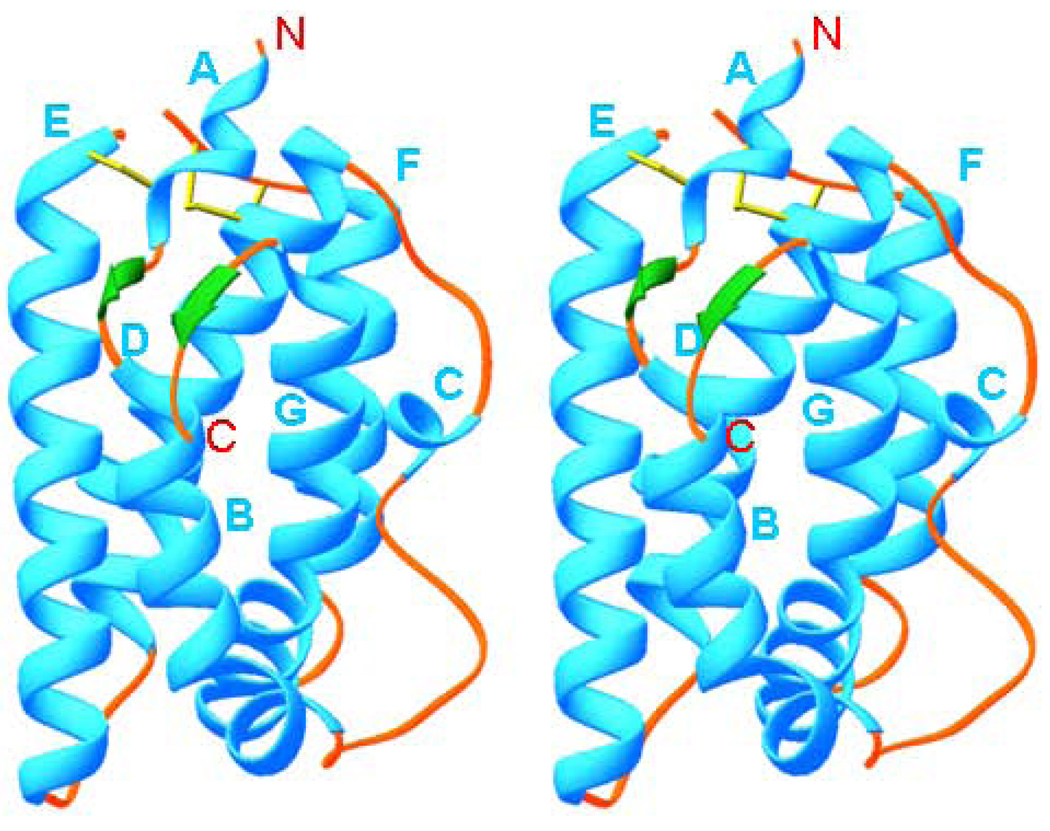
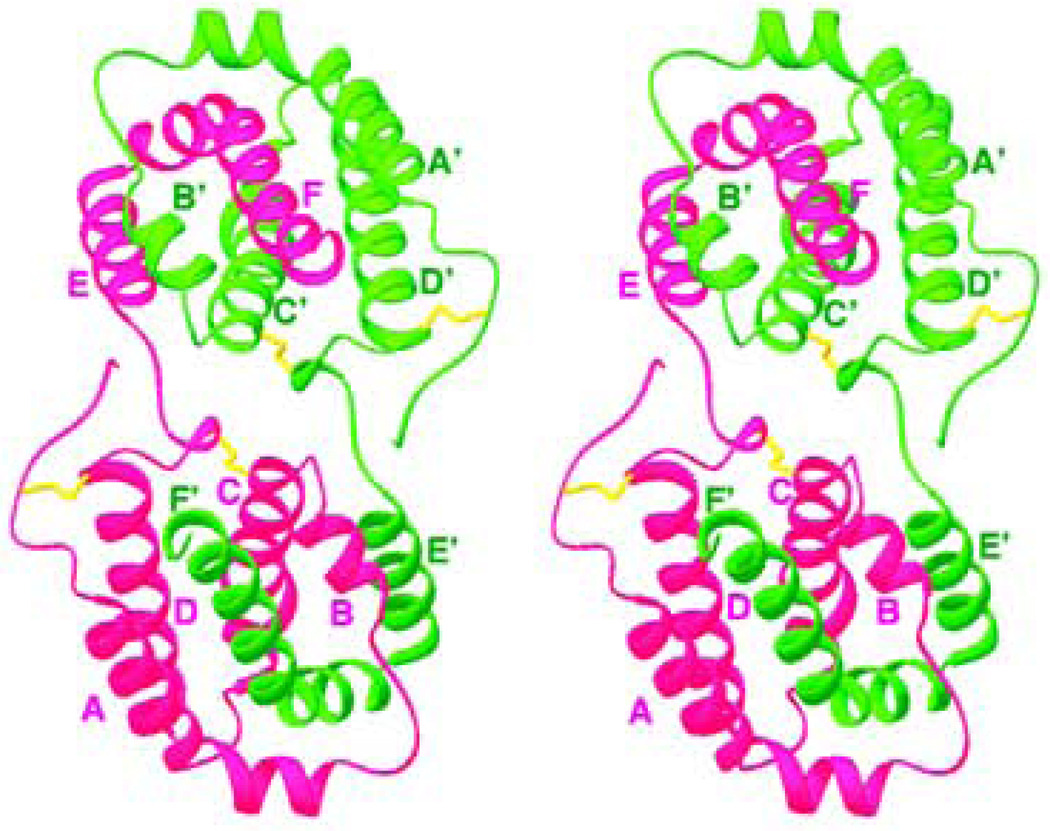
Helical cytokines are very compact molecules made of five to seven amphipathic helices, with a total helical content of 70–90%. All of the cellular homologs of IL-10 are monomers [51, 63–65] (Table 1), whereas IL-10 and its viral homologs are intercalated dimers (Fig. 2) [66, 67] composed of two identical domains. Each domain is a topological equivalent of a monomeric cytokine [64] (Fig. 2, 3, 4). The amphipathic helices are arranged in such a way that most of the hydrophobic amino acid residues are involved in the formation of the extended internal hydrophobic core inside the five-, six-or seven-helix bundle, stabilized by one to three disulfide bridges. These structures always include the ‘classical’ left-handed four-helix bundle [68], a signature element of all helical cytokines. An example of such a bundle can be seen in the structure of IL-10 [67] (helices A, C, D, F’ and A’, C’, D’, F) (Fig. 2). Crystal structures of IL-19 [64] and IL-22 [65, 69, 70] showed new structural features (Fig. 3), which marked a new IL-19 subfamily of cytokines [64, 71]. Helix A becomes short and is connected to helix B by a short β-strand, which in the structure of IL-19 (Fig. 3) participates, along with the C-terminal β-strand, in formation of a two strand β-sheet, never before seen in helical cytokines. An analysis of the amino acid sequence of human IL-20 strongly suggests that its structure must be very close to that of IL-19 [5, 64], although it lacks the C-terminal β-sheet. It is also likely that IL-26 exists and is active both as a dimer [36, 40, 72, 73] and a monomer (data are not shown). Crystal structure of IFN-λ3 [74] (Fig. 4) has shown complete lack of short helix A, found in IL-19 and IL-22 structures, even though the general topology is that of a member of the IL-10 family. Structural superposition of IFN-λ3 with IL-19 and IL-22 showed that it fits better fits to IL-22. That may be the reason why IFN-λ3 and IL-22 possess parallel functions, protecting epithelial tissue against viral and bacterial infections[74]. Members of IL-10 family of cytokines also belong to long-chain subfamily of helical cytokines, characterized by the conformation and the location of crossover polypeptide chain between the first and the second helix of the four-helix bundle [75, 76]. The known structures (Table I) of the binary complexes of cytokines with their appropriate soluble receptors showed that the first receptor chain binding site on the surface of the ligand includes the first and the last helix, as well as the loop between the first and the second helix of the four-helix bundle [68]. For example, in the case of IL-10 (Fig. 3), the receptor binding site includes [77, 78]: helix A, loop AB, and helix F’ of one domain, and helix A’, loop A’B’ and helix F of another domain. In the case of IL-19 (Fig. 4), an equivalent site includes helix B, loop BC, and helix G.
Fig.2.
Stereo diagram of the structure of IL-10 (PDB code 2ILK). The two monomers that form a domain-swapped dimer are shown in violet and green, respectively, and the disulfide bridges are in yellow. Figures 2–6 were generated with the program RIBBONS [93].
Fig. 3.
Stereo diagram of IL-19 (PDB code 1N1F). The helices are shown in cyan, loops in light-brown, β-strands in green, and disulfide bridges in yellow. The helices are labeled in cyan and the N-and C-termini are labeled in red.
Fig. 4.
Stereo diagram of IFN-λ3 (PDB code 3HHC). The helices shown in green, loops in magenta, and the disulfide bridges in yellow. The helices and the N-, C-termini are marked in green and red, respectively.
Receptors and binary complexes
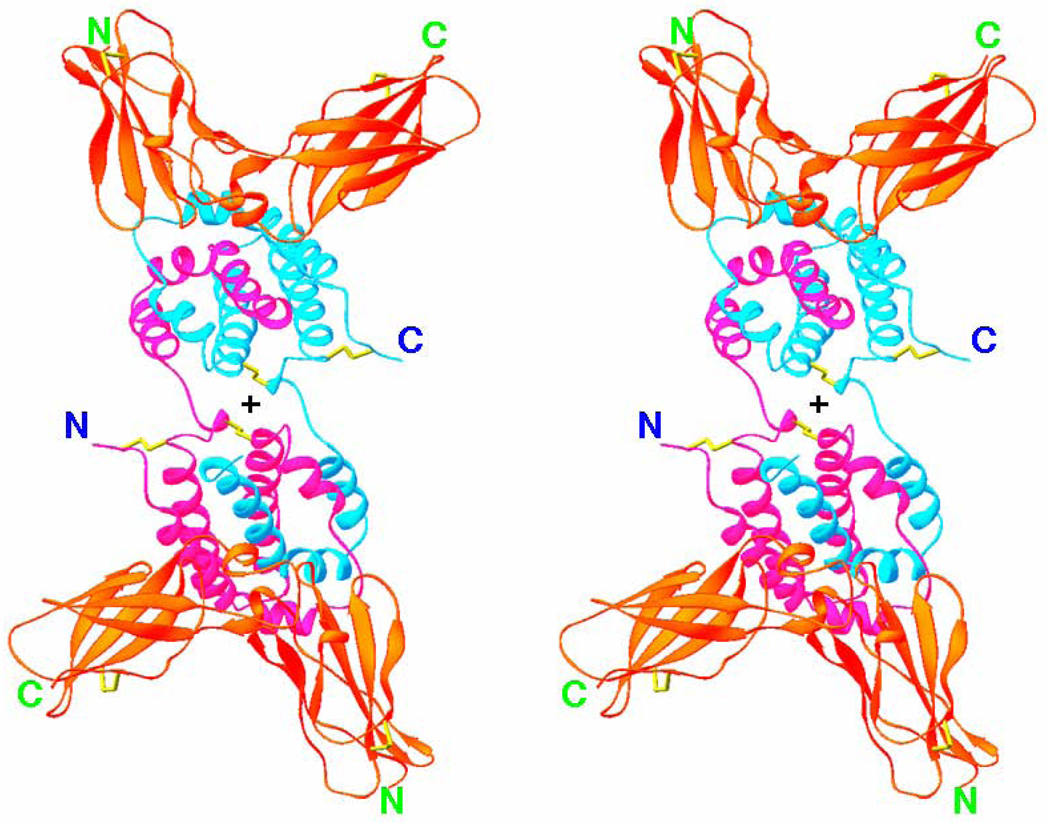
No structures of the unbound receptor chains, nor of the whole receptor complex or have been determined to date. However, structures of the binary complexes of IL-10R1 [78], a number of viral homologs, IL-22/IL22R1[79, 80], IL-22/IL-22BP[81] and IFN-λ1/IFN- λR1 (data not shown) are available (Table I). These structures showed that an ECD of class II cytokine receptor chains consist of two domains, each having fibronectin type III-like topology [47, 48, 82]. Fig. 5 shows the crystal structure of the complex of IL-10 with its soluble receptor chain, sIL-10R1 [78]. The molecule of the complex is a tetramer made up of an intercalated dimer of IL-10 and two sIL-10R1 molecules attached to each domain of IL-10. Two parts of the complex, including one domain of IL-10 and bound to it sIL-10R1. are related by a crystallographic two-fold axis, which is approximately perpendicular to the plane of Fig. 5 (its position is shown by a cross). Therefore, a 180° rotation around this axis perfectly superimposes both parts of the complex. Each domain of sIL-10R1 consists of one 3-stranded and one 4-stranded β-sheets (Fig. 5), packed one against another in the form of a sandwich. The domains are linked together by a short linkage consisting of one helical turn and a short β strand. Out of 14 loops connecting the β strands, five that are located in the vicinity of the receptor inter-domain junction were found to be the most important [78] because of their involvement in ligand-receptor interactions. The structure of the complex is quite rigid, with no conformational changes found between sIL-10R1 molecules bound to either IL-10 or to viral IL-10s [83, 84]. In spite of the low amino acid sequence identity (27%) between IL-10 and CMVIL-10, the interaction of either one with the receptor involves essentially the same areas on the surface of both the ligand and the receptor. In fact, a superposition of the CMVIL-10 domain bound to the sIL-10R1 with the structure of the IL-10 domain bound to the sIL-10R1 gives an r.m.s. deviation only 1.2 Å for 331 Cα pairs. Most of the ligand/receptor contacts are of polar character and can be clustered into two interacting sites [78, 83]. Binary complexes of other members of the IL-10 family have in general a similar topology, although the details of ligand/receptor interface are quite unique [79, 81, 85]. These specific differences in the interactions must define their high specificity.
Fig. 5.
Stereo diagram of IL-10/sIL-10R1 complex (PDB code 1J7V) viewed approximately along the crystallographic two-fold axis (its position is marked by cross). IL-10 is shown in violet and blue, the receptor chains are red, and the disulfide bridges are yellow.
Ternary complexes
As mentioned before, no crystal structure of a ternary or signaling complex has been determined so far for any member of the IL-10 family. However, an analysis of all known crystal structures of ternary complexes of other helical cytokines [46, 86] allows to mark the putative second receptor chain binding site as the first and the fourth helices of the four-helix bundle. In the case of IL-10 these are helices A and D of one domain and helices A’, D’ of another domain. Based on this information and on the assumption that the molecule should keep its two-fold symmetry, a homology model [86] of the ternary IL-10/sIL-10R1/sIL-10R2 complex was generated (Fig. 6). This model was later found to be in good agreement with both peptide scans [87] and with mutagenesis/surface plasmon resonance studies [88]. The receptor chains bind adjacent sides on the surface of each IL-10 domain. sIL-10R1 binding site or site I is exactly the same as in the binary IL-10/sIL-10R1 complex, sIL-10R2 binding site II is located on the surface of helices A and D, while sIL-10R1 and sIL-10R2, bound to the same IL-10 domain, interact with each other by their C-terminal domains, creating site III (Fig. 6). Most of the interactions found in all three interfaces are of polar nature, although hydrophobic interactions are also important, constituting 20%–35% of the total number of intermolecular contacts. The details of the ligand/receptor and receptor/receptor interactions can be found in the original papers [86] and in many reviews [5, 46, 89–91]. Both IL-10 receptor chains have a few glycosylation sites, which were found not to interfere with an overall organization of the IL-10/sIL-10R1/sIL-10R2 complex, and since the binary complexes of both IL-10 and its viral homologs with deglycosylated mutant of sIL-10R1 were also found quite stable [78, 83, 84, 92], it is clear that N-glycosylation is not that important for the formation of a biologically relevant ternary complex of IL-10/IL-10R1/IL-10R2. However, in some other instances the N- and O-linked olygosaccharides may be quite important [46]. In general, we may conclude that even though the role of glycosylation is not completely understood, it is likely that, in some cases, it may enhance the biological activity, as well as change the solubility and thermostability of both the cytokine and its receptor.
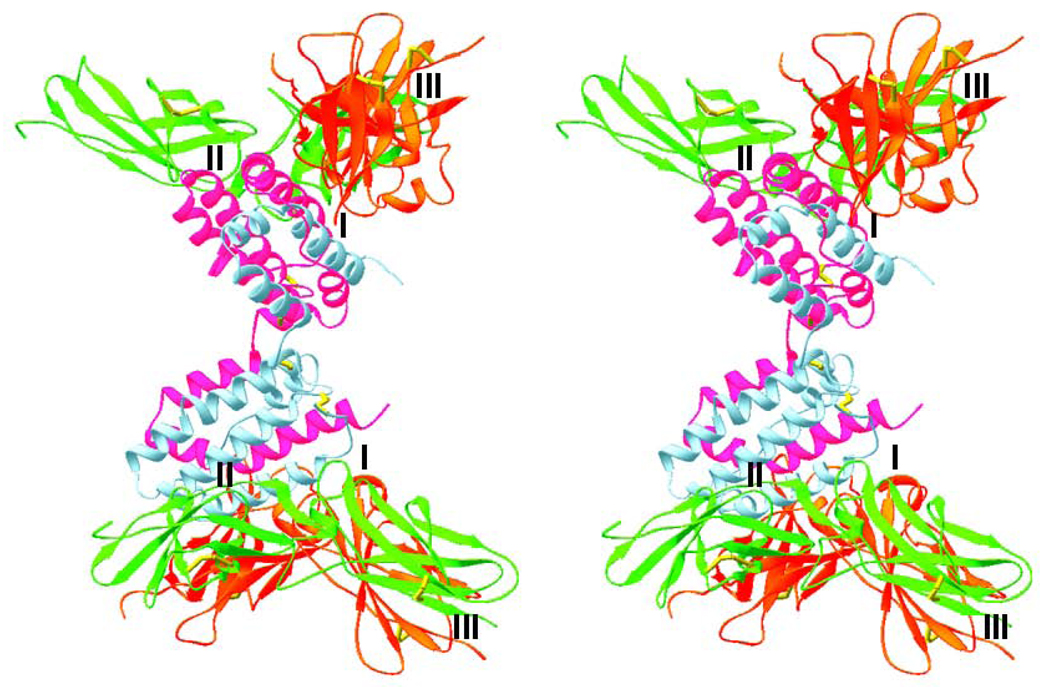
Fig. 6.
Stereo diagram of a model of the ternary complex of IL-10/sIL-10R1/sIL-10R2. IL-10 is shown in violet and blue, sIL-10R1 in red, sIL-10R2 in green, and the disulfide bridges are in yellow. A postulated position of the cell membrane is on the right hand side of the figure. The ligand/receptor and receptor/receptor interfaces are marked I–III in each half of the complex.
Discussion and concluding remarks
The IL-10 family of cytokines includes IL-10, viral homologs of IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, and three IFN-λs, the latter forming a unique subfamily of type III IFNs. Biologically active IL-10 and its viral homologs exist as intercalated dimers, whereas IL-19, IL-20, IL-22, and IL-24 are active as monomers. IL-26 is active as either a dimer or a monomer. The three-dimensional structures of the monomeric members of IL-10 family are topologically similar to the structure of a single domain of IL-10, which consists of a six-helix bundle that includes a classical four-helix bundle found in the structures of all helical cytokines. Published crystal structures of the complexes of IL-10/IL-10R1, its viral homologs, along with the recently solved structures of IL-22/IL-22R1, IL-22/IL-22BP and IFN-λ1/IFN-λ1R1 (data not shown) allowed detailed description of the ligand/receptor interface. The areas of the ligand creating the interface include helix A, loop AB and helix F (IL-10 notation), whereas the receptor utilizes three-four loops of domain D1, the interdomain junction, and two-three loops of domain D2. Topologically, structural elements involved in the interaction are similar, but at atomic level they are quite unique for each IL-10 family member, which certainly is necessary to provide high specificity of ligand/receptor interactions. The known crystal structures of the binary and ternary complexes allowed not only marking the receptor binding sites on the surface of IL-10 domains, but also provided guidance for generating a ternary model of IL-10/sIL-10R1/sIL-10R2 complex. This model is likely not very accurate in details, but this is the only model of this signaling complex which is currently available that shows how it could look on the surface of the cell.
Acknowledgment
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Special thanks go to Prof. Alexander Wlodawer for accurately proofreading the manuscript.
Biography

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bazan JF. Proc. Natl. Acad. Sci. USA. 1990;87:6934. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bazan JF. Immunol. Today. 1990;11:350. doi: 10.1016/0167-5699(90)90139-z. [DOI] [PubMed] [Google Scholar]
- 3.Schein CH. Curr. Pharm. Des. 2002;8:2113. doi: 10.2174/1381612023393161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims JE, Nicklin MJ, Bazan JF, Barton JL, Busfield SJ, Ford JE, Kastelein RA, Kumar S, Lin H, Mulero JJ, et al. Trends Immunol. 2001;22:536. doi: 10.1016/s1471-4906(01)02040-3. [DOI] [PubMed] [Google Scholar]
- 5.Zdanov A. Curr. Pharm. Des. 2004;10:3873. doi: 10.2174/1381612043382602. [DOI] [PubMed] [Google Scholar]
- 6.Ho ASY, Liu Y, Khan TA, Hsu D-H, Bazan JF, Moore KW. Proc. Natl. Acad. Sci. USA. 1993;90:11267. doi: 10.1073/pnas.90.23.11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. J. Leukoc. Biol. 2004;76:314. doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- 8.Renauld JC. Nat. Rev. Immunol. 2003;3:667. doi: 10.1038/nri1153. [DOI] [PubMed] [Google Scholar]
- 9.Kotenko SV, Langer JA. Int. Immunopharmacol. 2004;4:593. doi: 10.1016/j.intimp.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Langer JA, Cutrone EC, Kotenko S. Cytokine Growth Factor Rev. 2004;15:33. doi: 10.1016/j.cytogfr.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Krause CD, Pestka S. Pharmacol. Ther. 2005;106:299. doi: 10.1016/j.pharmthera.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Zdanov A. In: Class II cytokines. Zdanov A, editor. Kerala, India: Transworld Research Network; 2007. pp. 1–14. [Google Scholar]
- 13.Roberts RM, Liu L, Alexenko A. Prog. Nucleic Acid Res. Mol. Biol. 1997;56:287. doi: 10.1016/s0079-6603(08)61008-9. [DOI] [PubMed] [Google Scholar]
- 14.Lefevre F, Guillomot M, D'Andrea S, Battegay S, La Bonnardiere C. Biochimie. 1998;80:779. doi: 10.1016/s0300-9084(99)80030-3. [DOI] [PubMed] [Google Scholar]
- 15.Bekisz J, Schmeisser H, Hernandez J, Goldman ND, Zoon KC. Growth Factors. 2004;22:243. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- 16.Oritani K, Kanakura Y. J. Cell Mol. Med. 2005;9:244. doi: 10.1111/j.1582-4934.2005.tb00353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oritani K, Kincade PW, Zhang C, Tomiyama Y, Matsuzawa Y. Cytokine Growth Factor Rev. 2001;12:337. doi: 10.1016/s1359-6101(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 18.Pestka S, Krause CD, Walter MR. Immunol. Rev. 2004;202:8. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 19.Dunn GP, Koebel CM, Schreiber RD. Nat. Rev. Immunol. 2006;6:836. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 20.Young HA. Immunity. 2006;24:506. doi: 10.1016/j.immuni.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Moore KW, de Waal MR, Coffman RL, O'Garra A. Annu. Rev. Immunol. 2001;19:683. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 22.Moore KW, Vieira P, Fiorentino DF, Trounstine ML, Khan TA, Mosmann TR. Science. 1990;248:1230. doi: 10.1126/science.2161559. [DOI] [PubMed] [Google Scholar]
- 23.Hsu DH, de Waal Malefyt R, Fiorentino DF, Dang MN, Vieira P, de Vries J, Spits H, Mosmann TR, Moore KW. Science. 1990;250:830. doi: 10.1126/science.2173142. [DOI] [PubMed] [Google Scholar]
- 24.Vieira P, Malefyt RW, Dang M-N, Johnson KE, Kastelein R, Fiorentino DF, DeVries JE, Roncarolo M-G, Mosmann TR, Moore KW. Proc. Natl. Acad. Sci. USA. 1991;88:1172. doi: 10.1073/pnas.88.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rode HJ, Janssen W, Rosen-Wolff A, Bugert JJ, Thein P, Becker Y, Darai G. Virus Genes. 1993;7:111. doi: 10.1007/BF01702353. [DOI] [PubMed] [Google Scholar]
- 26.Fleming SB, McCaughan CA, Andrews AE, Nash AD, Mercer AA. J. Virol. 1997;71:4857. doi: 10.1128/jvi.71.6.4857-4861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleming SB, Haig DM, Nettleton P, Reid HW, McCaughan CA, Wise LM, Mercer A. Virus Genes. 2000;21:85. doi: 10.1023/b:viru.0000018443.19040.99. [DOI] [PubMed] [Google Scholar]
- 28.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Proc. Natl. Acad. Sci. USA. 2000;97:1695. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockridge KM, Zhou SS, Kravitz RH, Johnson JL, Sawai ET, Blewett EL, Barry PA. Virol. 2000;268:272. doi: 10.1006/viro.2000.0195. [DOI] [PubMed] [Google Scholar]
- 30.Gallagher G, Dickensheets H, Eskdale J, Izotova LS, Mirochnitchenko OV, Peat JD, Vazquez N, Pestka S, Donnelly RP, Kotenko SV. Genes Immun. 2000;1:442. doi: 10.1038/sj.gene.6363714. [DOI] [PubMed] [Google Scholar]
- 31.Blumberg H, Conklin D, Xu WF, Grossmann A, Brender T, Carollo S, Eagan M, Foster D, Haldeman BA, Hammond A, et al. Cell. 2001;104:9. doi: 10.1016/s0092-8674(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 32.Dumoutier L, Louahed J, Renauld JC. J. Immunol. 2000;164:1814. doi: 10.4049/jimmunol.164.4.1814. [DOI] [PubMed] [Google Scholar]
- 33.Xie MH, Aggarwal S, Ho WH, Foster J, Zhang Z, Stinson J, Wood WI, Goddard AD, Gurney AL. J. Biol. Chem. 2000;275:31335. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, Su ZZ, Lin JJ, Goldstein NI, Young CS, Fisher PB. Proc. Natl. Acad. Sci. USA. 1996;93:9160. doi: 10.1073/pnas.93.17.9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang H, Lin JJ, Su ZZ, Goldstein NI, Fisher PB. Oncogene. 1995;11:2477. [PubMed] [Google Scholar]
- 36.Knappe A, Hor S, Wittmann S, Fickenscher H. J. Virol. 2000;74:3881. doi: 10.1128/jvi.74.8.3881-3887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Annu. Rev. Immunol. 2004;22:929. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 38.Conti P, Kempuraj D, Frydas S, Kandere K, Boucher W, Letourneau R, Madhappan B, Sagimoto K, Christodoulou S, Theoharides TC. Immunol. Lett. 2003;88:171. doi: 10.1016/s0165-2478(03)00087-7. [DOI] [PubMed] [Google Scholar]
- 39.Kotenko SV. Cytokine Growth Factor Rev. 2002;13:223. doi: 10.1016/s1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 40.Fickenscher H, Hor S, Kupers H, Knappe A, Wittmann S, Sticht H. Trends Immunol. 2002;23:89. doi: 10.1016/s1471-4906(01)02149-4. [DOI] [PubMed] [Google Scholar]
- 41.Dumoutier L, Renauld JC. Eur. Cytokine Netw. 2002;13:5. [PubMed] [Google Scholar]
- 42.Ozaki K, Leonard WJ. J. Biol. Chem. 2002;277:29355. doi: 10.1074/jbc.R200003200. [DOI] [PubMed] [Google Scholar]
- 43.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. Nature Immunol. 2003;4:69. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 44.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. Nature Immunol. 2003;4:63. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 45.Ank N, West H, Paludan SR. J. Interferon Cytokine Res. 2006;26:373. doi: 10.1089/jir.2006.26.373. [DOI] [PubMed] [Google Scholar]
- 46.Zdanov A. Anti-Inflammatory & Anti-Allergy Agents in Medicinal Chemistry. 2006;5:207. [Google Scholar]
- 47.Leahy DM, Hendrickson WA, Aukhuil E, Erickson HP. Science. 1992;258:987. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 48.de Vos AM, Ultsch M, Kossiakoff AA. Science. 1992;255:306. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 49.Schindler C, Darnell JE., Jr Annu. Rev. Biochem. 1995;64:621. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 50.Decker T, Meinke A. Immunobiology. 1997;198:99. doi: 10.1016/S0171-2985(97)80031-9. [DOI] [PubMed] [Google Scholar]
- 51.Pletnev S, Magracheva E, Kozlov S, Tobin G, Kotenko SV, Wlodawer A, Zdanov A. Biochemistry. 2003;42:12617. doi: 10.1021/bi0354583. [DOI] [PubMed] [Google Scholar]
- 52.Sheikh F, Baurin VV, Lewis-Antes A, Shah NK, Smirnov SV, Anantha S, Dickensheets H, Dumoutier L, Renauld JC, Zdanov A, et al. J. Immunol. 2004;172:2006. doi: 10.4049/jimmunol.172.4.2006. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Wei SHY, Ho ASY, Malefyt RW, Moore KW. J. Immunol. 1994;152:1821. [PubMed] [Google Scholar]
- 54.Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. EMBO J. 1997;16:5894. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, Tan Z, Zhang R, Kotenko SV, Liang P. J. Biol. Chem. 2002;277:7341. doi: 10.1074/jbc.M106043200. [DOI] [PubMed] [Google Scholar]
- 56.Dumoutier L, Leemans C, Lejeune D, Kotenko SV, Renauld JC. J. Immunol. 2001;167:3545. doi: 10.4049/jimmunol.167.7.3545. [DOI] [PubMed] [Google Scholar]
- 57.Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. J. Biol. Chem. 2001;276:2725. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 58.Dumoutier L, Van Roost E, Colau D, Renauld JC. Proc. Natl. Acad. Sci. USA. 2000;97:10144. doi: 10.1073/pnas.170291697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramesh R, Mhashilkar AM, Tanaka F, Saito Y, Branch CD, Sieger K, Mumm JB, Stewart AL, Boquio A, Dumoutier L, et al. Cancer Res. 2003;63:5105. [PubMed] [Google Scholar]
- 60.Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. J. Immunol. 2001;166:7096. doi: 10.4049/jimmunol.166.12.7096. [DOI] [PubMed] [Google Scholar]
- 61.Dumoutier L, Lejeune D, Colau D, Renauld JC. J. Immunol. 2001;166:7090. doi: 10.4049/jimmunol.166.12.7090. [DOI] [PubMed] [Google Scholar]
- 62.Xu W, Presnell SR, Parrish-Novak J, Kindsvogel W, Jaspers S, Chen Z, Dillon SR, Gao Z, Gilbert T, Madden K, et al. Proc. Natl. Acad. Sci. USA. 2001;98:9511. doi: 10.1073/pnas.171303198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauane M, Gopalkrishnan RV, Sarkar D, Su ZZ, Lebedeva IV, Dent P, Pestka S, Fisher PB. Cytokine Growth Factor Rev. 2003;14:35. doi: 10.1016/s1359-6101(02)00074-6. [DOI] [PubMed] [Google Scholar]
- 64.Chang C, Magracheva E, Kozlov S, Fong S, Tobin G, Kotenko S, Wlodawer A, Zdanov A. J. Biol. Chem. 2003;278:3308. doi: 10.1074/jbc.M208602200. [DOI] [PubMed] [Google Scholar]
- 65.Nagem RAP, Colau D, Dumoutier L, Renauld J-C, Ogata C, Polikarpov I. Structure. 2002;10:1051. doi: 10.1016/s0969-2126(02)00797-9. [DOI] [PubMed] [Google Scholar]
- 66.Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, Weatherbee J, Wlodawer A. Structure. 1995;3:591. doi: 10.1016/s0969-2126(01)00193-9. [DOI] [PubMed] [Google Scholar]
- 67.Zdanov A, Schalk-Hihi C, Wlodawer A. Protein Sci. 1996;5:1955. doi: 10.1002/pro.5560051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Presnell SR, Cohen FE. Proc. Natl. Acad. Sci. USA. 1989;86:6592. doi: 10.1073/pnas.86.17.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagem RA, Lucchesi KW, Colau D, Dumoutier L, Renauld JC, Polikarpov I. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:529. doi: 10.1107/s0907444902001063. [DOI] [PubMed] [Google Scholar]
- 70.Xu T, Logsdon NJ, Walter MR. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:1295. doi: 10.1107/S0907444904010492. [DOI] [PubMed] [Google Scholar]
- 71.Zdanov A. Vitam. Horm. 2006;74:61. doi: 10.1016/S0083-6729(06)74003-1. [DOI] [PubMed] [Google Scholar]
- 72.Fickenscher H, Pirzer H. Int. Immunopharmacol. 2004;4:609. doi: 10.1016/j.intimp.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 73.Hor S, Pirzer H, Dumoutier L, Bauer F, Wittmann S, Sticht H, Renauld JC, de Waal MR, Fickenscher H. J. Biol. Chem. 2004;279:33343. doi: 10.1074/jbc.M405000200. [DOI] [PubMed] [Google Scholar]
- 74.Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. J Biol. Chem. 2009;284:20869. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rozwarski DA, Gronenborn AM, Clore GM, Bazan JF, Bohm A, Wlodawer A, Hatada M, Karplus PA. Structure. 1994;2:159. doi: 10.1016/s0969-2126(00)00018-6. [DOI] [PubMed] [Google Scholar]
- 76.Sprang SR, Bazan JF. Curr. Opin. Struct. Biol. 1993;3:815. [Google Scholar]
- 77.Josephson K, McPherson DT, Walter MR. Acta Crystallogr. D. Biol. Crystallogr. 2001;57:1908. doi: 10.1107/s0907444901016249. [DOI] [PubMed] [Google Scholar]
- 78.Josephson K, Logsdon NJ, Walter MR. Immunity. 2001;15:35. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- 79.Bleicher L, de Moura PR, Watanabe L, Colau D, Dumoutier L, Renauld JC, Polikarpov I. FEBS Lett. 2008;582:2985. doi: 10.1016/j.febslet.2008.07.046. [DOI] [PubMed] [Google Scholar]
- 80.Jones BC, Logsdon NJ, Walter MR. Structure. 2008;16:1333. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Moura PR, Watanabe L, Bleicher L, Colau D, Dumoutier L, Lemaire MM, Renauld JC, Polikarpov I. FEBS Lett. 2009;583:1072. doi: 10.1016/j.febslet.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 82.Bork P, Holm L, Sander C. J. Mol. Biol. 1994;242:309. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 83.Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, Walter MR. Proc. Natl. Acad. Sci. USA. 2002;99:9404. doi: 10.1073/pnas.152147499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoon SI, Jones BC, Logsdon NJ, Walter MR. Structure. (Camb.) 2005;13:551. doi: 10.1016/j.str.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 85.Jones BC, Logsdon NJ, Walter MR. Structure. 2008;16:1333. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pletnev S, Magracheva E, Wlodawer A, Zdanov A. BMC. Struct. Biol. 2005;5:10. doi: 10.1186/1472-6807-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wolk K, Witte E, Reineke U, Witte K, Friedrich M, Sterry W, Asadullah K, Volk HD, Sabat R. Genes Immun. 2005;6:8. doi: 10.1038/sj.gene.6364144. [DOI] [PubMed] [Google Scholar]
- 88.Yoon SI, Logsdon NJ, Sheikh F, Donnelly RP, Walter MR. J. Biol. Chem. 2006;281:35088. doi: 10.1074/jbc.M606791200. [DOI] [PubMed] [Google Scholar]
- 89.Walter MR. Adv. Protein Chem. 2004;68:171. doi: 10.1016/S0065-3233(04)68006-5. [DOI] [PubMed] [Google Scholar]
- 90.Walter MR. Immunol. Res. 2002;26:303. doi: 10.1385/ir:26:1-3:303. [DOI] [PubMed] [Google Scholar]
- 91.Walter MR. BioTechniques. 2002 Suppl:46. [PubMed] [Google Scholar]
- 92.Hoover DM, Schalk-Hihi C, Chou CC, Menon S, Wlodawer A, Zdanov A. Eur. J. Biochem. 1999;262:134. doi: 10.1046/j.1432-1327.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- 93.Carson M. J. Appl. Crystallogr. 1991;24:958. [Google Scholar]
- 94.Walter MR, Nagabhushan TL. Biochemistry. 1995;34:12118. doi: 10.1021/bi00038a004. [DOI] [PubMed] [Google Scholar]
- 95.Zdanov A, Schalk-Hihi C, Menon S, Moore KW, Wlodawer A. J. Mol. Biol. 1997;268:460. doi: 10.1006/jmbi.1997.0990. [DOI] [PubMed] [Google Scholar]
- 96.Xu T, Logsdon NJ, Walter MR. Acta Crystallogr. D. Biol. Crystallogr. 2005;61:942. doi: 10.1107/S0907444905009601. [DOI] [PubMed] [Google Scholar]
- 97.Josephson K, Jones BC, Walter LJ, DiGiacomo R, Indelicato SR, Walter MR. Structure. (Camb.) 2002;10:981. doi: 10.1016/s0969-2126(02)00791-8. [DOI] [PubMed] [Google Scholar]