SUMMARY
Ail is an outer membrane protein and virulence factor of Yersinia pestis, an extremely pathogenic, category A biothreat agent, responsible for precipitating massive human plague pandemics throughout history. Due to its key role in bacterial adhesion to host cells and bacterial resistance to host defense, Ail is a key target for anti-plague therapy. However, little information is available about the molecular aspects of its function and interactions with the human host, and the structure of Ail is not known. Here we describe the recombinant expression, purification, refolding, and sample preparation of Ail for solution and solid-state NMR structural studies in lipid micelles and lipid bilayers. The initial NMR and CD spectra show that Ail adopts a well-defined transmembrane β-sheet conformation in lipids.
1. INTRODUCTION
The Yersinia species of gram-negative bacteria include three strains that are pathogenic for humans, each causing diseases ranging from gastroenteritis (Y. pseudotuberculosis, Y. enterocolitica) to plague (Y. pestis), one of the most deadly human infectious diseases [1–4]. Of these, Y. pestis is extremely pathogenic, causing disease with only a few cells, and is easily transmittable from rats to humans through the bite of an infected flea, or from human to human through the air during widespread infection. Y. pestis has a long history of precipitating devastating human pandemics on a scale unmatched by any other infectious agent, including through its use in bioterrorism (e.g. during Black Death), and plague is still responsible for human outbreaks in endemic areas throughout the world, including the US, where more than 90% of human plague occurs in the Southwest, especially New Mexico, Arizona, California, and Colorado. Because Y. pestis spreads very easily and kills very quickly, it is classified as a “category A biothreat agent”, and plague is recognized as a re-emerging disease by the World Health Organization. The potential of bio-engineered antibiotic resistance and the lack of a vaccine providing protection from aerosolized Y. pestis, further underscore the need to identify new drugs [5].
Based on epidemiological observations and historical records, three Y. pestis biotypes have been associated with the three major human pandemics: biotype Antiqua with the Justinian Plague during the first millennium, biotype Medievalis with the Black Death from the 12th up to the 19th century, and biotype Orientalis with the third pandemic, which started during the 19th century and is still widespread and associated with modern plague [6]. The recently sequenced genomes of three Y. pestis strains, one from Orientalis (CO92) obtained from a clinical isolate in the United States, one from Medievalis (KIM), and one from Microtus (9100), which constitutes a fourth biotype, have provided a wealth of information about mechanisms of infection and pathogenesis [7–9].
The pathogenicity of Y. pestis stems from its striking ability to overcome the defenses of the mammalian host and overwhelm it with its massive growth, as well as its ability to survive in a variety of diverse environments (flea, mammalian host tissues, macrophages, and blood), which is critical for transmission from host to host. All three pathogenic Yersinia establish infection by adhering to the host and injecting cytotoxic Yersinia outer proteins (Yops) into host cells via a type III secretion system. Once in the host cell, Yops interfere with the host signaling pathways required for bacterial phagocytosis, actively blocking host-defense-mediated bacterial phagocytosis and lysis (reviewed in [2, 10, 11]). Establishing contact with the host cell is essential for engagement of secretion apparatus.
The outer membrane protein Ail (Attachment Invasion Locus; gene y1324) is a key virulence factor of Y. pestis, important for colonization, and pathogenesis [12–16]. Ail mediates bacterial adhesion to host epithelial cells through its association with the extracellular matrix component fibronectin [12–14, 16], facilitates Yop delivery to host cells [14, 15], and confers resistance to host serum [12, 13]. Deletion of the ail gene significantly attenuates the lethality of Y. pestis, increasing the pathogen’s median lethal dose by >3,000 fold. Furthermore, the ail deletion mutant is rapidly killed by exposure to animal sera, exhibits reduced binding to human-derived cell lines, and colonizes host tissue at much lower levels, compared to the wild-type strain [13, 14]. The expression and function of Ail are sensitive to temperature, reflecting sensitivity to the bacterial host environment [13, 17]. At 26°C and 37°C, the ambient temperatures of the flea vector and its mammalian hosts, Ail is expressed at high levels and is required for resistance to complement-mediated killing. However, expression is minimal at 6°C, i.e. outside the host. Furthermore, the serum-sensitive E. coli strain DH5α becomes serum resistant when it is transformed with an Ail expression vector, confirming that Ail confers serum resistance independent of other Y. pestis virulence factors [13].
Y. pestis, diverged from its closest ancestor Y. pseudotuberculosis only 1,500 to 20,000 years ago [18], and the Ail proteins from Y. pseudotuberculosis (gene YPTB2867) and Y. enterocolitica (gene YE1820) also mediate cell adhesion and invasion [16, 19] and confer serum resistance by binding to host complement regulatory proteins [20–23]. However, the single amino acid difference between Y. pestis Ail (Phe101) and Y. pseudotuberculosis Ail (Val101) is reported to reduce the adhesion and invasion activities in the latter [16].
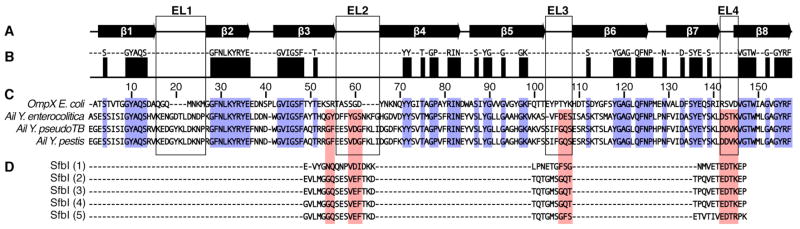
Ail belongs to a family of outer membrane proteins (Ail/Lom; pfam: PF06316) that function, at least in part, to protect bacteria from complement-mediated host defense [24–29]. The family members share amino acid sequence homology and are predicted to have similar membrane topologies, exemplified by the structure of E. coli OmpX [30], also a family member, which consists of eight transmembrane amphipathic β-strands and four extracellular loops (Fig. 1). The regions of greatest sequence homology are concentrated in the predicted membrane-spanning segments, while the extracellular loops of Ail are significantly longer than those of OmpX. The second and third extracellular loops (EL2, EL3) of Ail are reported to play a critical role in Y. enterocolitica cell adhesion [31] and are also predicted to be important for the functions of Ail from Y. pestis and Y. pseudotuberculosis. Interestingly, these loops exhibit significant amino acid sequence similarity to those in the unstructured/β-strand extracellular regions of bacterial fibronectin-binding proteins (Fig. 1D), which mediate bacterial adhesion to host by binding to the Fn type 1 (F1) domains in the N-terminus of mammalian fibronectin [32]. The sequence similarity is somewhat stronger for Ail from Y. pestis and Y. pseudotuberculosis than for Y. enterocolitica, suggesting a potential explanation for the somewhat weaker adhesion activity of the latter. Given the predicted integral membrane β-barrel structure of Ail, we anticipate that its mode of binding fibronectin will be different from that observed for other bacterial fibronectin-binding proteins [32].
Figure 1.
Multiple sequence alignment of Ail, OmpX, and fibronectin binding proteins. (A) Secondary structure of OmpX [30]. (B) Consensus sequence for Ail and OmpX. (C) Alignment of mature Ail from Y. pestis (gene y1324), Y. pseudotuberculosis (gene YPTB2867), and Y. enterocolitica (gene YE1820), and OmpX from E. coli (gene c0900). The Ail sequences from Y. pestis and Y. pseudotuberculosis are identical except for the F101V mutation before the predicted extracellular loop EL3. Amino acids identical in all four proteins are in blue. (D) Sequences of fibronectin F1 binding domains in the C-terminal region of the fibronectin-binding protein SfbI from Streptococcus pyogenes. Amino acids important for fibronectin binding, sharing similarity with the extracellular loops of Ail, are in red. Alignments were produced with ClustalW and rendered with Jalview.
Outer membrane proteins are often the first line of communication with the extracellular environment, with prominent roles in molecular transport, cellular homeostasis, and bacterial pathogenesis, and therefore, represent important targets for structural and functional characterization [33]. The structures of about twenty β-barrel membrane proteins have been determined in crystals by x-ray diffraction (reviewed in: [33–36]), the structures of five proteins (OmpX, OmpA, PagP, OmpG, VDAC) have been determined in micelles by solution NMR [37–41] and three proteins (OmpG, OmpA, OmpX) have been structurally characterized by solid-state NMR in lipid bilayers [42–45]. Lipid bilayers have the important advantage of providing an environment that closely resembles the cellular membrane, and structures determined in this environment are highly representative of the native in vivo conformations. Although samples of membrane proteins in lipid bilayers are too large for solution NMR structural studies, they are suitable for solid-state NMR studies where macroscopic alignment of the samples in the magnetic field provide very high-resolution, orientation-dependent restraints for the determination of both protein structure and protein orientation within the membrane [46–48].
The structure of Ail is not known, but is essential for understanding the molecular mechanism underlying its interaction with fibronectin and cell adhesion. Here we describe the expression, purification, refolding and sample preparation of Ail for solution and solid-state NMR structural studies of the protein in micelles and lipid bilayers.
2. MATERIALS AND METHODS
2.1 Protein expression and purification
The gene encoding mature Ail from Y. pestis KIM 10 (gene y1324, without the signal sequence) was cloned in the NdeI and BamHI restriction sites of the E. coli plasmid vector pET-3b (Novagen). Deletion of the signal sequence directs the expression of Ail into inclusion bodies. The Ail-bearing plasmid was transformed in E. coli C41(DE3) cells [49] (Lucigen) and positive clones were grown in M9 minimal medium, at 37°C, to a cell density of OD600 = 0.6, before induction with 1 mM IPTG (isopropyl 1-thio-β-D-galactopyranoside) for 6 hr. Cells were harvested by centrifugation (10,000 × g, 4°C, 30 min) and stored at −80°C overnight. For 15N and 13C isotopic labeling, the growth medium was prepared with (15NH4)2SO4 and 13C-glucose (Cambridge Isotope Laboratories) as the sole nitrogen and carbon sources.
Cells from 1 L of culture were suspended in 20 mL of buffer A (Table 1) and lysed with two passes through a French Press. The soluble fraction was removed by centrifugation (48,000 × g, 4°C, 30 min) and the resulting pellet, enriched in inclusion bodies, was thoroughly suspended in 30 mL of buffer B. The soluble fraction was removed by a second centrifugation step (48,000 × g, 4°C, 30 min) and the resulting pellet was dissolved in 30 mL of buffer C, transferred to a centrifuge bottle, and diluted 10-fold with water to precipitate Ail. The precipitate was collected by centrifugation (28,000 × g, 4°C, 1 hr).
Table 1.
List of buffers used for protein purification and sample preparation.
| buffer A: | (20 mM Tris, pH 8, 15% glycerol, 1 mM NaN3) |
| buffer B: | (20 mM Tris, pH 8, 1% deoxycholic acid, 1% igepal, 1 mM NaN3) |
| buffer C: | (20 mM Tris, pH 8, 500 mM NaCl, 6 M guanidinium hydrochloride) |
| buffer D: | (20 mM Na acetate, pH 5, 8 M urea) |
| buffer E: | (20 mM Tris, pH 8, 8 mM urea) |
| buffer F: | (20 mM glycine, pH 10.2, 200 mM KCl) |
| buffer G: | (20 mM Na phosphate, pH 6) |
Crude Ail protein from 5 L of cell culture was dissolved in 5–10 mL of buffer D, filtered through a 0.45 μM polypropylene membrane, and purified first by size exclusion chromatography (Sephacryl S-200 HR HiPrep 16/60 Column, GE Healthcare), also in buffer D, and then by cation exchange chromatography (HiTrap SP HP 5 ml column, GE Healthcare) in the same buffer with a NaCl gradient. Typically, 15–20 mg of purified AIL are obtained from a bacterial culture in 1 L of isotopically labeled (15N, 13C) M9 minimal media.
Purified Ail was precipitated by dialysis (10 kD cutoff) against water, lyophilized, and stored at −20°C. The protein mass was characterized by MALDI-TOF mass spectrometry. Protein expression and purification were monitored by SDS-PAGE performed with the Tris-Tricine system [50] and Coomassie Blue staining.
2.2 Screen for Ail refolding conditions
Refolding conditions were tested in a 96-well plate. Ail (20 μL, 5 mg/mL in buffer E) was added to each well, containing 200 μL of detergent (DPC, dodecylphosphocholine; DHPC, 1,2-dihexyl-sn-glycero-3-phosphocholine; 6-O-PC, 1,2-O-dihexyl-sn-glycero-3-phosphocholine; LMPG, 14:0-lysophosphatidylglycerol; LPPG, 16:0-lysophosphatidylglycerol; C8POE, n-octylpolyoxyethylene) in refolding buffer (Table 2). After incubating for 2 hr and 24 hr at room temperature, Protein aggregation was monitored by reading the absorbance from each well at 595 nm in a plate reader, and refolding was monitored by taking a 10 μL sample of the folding reaction for SDS-PAGE.
Table 2.
List of buffers used for protein refolding screen.
| pH 3–pH 5: | (20 mM Na acetate, ±100 mM KCl, ±600 mM Arginine) |
| pH 6–pH 8: | (20 mM Na phosphate, ±100 mM KCl, ±600 mM Arginine) |
| pH 6–pH 8: | (20 mM Tris, ±100 mM KCl, ±600 mM Arginine) |
| pH 9–pH 10: | (20 mM glycine Cl, ±100 mM KCl, ±600 mM Arginine) |
2.3 Reconstitution of Ail in lipid micelles
Pure, lyophilized Ail was dissolved in Buffer E at 5 mg/mL. Refolding in DHPC was obtained at room temperature by dropwise addition of 5 mg of Ail to 10 mL of buffer F, supplemented with 70 mM DHPC and 600 mM urea, while stirring, followed by overnight incubation with stirring. The refolded protein solution was dialyzed (10 kD cutoff, 10°C) for 45 minutes against buffer G supplemented with 500 mM urea, then for another 45 minutes against buffer G, and finally its volume was reduced in a spin concentrator (10 kD cutoff) to achieve a protein concentration of 300–500 μM (~500 μL). Small amounts of protein precipitate observed during this process, were removed by centrifugation.
The supernatant was dialyzed overnight, at 10°C, against 5 mL of buffer G supplemented with 100 mM DHPC, before transferring it to the NMR tube. The final sample, used for solution NMR studies, contained 0.3 mM Ail in 500 μL of 100 mM DHPC, 20 mM Na-phosphate, pH 6, and 5% D2O. The final concentration of DHPC was estimated by monitoring the intensity of the 1H NMR peak from the β-methylene protons of the choline headgroup at 3.15 ppm. The concentration of Ail was estimated by monitoring absorbance at 280 nm with an extinction coefficient of 1.641 ml•mg−1•cm−1). For mixed micelles, LPPG was further added to the Ail/DHPC sample from a stock solution of the lipid in water. All lipids were from Avanti Polar Lipids (www.avantilipids.com).
2.4 Reconstitution of Ail in lipid bicelles
Bicelles are lipid bilayers formed by mixtures of long and short chain phospholipids, where the parameter q corresponds to the molar ratio of long to short chain components. Small bicelles (q≤1.0) tumble rapidly and isotropically in solution and, therefore, are suitable for solution NMR experiments. In contrast, large (q=3.2) bicelles tumble slowly and have a net anisotropic diamagnetic susceptibility, which induces them to align spontaneously and homogeneously in the field of the NMR magnet, therefore, they are suitable for solid-state NMR experiments.
To prepare q=0.1 and q=3.2 bicelle samples, the appropriate amounts of DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) in chloroform solution were transferred to pear-shaped glass flasks, and the solvent was evaporated under a stream of N2 gas, followed by high vacuum, to generate a thin film of lipid on the flask interior. Refolded Ail, in 200 μL of DHPC-containing buffer G, was added to the dry DMPC film, and bicelles were formed by repeated freeze-thawing and vortexing as described previously [51, 52], then transferred to the NMR tube. The q=0.1 sample, used for solution NMR studies, contained 0.3 mM Ail in 500 μL of buffer G with 10 mM DMPC and 100 mM DHPC, and was transferred to a standard 5 mm OD NMR tube. The q=3.2 sample, used for solid-state NMR studies, contained 0.3 mM Ail in 300 μL of buffer G with 320 mM DMPC and 100 mM DHPC and was transferred to a flat-bottomed, 5 mm OD, NMR tube (New Era Enterprises).
2.5 Reconstitution of Ail in lipid bilayer vesicles
Pure lyophilized Ail (4 mg) was dissolved in 1 mL of 20 mM SDS in water, and added dropwise, at 45°C, to small unilamellar vesicles, prepared by sonicating 46.5 mg of 14-O-PC (1,2-O-ditetradecyl-sn-glycero-3-phosphocholine) in 10 mL of buffer F, as described [45]. The protein/detergent/lipid solution was incubated for ~3 hr at 40°C, and SDS was removed by dialysis (10 kD cutoff) against four 4 L changes of buffer F, followed by two 4 L changes of water. Protein folding was monitored by SDS-PAGE, where the appearance of a band with lower apparent molecular weight corresponds to folded Ail.
2.6 Circular dichroism experiments
CD (circular dichroism) spectra were recorded on a Jasco J-810 spectropolarimeter at room temperature in a 1 mm path length rectangular cuvette. For these experiments, Ail was refolded in DPC, as described above for DHPC, and diluted to obtain a final sample of 30 μM Ail, in, 10 mM KCl, 2 mM Tris, pH 9.6. buffer. The pH was adjusted by addition of HCl. The lower critical micelle concentration of DPC (1 mM), relative to DHPC (15 mM), permitted the use of samples with lower lipid concentration, which is beneficial for CD.
2.7 NMR experiments
Solution NMR experiments were performed on a Bruker AVANCE 600 spectrometer equipped with a cryoprobe, at 45° C, as described previously [45]. Resonance assignments were obtained using 1H/13C/15N triple resonance experiments with a 13C/15N-labeled Ail sample. Solid-state NMR experiments were performed on a Bruker AVANCE 500 spectrometer equipped with a 500/89 AS Magnex magnet, using double-resonance (1H/15N or 1H/31P) probes with a 5 mm inner diameter cylindrical coil for the bicelle samples. The probes were built at the UC San Diego NIH Resource for Molecular Imaging of Proteins (nmrresource.ucsd.edu). Bicelle samples were allowed to equilibrate in the magnetic field for at least 2 hr at 43°C before experiments. One-dimensional 15N and 31P chemical shift spectra were obtained as described previously [45]. The NMR data were processed using NMRPipe [53], and the spectra were analyzed with Sparky [54].
3. RESULTS AND DISCUSSION
3.1 Expression and purification of mature Ail
The production of proteins in bacteria is particularly useful for NMR studies because it allows milligram quantities of isotopically labeled proteins to be obtained relatively economically, and a variety of isotopic labeling schemes to be incorporated into the NMR experimental strategy. For Ail, expression was driven into inclusion bodies by deleting the ~20-residue leader sequence from the full-length gene. The formation of inclusion bodies often leads to increased yields by keeping the protein away from the bacterial membranes and limiting proteolytic degradation. It also facilitates purification since inclusion bodies are highly enriched in the target protein, virtually eliminating the need for engineered affinity tags (e.g. His tag) as in this case.
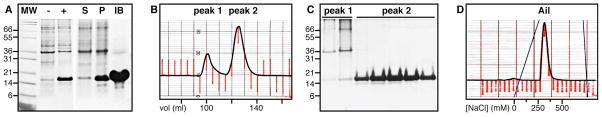
As shown in Figure 2A, very high levels of Ail expression were obtained upon induction with IPTG, and the protein purity, obtained after isolation of inclusion bodies from the soluble cellular fraction, was excellent. This facilitated the subsequent purification steps by size exclusion chromatography (Fig. 2B, 2C) and ion exchange chromatography (Fig. 2D).
Figure 2.
Expression and purification of Ail. (A) SDS-PAGE showing Ail expression and isolation. E. coli C41 cells transformed with Ail-bearing plasmid were grown on 15N-labeled M9 media and collected before (−) or after (+) induction with IPTG. Expressed Ail is visible between the 14 kD and 21 kD molecular weight markers (MW). The cell lysate supernatant (S) was separated from the pellet (P) by centrifugation, and inclusion bodies (IB) enriched in Ail were obtained after a second wash and separation from the resulting supernatant. (B) Purification by size exclusion chromatography in 8 M urea yields two major peaks. (C) SDS-PAGE of the two peaks from size exclusion chromatography shows that Ail elutes in peak 2. (D) Cation exchange chromatography with a NaCl gradient yields purified Ail.
The purified protein yielded a MALDI-TOF mass spectrum characterized by a single species, with a major peak with the exact molecular mass of mature 15N-labeled Ail (17,810.55 m/z) and a smaller peak at half mass from doubly charged species (Fig. 3A). The absence of signal at twice the mass reflects the absence of dimeric species. Importantly, the mass spectrum shows no evidence of degradation, chemical modifications, or impurities. SDS-PAGE further shows that purified Ail runs as a single band (Fig. 3B, lane 2), at an apparent molecular weight that is slightly higher than that expected from the amino acid sequence of the protein, as observed for many other β-barrel membrane proteins.
Figure 3.
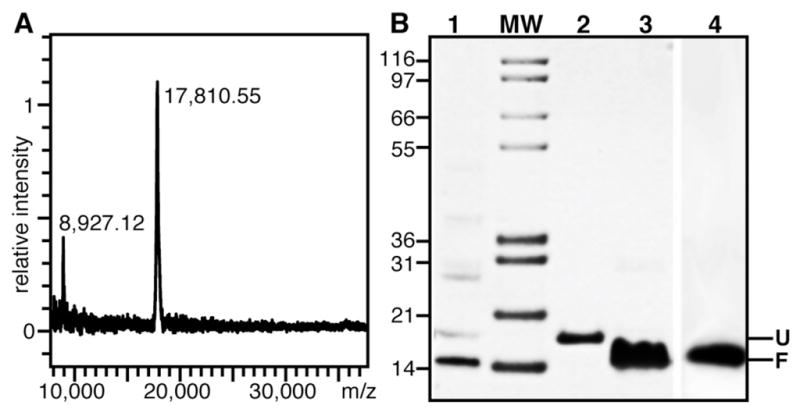
Refolding of purified Ail. (A) MALDI mass spectrum of purified Ail. The observed major peak at 17,810.55 m/z corresponds to the calculated molecular weight of 15N-labeled Ail. (B) SDS-PAGE showing: Ail folded in 14-O-PC (Lane 1); Ail dissolved in 8 M urea (Lane 2); Ail folded in 100 mM DHPC (Lane 3); Ail folded in 100 mM DHPC/10 mM DMPC (Lane 4). Unfolded (U) and folded (F) protein migrate at apparent molecular weights near 20 kD and 18 kD, respectively. Samples were loaded without boiling. (MW) Molecular weight markers.
3.2 Reconstitution of Ail in lipids is assisted by high pH
Quantitative refolding of Ail was obtained in four types of lipid environments: micelles, isotropic bicelles, large bicelles, and bilayer vesicles (Fig. 3B). These samples are key for all subsequent biochemical and NMR studies. The refolding efforts were guided by our previous studies with OmpX [44, 45] and by the extensive literature on β-barrel membrane proteins (reviewed in [36]).
The folded and unfolded states of β-barrel membrane proteins often migrate at different apparent molecular weights on SDS-PAGE and this can be used to monitor the folding process [30, 36]. Indeed, while Ail in 8 M urea has an apparent molecular weight near 20 kD (Fig. 3B, lane 2), Ail refolded in 14-O-PC vesicles (Fig. 3B, Lane 1) or in DHPC micelles (Fig. 3B, Lane 3) migrates closer to its calculated molecular weight of 17.6 kD. The near absence of a higher molecular weight band indicates that ~100% refolding is obtained. Notably, the fold of Ail in DHPC is maintained after addition of the long chain phospholipid DMPC to generate bicelles (Fig. 3B, Lane 4).
To screen for optimal refolding conditions, we used SDS-PAGE and visible light absorbance at 595 nm to monitor both the extent of refolding and the extent of protein aggregation over a wide range of pH (from pH 3 to pH 10), for several detergents, detergent concentrations, and buffer salts. High pH assists refolding of Ail in all of the detergents that we tested, as illustrated in Figure 4(A, B) for DHPC. This effect of pH had been noted previously for other outer membrane proteins with low isoelectric point (pI=5–6) and has been attributed to electrostatic repulsions from negatively charged protein in preventing aggregation during refolding [55]. However, the theoretical pI of Ail is close to 8, indicating that, at high pH, individual charges rather than net charge help keep the protein in solution. Indeed, we saw significantly less folding in DHPC below pH 6, and protein aggregation, was substantial at pH 5–8, but highly reduced at pH 9–10. Refolding in negatively charged LMPG further decreased aggregation. CD spectroscopy shows that once refolded, the structure of Ail resists denaturation over a wide range of pH (Fig. 4C). Addition of 6 M urea and sample heating up to 75°C also do not denature refolded Ail. In the CD spectra of folded Ail, the prominent minimum near 200 nm reflects a predominantly β-sheer conformation for the protein, as predicted based on its sequence homology to E. coli OmpX.
Figure 4.
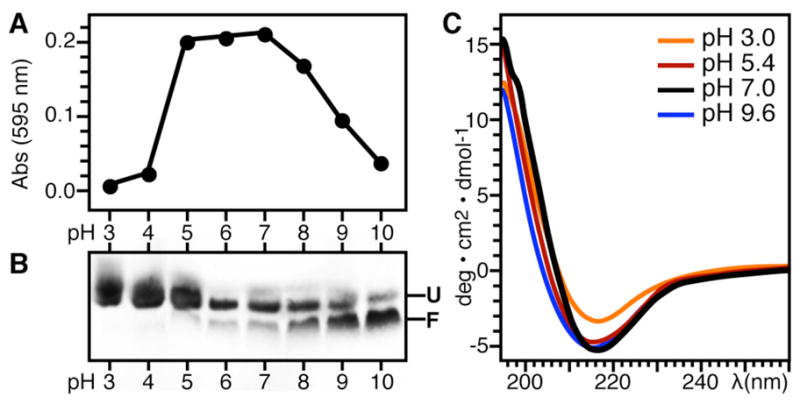
Ail adopts a β-sheet structure and refolding is assisted by high pH. (A) Protein aggregation at pH 3–10 monitored by visible light absorbance at 595 nm. (B) Corresponding SDS-PAGE showing Ail refolding in DHPC for each light absorbance measurement over the pH range from 3 to 10. The refolding reactions contained 70 mM DHPC, 100 mM KCl, and 600 mM Arginine, plus 20 mM of the appropriate buffer (Table 2). Folded (F) and unfolded (U) Ail migrate as different bands. Samples were loaded without boiling. (C) CD spectra of Ail refolded in DPC at pH 9.6 and then adjusted to pH 7-3 by the addition of HCl.
3.3 Ail adopts a transmembrane β-barrel in lipids
To further examine the conformation of Ail in lipids we obtained solution and solid-state NMR spectra of the protein reconstituted in micelles and bicelles. The solution NMR 1H/15N HSQC (heteronuclear single quantum correlation) spectra of Ail in DHPC, DHPC/DMPC, and DHPC/LPPG all exhibit well-resolved peaks over a wide range of 1H and 15N chemical shift frequencies (Fig. 5). This is typical of proteins with β-sheet structure and is in dramatic contrast to the narrow (<0.5 ppm) dispersion seen for unfolded Ail in urea (Fig. 5A). The spectra in lipids all have similar features with several overlapping peaks (Fig. S1), and the presence of a single peak for each residue indicates that Ail adopts a single well-defined conformation.
Figure 5.
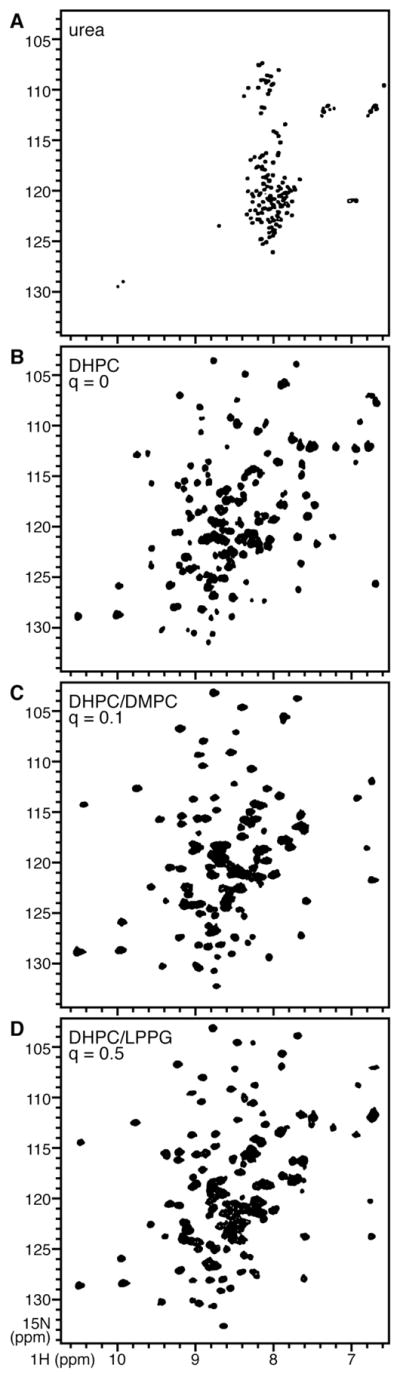
Solution NMR 1H/15N HSQC spectra of uniformly 15N labeled Ail in (A) 8 M urea; (B) 100 mM DHPC; (C) 100 mM DHPC plus 10 mM DMPC; (D) 100 mM DHPC plus 50 mM LPPG. The spectra are overlaid in Fig. S1.
Protein-containing bicelles can be used for both solution and solid-state NMR studies of membrane proteins by varying the parameter q corresponding to the molar ratio of the long to short chain lipids [51, 52, 56, 57]. Solution NMR spectra of proteins in small isotropic bicelles (q<1) and solid-state NMR spectra of proteins in large bicelles (q=3.2) contain complementary populations of resonances reflecting backbone dynamics, and comparisons of signal intensities observed in the HSQC spectra from micelles and isotropic bicelles over a range of q values provide insights about protein backbone dynamics [58]. The 1H/15N HSQC spectrum of Ail obtained after addition of the long chain lipid DMPC (Fig. 5C) demonstrates the feasibility of performing NMR spectroscopy on isotropic bicelles, which provide an environment that more closely resembles a membrane. This also sets the stage for bicelle titration experiments designed to examine details of protein dynamics.
Comparison of the spectra obtained for Ail/DHPC micelles in H2O with those obtained in D2O (Fig. 6) indicates that several residues have amide protons which resist exchange with the surrounding aqueous solvent and, therefore, have HSQC peaks that persist for days in D2O. In contrast several amide sites are highly susceptible to exchange and their peaks disappear after exchanging 1H for 2H. Preliminary resonance assignments indicate that the exchangeable amides are from residues in the predicted loops of the protein that are exposed to the bulk aqueous solvent, while resistant amides are from residues in the predicted transmembrane domain, which would be expected to participate in a tight β-sheet hydrogen bond network and to be buried within the hydrophobic interior of the micelle.
Figure 6.
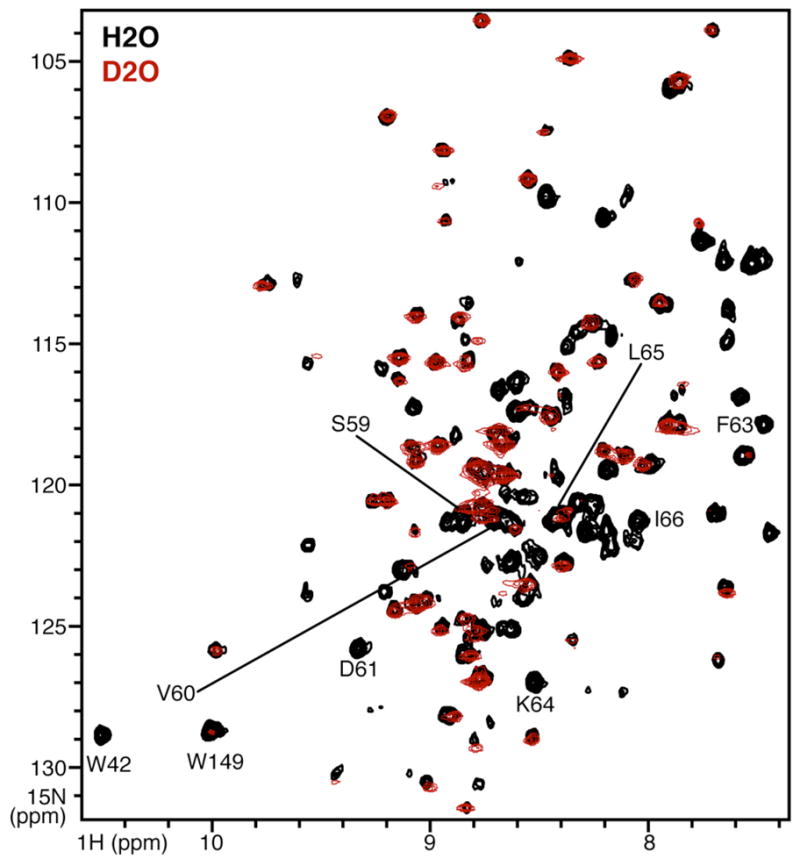
Expanded region of the solution NMR 1H/15N HSQC spectra of uniformly 15N labeled Ail refolded in 100 mM DHPC, obtained in H2O (black) or in D2O (red). Assigned peaks from the second extracellular loop (EL2) are labeled.
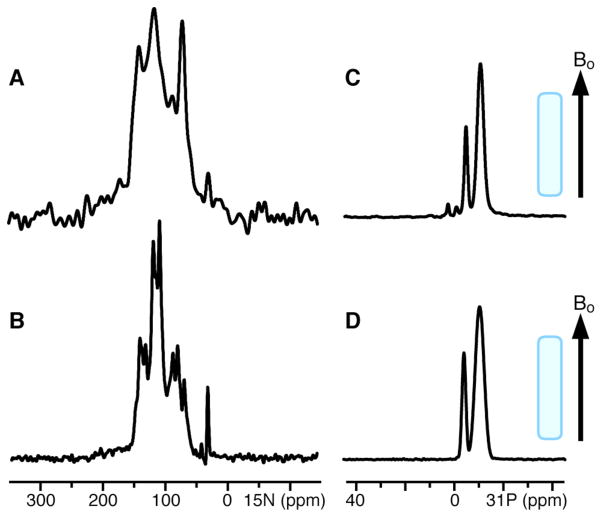
Ail could also be refolded directly in DMPC by mixing a solution of Ail in SDS with preformed DMPC vesicles (Fig. 3B), as we described previously for OmpX [45]. Alternatively, refolded Ail could be reconstituted in large lipid bicelles by adding the protein refolded in short chain lipid (DHPC) directly to long-chain lipid (14-O-PC, the ether-linked analog of DMPC). Bicelles formed in this way aligned spontaneously when placed in the field of the spectrometer magnet, and yielded high quality solid-state NMR 15N and 31P chemical shift spectra that manifest orientation dependent frequencies (Fig. 7A, 7C). These spectra are comparable to those obtained previously for OmpX (Fig. 7B, 7D) [44, 45], and demonstrate that Ail can be incorporated in lipid bicelles oriented with the bilayer plane parallel to the field of the NMR magnet (unflipped bicelles) for structural studies.
Figure 7.
Solid-state NMR 15N and 31P chemical shift spectra of uniformly 15N labeled (A, C) Ail and (B, D) OmpX, in q=3.2 magnetically aligned phospholipid bicelles (320 mM DMPC/100 mM DHPC). The lipid bilayer plane (gray) is oriented parallel to the magnetic field (Bo).
The 15N spectrum of Ail in unflipped bicelles shows some discrete peaks from amide sites over a range from about 70 to 160 ppm (Fig. 7A), indicating that the protein or protein/bicelle assembly undergoes rotational diffusion (> 105 sec−1) around an axis perpendicular to the plane of the lipid bilayer. This is similar to previous observations for helical membrane proteins [51, 52, 59], for the transmembrane domain of OmpA [43], and for OmpX [44, 45]. The chemical shift frequencies in these types of spectra reflect the orientations of individual peptide bonds rather than the local chemical environment and, thus, can be used as orientation restraints for structure determination [46–48]. High uniaxial alignment is also demonstrated by the 31P chemical shift spectra of the phospholipids (Fig. 7C), which display single lines at the expected frequencies corresponding to signals from 14-O-PC (−11 ppm) and DHPC (−4 ppm). Two additional peaks of low intensity are present in the 31P spectrum of Ail in bicelles (Fig. 7C). They may be due to an impurity that cannot be detected by SDS-PAGE (e.g. endogenous lipid which co-purifies with the protein) and/or an impurity that interferes with complete homogeneous alignment of the bicelles in the magnetic field. We are in the process of examining this issue.
4. CONCLUSIONS
In conclusion, we have described the expression and purification of Y. pestis Ail, as well as its reconstitution in both lipid micelles and lipid bilayers for solution and solid-state NMR structural studies. The CD and NMR spectra obtained in micelles indicate that the protein adopts a transmembrane β-barrel fold similar to E. coli OmpX. The observation of a strongly hydrogen bonded domain that resists exchange with aqueous 2H for days, indicates that the Ail β-barrel is folded in micelles, while peaks from residues in the predicted loops disappear upon exchange with 2H, indicating that these sites are more disordered and more accessible to the aqueous phase. The observation of discrete peaks in the spectra from magnetically aligned phospholipid bilayers demonstrates the presence of rapid rotational diffusion of the sample around the membrane normal, enabling solid-state NMR methods to be applied for Ail structure determination. Multi-dimensional spectroscopy and selectively labeled samples will be required to resolve and assign the spectrum of Ail in oriented lipid bilayers. However, the data presented in this report indicate that it will be possible to determine the structure, tilt, and rotation of Ail in the membrane, using the solid-state NMR methods that are currently being applied to α-helical membrane proteins.
Supplementary Material
Acknowledgments
We thank Greg Plano for providing the DNA encoding Ail, and Jinghua Yu for her assistance NMR experiments. This work was supported by the NIH Roadmap for Membrane Proteins (R21 GM075917). It utilized the NIH-supported Resource for NMR Molecular Imaging of Proteins (P41 EB002031) and the NIH-supported NMR Facility at the Sanford Burnham Medical Research Institute (P30 CA030199).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perry RD, Fetherston JD. Yersinia pestis--etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelis GR. Molecular and cell biology aspects of plague. Proc Natl Acad Sci U S A. 2000;97:8778–8783. doi: 10.1073/pnas.97.16.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brubaker RR. The recent emergence of plague: a process of felonious evolution. Microb Ecol. 2004;47:293–299. doi: 10.1007/s00248-003-1022-y. [DOI] [PubMed] [Google Scholar]
- 4.Huang XZ, Nikolich MP, Lindler LE. Current trends in plague research: from genomics to virulence. Clinical medicine & research. 2006;4:189–199. doi: 10.3121/cmr.4.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darling RG, Catlett CL, Huebner KD, Jarrett DG. Threats in bioterrorism. I: CDC category A agents. Emerg Med Clin North Am. 2002;20:273–309. doi: 10.1016/s0733-8627(02)00005-6. [DOI] [PubMed] [Google Scholar]
- 6.Wren BW. The yersiniae--a model genus to study the rapid evolution of bacterial pathogens. Nature reviews. 2003;1:55–64. doi: 10.1038/nrmicro730. [DOI] [PubMed] [Google Scholar]
- 7.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeno-Tarraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 8.Deng W, Burland V, Plunkett G, 3rd, Boutin A, Mayhew GF, Liss P, Perna NT, Rose DJ, Mau B, Zhou S, Schwartz DC, Fetherston JD, Lindler LE, Brubaker RR, Plano GV, Straley SC, McDonough KA, Nilles ML, Matson JS, Blattner FR, Perry RD. Genome sequence of Yersinia pestis KIM. J Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song Y, Tong Z, Wang J, Wang L, Guo Z, Han Y, Zhang J, Pei D, Zhou D, Qin H, Pang X, Zhai J, Li M, Cui B, Qi Z, Jin L, Dai R, Chen F, Li S, Ye C, Du Z, Lin W, Yu J, Yang H, Huang P, Yang R. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 2004;11:179–197. doi: 10.1093/dnares/11.3.179. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis GR. Yersinia type III secretion: send in the effectors. J Cell Biol. 2002;158:401–408. doi: 10.1083/jcb.200205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolodziejek AM, Sinclair DJ, Seo KS, Schnider DR, Deobald CF, Rohde HN, Viall AK, Minnich SS, Hovde CJ, Minnich SA, Bohach GA. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology. 2007;153:2941–2951. doi: 10.1099/mic.0.2006/005694-0. [DOI] [PubMed] [Google Scholar]
- 13.Bartra SS, Styer KL, O’Bryant DM, Nilles ML, Hinnebusch BJ, Aballay A, Plano GV. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infect Immun. 2008;76:612–622. doi: 10.1128/IAI.01125-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felek S, Krukonis ES. The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect Immun. 2009;77:825–836. doi: 10.1128/IAI.00913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felek S, Tsang TM, Krukonis ES. Three Yersinia pestis adhesins facilitate Yop delivery to eukaryotic cells and contribute to plague virulence. Infect Immun. 2010 doi: 10.1128/IAI.00167-10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsang TM, Felek S, Krukonis ES. Ail binding to fibronectin facilitates Yersinia pestis binding to host cells and Yop delivery. Infect Immun. 2010;78:3358–3368. doi: 10.1128/IAI.00238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, Perry RD, Fleischmann RD, Peterson SN. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology. 2009;155:498–512. doi: 10.1099/mic.0.022160-0. [DOI] [PubMed] [Google Scholar]
- 18.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller VL, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bliska JB, Falkow S. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc Natl Acad Sci U S A. 1992;89:3561–3565. doi: 10.1073/pnas.89.8.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierson DE, Falkow S. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect Immun. 1993;61:1846–1852. doi: 10.1128/iai.61.5.1846-1852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirjavainen V, Jarva H, Biedzka-Sarek M, Blom AM, Skurnik M, Meri S. Yersinia enterocolitica serum resistance proteins YadA and ail bind the complement regulator C4b-binding protein. PLoS Pathog. 2008;4:e1000140. doi: 10.1371/journal.ppat.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Merriam JJ, Mueller JP, Isberg RR. The psa locus is responsible for thermoinducible binding of Yersinia pseudotuberculosis to cultured cells. Infect Immun. 1996;64:2483–2489. doi: 10.1128/iai.64.7.2483-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barondess JJ, Beckwith J. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature. 1990;346:871–874. doi: 10.1038/346871a0. [DOI] [PubMed] [Google Scholar]
- 25.Miller VL, Bliska JB, Falkow S. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J Bacteriol. 1990;172:1062–1069. doi: 10.1128/jb.172.2.1062-1069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pulkkinen WS, Miller SI. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J Bacteriol. 1991;173:86–93. doi: 10.1128/jb.173.1.86-93.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heffernan EJ, Reed S, Hackett J, Fierer J, Roudier C, Guiney D. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella typhimurium virulence plasmid gene rck. J Clin Invest. 1992;90:953–964. doi: 10.1172/JCI115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heffernan EJ, Wu L, Louie J, Okamoto S, Fierer J, Guiney DG. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect Immun. 1994;62:5183–5186. doi: 10.1128/iai.62.11.5183-5186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mecsas J, Welch R, Erickson JW, Gross CA. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol. 1995;177:799–804. doi: 10.1128/jb.177.3.799-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt J, Schulz GE. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure. 1999;7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 31.Miller VL, Beer KB, Heusipp G, Young BM, Wachtel MR. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol Microbiol. 2001;41:1053–1062. doi: 10.1046/j.1365-2958.2001.02575.x. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz-Linek U, Werner JM, Pickford AR, Gurusiddappa S, Kim JH, Pilka ES, Briggs JA, Gough TS, Hook M, Campbell ID, Potts JR. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature. 2003;423:177–181. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- 33.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 35.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 36.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez C, Hilty C, Wider G, Guntert P, Wuthrich K. NMR structure of the integral membrane protein OmpX. J Mol Biol. 2004;336:1211–1221. doi: 10.1016/j.jmb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 39.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci U S A. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang B, Tamm LK. Structure of outer membrane protein G by solution NMR spectroscopy. Proc Natl Acad Sci U S A. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiller M, Krabben L, Vinothkumar KR, Castellani F, van Rossum BJ, Kuhlbrandt W, Oschkinat H. Solid-state magic-angle spinning NMR of outer-membrane protein G from Escherichia coli. Chembiochem. 2005;6:1679–1684. doi: 10.1002/cbic.200500132. [DOI] [PubMed] [Google Scholar]
- 43.Triba MN, Zoonens M, Popot JL, Devaux PF, Warschawski DE. Reconstitution and alignment by a magnetic field of a beta-barrel membrane protein in bicelles. Eur Biophys J. 2006;35:268–275. doi: 10.1007/s00249-005-0014-x. [DOI] [PubMed] [Google Scholar]
- 44.Mahalakshmi R, Marassi FM. Orientation of the Escherichia coli outer membrane protein OmpX in phospholipid bilayer membranes determined by solid-State NMR. Biochemistry. 2008;47:6531–6538. doi: 10.1021/bi800362b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahalakshmi R, Franzin CM, Choi J, Marassi FM. NMR structural studies of the bacterial outer membrane protein OmpX in oriented lipid bilayer membranes. Biochim Biophys Acta. 2007;1768:3216–3224. doi: 10.1016/j.bbamem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marassi FM. NMR of peptides and proteins in membranes. Concepts Magn Resonance. 2002;14:212–224. [Google Scholar]
- 47.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Page RC, Li C, Hu J, Gao FP, Cross TA. Lipid bilayers: an essential environment for the understanding of membrane proteins. Magn Reson Chem. 2007;45:S2–S11. doi: 10.1002/mrc.2077. [DOI] [PubMed] [Google Scholar]
- 49.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 50.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 51.De Angelis AA, Jones DH, Grant CV, Park SH, Mesleh MF, Opella SJ. NMR experiments on aligned samples of membrane proteins. Methods Enzymol. 2005;394:350–382. doi: 10.1016/S0076-6879(05)94014-7. [DOI] [PubMed] [Google Scholar]
- 52.De Angelis AA, Nevzorov AA, Park SH, Howell SC, Mrse AA, Opella SJ. High-resolution NMR spectroscopy of membrane proteins in aligned bicelles. J Am Chem Soc. 2004;126:15340–15341. doi: 10.1021/ja045631y. [DOI] [PubMed] [Google Scholar]
- 53.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 54.Goddard TD, Kneller DG. SPARKY. Vol. 3. University of California; San Francisco: 2004. [Google Scholar]
- 55.Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vold RR, Prosser RS, Deese AJ. Isotropic solutions of phospholipid bicelles: a new membrane mimetic for high-resolution NMR studies of polypeptides. J Biomol NMR. 1997;9:329–335. doi: 10.1023/a:1018643312309. [DOI] [PubMed] [Google Scholar]
- 57.Prosser RS, Evanics F, Kitevski JL, Al-Abdul-Wahid MS. Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry. 2006;45:8453–8465. doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 58.Opella SJ, Zeri AC, Park SH. Structure, dynamics, and assembly of filamentous bacteriophages by nuclear magnetic resonance spectroscopy. Annu Rev Phys Chem. 2008;59:635–657. doi: 10.1146/annurev.physchem.58.032806.104640. [DOI] [PubMed] [Google Scholar]
- 59.Park SH, Mrse AA, Nevzorov AA, De Angelis AA, Opella SJ. Rotational diffusion of membrane proteins in aligned phospholipid bilayers by solid-state NMR spectroscopy. J Magn Reson. 2006;178:162–165. doi: 10.1016/j.jmr.2005.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.