Abstract
Mutations in FUS have been reported to cause a subset of familial amyotrophic lateral sclerosis (ALS) cases. Wild-type FUS is mostly localized in the nuclei of neurons, but the ALS mutants are partly mislocalized in the cytoplasm and can form inclusions. Little is known about the regulation of FUS subcellular localization or how the ALS mutations alter FUS function. Here we demonstrate that the C-terminal 32 amino acid residues of FUS constitute an effective nuclear localization sequence (NLS) as it targeted beta-galactosidase (LacZ, 116 kDa) to the nucleus. Deletion of or the ALS point mutations within the NLS caused cytoplasmic mislocalization of FUS. Moreover, we identified the poly-A binding protein (PABP1), a stress granule marker, as an interacting partner of FUS. PABP1 formed large cytoplasmic foci that co-localized with the mutant FUS inclusions. No such foci, which resemble stress granules, were observed in the presence of wild-type FUS. In addition, processing bodies, which are functionally related to stress granules, were adjacent to but not co-localized with the mutant FUS inclusions. Our results suggest that the ALS mutations in the C-terminal NLS of FUS can impair FUS nuclear localization and induce cytoplasmic mislocalization, inclusion formation, and potential perturbation of RNA metabolism.
Keywords: ALS, FUS/TLS, nuclear localization sequence (NLS), poly-A binding protein 1 (PABP1), stress granules, processing bodies, RNA metabolism
Introduction
Amyotrophic lateral sclerosis (ALS, also known as Lou Gehrig's disease) is a progressive and fatal neurodegenerative disease. A general symptom of ALS is muscle weakness and wasting triggered by the loss of innervation by motor neurons. The majority of ALS cases are sporadic, and approximately 10% are familial. Several ALS genes have been identified as their mutation can lead to familial ALS. Mutations in Cu/Zn superoxide dismutase (SOD1) were first discovered (Rosen, et al., 1993), which account for about 20% of the familial cases. Most recently, mutations in an RNA processing protein named fused in sarcoma/translocated in liposarcoma (FUS/TLS) were found to cause type 6 familial ALS (Kwiatkowski, et al., 2009,Vance, et al., 2009). In addition, FUS mutations have also been reported in sporadic ALS cases (Belzil, et al., 2009,Corrado, et al., 2009,Dejesus-Hernandez, et al., 2010). Interestingly, mutations in another RNA metabolism protein named TAR DNA binding protein (TDP-43) were also reported in familial ALS in recent years (Kabashi, et al., 2008,Sreedharan, et al., 2008,Van Deerlin, et al., 2008). In addition, perikaryal ubiquitinated TDP-43 proteinopathy is found in a large portion of familial and sporadic ALS cases (Kwong, et al., 2007,Mackenzie, et al., 2007,Neumann, et al., 2006). While TDP-43 has been intensely studied in the past several years, the role of FUS in ALS is largely unknown and is the focus of this study.
FUS is a ubiquitously expressed multi-domain protein belonging to the FET family of RNA-binding proteins along with EWS (Ewing sarcoma) (Bertolotti, et al., 1996). In neurons and glial cells, FUS is almost exclusively localized to the nucleus (Aman, et al., 1996,Andersson, et al., 2008). FUS engages in rapid nucleocytoplasmic shuttling, binds RNA both in the nucleus and the cytoplasm and likely shuttles RNA between the nucleus and the cytoplasm (Zinszner, et al., 1997). The FUS protein also plays a role in the transport of mRNA for local translation in dendrites in neurons (Fujii, et al., 2005,Fujii and Takumi, 2005). In addition, FUS has also been reported to bind single- and double-stranded DNA (Baechtold, et al., 1999) and to play a role in a variety of processes including DNA repair (Baechtold, et al., 1999), pairing of homologous DNA and cell proliferation (Bertrand, et al., 1999), transcriptional regulation (Prasad, et al., 1994,Tan and Manley, 2010) and mRNA splicing (Yang, et al., 1998).
Protein inclusions in degenerating motor neurons are a hallmark of ALS (Piao, et al., 2003). In the familial ALS cases caused by FUS mutations, aberrant FUS mislocalization to the cytoplasm and FUS-positive cytoplasmic inclusions were detected (Belzil, et al., 2009,Corrado, et al., 2009,Kwiatkowski, et al., 2009,Vance, et al., 2009). However, the mechanism for such altered localization is unknown. Thus, in this study we set forth to determine the underlying mechanism and potential consequences of mutant FUS mislocalization.
The regulation of FUS subcellular localization is not fully understood. The calculated molecular weight of human FUS protein is 53,426 daltons although it migrates as a band at approximately 70 kDa in SDS-PAGE. The cutoff for passive diffusion through the nuclear pore complex is estimated to vary between 40-60 kDa (Gerace, 1995,Peters, 2009,Rout, et al., 2003). Thus, it is theoretically possible but likely difficult for FUS to migrate across the nuclear pore complex with passive diffusion. There is a predicted nuclear export sequence (NES) in FUS (Lagier-Tourenne and Cleveland, 2009), whose sequence shows similarity but does not match well with the consensus NES sequence. There is no classic nuclear localization sequence (NLS) reported or predicted in FUS. Bioinformatic attempts revealed no classically predictable NLS (e.g., PredictNLS server at http://cubic.bioc.columbia.edu/services/predictNLS, (Cokol, et al., 2000)). However, a potential NLS has been reported within EWS, which is homologous to the C-terminus of FUS (Lee, et al., 2006,Zakaryan and Gehring, 2006). It is also noted that the majority of the FUS mutations identified to date are clustered in the C-terminus of the protein (Kwiatkowski, et al., 2009,Vance, et al., 2009). We thus tested the hypothesis that the C-terminus of FUS played a critical role in the nuclear localization of FUS by functioning as an NLS.
Sequence analysis led us first to test the C-terminal 17 residues, which contain several positively charged residues and likely constitute a distinct motif from the preceding sequences. Deletion of the 17 residues caused a dramatic mislocalization to cytoplasm and formation of cytoplasmic inclusions. Fusion of the 17 residues to GFP was sufficient to cause accumulation of GFP in the nucleus, however tagging the 17 residues to a large protein beta-galactosidase (LacZ) had marginal effect in targeting LacZ to the nucleus. Our results suggest that the C-terminal 17 residues are essential for FUS nuclear localization but may not be sufficient to function as an effective NLS. Based on the most recently discovered ALS truncation mutant FUS lacking the C-terminal 32 residues, we tested and demonstrated that the 32-residue fragment was highly effective in targeting LacZ to the nucleus. Moreover, the ALS point mutations in the C-terminal fragment significantly impaired its ability of targeting LacZ and GFP to the nucleus. These results support that the C-terminus of FUS indeed plays a critical role in its nuclear localization. Using proteomic techniques, we identified a stress granule component, the poly-A binding protein 1 (PABP1) as a FUS interacting partner. Confocal microscopic analysis showed that the mutant FUS inclusions were co-localized with stress granules using PABP-1 and TIA-1 as markers, whereas no stress granules were induced in cells expressing wild-type FUS. Moreover, the mutant FUS inclusions were adjacent to but not co-localized with processing bodies that are functionally related to stress granules.
Materials and Methods
Plasmid construction
The wild-type (WT) or mutant FUS sequence was PCR amplified using the pDEST53-WT FUS plasmid (generous gift from Dr. Lawrence J. Hayward, University of Massachusetts Medical School). The amplification products were inserted into p3XFLAG-CMV10 (Sigma), pEGFP-C3 (Clontech) and p5xMyc-UAST (Liu, et al., 2007) vectors using the BglII and KpnI sites, or the pEBG glutathione S-transferase (GST) fusion vector (Tanaka, et al., 1995) using the BamHI and KpnI sites. The FUS C-terminal segments K510-Y526, R495-Y526 and R495-G509 were fused to the C-terminus of the GFP and LacZ vectors by inserting double-stranded oligonucleotides between the BglII and KpnI sites of pEGFP-C1 (Clontech) or the NheI and XhoI sites of pHM829 (Addgene Plasmid 20703) (Sorg and Stamminger, 1999), The R518K, R521G and R521H mutations were introduced into the LacZ-FUS (R495-Y526) construct using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene).
Primary mouse motor neuron culture
Primary mouse motor neurons were purified and cultured following a previously reported protocol (Gingras, et al., 2007). Briefly, spinal cords were obtained from embryonic day 13 mice by careful dissection. Spinal cord cells were then dissociated by trituration and motor neurons were purified by differential centrifugation using a step gradient of NycoPrep (Accurate Chemical) solutions. The motor neurons were then plated on poly-Lysine treated glass coverslips and incubated at 37°C, 5% CO2 in growth-factor supplemented medium. The cells were transfected using Lipofectamine LTX (Invitrogen) on the sixth day in vitro (DIV6) and fixed on DIV 10 for confocal microscopy.
Cell culture and transfection
The NSC34, N2A and HEK293 cells were cultured in DMEM medium (Invitrogen) supplemented with 10% FBS and penicillin/streptomycin in a 37°C incubator in 5% CO2/95% air as previously described (Strom, et al., 2008). The cells were transfected in a 6-well format using the Lipofectamine transfection reagent (Invitrogen). The S2 cells were cultured in Schneider's Drosophila Medium (Invitrogen) with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Standard protocols were used for transfection and immunostaining as described previously (Liu, et al., 2007).
Nuclear-cytosolic fractionation
The nuclear fractionation was performed as described in (Vance, et al., 2009). Briefly, the transfected cells were rinsed with phosphate-buffered saline (PBS), resuspended in PBS, pelleted at 1,000 RCF, and resuspended in lysis buffer (50 mM K-HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, Sigma P-8340 protease inhibitor cocktail at 1:1,000 dilution and 0.2 mM PMSF). After incubation at room temperature for 2 min and on ice for 10 min, Triton-X100 was added to the suspensions to the final concentration of 0.5%. The suspension was incubated on ice for additional 10 min, and pelleted at 13,000 RCF for 1 minute. The supernatant and the pellet was separately collected, boiled with SDS loading buffer and used as the cytosolic and nuclear fraction, respectively. SOD1 and histone H4 was used as the cytosolic and nuclear marker, respectively.
Western blotting
The cell culture extracts were prepared two days after transfection. The primary antibodies used in Western blotting were anti-actin (Santa Cruz, sc-1616), anti-GFP (Santa Cruz, sc-8334), anti-GST (Santa Cruz, sc-459), anti-FUS (Santa Cruz, sc-47711), anti-SOD1 (Santa Cruz, sc-11407), anti-PABP1 (Santa Cruz, sc-32318) and anti-acetyl-histone H4 (Upstate 17-630). The protein bands were visualized using ECL reagents (Pierce).
Fluorescence microscopy
The cells were cultured and transfected on sterile gelatin-coated coverslips and subjected to fluorescence microscopy analysis as previously described (Gal, et al., 2009). Briefly, the cells were washed with PBS 48 hours after transfection and fixed in 4% paraformaldehyde in PBS at 37°C for 15 min. The cells with GFP fluorescence were incubated with 4’,6-diamidino-2-phenylindole (DAPI, Invitrogen) in PBS/ 0.1% Triton-X100, then mounted on glass slides with Vectashield mounting medium (Vector Laboratories). The cells for immunofluorescence staining were fixed as above, permeabilized with PBS/0.25% Triton-X100, blocked with 10% BSA in PBS, and incubated with primary antibodies diluted in 3% BSA/PBS. The primary antibodies were anti-FLAG (Sigma F3165), anti-beta-galactosidase (Promega Z3781), anti-PABP (Santa Cruz sc-32318), anti-TIA-1 (Santa Cruz sc-28237) and anti-GE-1 (Santa Cruz sc-8418). The slides were subsequently washed with PBS and incubated with DAPI and Alexa Fluor 488 anti-mouse (Invitrogen A21202), Alexa Fluor 594 anti-mouse (Invitrogen A21203) or Alexa Fluor 594 anti-rabbit (Invitrogen A11012) secondary antibodies at 1:400 dilution in 3% BSA/PBS. The coverslips were then washed with PBS and mounted using Vectashield mounting medium. Fluorescence microscopy was carried out using a Leica SP5 inverted confocal microscope with a 63x or 100x objective.
Image analysis was performed using the ImageJ program. The quantitative analysis of the fluorescence intensities in the nucleus and cytoplasm was performed using confocal microscopic images of randomly selected cells. For each individual cell, the boundary of the nucleus was identified by tracing the acquired DAPI-channel image. The boundary of the cell was similarly traced using the GFP or LacZ immunofluorescence image. Cytoplasm was the area between the nuclear and cellular boundaries as defined above. An ImageJ macro then calculated the average intensity of GFP or LacZ immunofluorescence from pixels within the nucleus (N) and within the cytoplasm (C) using these traced areas. The ratio of these average intensities (N/C) was calculated from at least 20 randomly selected cells in each group. The statistical analysis was performed using Student's t-test.
Identification of PABP1 as a FUS interaction partner
The GST-FUS (WT and R521G) constructs were transfected into HEK293 cells. Two days post-transfection, cell lysates were prepared in RIPA buffer (Millipore) and subjected to pulldown with Glutathione Sepharose 4B (GE Healthcare 17-0756-01) as previously published (Zhang, et al., 2007). The eluted proteins were resolved by SDS-PAGE, and individual gel slices of interest were subjected to trypsin digestion. The extracted peptide mixtures were analyzed by LC-MS/MS using a QSTAR XL (Applied Biosystems) as previously described (Fukada, et al., 2004). The LC-MS/MS results were processed with a local Mascot server (Matrix Science) for protein identification.
Results
The in vivo pathology of FUS mutants is recapitulated in cultured cells
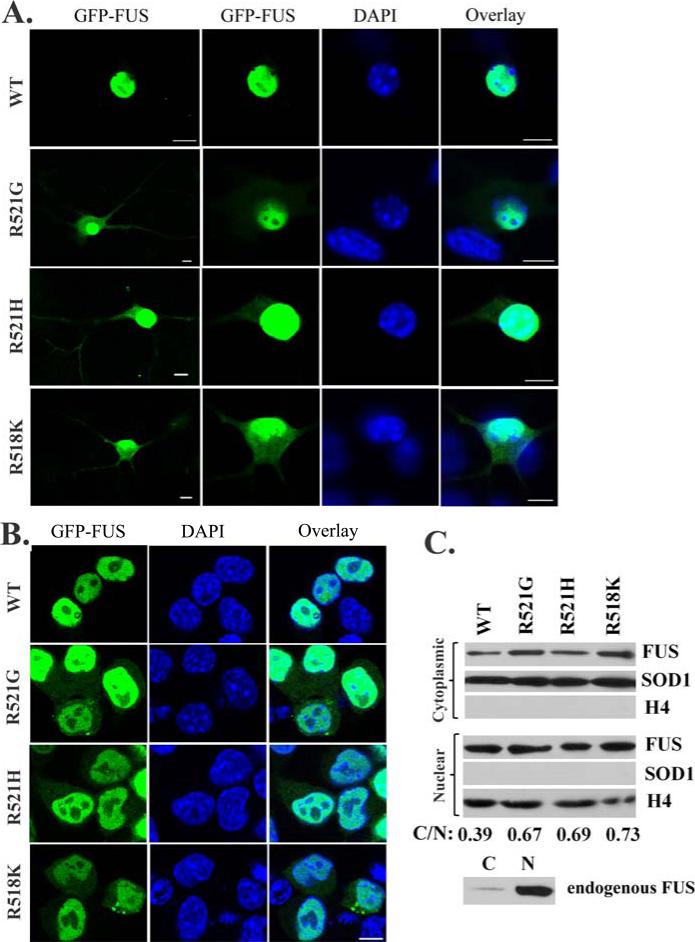
We first tested whether the pathological cytoplasmic localization of FUS mutants were replicated in primary motor neurons using the GFP-tagged wild-type and familial ALS mutant FUS constructs. R518K, R521G and R521H were chosen since they are the more prevailing mutations in the reports (Kwiatkowski, et al., 2009,Vance, et al., 2009). As shown in Fig 1A, all three mutants were evident in the perikaryon and neurites in addition to the nucleus, clearly suggesting cytoplasmic mislocalization. In contrast, WT FUS was localized in the nucleus with nearly undetectable cytoplasmic signal.
Figure 1. Cytoplasmic mislocalization of the familial ALS mutant FUS in primary motor neurons and N2A cells.
(A). Confocal microscopic images of mouse embryonic primary motor neurons transfected with GFP-tagged WT or mutant FUS. Partial localization in the cytoplasm was observed for the ALS mutants R521G, R521H and R518K, but WT FUS was mostly localized in the nucleus. (B). Confocal microscopic images of N2A cells transfected with GFP-tagged WT or mutant FUS. Similarly, cytoplasmic localization of mutant FUS was observed. In addition, inclusions were also observed in cells expressing mutant FUS. (C). Western blot of the cytoplasmic and nuclear fractions of cells expressing WT or mutant FUS. Increased levels of mutant FUS were found in the cytoplasmic fraction as compared to WT FUS. The quantification of the relative abundance of the cytoplasmic FUS normalized against the nuclear FUS was shown as C/N values at the bottom of the figure. A nuclear protein histone 4 (H4) and a cytoplasmic protein superoxide dismutase 1 (SOD1) were used to demonstrate the purity of the fractions. Western blot of the endogenous FUS in N2A cells showed that the endogenous WT FUS was largely in the nuclear fraction while a weak band was also found in the cytoplasmic fraction. All scale bars are 10 μm.
We then tested whether the pathological cytoplasmic mislocalization and inclusion formation of FUS mutants were also observed in several different cell lines including NSC34, N2A, HEK293 and Drosophila S2 cells. As shown in Fig 1B, cytoplasmic mislocalization of the mutants was also apparent in the N2A mouse neuroblastoma cells. The nuclear-cytosolic fractionation of transfected N2A cells showed that higher amounts of mutant FUS proteins were fractionated to the cytosol than the WT protein (Fig 1C). At the same time, the endogenous FUS was detected almost exclusively in the nuclear fraction. In addition, cytoplasmic inclusions were observed in some cells expressing mutant FUS (Fig 1B). The cytoplasmic mislocalization of mutant FUS was also observed in the motor neuron-like NSC34 cells, HEK293 cells and Drosophila S2 cells (Supplementary Fig S1). Inclusions of mutant FUS were also observed in some NSC34 and S2 cells, but were more prominent in HEK293 cells. Our results show that the pathological features of the familial ALS mutants of FUS could be replicated in the cell culture systems tested, including primary motor neurons.
Sequence analysis of FUS
FUS is a multi-domain RNA-binding protein (Bertolotti, et al., 1996,Lagier-Tourenne and Cleveland, 2009). It contains an N-terminal domain rich in Gln, Gly, Ser and Tyr residues, followed by a Gly-rich region, an RNA Recognition Motif (RRM) domain, two Gly-rich regions separated by a RanBP2-type zinc finger, and a C-terminal domain. The Gly-rich regions contain several embedded Arg-Gly motifs with the sequence “RG”, “RGG” or “RGGG” (referred to as the RGG motif in this study).
The sequence alignment of the C-terminal regions of FUS from multiple organisms, Drosophila CaZ protein, and several human proteins closely related to FUS is shown in Fig 2A. The C-terminal 17-residue segment of FUS is clearly distinct from the preceding region containing the RGG motifs. The C-terminal 17 residues are separated from the RGG motifs by a Gly-Pro-Gly sequence, which has a high turn and loop propensity (Leszczynski and Rose, 1986,Rose, et al., 1985). Thus, this C-terminal segment is likely to be a separate motif from the structural perspective. Moreover, the majority of reported familial ALS mutations of FUS are point mutations clustered within the C-terminus (Fig 2A) (Kwiatkowski, et al., 2009,Vance, et al., 2009). In addition, the C-terminus of EWS has been reported to function as an NLS (Zakaryan and Gehring, 2006). We first hypothesized that, despite the lack of a classic NLS in FUS, the C-terminal 17 residues may play a crucial role in the nuclear localization of FUS by functioning as an NLS, or a nuclear retention sequence (NRS).
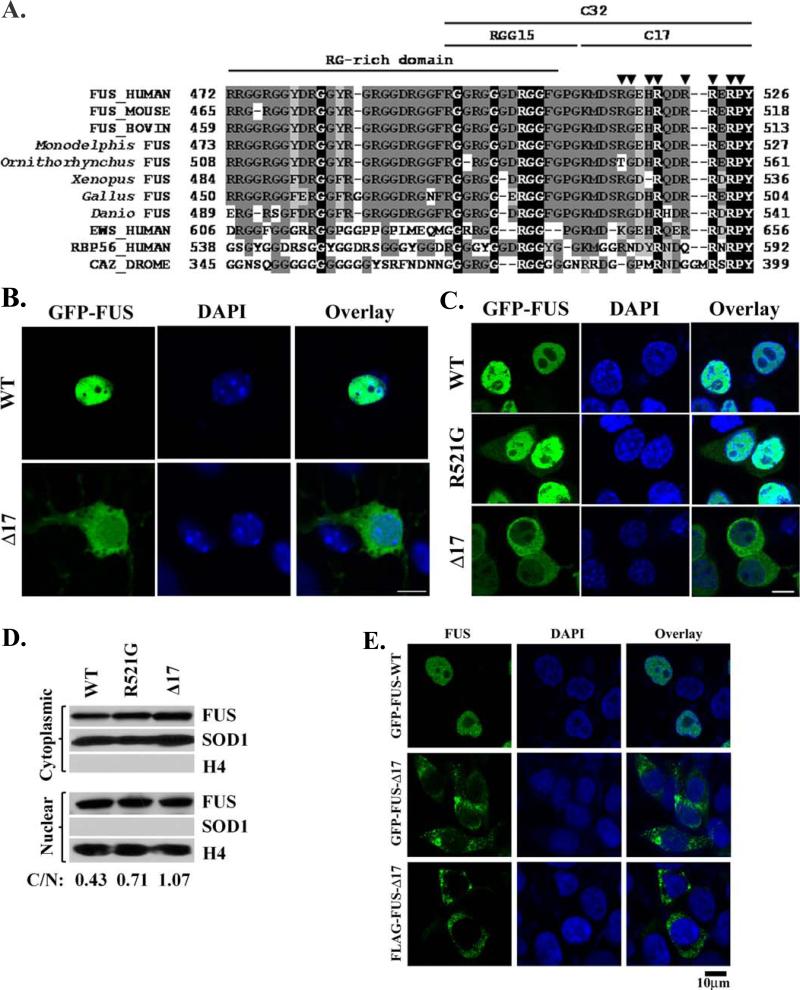
Figure 2. The C-terminal 17 residues are essential to the nuclear localization of FUS.
(A). The sequence alignment of the C-terminal regions of FUS from multiple organisms, Drosophila CaZ and several human proteins closely related to FUS. X: identical residues; X: strong conservation; X: weaker conservation/similarity. ▼ indicates the positions of known familial ALS mutations. The RG-rich domain, RGG15 motif, C-terminal 17-residue sequence (C17) and C-terminal 32-residue segment (C-32) are marked. (B). Confocal microscopic images of mouse embryonic primary motor neurons transfected with GFP-tagged WT or C17 deletion mutant FUS. FUS-Δ17 was observed in the perikaryon, neurites and nucleus while WT FUS was mostly localized in the nucleus. (C). Similar cytoplasmic localization of FUS-Δ17 was also observed in N2A cells. In addition, inclusions were also observed in cells expressing mutant FUS. (D). Western blot of the cytoplasmic and nuclear fractions of cells expressing WT FUS, R521G or Δ17 mutants. Increased levels of mutant FUS were found in the cytoplasmic fraction as compared to WT FUS. The quantification of the relative abundance of the cytoplasmic FUS normalized against the nuclear FUS was shown as C/N values at the bottom of the figure. H4 and SOD1 were used as nuclear and cytoplasmic markers, respectively. (E). Similar cytoplasmic localization and inclusions of FUS-Δ17 were also observed in HEK293 cells using GFP-tagged FUS or immunofluorescence staining of FLAG-tagged FUS. All scale bars are 10 μm.
The effect of the deletion of the C-terminal 17 residues of FUS
To examine the role of the C-terminal 17 residues of FUS in the nuclear localization of the protein, we generated a truncation mutant of FUS lacking the C-terminal 17 residues (FUS-Δ17). Deletion of the 17 residues dramatically changed the subcellular localization of the FUS protein. In primary motor neurons, the GFP-tagged FUS-Δ17 fusion protein was apparent in the neurites, perikaryon, in addition to nucleus (Fig 2B). In N2A cells, GFP-tagged FUS-Δ17 was localized either predominantly in the cytoplasm or evenly throughout both nucleus and cytoplasm (Fig 2C). In addition, GFP-FUS-Δ17 inclusions were evident in many transfected cells (Fig 2C). The fractionation of transfected N2A cells also showed that FUS-Δ17 localized to the cytoplasm even more than the R521G mutant (Fig 2D). Similar results were obtained in HEK293 cells (Fig 2E) and NSC34 cells (Supplemental Fig S2). To eliminate the potentially fortuitous effect of the GFP tag itself, a 3xFLAG-tagged FUS-Δ17 fusion protein was also generated and tested. Anti-FLAG immunofluorescence studies showed that the localization of 3xFLAG-FUS-Δ17 was also predominantly cytoplasmic with inclusions in transfected HEK293 cells (Fig 2E). Taken together, the results suggest that the C-terminal 17 residues of FUS play an essential role in the nuclear localization of the protein.
The effect of the C-terminal 17 residues on nuclear localization
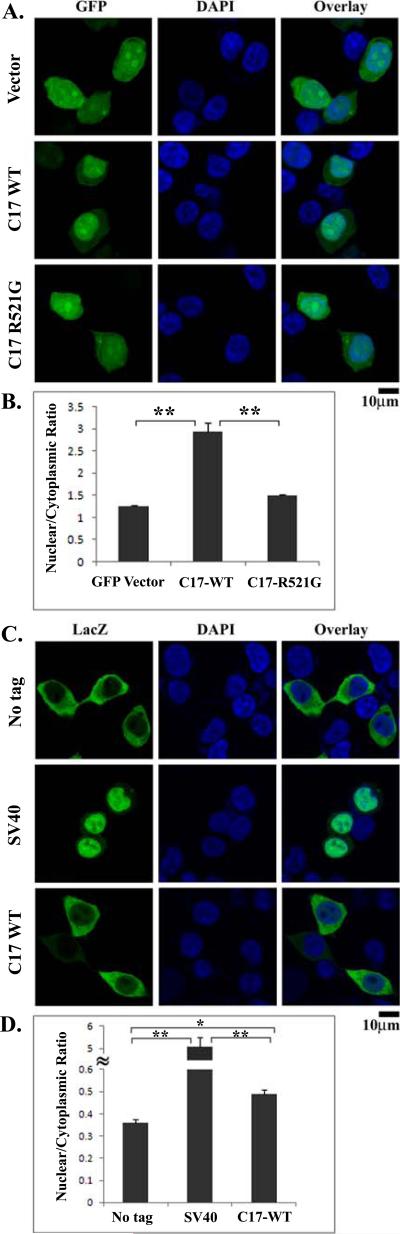
We fused the C-terminal 17 residues (K510-Y526) to the C-terminus of enhanced GFP (GFP-C17) and tested its effect on the localization of GFP. The GFP-C17 protein apparently accumulated in the nucleus in contrast to the almost equal distribution of GFP control in the cytoplasm and nucleus of HEK293 (Fig 3A). We also generated a mutation within the C-terminal C17 residues corresponding to the R521G mutation in full-length FUS (GFP-C17-R521G). The R521G mutation severely impaired the ability of the C-terminal 17 residues of FUS to drive the nuclear accumulation of GFP (Fig 3A). The results were quantified by measuring the nuclear/cytoplasmic fluorescence intensity ratios (N/C). As shown in Fig 3B, the N/C ratio of GFP-C17 protein (2.94) was more than two-fold higher than that of the GFP control protein (1.24). Compared to GFP-C17, the R521G mutation significantly reduced the N/C ratio of the GFP-C17-R521G protein (1.49). Statistical analysis showed that the differences in the above two pairs were statistically significant (p < 10-6). The results clearly support that the C-terminal 17 residues can facilitate the nuclear accumulation of GFP while the R521G mutation in the segment can impair nuclear accumulation.
Figure 3. The effect of the C-terminal 17 residues on the nuclear localization of GFP and LacZ.
(A). Confocal images of HEK293 cells transfected with GFP vector control, GFP-C17, or GFP-C17-R521G. Tagging the C-terminal 17 residues of FUS to GFP caused significant accumulation of GFP-C17 in the nucleus. The ALS mutation R521G within the C-17 segment caused nearly even distribution of GFP-C17-R521 in the entire cell. (B). The average ratios of nuclear and cytoplasmic fluorescence intensities (N/C) were quantified from at least 20 randomly selected cells, see Materials and Methods. (C). Confocal images of HEK293 cells transfected with LacZ alone (no tag), LacZ-SV40, or LacZ-C17. Tagging LacZ with the NLS of the large T antigen of SV40 caused significant relocalization of LacZ-SV40 to the nucleus. In contrast, LacZ-C17 largely remained in the cytoplasm with relatively weak fluorescence in the nucleus. (D). Quantification of the nuclear and cytoplasmic fluorescence intensities as the N/C ratios. The C-terminal 17 residues caused a mild but statistically significant increase of the N/C ratio of LacZ-C17 as compared to LacZ with no tag. SEM were calculated as error bars and Student's t-test was performed to obtain p values in (B) and (D). *, p < 0.0001; **, p < 10-6. All scale bars are 10 μm.
A caveat of this set of experiments is that the molecular weight of GFP is under 30kDa and it can enter the nucleus with passive diffusion (Peters, 2009,Sorg and Stamminger, 1999), consistent with the observation that GFP was localized in both the nucleus and cytoplasm (Fig 3A). To overcome this caveat, the bacterial beta-galactosidase (LacZ) with a molecular weight of 116 kDa was chosen to further evaluate the effect of the C-terminal 17 residues. When expressed in mammalian cells, LacZ is a cytoplasmic protein and can be imported into the nucleus if it is tagged with an NLS (Sorg and Stamminger, 1999). We fused the C-terminal 17 residues of FUS to the C-terminus of LacZ (LacZ-C17).We also generated a positive control construct with the well known SV40 large T antigen NLS (PKKKRKV) (Kalderon, et al., 1984) fused to the C-terminus of LacZ (LacZ-SV40).
The subcellular localization of the LacZ proteins examined by immunofluorescence microscopy is shown in Fig 3C. The untagged LacZ was largely in the cytoplasm and the LacZ-SV40 protein was largely in the nucleus, serving as negative and positive controls, respectively. Surprisingly, the C-terminal 17 residues of FUS were not nearly as effective as the SV40 large T antigen NLS in targeting LacZ to the nucleus (Fig 3C). Careful quantification (see Fig 3D) of the nuclear/cytosolic fluorescence intensity ratios showed that the N/C ratio of the LacZ-C17 protein (0.49) was mildly but statistically (p < 0.0001) increased compared to that of the untagged LacZ (0.36). The p values comparing LacZ-SV40 to LacZ control or LacZ-C17 were less than 10-6 (Fig 3D). The results in Fig 2 and 3 collectively show that although the C-terminal 17 residues of FUS are essential for the nuclear localization of the FUS protein, the segment is not an effective NLS.
The effect of the C-terminal 32 residues of FUS on the localization of LacZ
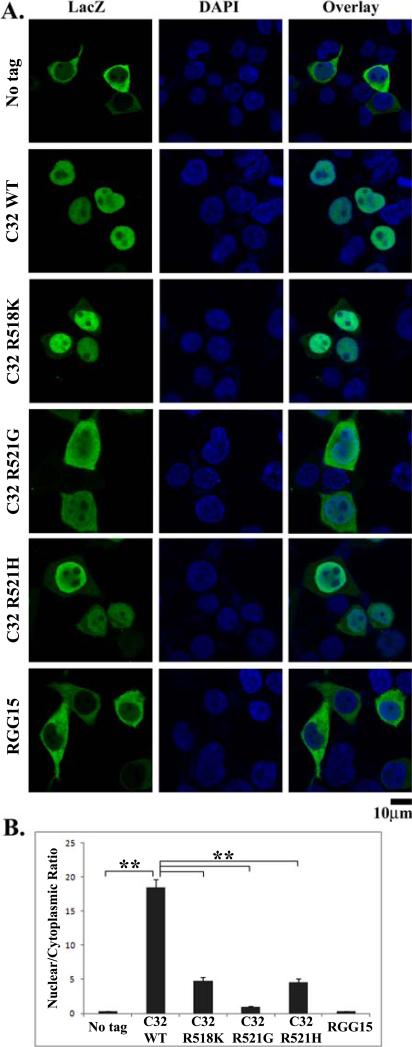
In light of the above results, we speculated that a larger segment of the C-terminus of FUS could function as a more competent NLS. Based on the most recently discovered R495X nonsense mutation (lacking residues R495-Y526) in familial ALS patients (personal communication with Dr. Lawrence Hayward), we tested whether the C-terminal 32 residues can function as an NLS. Fig 4A shows the representative confocal images and Fig 4B shows the quantitative analysis of the N/C ratios of different LacZ constructs examined. The C-terminal 32 residues of FUS were fused to the C-terminus of LacZ (LacZ-C32). When expressed in HEK293 cells, the LacZ-C32 protein was dramatically relocalized to the nucleus (p < 10-6). Introduction of the R518K, R521G and R521H ALS single point mutations in the C32 segment all resulted in significant reduction of the nuclear localization of LacZ-C32 (p < 10-6). Of note, the R521G mutation caused the most significant reduction of nuclear localization of LacZ-C32 among the three point mutations tested. As the C-terminal 17 residues could not work as an effective NLS, we further tested whether the first 15-residue segment of the C-terminal 32 residues alone might have acted as an NLS. We fused the 15-residue RGG motif (R495-G509) to the C-terminus of LacZ (LacZ-RGG15). LacZ-RGG15 was largely confined in the cytoplasm, which was indistinguishable from the untagged LacZ (Fig 4A), suggesting that the RGG15 motif alone is not an NLS. Taken together, our results suggest that the C-terminal 32 residues of FUS can act as a potent nuclear localization sequence.
Figure 4. The C-terminal 32 residues are an effective nuclear localization sequence (NLS).
(A). Confocal images of HEK293 cells transfected with LacZ (no tag), LacZ-C32, LacZ-C32-R521G, LacZ-C32-R518K, LacZ-C32-R521H, or LacZ-RGG15. LacZ-C32 was highly concentrated in the nucleus. The ALS mutations (R521G, R518K or R521H) in the C-32 sequence all increased the cytoplasmic localization of the fusion protein. The R521G mutation caused approximately even distribution of LacZ-C32-R521G between the cytoplasm and nucleus while the cytoplasmic fluorescence intensity of LacZ-C32-R518K or LacZC32-R521H was weaker. (B). Quantification of the nuclear and cytoplasmic fluorescence intensities as the N/C ratios. The C-terminal 32 residues were highly effective in increasing the N/C ratio. All three ALS mutations caused significant decrease in the N/C ratios, among which R521G caused the largest decrease. Tagging the RGG15 motif to LacZ did not induce significant change in the N/C ratio compared to the LacZ with no tag. Errors bars represent SEM and p values were calculated using Student's t-test. **, p < 10-6. All scale bars are 10 μm.
The cytoplasmic inclusions of mutant FUS co-localize with stress granules
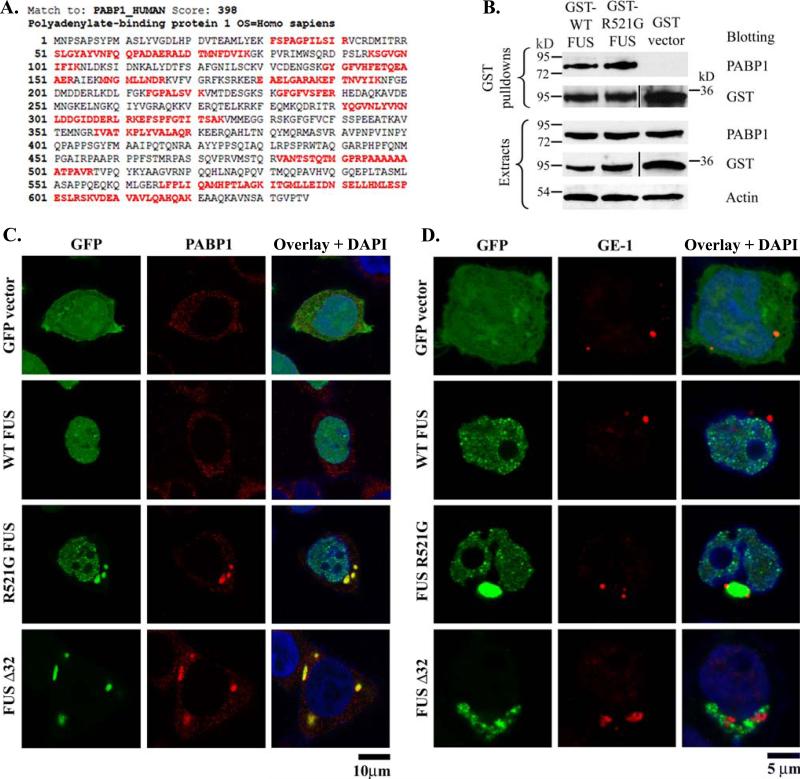
To better understand the differential functions of WT and mutant FUS, we used a mass spectrometry-based proteomic approach to identify FUS interacting partners. We generated mammalian expression constructs for GST-tagged WT and R521G FUS and carried out GST pulldown experiment as described previously (Zhang, et al., 2007). The poly-A binding protein 1 (PABP1) was identified as one of the interacting partners of WT and R521G FUS. The sequences identified by tandem MS/MS are in red in Fig 5A, and a MASCOT score of 398 also support a high-confidence identification. Furthermore, the interaction between PABP1 and FUS was validated by GST pulldown followed by PABP1 Western blot. As shown in Fig 5B, the endogenous PABP1 was co-precipitated with WT and R521G FUS (lanes 1 and 2) but not with the GST control (lane 3).
Figure 5. The familial ALS mutant FUS co-localized with stress granules.
(A). Identification of polyadenylate-binding protein 1 (PABP-1) in the GST-FUS pull-down sample by LC-MS/MS. The identified tryptic peptides within PABP-1 by tandem mass spectrometry (MS/MS) are shown in red. The MASCOT score of 398 suggests a high-confidence identification. (B). GST vector control, GST-WT-FUS or GST-R521G-FUS was transfected to HEK293 cells. The GST pull-down samples were subjected to PABP-1 Western blot. PABP-1 was co-precipitated with GST-FUS but not with the GST vector control. (C). Confocal images of GFP, PABP-1 immunofluorescence and DAPI were obtained from HEK293 cells transfected with GFP vector control, GFP-WT-FUS, GFP-R521G-FUS, or GFP-FUS-Δ32. PABP-1 showed dispersed puncta throughout the cytoplasm in cells transfected with GFP vector or WT FUS. In the cells expressing the R521G or C32 deletion mutant FUS, PABP-1 formed large cytoplasmic foci that resembled stress granules. The stress granules were co-localized with the mutant FUS inclusions. (D). Confocal images of GFP, GE-1 immunofluorescence and DAPI were obtained from HEK293 cells transfected with GFP vector control, GFP-WT-FUS, GFP-R521G-FUS, or GFP-FUS-Δ32. The processing bodies marker GE-1 was adjacent to, but not co-localized with the cytoplasmic inclusions of mutant FUS.
The PABP1 protein has been reported to be a component of stress granules (Kedersha, et al., 2007,Kedersha, et al., 1999), the cytoplasmic structures in which a population of silenced mRNAs are held in translationally inactive 48S ribosomal complexes (Anderson and Kedersha, 2008). PABP1 also plays a role in regulatory processes of mRNA metabolism, translational initiation regulation and translationally-coupled mRNA turnover (Mangus, et al., 2003). We hypothesize that the cytoplasmic inclusions containing mislocalized mutant FUS might facilitate stress granule formation and perturb translational regulation, which could contribute to the pathogenesis of ALS.
To test this hypothesis, we examined the subcellular localization of PABP1 in HEK293 cells transfected with GFP vector, GFP-WT-FUS, GFP-R521G-FUS or GFP-FUS-Δ32, and in the untransfected control cells. In the untransfected cells and the cells transfected with GFP vector control or GFP-WT-FUS, anti-PABP1 immunofluorescence microscopy revealed a dispersed punctate pattern throughout the cytoplasm, which was characteristic PABP-1 distribution as described in literature (Kedersha, et al., 1999) (Fig 5C). In contrast, in cells expressing GFP-R521G-FUS or GFP-FUS-Δ32, PABP-1 formed large cytoplasmic foci that appeared co-localized with the mutant FUS inclusions. These foci represented highly clustered PABP-1 positive puncta and resembled stress granules (Kedersha, et al., 2007,Kedersha, et al., 1999). No such cytoplasmic foci/stress granules were observed in cells expressing WT-FUS.
We further examined the co-localization of FUS and another stress granule marker TIA-1 (Anderson and Kedersha, 2008,Kedersha, et al., 2002). Similarly, while the TIA-1 positive puncta were evenly distributed throughout the cytoplasm in HEK293 cells transfected with GFP-WT-FUS, TIA-1 positive granules were clearly observed and co-localized with the GFP-R521G-FUS inclusions (Supplementary Fig S3).
To eliminate the possibility that the mutant FUS inclusions were incidentally co-localized with stress granules, a negative control experiment was performed to test whether the mutant FUS inclusions were co-localized with the processing bodies marker GE-1 (Anderson and Kedersha, 2008). As shown in Fig 5D, the processing bodies were evident in cells regardless whether FUS (WT or mutant) was over-expressed. When FUS-R521G point mutant and FUS-Δ32 truncation mutant formed the cytoplasmic inclusions, the processing bodies were adjacent to but not co-localized with the inclusions. The results suggest that the co-localization of mutant FUS inclusions and stress granule markers was not non-specific. Taken together, these results suggest that the cytoplasmic mislocalization and inclusion formation of ALS mutant FUS may induce stress granule formation, and subsequently perturb RNA metabolism and translation regulation.
Discussion
Since the initial report of FUS/TLS mutations in type 6 familial ALS (Kwiatkowski, et al., 2009,Vance, et al., 2009), the FUS pathology has also been found in sporadic ALS cases (Belzil, et al., 2009,Corrado, et al., 2009) and frontotemporal lobar degeneration (Neumann, et al., 2009,Seelaar, et al., 2009). In addition, cytoplasmic mislocalization and inclusion formation of mutant FUS protein have been shown in familial ALS tissues (Belzil, et al., 2009,Corrado, et al., 2009,Kwiatkowski, et al., 2009,Vance, et al., 2009). We first demonstrated that the molecular pathology of FUS mutants can be recapitulated in cell culture models. When mutant FUS was expressed in primary motor neurons and four cell lines, increased cytoplasmic localization of mutant FUS was observed in all five cell cultures and varying degrees of inclusion formation was seen in the four cell lines (Fig 1 and S1). The lack of apparent cytoplasmic inclusions in primary motor neurons is likely due to the relatively low expression level of exogenous FUS as compared to the cell lines. Using the cell culture model, we strived in this study to understand how WT FUS is localized to the nucleus, how ALS mutations interfere with this process, and how mutant FUS inclusions may disrupt other cellular processes.
Sequence analysis of the C-terminal region of FUS from multiple organisms suggested that the C-terminal 17 residues may function as an NLS (Fig 2A). Deletion of the 17 residues resulted in significant relocalization of the truncated protein to the cytoplasm and formation of cytoplasmic inclusions (Fig 2 and S2). Appending the 17 residues of FUS to the C-terminus of GFP caused significant nuclear accumulation of GFP-C17 (Fig 3A and 3B), but appending the 17 residues to a larger protein LacZ only caused a minimal nuclear relocalization of LacZ-C17 (Fig 3C and 3D). The results show that, although the C-terminal 17 residues are essential to the nuclear localization of FUS, this segment alone is not sufficient to target a large protein like LacZ into the nucleus. The C-terminal 17 residues may be part of an NLS that requires additional sequences, thus behaves as a very weak NLS on its own. Alternatively, this segment may facilitate the nuclear retention of FUS by functioning as an NRS as described (Kim, et al., 2007,Nakielny and Dreyfuss, 1996).
To determine whether a longer sequence of the C-terminus of FUS can function as an effective NLS, we chose to test the C-terminal 32 residues that are absent in the R495X mutant recently found in familial ALS patients. The results in Fig 4 strongly suggest that neither the 15-residue RGG motif nor the C-terminal 17-residue segment alone is sufficient to ensure nuclear localization. Rather, the entire 32-residue C-terminal motif is a highly effective NLS. Mutations (point mutations or deletion) in the NLS can significantly reduce the nuclear localization of the protein. Since this NLS within FUS evaded the prediction algorithms of classic NLS that is mediated by importin α or β, it is likely it functions via a non-classic mechanism, possibly involve the nuclear importer protein called karyopherin-β2 (Bonifaci, et al., 1997,Lee, et al., 2006,Pollard, et al., 1996).
Our finding that the C-terminal 17 residues of FUS facilitated the nuclear accumulation of GFP but not that of LacZ is reminiscent of an earlier report on the NLS of the FET family member EWS (Zakaryan and Gehring, 2006). The study showed that the C-terminal motif of 18 residues of EWS redirected the YFP protein to the nucleus, but acted rather inconsistently on a larger GST-YFP double-tag protein (Zakaryan and Gehring, 2006). The FUS and EWS proteins are closely related and our sequence alignment shows that EWS also contains an RGG repeat motif preceding the C-terminal 18 residues (Fig 2A). We expect that the RGG motif along with the C-terminal 18 residues together in EWS would also function as a consistently effective NLS. The sequence similarity between FUS and EWS enhances the notion that the entire C-terminal segment including the RGG motif is essential for ensuring nuclear localization of the proteins.
Interestingly, to the best of our knowledge, no ALS mutations have been identified in the 15-residue RGG motif whereas the C-terminal 17-residue segment is populated with the ALS mutations. We postulate that, although the 15-residue RGG motif is important for the nuclear localization of FUS, it might be more tolerant to missense mutations than the C-terminal 17 residues because additional RGG repeats exist in the FUS sequence.
It is noted that the three point mutations, R518K, R521G and R521H, had differential degrees of impairment of the nuclear localization of LacZ-C32 (Fig 4). The R521G mutation caused the most drastic reduction of the N/C ratio of LacZ-C32. Since Arg, Lys and His are all positively charged under the physiological pH, the substitution of Arg with Gly is obviously a more significant change compared to the substitution of Arg with Lys or His. This provides a likely rationale for the observation that R521G impaired the nuclear localization most severely among the three mutations. Moreover, the average duration of survival of the 17 familial ALS patients carrying the R521G mutation was between 24.0 to 28.3 months. The average survival duration of the 3 patients carrying the R521H mutation was 54.1 months (Kwiatkowski, et al., 2009). This suggests that R521G can potentially generate more severe phenotypes. In addition, the FUS-Δ32 truncation mutant lacking the NLS is reported to be associated with a severe early-onset clinical phenotype (personal communication with Dr. Lawrence Hayward). This correlates with the higher degree of cytoplasmic miclocalization of FUS-Δ32 mutant. Given the scarcely available clinical data from mutant FUS-mediated familial ALS patients, the above correlation could be premature. More mechanistic studies and clinical data are needed to better understand the relationship between the mutations and their clinical manifestation in human patients.
The FUS protein has been reported to relocalize to stress granules upon heat shock and oxidative stress (Andersson, et al., 2008). Stress granules are cytoplasmic structures containing translationally silent mRNAs in inactive 48S ribosomal complexes (Anderson and Kedersha, 2008). We independently identified poly A-binding protein 1 (PABP1), which is a stress granule marker, as a FUS interacting partner using proteomic approaches (Fig 5A) and validated it (Fig 5B). Confocal microscopic analysis showed that the PABP1 positive stress granules were co-localized with the cytoplasmic inclusions of mutant FUS whereas PABP-1 was dispersed throughout the cytoplasm in cells expressing WT FUS (Fig 5C). Similar results were obtained when we used another stress granule marker TIA-1 (Fig S3). In addition, the mutant FUS inclusions were adjacent to, but not co-localized with processing bodies, suggesting that the co-localization of stress granules with the mutant FUS inclusions was not incidental. The results suggest that, in the absence of additional insults, mutant FUS alone can induce stress granules. Stress granules and processing bodies are often adjacent to each other and functionally related (Anderson and Kedersha, 2008,Kedersha, et al., 2005). Thus, the observation of the adjacent processing bodies suggests that stress granules induced by the mutant FUS are likely to be functionally relevant.
Similarities between the pathologies caused by TDP-43 and FUS mutants are noted. Although the two proteins are not closely related, they share similar sequence elements such as the RRM domain and glycine-rich regions, which are common to RNA/DNA binding proteins (Lagier-Tourenne and Cleveland, 2009). Similar to FUS, TDP-43 has been reported to play roles in RNA transcription, splicing and transport (Buratti and Baralle, 2008,Volkening, et al., 2009). Both proteins are normally mostly nuclear in neurons, and their molecular features in ALS encompass their mislocalization to the cytoplasm and the formation of protein inclusions (Arai, et al., 2006,Kwiatkowski, et al., 2009,Kwong, et al., 2007,Neumann, et al., 2006,Sreedharan, et al., 2008,Vance, et al., 2009). TDP-43 has been recently reported to localize to cytoplasmic foci called stress granules (Andersson, et al., 2008,Colombrita, et al., 2009,Moisse, et al., 2009,Volkening, et al., 2009), which is similar to mutant FUS in this study (Fig 5). Stress granules represent a potentially protective mechanism in response to a variety of insults. When stress granules are formed, with the exception of certain essential transcripts, the majority of mRNAs are held silent in translationally inactive 48S ribosomal complexes (Anderson and Kedersha, 2008). Our results demonstrate that mutant FUS alone, without the induction of heat shock or oxidative stress, can induce stress granules (Fig 5 and S3). The above evidence support the emerging hypothesis that altered RNA metabolism and processing may play a central role in many familial, and possibly sporadic, forms of ALS (Lagier-Tourenne and Cleveland, 2009,Strong, 2010). Future studies will determine what role stress granule formation and impaired translational control might play in the pathogenesis of ALS.
This study identifies the C-terminus of FUS as a functional NLS, suggesting that the primary reason for the cytoplasmic mislocalization of most reported familial ALS FUS mutants is a damaged or missing NLS. Mutant FUS was also demonstrated to induce stress granules. However, more studies are required to determine whether the ALS mutants of FUS cause the disease due to a loss-of-function or toxic gain-of-function mechanism. Both hypotheses appear to be possible at this time. Although the R521G mutant retained the ability of binding with RNAs containing GGUG motif (Kwiatkowski, et al., 2009,Lerga, et al., 2001), reduced nuclear localization can possibly impair the essential processes that FUS normally participates in inside the nucleus, providing a possible “loss-of-function” mechanism. At the same time, the cytoplasmic accumulation of mutant FUS leads to the aberrant clustering of TIA-1 and PABP1 positive granules. The alterations in the stress granules can potentially lead to aberrant RNA processing and perturb or likely inhibit RNA translation, providing a possible “gain-of-function” mechanism. A recent study of TDP-43 suggested that both gain and loss of function may contribute to the toxicity of the ALS-related TDP-43 mutants in the cell culture and zebrafish models (Kabashi, et al., 2010). Several TDP-43 animal models have been reported recently (Hanson, et al., 2010,Kabashi, et al., 2010,Li, et al., 2010,Wegorzewska, et al., 2009,Wils, et al., 2010) whereas FUS animal models are yet to be developed. More studies are needed to delineate the loss or gain of function for the FUS mutants and determine how the mutations in FUS cause familial ALS.
Supplementary Material
Acknowledgments
We gratefully thank Dr. Lawrence J. Hayward for wild-type FUS plasmid. We are grateful to Drs. Edward Kasarskis and Qingjun Wang for helpful discussions and reading the manuscript. This work was in part supported by the National Institutes of Health grants R01NS049126 and R21AG032567 to HZ. The support from the Center of Biomedical Research Excellence in the Molecular Basis of Human Disease (COBRE, P20RR020171) funded by National Center for Research Resources is acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Hoglund M, Forster A, Rabbitts TH, Ron D, Mandahl N, Mitelman F. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics. 1996;37(1):1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33(3):141–50. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351(3):602–11. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Baechtold H, Kuroda M, Sok J, Ron D, Lopez BS, Akhmedov AT. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol. Chem. 1999;274(48):34337–42. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- Belzil VV, Valdmanis PN, Dion PA, Daoud H, Kabashi E, Noreau A, Gauthier J, Hince P, Desjarlais A, Bouchard JP, Lacomblez L, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73(15):1176–9. doi: 10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. Embo J. 1996;15(18):5022–31. [PMC free article] [PubMed] [Google Scholar]
- Bertrand P, Akhmedov AT, Delacote F, Durrbach A, Lopez BS. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene. 1999;18(31):4515–21. doi: 10.1038/sj.onc.1203048. [DOI] [PubMed] [Google Scholar]
- Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci U S A. 1997;94(10):5055–60. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–78. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1(5):411–5. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111(4):1051–61. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- Corrado L, Del Bo R, Castellotti B, Ratti A, Cereda C, Penco S, Soraru G, Carlomagno Y, Ghezzi S, Pensato V, Colombrita C, Gagliardi S, Cozzi L, Orsetti V, Mancuso M, Siciliano G, Mazzini L, Comi GP, Gellera C, Ceroni M, D'Alfonso S, Silani V. Mutations of FUS Gene in Sporadic Amyotrophic Lateral Sclerosis. J Med Genet. 2009 doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- Dejesus-Hernandez M, Kocerha J, Finch N, Crook R, Baker M, Desaro P, Johnston A, Rutherford N, Wojtas A, Kennelly K, Wszolek ZK, Graff-Radford N, Boylan K, Rademakers R. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum Mutat. 2010 doi: 10.1002/humu.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15(6):587–93. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- Fujii R, Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci. 2005;118(Pt 24):5755–65. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- Fukada K, Zhang F, Vien A, Cashman NR, Zhu H. Mitochondrial Proteomic Analysis of a Cell Line Model of Familial Amyotrophic Lateral Sclerosis. Mol Cell Proteomics. 2004;3(12):1211–23. doi: 10.1074/mcp.M400094-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, Strom AL, Kwinter DM, Kilty R, Zhang J, Shi P, Fu W, Wooten MW, Zhu H. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111(4):1062–73. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82(3):341–4. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- Gingras M, Gagnon V, Minotti S, Durham HD, Berthod F. Optimized protocols for isolation of primary motor neurons, astrocytes and microglia from embryonic mouse spinal cord. J Neurosci Methods. 2007;163(1):111–8. doi: 10.1016/j.jneumeth.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Hanson KA, Kim SH, Wassarman DA, Tibbetts RS. Ubiquilin modifies toxicity of the 43 kilodalton TAR-DNA binding protein (TDP-43) in a Drosophila model of amyotrophic lateral sclerosis (ALS). J Biol. Chem. 2010 doi: 10.1074/jbc.C109.078527. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Lin L, Tradewell ML, Dion PA, Bercier V, Bourgouin P, Rochefort D, Bel Hadj S, Durham HD, Velde CV, Rouleau GA, Drapeau P. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet. 2010;19(4):671–83. doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40(5):572–4. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311(5981):33–8. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P, Jon L. Methods in Enzymology. Academic Press; 2007. Mammalian Stress Granules and Processing Bodies. pp. 61–81. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13(1):195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169(6):871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147(7):1431–42. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Kim SY, Lee H. Identification and characterization of human cdc7 nuclear retention and export sequences in the context of chromatin binding. J Biol. Chem. 2007;282(41):30029–38. doi: 10.1074/jbc.M703705200. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwong LK, Neumann M, Sampathu DM, Lee VM, Trojanowski JQ. TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol. 2007;114(1):63–70. doi: 10.1007/s00401-007-0226-5. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136(6):1001–4. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126(3):543–58. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerga A, Hallier M, Delva L, Orvain C, Gallais I, Marie J, Moreau-Gachelin F. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol. Chem. 2001;276(9):6807–16. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- Leszczynski JF, Rose GD. Loops in globular proteins: a novel category of secondary structure. Science. 1986;234(4778):849–55. doi: 10.1126/science.3775366. [DOI] [PubMed] [Google Scholar]
- Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, Fushimi K, Wu JY. A Drosophila model for TDP-43 proteinopathy. PNAS. 2010;107(7):3169–74. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao X, Jiang J, Jia J. Fused–Costal2 protein complex regulates Hedgehog-induced Smo phosphorylation and cell-surface accumulation. Genes & Development. 2007;21(15):1949–63. doi: 10.1101/gad.1557407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, Kwong LK, Forman MS, Ravits J, Stewart H, Eisen A, McClusky L, Kretzschmar HA, Monoranu CM, Highley JR, Kirby J, Siddique T, Shaw PJ, Lee VM, Trojanowski JQ. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61(5):427–34. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4(7):223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisse K, Volkening K, Leystra-Lantz C, Welch I, Hill T, Strong MJ. Divergent patterns of cytosolic TDP-43 and neuronal progranulin expression following axotomy: implications for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009;1249:202–11. doi: 10.1016/j.brainres.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134(6):1365–73. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132(Pt 11):2922–31. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Peters R. Translocation through the nuclear pore: Kaps pave the way. Bioessays. 2009;31(4):466–77. doi: 10.1002/bies.200800159. [DOI] [PubMed] [Google Scholar]
- Piao YS, Wakabayashi K, Kakita A, Yamada M, Hayashi S, Morita T, Ikuta F, Oyanagi K, Takahashi H. Neuropathology with clinical correlations of sporadic amyotrophic lateral sclerosis: 102 autopsy cases examined between 1962 and 2000. Brain Pathol. 2003;13(1):10–22. doi: 10.1111/j.1750-3639.2003.tb00002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86(6):985–94. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Prasad DD, Ouchida M, Lee L, Rao VN, Reddy ES. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9(12):3717–29. [PubMed] [Google Scholar]
- Rose GD, Gierasch LM, Smith JA. Turns in peptides and proteins. Adv Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13(12):622–8. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Seelaar H, Klijnsma KY, de Koning I, van der Lugt A, Chiu WZ, Azmani A, Rozemuller AJ, van Swieten JC. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J Neurol. 2009 doi: 10.1007/s00415-009-5404-z. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg G, Stamminger T. Mapping of nuclear localization signals by simultaneous fusion to green fluorescent protein and to beta-galactosidase. Biotechniques. 1999;26(5):858–62. doi: 10.2144/99265bm12. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–72. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AL, Shi P, Zhang F, Gal J, Kilty R, Hayward LJ, Zhu H. Interaction of ALS-related mutant copper-zinc superoxide dismutase with the dynein-dynactin complex contributes to inclusion formation. J Biol. Chem. 2008;283(33):22795–805. doi: 10.1074/jbc.M800276200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong MJ. The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS). J Neurol Sci. 2010;288(1-2):1–12. doi: 10.1016/j.jns.2009.09.029. [DOI] [PubMed] [Google Scholar]
- Tan AY, Manley JL. TLS Inhibits RNA Polymerase III Transcription. Mol Cell Biol. 2010;30(1):186–96. doi: 10.1128/MCB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Gupta R, Mayer BJ. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol. 1995;15(12):6829–37. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, Steinbart E, McCluskey L, Grossman M, Neumann M, Wu IL, Yang WS, Kalb R, Galasko DR, Montine TJ, Trojanowski JQ, Lee VM, Schellenberg GD, Yu CE. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7(5):409–16. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkening K, Leystra-Lantz C, Yang W, Jaffee H, Strong MJ. Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res. 2009;1305:168–82. doi: 10.1016/j.brainres.2009.09.105. [DOI] [PubMed] [Google Scholar]
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. PNAS. 2009;106(44):18809–14. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, Cuijt I, Smits V, Ceuterick-de Groote C, Van Broeckhoven C, Kumar-Singh S. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. PNAS. 2010;107(8):3858–63. doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serinearginine proteins involved in RNA splicing. J Biol. Chem. 1998;273(43):27761–4. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- Zakaryan RP, Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J Mol Biol. 2006;363(1):27–38. doi: 10.1016/j.jmb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Zhang F, Strom AL, Fukada K, Lee S, Hayward LJ, Zhu H. Interaction between familial amyotrophic lateral sclerosis (ALS)-linked SOD1 mutants and the dynein complex. J Biol. Chem. 2007;282(22):16691–9. doi: 10.1074/jbc.M609743200. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110(Pt 15):1741–50. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.