Abstract
Many drug candidates fail in clinical development due to their insufficient selectivity that may cause undesired side effects. Therefore, modern drug discovery is routinely supported by computational techniques, which can identify alternate molecular targets with a significant potential for cross-reactivity. In particular, the development of highly selective kinase inhibitors is complicated by the strong conservation of the ATP-binding site across the kinase family. In this paper, we describe X-ReactKIN, a new machine learning approach that extends the modeling and virtual screening of individual protein kinases to a system level in order to construct a cross-reactivity virtual profile for the human kinome. To maximize the coverage of the kinome, X-ReactKIN relies solely on the predicted target structures and employs state-of-the-art modeling techniques. Benchmark tests carried out against available selectivity data from high-throughput kinase profiling experiments demonstrate that for almost 70% of the inhibitors, their alternate molecular targets can be effectively identified in the human kinome with a high (>0.5) sensitivity at the expense of a relatively low false positive rate (<0.5). Furthermore, in a case study, we demonstrate how X-ReactKIN can support the development of selective inhibitors by optimizing the selection of kinase targets for small-scale counter-screen experiments. The constructed cross-reactivity profiles for the human kinome are freely available to the academic community at http://cssb.biology.gatech.edu/kinomelhm/
Keywords: X-ReactKIN, human kinome, kinase functional space, kinome structural coverage, kinase inhibitors, drug development, drug off-targets, Chemical Systems Biology
Introduction
The Human kinome, one of the largest families in the human proteome, comprises >500 genes 1. The pivotal function of kinases is the signal transduction through a reversible phosphorylation of tyrosine, threonine and serine residues in other proteins 2, 3. The strong implication of kinase activity in numerous disease states such as cancer 4, diabetes 5, inflammation 6, multiple sclerosis 7, cardiovascular disease 8 and neurological dysfunctions 9 makes them very important drug targets. Consequently, there is a growing interest in the development of novel compounds with kinase inhibition as their mode of action 10–12; this has resulted in over a hundred of kinase crystal structures complexed with low-molecular-weight inhibitors reported in the public domain 13.
Many therapeutic strategies have been developed to modulate kinase activity 14. The most prevalent is kinase inhibition by targeting the catalytic site of kinases with ATP-competitive inhibitors 15. The ATP-binding site provides a compelling environment for binding a diverse range of organic molecules devised to compete with ATP, mostly by mimicking the binding interactions of the adenosine moiety 16. Indeed, ATP-binding pockets are the primary target sites for the majority of the currently available kinase inhibitors 17. However, the structural and chemical features of the ATP-binding site as well as the catalytic mechanism are highly conserved across the kinase family, which significantly complicates the development of kinase inhibitors with sufficient target selectivity.
To address this significant issue, a number of computational techniques have been developed to support experimental efforts directed towards the development of selective kinase inhibitors. Most employ various classification schemas for the kinase space with the underlying assumption that kinases belonging to a common category have higher potential to bind similar compounds, which may give rise to undesired cross-reactivity effects. The most straightforward approach to the classification of kinases is based on the global sequence or/and structure similarity. A comprehensive survey carried out for all available kinase sequences classified them into 30 distinct families, with 19 of them covering nearly 98% of all sequences and representing seven general structural folds 18. Nevertheless, it has been demonstrated that a high probability of being inhibited by the same groups of compounds requires very high sequence identity thresholds, typically more than 50–60% 19–21. However, the average pairwise global sequence identity in the human kinome is ~25%; those kinase pairs with a sequence identity of 50–60% and less, might or might not have similar pharmacological profiles.
In that regard, alternative approaches are required. A new method was proposed to classify the medicinally relevant kinase space based on structure-activity relationship, SAR, profiles 22. Results obtained for 38 crystal structures of protein kinases and available small molecule inhibition data showed that the SAR-based dendograms differ significantly from the sequence-based clustering for distantly homologous targets. Another approach exploits structure comparison of kinases based on a feature-similarity matrix 23. This new metric is well correlated with a pharmacological distance generated by comparing affinity fingerprints constructed from experimental cross-reactivity profiles. An interesting study reported recently employs the QSAR analysis of residue contributions to the kinase inhibition profile 24. Using various experimental data sets, binding profiles are constructed based on the properties of 29 residues in the active site, which can be applied to predict binding similarities for untested kinases. Other chemical/structure-based classifications of ATP-binding sites in protein kinases are based on target family landscapes constructed using molecular interaction field analysis 25, exposed physicochemical properties of the active sites calculated by Cavbase 26, geometric hashing algorithms 27 and binding site signatures created from “hot spot” residues 28. These techniques have been shown to be relatively successful in the identification of protein kinase binding sites known experimentally to bind the same compound; however, they require high-resolution crystallographic structures of the target kinase proteins, preferably complexed with inhibitors. As a consequence, the covered kinase space remains incomplete because it is limited by the availability of experimentally solved crystal structures; this corresponds to only about 20% of the human kinome.
This gap can be bridged by protein structure prediction, particularly comparative modeling 29, 30. Current state-of-the-art protein structure prediction approaches have reached the level where they can construct protein models whose quality is often comparable to that of low-resolution experimentally determined structures 31. Nevertheless, theoretically predicted protein structures may still have significant structural inaccuracies in their ligand binding regions 32, 33; this requires appropriate computational techniques that are different from those applicable to the crystal structures and which can accommodate structural distortions without significant loss in accuracy.
In our previous study, we described the results of the first proteome-scale structure modeling and virtual screening of the entire human kinome 34. Using a template-based modeling procedure 35, 36, we constructed structural models for all kinase domains in humans. Subsequently, we applied a structure/evolution-based approach 37 to precisely detect target sites. These were then subject to large-scale virtual screening against a large collection of commercially available compounds using a novel hierarchical approach that combines ligand- and structure-based filters 38, 39. Retrospective benchmarks against several commonly used ligand libraries demonstrate that predicted molecular interactions between kinases and small ligands substantially overlap with available experimental data. In this paper, we attempt to extend the modeling and virtual screening of individual protein kinases to the system level in order to construct a cross-reactivity virtual profile for the entire human kinome. To achieve this goal, we develop X-ReactKIN, a machine learning approach that estimates the potential for cross-reactivity from sequence, structure and binding properties of the ATP-binding sites in protein kinases. We validate the results against available selectivity data from high-throughput kinase profiling experiments. Finally, we demonstrate how X-ReactKIN can support the development of selective inhibitors by suggesting alternate targets for small-scale counter-screen experiments. The constructed cross-reactivity profiles for the human kinome are freely available to the academic community via a user-friendly web interface that can be accessed from http://cssb.biology.gatech.edu/kinomelhm/
Methods
X-ReactKIN overview
Here, we use the concept of kinase family virtual profiling and compute the complete map of putative cross-interactions within the human kinome. We develop X-ReactKIN, a machine learning approach that combines sequence, structure and ligand binding similarities of the ATP-binding sites in protein kinases to estimate the potential for cross-interactions. We note that these similarities are calculated using modeled protein structures and virtual screening ranking. We train a Naive Bayes classifier on the available inhibitor selectivity data to calculate a new probabilistic cross-reactivity score, called a CR-score. Based on the estimated similarities expressed by the CR-score values, we construct a cross-reactivity virtual profile that corresponds to the matrix of pairwise interactions within the complete human kinase family. Below, we describe the scoring functions used to construct the cross-reactivity probabilistic score, the details of the datasets and machine learning implementation including training and validation protocols.
Sequence-based score
For each kinase domain in the human proteome, we constructed its structural model using a state-of-the-art template-based structure prediction approach. This procedure, described in detail in 34, involves the identification of evolutionary related templates in the PDB 40 using the PROSPECTOR_3 threading algorithm 36, followed by structure refinement/assembly by TASSER, a coarse-grained procedure guided by tertiary restraints extracted from the template structures 35. Subsequently, modeled kinase structures were taken as targets for the prediction of ATP-binding sites by FINDSITE, a structure/evolution-based method that identifies ligand-binding sites based on binding site similarity among superimposed groups of functionally and structurally related template structures 37. The sequence-based score corresponds to the sequence identity (a fraction of identical residues) of binding residues between two protein kinases calculated using FINDSITE identified residues and structure alignments generated by TM-align 41.
Structure-based score
In addition to the sequence-based scoring function, we also use a more structure-oriented measure of binding site similarity. Here, we employ a modified version of a PocketMatch score, PM-score, developed to provide a normalized similarity metric for binding site comparisons 42. PocketMatch applies a geometric hashing algorithm to Cα atoms and side-chain geometrical centers of ligand binding residues extracted from the crystal structures of protein-ligand complexes. Each binding site is represented by a set of 90 predefined distance bins, whose populations capture its shape and chemical features. The original PocketMatch approach uses residues, one or more of whose atoms are within a distance of 4A from the crystallographic ligand position 42. In our modified implementation, we use the consensus binding residues identified by FINDSITE in modeled kinase structures to populate the hash bins and calculate the PM-score.
Ligand-based score
Next, we introduce a new measure of binding site similarity that uses virtual screening ranks to calculate a chemical correlation. In the previous study, we carried out a large-scale virtual screening experiment for the complete human kinome 34. Here, we use this data to calculate the correlation between compound ranks obtained for two binding pockets. The chemical correlation corresponds to the Kendall τ rank correlation coefficient 43 calculated for the average top ranked set of 10,000 ZINC compounds 44 ranked for individual target sites of the entire human kinome by structure-based virtual screening using Q-DockLHM 39, 45. Details on the docking/screening protocol are given in 34. Retrospective benchmarks carried out against several ligand libraries demonstrate that this collection of compounds is likely to be significantly enriched in ATP-competitive kinase inhibitors 34. A high Kendall τ indicates that the pockets not only exhibit specific binding affinity toward similar compounds, but also do not bind similar ligands. This new measure based on the similarity of virtual screening ranks complements sequence-and structure-based similarities between binding pockets.
Bioassay data
We use three publicly available bioassay datasets to train and validate X-ReactKIN: 28 commercially available compounds examined against a panel of 20 protein kinases (Bioassay #1) 46, 38 kinase inhibitors assessed across a panel of 317 kinases representing >60% of the predicted human kinome (Bioassay #2) 47 and 20 kinase inhibitors including 16 approved drugs or those in clinical development screened against a panel of 119 protein kinases (Bioassay #3) 48. Bioassay #1 reports inhibitor potency as a percentage of kinase activity with respect to that in control incubations at an ATP concentration of 0.1 mM. Bioassays #2 and #3 use ATP site-dependent competition binding with each compound screened against the kinase targets at a single concentration of 10 μM and the binding efficacy reported in terms of quantitative dissociation constants, Kd. First, primary kinase targets(one per compound)are selected based on the strongest inhibition (Bioassay #1) or the lowest dissociation constant (Bioassays #2 and #3). Then, for each compound, we define alternate targets as kinases whose activity was inhibited to ≤25% of the control for Bioassay #1and those with Kd ≤10 μM for Bioassays #2 and #3. Remaining kinases are classified as non-targets. In this study, we use only compounds with at least one alternate kinase target. The list of compounds, primary target kinases and the number of alternate targets as well as non-targets is provided in Supplementary Information, SI Table 1.
Activity-based SAR profiles
In addition to the bioassay data described above, we comparethe virtual profiles constructed by X-ReactKIN to the experimentally derived activity-based SAR similarities on an orthogonal dataset of 577 diverse compounds screened across a panel of 203 protein kinases 21. Here, we use similarity scores expressed by a Tanimoto coefficient calculated for binding affinity fingerprints generated using an affinity threshold of 10%. Similarly to the CR-score values, kinase SAR similarity scores also range from 0(dissimilar)to 1(identical). For each kinase target, we assess the quality of X-ReactKIN virtual profiles calculated against the remaining kinases using the Pearson correlation coefficient between the SAR similarities and the CR-score values.
Machine learning
In X-ReactKIN, we use a Naive Bayes classifier to combine individual scoring functions: sequence-, structure- and ligand-based into a single probabilistic score. A classical Naive Bayes classification is based on estimating P(X|Y), the probability or probability density of a qualitative attribute X given class Y. In our classifier, the real-value attributes are modeled by a Gaussian distribution, i.e. the classifier first estimates a normal distribution for each class by computing the mean and standard deviation of the training data in that class, which is then used to estimate P(X|Y) during classification 49. For a given pair of protein kinases, the probabilistic score from the classifier, called a CR-score, estimates the chances of the cross-reactivity from sequence, structure and binding similarities. X-ReactKIN was validated using the following leave-one-out procedure: In each round, one inhibitor and its close analogs are removed from the dataset that consists of the bioassay data described above and the classifier is trained on the remaining compounds. Here, we define a close analog as a compound that has a Tanimo to coefficient calculated using SMILES strings ≥0.7 50. Then, for the excluded inhibitor and its primary target, the kinase proteins are ranked by the predicted CR-score, with the top-ranked kinases assumed to be alternate targets. We assess the accuracy of the off-target identification by a receiver operating characteristic (ROC)analysis with the CR-score used as a variable parameter. In addition to the standard ROC curves, we also calculate their distribution-free confidence bounds 51.
Virtual map of kinase cross-reactivity
Finally, X-ReactKIN was re-trained on all bioassay data and the complete map of putative cross-interactions within the human kinome was calculated. Moreover, we constructed a statistical model by fitting the distribution of the random CR-score values to a Normal Inverse Gaussian distribution 52 in order to calculate the associated p-values. The fitting procedure was done in R 53 using the ghyp package. The virtual cross-reactivity map is visualized using matrix2png 54, with the kinase proteins grouped according to the subfamily classification and clustered by sequence identity using CLUTO 55.
Results
X-ReactKIN validation
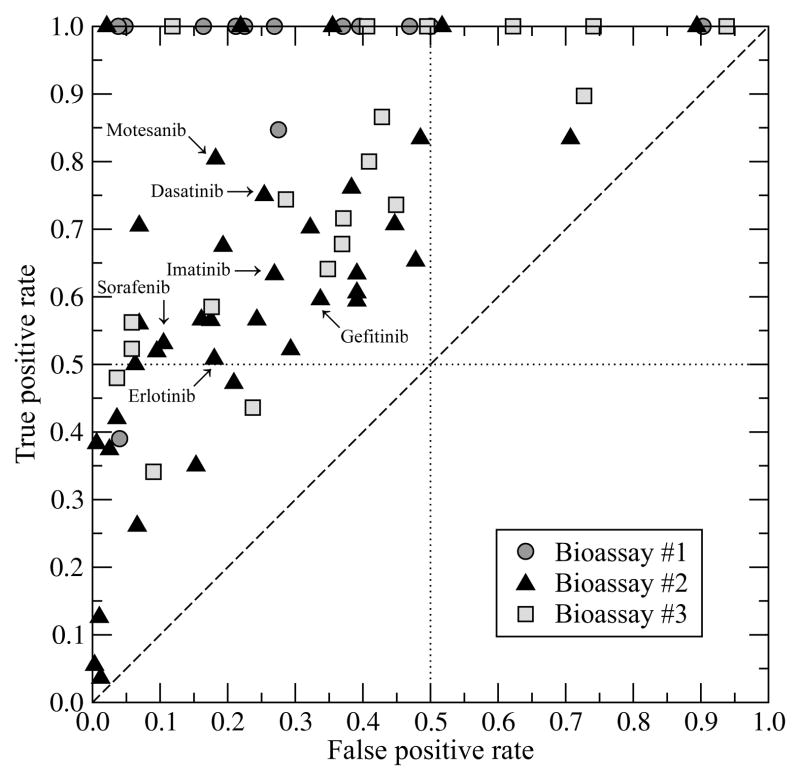
Here, we use the available selectivity data from high-throughput kinase profiling experiments to train and validate X-ReactKIN in the off-target prediction. As described in the Methods section, for each kinase inhibitor and the corresponding primary target, the remaining kinases are assessed with respect to the estimated potential for cross-reactivity, i.e. ability to bind similar compounds. The results of leave-one-out validation are presented as a ROC plot in Figure 1. Encouragingly, in all cases the performance of X-ReactKIN is better than random, with a true positive rate >0.5 and a false positive rate <0.5 for almost 70% of the benchmark inhibitors. Particularly the results obtained for Bioassay #2 are very promising since this panel of kinases covers >60% of the human kinome 47. In addition, individual ROC plots for six selected compounds that include approved drugs such as Gleevec (imatinib), Iressa (gefitinib), Nexavar (sorafenib), Sprycel (dasatinib) and Tarceva (erlotinib) are presented in Figure 2. In all cases, the cross-validated performance of X-ReactKIN is significantly better than random, with tight confidence bounds particularly for dasatinib (Figure 2A), erlotinib (Figure 2B), motesanib (Figure 2E) and sorafenib (Figure 2F). The calculated cut-off points (displayed in Figure 2), which maximize the sensitivity and specificity show that most of the cross-interacting kinases are identified at the expense of a relatively low false positive rate; the true (false) positive rate is 0.75 (0.25), 0.51 (0.18), 0.60 (0.34), 0.63 (0.27), 0.80 (0.18) and 0.53 (0.11) for dasatinib, erlotinib, gefitinib, imatinib, motesanib and sorafenib, respectively.
Figure 1.
ROC plot for the prediction of kinase inhibitor cross-reactivity using X-ReactKIN. Compounds from Bioassays #1, #2 and #3 are shown as dark gray circles, black triangles and light gray squares, respectively.
Figure 2.
Individual ROC plots for selected inhibitors: (A) dasatinib, (B) erlotinib, (C) gefitinib, (D) imatinib, (E) motesanib and (F) sorafenib. In each graph, the solid black line, the gray area and the dashed line show the ROC curve for the CR-score, its 95% confidence bounds and the accuracy of a random classifier, respectively. The cut-off point that maximizes the sensitivity and specificity is represented by a black triangle. Chemical structures of the inhibitors are also displayed.
Human kinome cross-reactivity profile
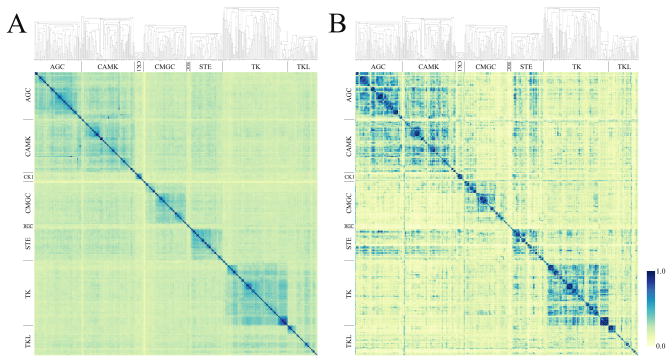
Encouraged by the satisfactory performance of X-ReactKIN in benchmark tests, we re-trained the model on all bioassay data and constructed a complete map of putative cross-reactions within the entire human kinome. The details on the trained classifier used in X-ReactKIN are provided in Supplementary Information, SI Table 2. In Figure 3, for the human kinome, we compare the cross-interaction potential expressed by a sequence-based classification (Figure 3A) to the CR-score based classification (Figure 3B). In both Figures 3A and B, the kinases are clustered using sequence identity and the resulting dendograms are shown on the top of each plot. Comparing the sequence identity score to the CR-score, we observe many off-diagonal interactions pointed out by high CR-values (Figure 3B, blue spots). These non-trivial similarities, which are clearly the most interesting, indicate the possibility to bind similar compounds by remotely related protein kinases that belong to different groups. In particular, many potential cross-interactions are observed between kinases that belong to AGC (containing PKA, PKC and PKG protein kinases), CAMK (calcium/calmodulin-dependent protein kinases) and STE (the homologues of yeast Sterile kinases) groups. We note that whereas the average pairwise sequence identity within these groups is relatively high: 38%, 34% and 36%, respectively, the inter-group sequence identity is notably lower: 29%, 26% and 26% for AGC/CAMK, AGC/STE and CAMK/STE, respectively. Even lower average sequence identity is seen between the TK (tyrosine kinases) group and those kinases that belong to AGC (23%), CAMK (24%) and CMGC (22%). The functional similarities indicated by the high CR-score values between these kinase proteinsare undetectable on the basis of the sequence similarity alone. We have also constructed a statistical model for the CR-score distribution in order to assign statistical significance values. Here, we use a Normal Inverse Gaussian distribution, which fits well to the data; this is shown as histograms as well as a quantile-quantile plot in Supplementary Information, SI Figure 1.
Figure 3.
Classification of the human kinome by X-ReactKIN: (A) sequence similarity matrix and (B) cross-reactivity matrix. In both plots, kinase proteins are grouped according to the subfamily classification displayed on both axes. Within each group, kinase members are clustered using sequence identity and the resulting dendograms are shown on the top of each graph. Color scale expressing the sequence similarity (A) as well as the potential cross-reactivity (B) is displayed on the right.
Comparison to SAR profiles
For a subset of 203 protein kinases, activity-based SAR similarities have been previously reported 21. These similarities were calculated directly from the experimental data obtained by screening the target kinases against a diverse set of >500 compounds, intended to represent kinase inhibitor chemical space. This large-scale kinase profiling provides an orthogonal dataset to validate the potential for cross-reactivity predicted by X-ReactKIN. The results are presented in Figure 4. The direct comparison of the similarity between pairs of kinases according to the SAR profiles and the CR-score values isshown in Figure 4A. In both cases, the joint inhibition of many of these kinase pairs is observed within the TK subfamily. Moreover, good agreement between both approaches is seen for the STE subfamily, for which many predicted cross-interactions with kinases that belong to other, particularly AGC and CAMK, groups are confirmed experimentally. The distribution of the Pearson correlation coefficients between SAR similarities and CR-score values calculated for 203 kinase targets is presented in Figure 4B. This distribution is clearly shifted toward high (>0.5) values, which indicate a good overlap between experimental SAR and virtual CR-score profiles for the majority of kinase targets. The average Pearson correlation coefficient calculated across this dataset is 0.53 ± 0.14. The qualitative agreement between the activity-based SAR similarities and the CR-score profiles provides significant validation of the X-ReactKIN approach.
Figure 4.
Comparison of the X-ReactKIN virtual profiles to the SAR similarities on a set of 203 protein kinases. (A) Similarity between pairs of kinases ordered according to the Sugen phylogenetic tree (available at http://kinase.com). Upper right and lower left triangles represent the CR-score values and SAR similarities, respectively. The color scale expressing both similarities is displayed in the right corner. (B) Histogram of the distribution of the Pearson correlation coefficients between SAR similarities and CR-score values calculated for 203 kinase targets. Inset: Correlation between SAR similarities and CR-score values for the leukocyte-specific protein tyrosine kinase, Lck.
Below, in a case study, we present a simple application of the human kinome cross-reactivity virtual profile constructed by X-ReactKIN to demonstrate how it can be used to optimize the selection of kinase targets for small-scale selectivity counter-screens in kinase inhibitor development.
Case study: Inhibitors of Lck
2-Aminopyrimidine carbamates are a new class of compounds with potent and selective inhibition of the leukocyte-specific protein tyrosine kinase, Lck. Structure-activity relationship studies and extensive pharmacological tests carried out for a series of substituted 2-aminopyrimidine carbamates identified 2,6-dimethylphenyl-2-((3,5-bis(methyloxy)-4-((3-(4-methyl-1-piperazinyl)propyl)oxy) phenyl)amino)-4-pyrimidinyl(2,4-bis(methyloxy)phenyl)carbamateas a potent inhibitor of Lck, with an IC50 of 0.6 nM (compound 43 in the original paper) 56. Subsequently, a counter-screen against 15 other kinases that belong to TK, CMGC and AGC groups was carried out in order to characterize the selectivity profile of this compound. Here, we compare the experimental inhibition data to the in silico profile of Lck and demonstrate that the map of putative cross-interactions within the human kinome constructed by X-ReactKIN can be used to suggest alternate kinase targets for the selectivity counter-screens. Figure 5 shows the selectivity profile for the pyrimidine carbamate inhibitor. Experimentally, this inhibitor was found to be highly selective with regards to the non-binding of JAK3(Kin. Dom. 2), MET, JNK3, PKCt, IGF1R and CDK2 (Figure 5A). With the exception of JAK3 (Kin. Dom. 2), the CR-score values (p-values) between Lck and these kinases are statistically insignificant: 0.483 (3.46×10−2), 0.126 (7.03×10−1), 0.182 (4.36×10−1), 0.267 (1.96×10−1), 0.229 (2.81×10−1) and 0.162 (5.23×10−1), respectively (Figure 5B). For another 8 kinase targets, the experimental IC50 values are in the range of 100 nM −1 μM; here the CR-scores are higher (~0.3, p-values ~0.1 or better), with p-values <0.05 for BTK (1.39×10−2) and JAK2 (Kin. Dom. 2, 4.07×10−2). No selectivity was shown against SRC kinase, for which the CR-score (p-value) is 0.961 (1.55×10−3).
Figure 5.
Selectivity profile for the pyrimidine carbamate inhibitor reported in 56: (A) experimental inhibition constant values in μM with the IC50≤1 μM (>1 μM) in turquoise (yellow); (B) pairwise CR-score matrix for the tested kinases, CR-score scale is given at the bottom; (C) chemical structure of the inhibitor. In B, kinase pairs with a pairwise sequence identity of >60% are marked with an X.
Furthermore, the map of putative cross-interactions reveals other similarities between e.g. FGFR1 and TIE2 (CR-score=0.856, p-value=2.92×10−3), JAK2 (Kin. Dom. 2)and TIE2 (CR-score=0.663, p-value=9.96×10−3), BTK and ZAP70 (CR-score=0.544, p-value=2.23×10−2), JNK3 and p38a (CR-score=0.532, p-value=2.43×10−2) or JAK3(Kin. Dom. 2)and SYK (CR-score=0.603, p-value=1.49×10−2), which indicate a high probability of inhibition by similar compounds. In fact, the joint inhibition of many of these kinase pairs has been already confirmed experimentally. We note that none of this information was used for the construction of the CR-score matrix; indeed we were unaware of the experimental results until after the predictions were made and we did a literature search. For example, an oral kinase inhibitor ACTB-1003 with multiple modes of action, targeting cancer mutations via FGFR1 inhibition (IC50=6 nM)and angiogenesis through inhibition of VEGFR2 (2 nM) and TIE2 (4 nM) has been recently reported 57. Several inhibitors (compounds 10, 11, 12, 13 and 14 in the original paper) were found to non-selectively inhibit JAK2 (TIE2) with the percent of enzyme activity at 1μM concentration of 6 (35), 5 (0), 0 (1), 30 (1) and 27 (7), respectively 58. Moreover, compound 7 in the original paper was found to be the most selective against JAK2 and TIE2 (3% and 26%) across a panel of 59 recombinant serine/threonine and tyrosine kinases. Many JNK3 inhibitors are known to also inhibit p38a; e.g. two compounds with a nanomolar activity against JNK3 (IC50 of 7 and 1 nM) have been reported as potent p38a inhibitors as well, with the IC50 of 0.2 and 4 nM, respectively 59. Finally, in vitro enzymatic assays of the novel JAK3 inhibitor R348 showed potent inhibition of JAK3-and SYK-dependent pathways 60.
Lck was also included in the large-scale assessment of the chemical coverage of the kinome space using activity-based SAR profiles 21. In Figure 4B (inset), we compare the experimentally derived SAR similarities to the CR-score values calculated against the remaining 202 protein kinases used as targets in the high-throughput binding assay. Here, the Pearson correlation coefficient between the SAR similarities and the CR-score values is 0.73. This high correlation additionally confirms the good agreement between the potential for cross-reactivity predicted by X-ReactKIN and the experimentally observed joint inhibition of protein kinases.
Of course, a high probability of inhibition by the same groups of compounds does not preclude a successful design of selective inhibitors. Rather, it should support the counter-screen selectivity experiments by optimizing the selection of possible off-targets, whose binding sites have the highest potential for cross-reactivity.
Discussion
Many drug candidates fail in clinical development due to their poor pharmacokinetic characteristics and because of intolerable adverse effects, which may sometimes originate in their insufficient selectivity 61. The physicochemical similarity between highly conserved ATP-binding sites in protein kinases, one of the most important drug targets, has rendered the challenge of designing selective inhibitors difficult. Nevertheless, the discovery of selective kinase inhibitors demonstrate that there is enough conformational and chemical diversity in and around the active site that can be explored to design compounds with sufficient selectivity 14, 15. Thus, particularly in the early stages of drug development, the knowledge of alternate kinase targets with significant potential for cross-reactivity is critical. One common strategy in inhibitor design involves differential lead optimization to increase the selectivity toward a particular drug target; such efforts are typically oriented towards the development of highly specific inhibitors acting on single protein kinases. Later on, with the approval of multi-target inhibitors, such as imatinib, sunitinib or lapatinib, an alternate strategy has emerged, where drug-resistance can be overcome by simultaneously targeting multiple kinase pathways 62. Multikinase inhibitors with highly tuned selectivity profiles are currently of particular interest in pharmaceutical research 63. The functional classification of the entire human kinome is of paramount importance in the development of both highly selective as well as selectively unselective novel inhibitors.
Due to the sparse and non-uniformly distributed structural data 64, cross-interactions are still poorly defined atthe kinome level. To maximize the coverage of kinase functional space, we developed X-ReactKIN, a Chemical Systems Biology approachfor in silico cross-reactivity profiling that does not require high-resolution structural data. X-ReactKIN employs a state-of-the-art protein structure prediction algorithm followed by the recently developed Ligand Homology Modeling approach to model kinase-drug interactions 34. Subsequently, the modeling of individual kinase members is now extended to construct a cross-reactivity virtual profile for the entire human kinome. This proteome-wide analysis represents a significant improvement over other methods, which are generally confined to high-resolution structures solved by protein crystallography.
In addition to the traditional sequence and structure similarity measures, our method also uses a novel type of the binding site comparison by means of virtual screening ranks. A high correlation between ligand rankings for two binding sites, referred to as a chemical correlation, indicates that these sites not only exhibit specific binding affinity toward similar molecules, but also do not bind similar compounds. Here, the accuracy of ligand docking and ranking is essential. Particularly, using predicted receptor structures requires reliable docking techniques capable of dealing with structural inaccuracies in protein models. It has been demonstrated that even moderate structural distortions of the modeled binding pockets drastically interfere with the ability of the all-atom docking approaches to identify correct docking geometries and to rank ligands 39, 65. Our virtual screening protocol that provides compound ranking for the estimation of the chemical correlation employs evolution-based ligand docking 38 followed by low-resolution binding pose refinement 39, 45. Such a docking/ranking procedure is well suited for virtual screening applications using modeled receptor structures since it exhibits significant tolerance to receptor structure deformation 39.
Modern drug discovery is routinely supported by computational techniques, such as virtual screening, which prioritize drug candidates and increase the hit rate by restricting screening libraries to compounds that likely exhibit the desired bioactivity. At the system level, the functional classification of the human kinome expands our understanding of the structural, chemical and pharmacological aspects of the kinase space and provides a practical strategy that should prove useful for the design of more selective therapeutics.
Availability
The cross-reactivity virtual profile of the human kinase space is available at http://cssb.biology.gatech.edu/kinomelhm/
Supplementary Material
Fitting of the asymmetric normal inverse Gaussian (NIG) to the distribution of CR-scores in the human kinome: (A) histogram of the empirical distribution (gray bars) compared to the fitted NIG and normal distribution, (B) same as A but in log scale, (C) quantile-quantile plot.
Bioassay data used in this study. Primary targets are selected based on the strongest inhibition (Bioassay #1) or the lowest dissociation constant Kd (Bioassays #2 and #3). #P and #N correspond to the number of alternate targets (≤25% for Bioassay #1, and ≤10 μM for Bioassays #2 and #3) and non-targets (>25% for Bioassay #1, and >10 μM for Bioassays #2 and #3) for a given compound, respectively.
Trained Naive Bayes classifier used by X-ReactKIN to calculate the CR- score.
Acknowledgments
This work was supported in part by grant Nos. GM-48835 and GM-37408 of the Division of General Medical Sciences of the National Institutes of Health.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298 (5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9 (8):576–96. [PubMed] [Google Scholar]
- 3.Kennelly PJ. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol Lett. 2002;206 (1):1–8. doi: 10.1111/j.1574-6968.2002.tb10978.x. [DOI] [PubMed] [Google Scholar]
- 4.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411 (6835):355–65. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 5.Sasase T. PKC -a target for treating diabetic complications. Drugs of the Future. 2006;31 (6):503–11. [Google Scholar]
- 6.Muller S, Knapp S. Targeting kinases for the treatment of inflammatory diseases. Expert Opin Drug Discov. 2010;5 (9):867–81. doi: 10.1517/17460441.2010.504203. [DOI] [PubMed] [Google Scholar]
- 7.Saarela J, Kallio SP, Chen D, Montpetit A, Jokiaho A, Choi E, Asselta R, Bronnikov D, Lincoln MR, Sadovnick AD, Tienari PJ, Koivisto K, Palotie A, Ebers GC, Hudson TJ, Peltonen L. PRKCA and multiple sclerosis: association in two independent populations. PLoS Genet. 2006;2 (3):e42. doi: 10.1371/journal.pgen.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bynagari-Settipalli YS, Chari R, Kilpatrick L, Kunapuli SP. Protein Kinase C -Possible Therapeutic Target to Treat Cardiovascular Diseases. Cardiovasc Hematol Disord Drug Targets. 2010 doi: 10.2174/187152910793743869. [DOI] [PubMed] [Google Scholar]
- 9.Mueller BK, Mack H, Teusch N. Rho kinase, a promising drug target for neurological disorders. Nat Rev Drug Discov. 2005;4 (5):387–98. doi: 10.1038/nrd1719. [DOI] [PubMed] [Google Scholar]
- 10.Cohen P. The development and therapeutic potential of protein kinase inhibitors. Curr Opin Chem Biol. 1999;3 (4):459–65. doi: 10.1016/S1367-5931(99)80067-2. [DOI] [PubMed] [Google Scholar]
- 11.Johnson L. Protein kinases and their therapeutic exploitation. Biochem Soc Trans. 2007;35 (Pt 1):7–11. doi: 10.1042/BST0350007. [DOI] [PubMed] [Google Scholar]
- 12.Weinmann H, Metternich R. Drug discovery process for kinase inhibitors. Chembiochem. 2005;6 (3):455–9. doi: 10.1002/cbic.200500034. [DOI] [PubMed] [Google Scholar]
- 13.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303 (5665):1800–5. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 14.McInnes C, Fischer PM. Strategies for the design of potent and selective kinase inhibitors. Curr Pharm Des. 2005;11 (14):1845–63. doi: 10.2174/1381612053764850. [DOI] [PubMed] [Google Scholar]
- 15.Sawa M. Strategies for the design of selective protein kinase inhibitors. Mini Rev Med Chem. 2008;8 (12):1291–7. doi: 10.2174/138955708786141043. [DOI] [PubMed] [Google Scholar]
- 16.Liao JJ. Molecular recognition of protein kinase binding pockets for design of potent and selective kinase inhibitors. J Med Chem. 2007;50 (3):409–24. doi: 10.1021/jm0608107. [DOI] [PubMed] [Google Scholar]
- 17.Stout TJ, Foster PG, Matthews DJ. High-throughput structural biology in drug discovery: protein kinases. Curr Pharm Des. 2004;10 (10):1069–82. doi: 10.2174/1381612043452695. [DOI] [PubMed] [Google Scholar]
- 18.Cheek S, Zhang H, Grishin NV. Sequence and structure classification of kinases. J Mol Biol. 2002;320 (4):855–81. doi: 10.1016/s0022-2836(02)00538-7. [DOI] [PubMed] [Google Scholar]
- 19.Vieth M, Higgs RE, Robertson DH, Shapiro M, Gragg EA, Hemmerle H. Kinomics-structural biology and chemogenomics of kinase inhibitors and targets. Biochim Biophys Acta. 2004;1697 (1–2):243–57. doi: 10.1016/j.bbapap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 20.Vieth M, Sutherland JJ, Robertson DH, Campbell RM. Kinomics: characterizing the therapeutically validated kinase space. Drug Discov Today. 2005;10 (12):839–46. doi: 10.1016/S1359-6446(05)03477-X. [DOI] [PubMed] [Google Scholar]
- 21.Bamborough P, Drewry D, Harper G, Smith GK, Schneider K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J Med Chem. 2008;51 (24):7898–914. doi: 10.1021/jm8011036. [DOI] [PubMed] [Google Scholar]
- 22.Frye SV. Structure-activity relationship homology (SARAH): a conceptual framework for drug discovery in the genomic era. Chem Biol. 1999;6 (1):R3–7. doi: 10.1016/S1074-5521(99)80013-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Fernandez A. In silico drug profiling of the human kinome based on a molecular marker for cross reactivity. Mol Pharm. 2008;5 (5):728–38. doi: 10.1021/mp800010p. [DOI] [PubMed] [Google Scholar]
- 24.Sheridan RP, Nam K, Maiorov VN, McMasters DR, Cornell WD. QSAR models for predicting the similarity in binding profiles for pairs of protein kinases and the variation of models between experimental data sets. J Chem Inf Model. 2009;49 (8):1974–85. doi: 10.1021/ci900176y. [DOI] [PubMed] [Google Scholar]
- 25.Naumann T, Matter H. Structural classification of protein kinases using 3D molecular interaction field analysis of their ligand binding sites: target family landscapes. J Med Chem. 2002;45 (12):2366–78. doi: 10.1021/jm011002c. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn D, Weskamp N, Hullermeier E, Klebe G. Functional classification of protein kinase binding sites using Cavbase. ChemMedChem. 2007;2 (10):1432–47. doi: 10.1002/cmdc.200700075. [DOI] [PubMed] [Google Scholar]
- 27.Kinnings SL, Jackson RM. Binding site similarity analysis for the functional classification of the protein kinase family. J Chem Inf Model. 2009;49 (2):318–29. doi: 10.1021/ci800289y. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian G, Sud M. Computational Modeling of Kinase Inhibitor Selectivity. ACS Med Chem Lett. 2010 doi: 10.1021/ml1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009;77 (Suppl 9):18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ginalski K. Comparative modeling for protein structure prediction. Curr Opin Struct Biol. 2006;16 (2):172–7. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15 (3):285–9. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 32.DeWeese-Scott C, Moult J. Molecular modeling of protein function regions. Proteins. 2004;55 (4):942–61. doi: 10.1002/prot.10519. [DOI] [PubMed] [Google Scholar]
- 33.Piedra D, Lois S, de la Cruz X. Preservation of protein clefts in comparative models. BMC Struct Biol. 2008;8:2. doi: 10.1186/1472-6807-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brylinski M, Skolnick J. Comprehensive structural and functional characterization of the human kinome by protein structure modeling and ligand virtual screening. J Chem Inf Model. 2010 doi: 10.1021/ci100235n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Skolnick J. Tertiary structure predictions on a comprehensive benchmark of medium to large size proteins. Biophys J. 2004;87 (4):2647–55. doi: 10.1529/biophysj.104.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skolnick J, Kihara D, Zhang Y. Development and large scale benchmark testing of the PROSPECTOR_3 threading algorithm. Proteins. 2004;56 (3):502–18. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- 37.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105 (1):129–34. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brylinski M, Skolnick J. FINDSITE(LHM): a threading-based approach to ligand homology modeling. PLoS Comput Biol. 2009;5(6):e1000405. doi: 10.1371/journal.pcbi.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brylinski M, Skolnick J. Q-Dock(LHM): Low-resolution refinement for ligand comparative modeling. J Comput Chem. 2010;31 (5):1093–105. doi: 10.1002/jcc.21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28 (1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33 (7):2302–9. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeturu K, Chandra N. PocketMatch: a new algorithm to compare binding sites in protein structures. BMC Bioinformatics. 2008;9:543. doi: 10.1186/1471-2105-9-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kendall MG. A new measure of rank correlation. Biometrika. 1938;30 (Pt 1–2):81–9. [Google Scholar]
- 44.Irwin JJ, Shoichet BK. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45 (1):177–82. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brylinski M, Skolnick J. Q-Dock: Low-resolution flexible ligand docking with pocket-specific threading restraints. J Comput Chem. 2008;29 (10):1574–1588. doi: 10.1002/jcc.20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351 (Pt 1):95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26 (1):127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 48.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23 (3):329–36. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 49.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2. Morgan Kaufmann Publishers; San Francisco: 2005. [Google Scholar]
- 50.Willett P, Barnard JM, Downs GM. Chemical Similarity Searching. J Chem Inf Comput Sci. 1998;38 (6):983–996. [Google Scholar]
- 51.Kestler HA. ROC with confidence -a Perl program for receiver operator characteristic curves. Comput Methods Programs Biomed. 2001;64 (2):133–136. doi: 10.1016/s0169-2607(00)00098-5. [DOI] [PubMed] [Google Scholar]
- 52.Dagpunar JS. An Easily Implemented Generalised Inverse Gaussian Generator. Communications in Statistics -Simulation and Computation. 1989;18 (2):703 –710. [Google Scholar]
- 53.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: 2008. [Google Scholar]
- 54.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19 (2):295–6. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 55.Karypis G. CLUTO: A Clustering Toolkit, 2.1.1. 2003. [Google Scholar]
- 56.Martin MW, Newcomb J, Nunes JJ, McGowan DC, Armistead DM, Boucher C, Buchanan JL, Buckner W, Chai L, Elbaum D, Epstein LF, Faust T, Flynn S, Gallant P, Gore A, Gu Y, Hsieh F, Huang X, Lee JH, Metz D, Middleton S, Mohn D, Morgenstern K, Morrison MJ, Novak PM, Oliveira-dos-Santos A, Powers D, Rose P, Schneider S, Sell S, Tudor Y, Turci SM, Welcher AA, White RD, Zack D, Zhao H, Zhu L, Zhu X, Ghiron C, Amouzegh P, Ermann M, Jenkins J, Johnston D, Napier S, Power E. Novel 2-aminopyrimidine carbamates as potent and orally active inhibitors of Lck: synthesis, SAR, and in vivo antiinflammatory activity. J Med Chem. 2006;49 (16):4981–91. doi: 10.1021/jm060435i. [DOI] [PubMed] [Google Scholar]
- 57.Patel K, Fattaey A, Burd A. ACTB-1003: An oral kinase inhibitor targeting cancer mutations (FGFR), angiogenesis (VEGFR2, TEK), and induction of apoptosis (RSK and p70S6K) J Clin Oncol (Meeting Abstracts) 2010;28 (15_suppl):e13665. [Google Scholar]
- 58.Okram B, Nagle A, Adrian FJ, Lee C, Ren P, Wang X, Sim T, Xie Y, Xia G, Spraggon G, Warmuth M, Liu Y, Gray NS. A general strategy for creating "inactive-conformation" abl inhibitors. Chem Biol. 2006;13 (7):779–86. doi: 10.1016/j.chembiol.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 59.Scapin G, Patel SB, Lisnock J, Becker JW, LoGrasso PV. The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity. Chem Biol. 2003;10 (8):705–12. doi: 10.1016/s1074-5521(03)00159-5. [DOI] [PubMed] [Google Scholar]
- 60.Deuse T, Velotta JB, Hoyt G, Govaert JA, Taylor V, Masuda E, Herlaar E, Park G, Carroll D, Pelletier MP, Robbins RC, Schrepfer S. Novel immunosuppression: R348, a JAK3-and Syk-inhibitor attenuates acute cardiac allograft rejection. Transplantation. 2008;85 (6):885–92. doi: 10.1097/TP.0b013e318166acc4. [DOI] [PubMed] [Google Scholar]
- 61.Bleicher KH, Bohm HJ, Muller K, Alanine AI. Hit and lead generation: beyond high-throughput screening. Nat Rev Drug Discov. 2003;2 (5):369–78. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 62.Petrelli A, Giordano S. From single-to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Curr Med Chem. 2008;15 (5):422–32. doi: 10.2174/092986708783503212. [DOI] [PubMed] [Google Scholar]
- 63.Morphy R. Selectively nonselective kinase inhibition: striking the right balance. J Med Chem. 2010;53 (4):1413–37. doi: 10.1021/jm901132v. [DOI] [PubMed] [Google Scholar]
- 64.Marsden BD, Knapp S. Doing more than just the structure-structural genomics in kinase drug discovery. Curr Opin Chem Biol. 2008;12 (1):40–5. doi: 10.1016/j.cbpa.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 65.Verdonk ML, Mortenson PN, Hall RJ, Hartshorn MJ, Murray CW. Protein-Ligand Docking against Non-Native Protein Conformers. J Chem Inf Model. 2008;48 (11):2214–25. doi: 10.1021/ci8002254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fitting of the asymmetric normal inverse Gaussian (NIG) to the distribution of CR-scores in the human kinome: (A) histogram of the empirical distribution (gray bars) compared to the fitted NIG and normal distribution, (B) same as A but in log scale, (C) quantile-quantile plot.
Bioassay data used in this study. Primary targets are selected based on the strongest inhibition (Bioassay #1) or the lowest dissociation constant Kd (Bioassays #2 and #3). #P and #N correspond to the number of alternate targets (≤25% for Bioassay #1, and ≤10 μM for Bioassays #2 and #3) and non-targets (>25% for Bioassay #1, and >10 μM for Bioassays #2 and #3) for a given compound, respectively.
Trained Naive Bayes classifier used by X-ReactKIN to calculate the CR- score.