Abstract
Gα12/13-mediated pathways have been shown to be involved in various fundamental cellular functions in mammalian cells such as axonal guidance, apoptosis, and chemotaxis. Here, we identified a homologue of Rho-guanine nucleotide exchange factor (GEF) in Caenorhabditis elegans (CeRhoGEF), which functions downstream of gpa-12, the C. elegans homologue of Gα12/13. CeRhoGEF contains a PSD-95/Dlg/ZO-1 domain and a regulator of G protein signaling (RGS) domain upstream of the Dbl homology–pleckstrin homology region similar to mammalian RhoGEFs with RGS domains, PSD-95/Dlg/ZO-1–RhoGEF and leukemia-associated RhoGEF. It has been shown in mammalian cells that these RhoGEFs interact with activated forms of Gα12 or Gα13 through their RGS domains. We demonstrated by coimmunoprecipitation that the RGS domain of CeRhoGEF interacts with GPA-12 in an 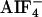 activation-dependent manner and confirmed that the Dbl homology–pleckstrin homology domain of CeRhoGEF was capable of Rho-dependent signaling. These results proved conservation of the Gα12–RhoGEF pathway in C. elegans. Expression of DsRed or GFP under the control of the promoter of CeRhoGEF or gpa-12 revealed an overlap of their expression patterns in ventral cord motor neurons and several neurons in the head. RNA-mediated gene interference for CeRhoGEF and gpa-12 resulted in similar phenotypes such as embryonic lethality and sensory and locomotive defects in adults. Thus, the Gα12/13–RhoGEF pathway is likely to be involved in embryonic development and neuronal function in C. elegans.
activation-dependent manner and confirmed that the Dbl homology–pleckstrin homology domain of CeRhoGEF was capable of Rho-dependent signaling. These results proved conservation of the Gα12–RhoGEF pathway in C. elegans. Expression of DsRed or GFP under the control of the promoter of CeRhoGEF or gpa-12 revealed an overlap of their expression patterns in ventral cord motor neurons and several neurons in the head. RNA-mediated gene interference for CeRhoGEF and gpa-12 resulted in similar phenotypes such as embryonic lethality and sensory and locomotive defects in adults. Thus, the Gα12/13–RhoGEF pathway is likely to be involved in embryonic development and neuronal function in C. elegans.
Members of the Rho family GTPases (Rho, Rac, and Cdc42) regulate a variety of cellular activities by controlling actin cytoskeletal rearrangements or gene expression (1). Activation of Rho family GTPases is catalyzed by a large number of guanine nucleotide exchange factors (GEFs). These GEFs share a Dbl homology (DH) domain, which is responsible for GDP–GTP exchange activity toward Rho GTPases, and an adjacent pleckstrin homology (PH) domain (2, 3). Except for this DH–PH structure, GEFs contain various different protein motifs that are implicated in signal transduction. In addition, mutants of several GEFs have been isolated as oncogenes or disease-related genes. Thus, it is expected that the activation of Rho family GTPases by these GEFs is tightly regulated through a variety of signaling mechanisms.
Heterotrimeric G proteins G12 and G13 have been shown to mediate signals from G protein-coupled receptors such as thrombin receptor or lysophosphatidic acid receptor to RhoGTPase activation (4–6). Possible involvement of the Gα12/13-mediated pathway in physiological processes such as tumorgenesis, angiogenesis, or neurite retraction has been reported (7–9). We recently identified that p115RhoGEF, one of the GEFs for Rho, acts as a direct link between heterotrimeric G13 and Rho activation (10, 11). p115RhoGEF contains a regulator of G protein signaling (RGS) domain at its N-terminal region that specifically interacts with the activated form of Gα13 or Gα12. It was demonstrated that activated Gα13 stimulated RhoGEF activity of p115 through the interaction with this RGS domain. Recently, two other mammalian RhoGEFs with the RGS domain (RGS-RhoGEFs), PSD-95/Dlg/ZO-1 (PDZ)-RhoGEF, and leukemia-associated RhoGEF (LARG), have been shown to mediate Rho activation through Gα12/13 (12–14).
In Drosophila, DRhoGEF2, a putative Rho exchange factor, was identified as a critical signaling component for gastrulation (15, 16). Embryos lacking DRhoGEF2 failed to gastrulate because of an inability of cells to undergo shape changes required for tissue invagination. Expression of a dominant-negative Rho in early embryos caused a similar defect. In addition, the gastrulation defects seen in embryos lacking DRhoGEF2 closely resemble those seen in embryos lacking an extracellular ligand, folded gastrulation (Fog), or the Drosophila Gα12/13 homologue concertina (Cta) (17, 18). Further genetic studies led to the hypothetical model that the signal from Fog activated Cta and that Cta stimulated the exchange activity of DRhoGEF2 for RhoGTPase (15, 16). Furthermore, an RGS domain that is highly homologous to that of mammalian RGS–RhoGEFs was identified at the N terminus of the DH–PH domain in DRhoGEF2 (10). These results led to the conclusion that the G12/13–RhoGEF–Rho pathway plays a critical role in early embryogenesis in Drosophila and has been highly conserved during evolution.
Caenorhabditis elegans is an excellent model organism for studying the function of genes at the animal level. In particular, G protein-mediated signaling pathways such as Go- or Gq-mediated pathways have been well characterized in C. elegans (19, 20). In addition, analysis of Gα12 expression in C. elegans (21) and its signaling via tpa-1, a homologue of protein kinase C in pharyngeal muscle cells, have recently been reported (22). However, the Gα12–RGS-RhoGEF signaling pathway has not yet been identified. In this study, we identified a homologue of mammalian RGS-RhoGEF in C. elegans and characterized its involvement in Gα12-mediated signaling pathway.
Methods
Nematode Strains and Culture. WT Bristol nematode strain N2 was obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis). Nematode strain rrf-3 (pk1426) (23), which is hypersensitive to RNA-mediated genetic interference (RNAi), was obtained from Shin Takagi (Nagoya University Graduate School of Science, Nagoya, Japan). The worms were maintained at 20°C on NGM plates seeded with Escherichia coli OP50 according to standard methods (24).
Cloning of CeRhoGEF and gpa-12 cDNAs. A 3.8-kb fragment encoding for CeRhoGEF (F13E6.6) was amplified from the C. elegans cDNA library by using the set of primers of F13E6-D1F (5′-TAGTAGTTCAACGACAACCAGATG-3′) and F13E6-D4R (5′-TCGGAACGATTTTTCGATCT-3′). To avoid mutations during PCR, the C. elegans cDNA library was screened with this 3.8-kb DNA fragment as a probe. Several positive clones were isolated, and the full-length CeRhoGEF cDNA was constructed. Gpa-12 (F18G5.3) was PCR-amplified from the C. elegans cDNA library by using the set of primers of F18G5-F (5′-TCAACATGGTATGCTGTTTCG-3′) and F18G5-R (5′-GGAAACATTTGAGCAACAACAA3′). The PCRs were performed with a Supermix High Fidelity kit (GIBCO) as follows: (i)94°C 2 min; (ii)94°C30s,58°C30s,68°C 5 min, 35 times; and (iii) 68°C 5 min. The PCR products were subcloned into pCR4-TOPO (Invitrogen). The nucleotide sequence of CeRhoGEF or gpa-12 was identical to the predicted sequence for the respective gene.
Construction of Expression Plasmids. The RGS domain of CeRhoGEF (CeRGS-RhoGEF, 670 bp, amino acids 181–403) was amplified from CeRhoGEF cDNA by PCR using the HindIII-CeRGS-F forward primer (5′-ATAAAGCTTATGGACATTGATAGTGACGAAGAG-3′) and CeRev2-R reverse primer (5′-TAATCTAGAACTCGTTAGACGACGACGATG-3′). The product was subcloned into pCR4-TOPO. CeRGS-RhoGEF/pCR4-TOPO was digested with HindIII and XbaI and subcloned into 3× flag-pCMV7 (Sigma) (3×flag-CeRGS-RhoGEF). For the expression vector for the DH–PH domain of CeRhoGEF, CeRhoGEF/pBS II KS(–) was digested with BglII and EcoRI and subcloned into myc-pcDNA at the BamHI and EcoRI sites (myc-CeRhoGEF DH–PH). Gpa-12/pBSII KS(–) cDNA (1.1 kb) was digested with EcoRI and subcloned into myc-pcDNA (myc-gpa-12). The DH–PH domain of LARG in myc-pcDNA (myc-LARG DH–PH) was kindly provided by Nobuchika Suzuki (University of Illinois, Chicago). SRE.L-luciferase reporter plasmid was kindly provided by Paul C. Sternweis (University of Texas Southwestern Medical Center, Dallas).
Cell Culture. COS-7 cells were maintained in DMEM (Invitrogen) supplemented with 10% heat-inactivated FBS (Invitrogen). Transfections were performed with Lipofectamine 2000 (Life Technologies, Gaithersburg, MD).
Coimmunoprecipitation. COS-7 cells (3.3 × 106) were plated onto 100-mm dishes 1 day before transfection. Cells were cotransfected with myc-gpa-12 (15 μg) or myc-pcDNA (15 μg) and flag-CeRGS-RhoGEF (5 μg) in OPTI-MEM (Invitrogen). After 4 h, FBS was supplemented into the culture to a final concentration of 10%. Cells were harvested after 20 h. Immunoprecipitation by myc antibody in the presence or absence of 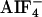 was performed as described (14). The immunoprecipitates were loaded onto 10% SDS/polyacrylamide gel for electrophoresis and immunoblotted with anti-myc antibody (Sigma) or anti-flag M2 antibody (Sigma). Immunoblots were developed by using an ECL+plus Western blotting detection kit (Amersham Pharmacia Biosciences).
was performed as described (14). The immunoprecipitates were loaded onto 10% SDS/polyacrylamide gel for electrophoresis and immunoblotted with anti-myc antibody (Sigma) or anti-flag M2 antibody (Sigma). Immunoblots were developed by using an ECL+plus Western blotting detection kit (Amersham Pharmacia Biosciences).
Serum Response Element (SRE)-Luciferase Assay. COS-7 cells (3 × 105) were plated onto six-well plates 1 day before transfection. Cells were cotransfected with SRE.L-luciferase reporter plasmid (0.5 μg), pCMV-β-galactosidase (0.5 μg), and the indicated constructs. Total amounts of transfected DNA were kept constant among wells with the supplementation of empty vector DNA. β-Galactosidase activities of cell lysates were used to normalize the transfection efficiency. pEGFP-C3 expression vector for C3 exoenzyme was kindly provided by Shuh Narumiya (Kyoto University, Kyoto). Luciferase activities in cell extracts were measured 24 h after transfection according to the manufacturer's instruction (Promega).
Construction of Promoter-GFP Reporter Plasmids. GFP reporter plasmids were constructed by inserting genomic DNA fragments from CeRhoGEF (F13E6.6) or gpa-12 (F18G5.3) into pFXneo-EGFP (enhanced GFP) vector or pPD122.22 nuclear localization signal (NLS) GFP vector (a gift from Shohei Mitani, Tokyo Women's Medical University, Tokyo, or Andrew Fire, Carnegie Institute of Washington, Baltimore, respectively). The NLS-containing vector (pPD122.22 NLS GFP) was used to assist in the identification of cells expressing GFP. CeRhoGEFp::EGFP was constructed by subcloning a 6.5-kb genomic fragment that contained the promoter region of CeRhoGEF and exon 1 to part of exon 10 (BamHI–XhoI) at the BamHI–SalI sites of pFXne-EGFP. CeRhoGEFp::NLS::GFP was constructed by subcloning a 2.7-kb genomic fragment that contained the promoter region of CeRhoGEF and exon 1 to part of exon 2 into NLS::GFP at the XmaI–EcoRV sites. Gpa-12p::EGFP and Gpa-12p::NLS::GFP were constructed by subcloning a 3.7-kb genomic fragment that contained the promoter region of gpa-12 including exon 1 to part of exon 2 into BamHI site of pFXneEGFP vector and pPD 122.22 NLS GFP, respectively. Each GFP construct (200 ng/μl) was mixed with pRF4 (200 ng/μl) containing rol-6 (su1006) and injected into N2 worms. F1 progeny exhibiting the rolling phenotype induced by rol-6 were isolated for observation (25). The staining with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine (DiI) dye (0.1 mg/ml, Molecular Probes) was performed as described (26). For colocalization studies, CeRhoGEFp::DsRed was constructed by subcloning the previously described 6.5-kb genomic fragment of CeRhoGEF (BamHI–XhoI) into the BamHI–SalI sites of pFXneDsRedXT (a gift from Shohei Mitani). CeRhoGEFp::DsRed (100 ng/μl) and Gpa-12p::NLS::GFP (100 ng/μl) were injected into N2 worms along with pRF4 (100 ng/μl). The F1 progeny were analyzed.
RNAi. CeRhoGEF (1.4-kb C-terminal region, amino acids 807-1293) or gpa-12 (1.1 kb full length) in pBS KS(–) was used as a template for in vitro RNA synthesis with T7 or T3 RNA polymerase by using a MAXIscript T7/T3 kit (Ambion). PpD 79.44 gfp vector (from 1995 Fire Vector Kit, a gift from Peter Okkema, University of Illinois, Chicago) was used as a control template for RNAi. RNA was purified and dissolved in TE (10 mM Tris·HCl, pH 8.0/1 mM EDTA). Equal amounts of sense and antisense RNA were mixed, denatured (10 min at 68°C), and annealed (30 min at 37°C) to prepare double-stranded RNA (dsRNA) with a concentration between 1.0 and 1.3 μg/μl. dsRNA was mixed with 10× injection buffer [20% polyethylene glycol (MW 6,000–8,000), 200 mM potassium phosphate (pH 7.5), and 30 mM potassium citrate (pH 7.5)] to make a final concentration of 0.5–0.8 μg/μl in 1× injection buffer. The dsRNA was injected into the gonad of WT N2 or rrf-3 hermaphrodites, and the phenotypes of the F1 progeny were examined 3 days postinjection.
Results
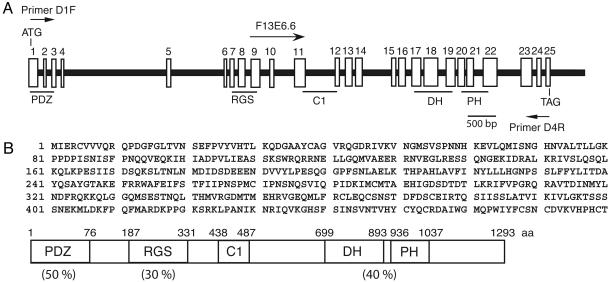
Molecular Cloning of CeRhoGEF cDNA. Based on the conservation of the G12/13–RhoGEF pathway in mammalian cells and Drosophila, we predicted that a similar signal transduction pathway might also exist in C. elegans. Thus, we carried out a blast search to look for the homologue of RGS-RhoGEF in the C. elegans genome by using the DH–PH domain of p115RhoGEF. F13E6.6 was identified to contain the DH–PH domain that is most homologous to that of p115RhoGEF (40% identity in amino acid sequence). The database predicted that F13E6.6 cDNA encodes a protein with 1,006 amino acid residues with 17 exons. Although, we could not detect PDZ or RGS domains at the N-terminal region of the DH–PH domain in F13E6.6, further examination of the nucleotide sequence of the 5′ region of the genome revealed eight additional exons that could encode PDZ and RGS domains (Fig. 1A). To confirm the existence of a cDNA containing PDZ, RGS, and DH–PH domains, we synthesized primers D1F (5′-TAG TAG TTC AAC GAC AAC CAG ATG–3′) and D4R (5′-TCG GAA CCA TTT TTC GAT CT–3′), which correspond to the 5′ and 3′ ends of the predicted sequence of the RhoGEF cDNA, respectively. PCR analysis of the C. elegans cDNA library using D1F and D4R generated a 3.8-kb product that matched the size of the predicted cDNA. Using this 3.8-kb fragment as a probe, the C. elegans cDNA library was screened to obtain the full-length cDNA. This cDNA encodes a protein with 1,293 aa containing PDZ, RGS, and DH–PH domains. These domains share 35–50% amino acid sequence identity to the corresponding domains of LARG (Fig. 1B). We thus considered this cDNA as a homologue of mammalian RGS-RhoGEF, and it will be referred to as CeRhoGEF. Like DRhoGEF2, CeRhoGEF contains a C1 domain, a putative phorbol ester/diacylglycerol binding site between the RGS and DH–PH domains.
Fig. 1.
Molecular cloning of CeRhoGEF.(A) The structure of the CeRhoGEF gene is schematically represented. White boxes with numbers indicate exons. The F13E6.6 cDNA starting site and D1F and D4R primer positions are indicated. Exons encoding PDZ, RGS, C1, or the DH–PH domain are underlined. (B) The amino acid sequence of the amino-terminal region of CeRhoGEF (amino acids 1–480) and the domain structure of full-length CeRhoGEF are shown. Amino acid identity of each region of CeRhoGEF with the corresponding domain of LARG is indicated in parentheses.
The C. elegans homologue of Gα12 (gpa-12) has already been identified as a product of F18G5.3 (21, 22). GPA-12 is 50% identical in amino acid sequence to the mammalian Gα12. In particular, the sequences in three switch regions of GPA-12 are most homologous to those of mammalian Gα12 or Gα13 (data not shown). In contrast to mammalian cells, only one member of the G12 subfamily exists in C. elegans.
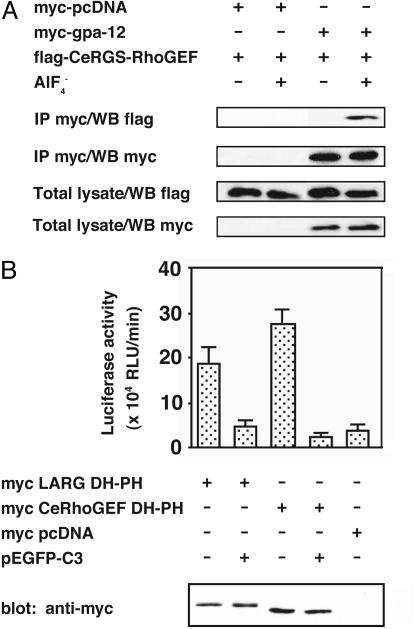
The Biochemical Interaction of the GPA-12–CeRhoGEF Pathway. It has been shown in mammalian cells that three RhoGEFs with a RGS domain (RGS-RhoGEFs) specifically interacted with active forms of Gα12 or Gα13 through their RGS domains (10, 12, 13). To analyze the interaction between the RGS domain of CeRhoGEF and GPA-12, we coexpressed them in COS-7 cells and examined their interaction by immunoprecipitation. COS-7 cells were cotransfected with myc-tagged gpa-12 and the flag-tagged RGS domains of CeRhoGEF. The cell lysates were incubated with or without 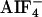 , a reversible activator of the Gα subunit. Then, GPA-12 was immunoprecipitated with anti-myc antibody from the lysates. Fig. 2A shows that the RGS domain of CeRhoGEF immunoprecipitated with GPA-12 only in the presence of
, a reversible activator of the Gα subunit. Then, GPA-12 was immunoprecipitated with anti-myc antibody from the lysates. Fig. 2A shows that the RGS domain of CeRhoGEF immunoprecipitated with GPA-12 only in the presence of 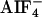 , suggesting that CeRhoGEF specifically interacts with the active form of GPA-12 through its RGS domain.
, suggesting that CeRhoGEF specifically interacts with the active form of GPA-12 through its RGS domain.
Fig. 2.
Biochemical interaction of the GPA-12–CeRhoGEF pathway. (A) CeRGS-RhoGEF interacts with activated GPA-12 in the presence of 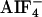 . Myc-tagged gpa-12 was cotransfected with the flag-tagged RGS domain of CeRhoGEF (CeRGS-RhoGEF) in COS-7 cells. GPA-12 was immunoprecipitated (IP) with anti-myc antibody from the cell lysates in the presence or absence of
. Myc-tagged gpa-12 was cotransfected with the flag-tagged RGS domain of CeRhoGEF (CeRGS-RhoGEF) in COS-7 cells. GPA-12 was immunoprecipitated (IP) with anti-myc antibody from the cell lysates in the presence or absence of 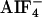 . The immunoprecipitates were separated on SDS/PAGE and immunoblotted with anti-flag (Top) or anti-myc (second panel from Top) antibody. Expression of gpa-12 (Bottom, with anti-myc antibody) or CeRGS-RhoGEF (second panel from Bottom, with anti-flag antibody) in lysates is shown. (B) Stimulation of SRF activity by the DH–PH domain of CeRhoGEF. COS-7 cells were transfected with SRE.L-luciferase reporter (0.5 μg), pCMV-β-galactosidase (0.5 μg), and the indicated expression plasmids, myc-LARG DH–PH (0.5 μg), myc-CeRhoGEF DH–PH (6 μg), or pEGFP-C3 (0.5 μg). SRF activities of cell lysates were measured as described in Methods. The expression of LARG DH–PH or CeRhoGEF DH–PH in lysates was detected by immunoblotting with anti-myc antibody (Bottom).
. The immunoprecipitates were separated on SDS/PAGE and immunoblotted with anti-flag (Top) or anti-myc (second panel from Top) antibody. Expression of gpa-12 (Bottom, with anti-myc antibody) or CeRGS-RhoGEF (second panel from Bottom, with anti-flag antibody) in lysates is shown. (B) Stimulation of SRF activity by the DH–PH domain of CeRhoGEF. COS-7 cells were transfected with SRE.L-luciferase reporter (0.5 μg), pCMV-β-galactosidase (0.5 μg), and the indicated expression plasmids, myc-LARG DH–PH (0.5 μg), myc-CeRhoGEF DH–PH (6 μg), or pEGFP-C3 (0.5 μg). SRF activities of cell lysates were measured as described in Methods. The expression of LARG DH–PH or CeRhoGEF DH–PH in lysates was detected by immunoblotting with anti-myc antibody (Bottom).
We then performed SRE-luciferase reporter assay to examine whether CeRhoGEF can activate Rho through its DH–PH domain. As previously shown, Rho activation in cells can be monitored by serum response factor (SRF) activity with the SRE-luciferase reporter system (27). The expression of the DH–PH domain of CeRhoGEF in COS-7 cells stimulated SRF activation to a similar degree as the DH–PH domain of LARG (Fig. 2B). This SRF activation was almost completely inhibited by coexpression of C3 botulinus toxin, which specifically inactivates Rho by ADP ribosylation. These data indicate that CeRhoGEF can activate Rho through its DH–PH domain in mammalian cells. Thus, it is highly likely that CeRhoGEF functions as RhoGEF in C. elegans. Taken together, the results shown here support the existence of the conserved Gα12–RGS-RhoGEF pathway in C. elegans.
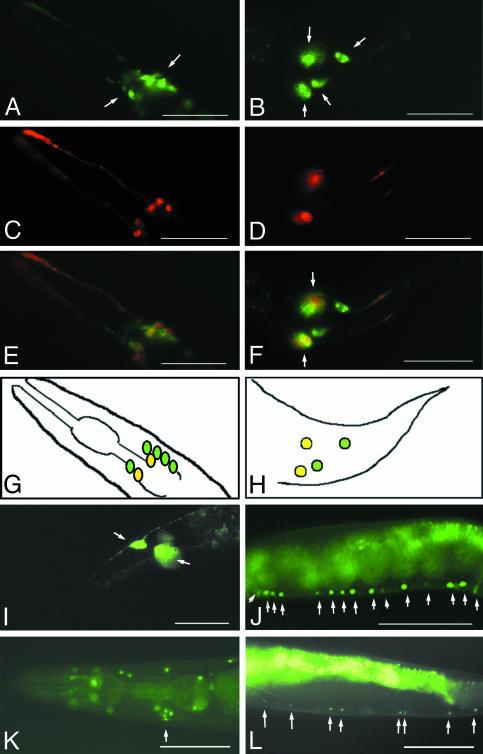
Expression of CeRhoGEF and gpa-12. We determined the expression patterns of CeRhoGEF and gpa-12 in vivo by using promoter-gfp fusion constructs as described in Methods. The CeRhoGEF expression was observed primarily in neurons, including several neurons in the head (Fig. 3A) and the bilaterally symmetrical lumbar ganglia in the tail (Fig. 3B). Identity of the subset of chemosensory neurons was further confirmed by double labeling with DiI staining of amphid and phasmid neurons (26) (Fig. 3 C and D). Overlaying the GFP and DiI images of the head and tail revealed their colocalization among the set of amphid and phasmid neurons (Fig. 3 E and F, shown in yellow-orange). A diagram of colocalization is displayed in Fig. 3 G and H. CeRhoGEF expression was also observed in several additional neurons in the head and tail (Fig. 3 E and F, shown in green). Based on cell position and pattern of bilaterally directed processes, we found that PLML and PLMR touch neurons also express GFP in the lumbar ganglia (Fig. 3I). CeRhoGEF expression was also observed in the motor neurons of the ventral nerve cord (Fig. 3J).
Fig. 3.
Expression of CeRhoGEF or Gpa-12 in C. elegans. (A) Expression of CeRhoGEFp::EGFP in the neurons in the head (white arrows). (B) Expression of CeRhoGEFp::EGFP in lumbar ganglia in the tail (white arrows). (C and D) DiI staining (shown in red) of amphid neurons in the head (C) or phasmid neurons in the tail (D). (E and F) GFP and DiI merged images show colocalization of CeRhoGEFp::EGFP at the amphid neurons in the head (E) or the phasmid neurons in the tail (F). (G and H) Diagram of CeRhoGEFp::EGFP expression in the head region (G) or tail region (H). (I) CeRhoGEFp::EGFP expression in the PLML and PLMR touch neurons (white arrows). (J) CeRhoGEFp::NLS::GFP expression in the ventral cord motor neurons (white arrows). (K) Possible expression in a cluster of neurons in the head (white arrow). (L) Gpa-12p::NLS::GFP expression in the ventral cord motor neurons (white arrows). (Scale bar: 100 μm.)
In contrast to CeRhoGEF, which shows GFP expression primarily in neural tissues, gpa-12p::EGFP expression was observed in the hypodermis, muscle tissue, intestinal cells, and pharynx (Fig. 6 A and C, which is published as supporting information on the PNAS web site) throughout the entire development of the animal, which is consistent with previously reported results (21, 22). In addition, we observed a weak GFP expression in a set of neurons in the tail and the primary excretory H cell (Fig. 6 B and D). We used another GFP construct that included the NLS to facilitate the identification of cells expressing gpa-12. Expression of gpa-12p::NLS::GFP was observed in the hypodermal nuclei, which can be seen in the head region, along the entire body length, and in the tail region (Fig. 6 E and F). In addition, we observed GFP expression in several neurons in the head (Fig. 3K) and in a subset of ventral cord motor neurons (Fig. 3L).
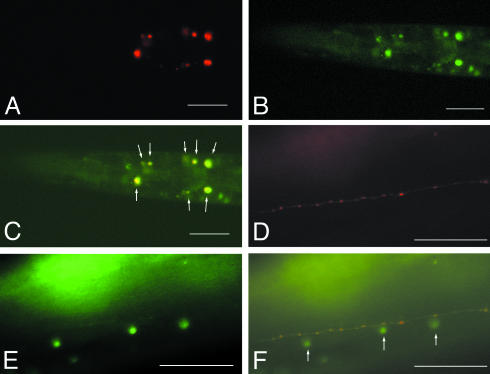
Coexpression of CeRhoGEF and gpa-12 was further confirmed by using CeRhoGEFp::DsRed and gpa-12p::NLS::GFP constructs. As shown in Fig. 4 A–C, the overlap of their expression was detected in a subset of neurons in the head, which could be the interneurons at the second pharyngeal bulb and the sensory neurons in the nerve ring area. In addition, the expression of CeRhoGEFp::DsRed or gpa-12p::NLS::GFP was observed in neuronal processes or nuclei of ventral cord motor neurons, respectively (Fig. 4 D–F).
Fig. 4.
Coexpression of CeRhoGEF and gpa-12 in C. elegans. Transgenic animals carrying CeRhoGEF-DsRed and gpa-12-NLS GFP were examined under a fluorescence microscope. (A) Expression of CeRhoGEFp::DsRed in neurons in the head. (B) Expression of gpa-12p::NLS GFP in hypodermal cells and in neurons in the head. (C) Merged image of A and B shows their coexpression in a subset of neurons in the head (white arrows). (D) Expression of CeRhoGEFp::DsRed in neuronal processes of ventral cord motor neurons. (E) Expression of gpa-12p::NLS GFP in nuclei of ventral cord motor neurons. (F) Merged image of E and F shows their coexpression in ventral cord motor neurons. (Scale bar: 50 μm.)
RNAi of CeRhoGEF and gpa-12. The role of CeRhoGEF and gpa-12 in development was investigated by RNAi (28). dsRNA of CeRhoGEF or gpa-12 was injected into rrf-3 young adult hermaphrodites as described in Methods. Table 1 summarizes the results. RNAi of CeRhoGEF resulted in only a marginal reduction in the fecundity of the treated animals. However, most of these eggs were not viable, suggesting that CeRhoGEF plays a critical role in embryonic development. The egg laying defect in the F1 hermaphrodites may be caused by abnormal vulva development, hypodermal defect, or neural function. CeRhoGEF RNAi also affected coordinated locomotion and touch sensitivity to a varying degree. For the progeny that did display locomotion defect or vulva defect, they were also touch insensitive.
Table 1. Characterization of CeRhoGEF and gpa-12 (RNAi) phenotypes.
| Gene | Brood size | RNAi phenotype |
|---|---|---|
| Control (injection buffer) | 73 ± 24 | No abnormal phenotype |
| Control (gfp) | 64 ± 28 | No abnormal phenotype |
| CeRhoGEF | 12 ± 9 | Egg laying defect, embryonic arrest, touch insensitivity, locomotory defect |
| gpa-12 | 6 ± 3 | Low egg production, egg laying defect, embryonic arrest, touch insensitivity, locomotory defect |
dsRNA of CeRhoGEF, gpa-12, or gfp (mock) was injected into rrf-3 young adult and N2 (data not shown) hermaphrodites. The progeny count and phenotypes were recorded 3 days postinjection.
Similar RNAi analysis was performed for gpa-12. Compared with CeRhoGEF dsRNA, gpa-12 dsRNA caused more severe defects in embryogenesis and oogenesis. There was a low egg production in the gpa-12 dsRNA-treated worms, with the observation of a few to none seen inside the gonad arms. Many of the eggs that were laid were developmentally arrested. The average number of surviving progeny was significantly lower than that of control (Table 1), which suggests that gpa-12 also plays a critical role in embryonic development. The progeny that survived embryogenesis demonstrated a variable degree of expression of abnormal phenotypes in locomotion or touch sensitivity. These phenotypes were similar to animals injected with CeRhoGEF dsRNA.
Discussion
In this study, we identified and characterized the function of CeRhoGEF, the RGS-RhoGEF homologue in C. elegans. CeRhoGEF is a unique RhoGEF that contains both PDZ and RGS domains upstream of the DH–PH domain with structural similarity to the previously characterized RGS-RhoGEFs, LARG, PDZ-RhoGEF, and DRhoGEF2. All of these RGS-RhoGEFs have been shown to function as downstream effectors for Gα12/13 in mammalian cells or Drosophila. From biochemical analysis, we found that CeRhoGEF specifically bound to the activated form of GPA-12 through its RGS domain and that it activated Rho through its DH–PH domain in COS-7 cells. Coexpression of gpa-12 and CeRhoGEF in a subset of neurons and several similar phenotypes induced by RNAi of these two genes were also observed. These results suggest that the CeRhoGEF function as a downstream target for Gα12 in C. elegans, especially in the nervous system (Fig. 5).
Fig. 5.
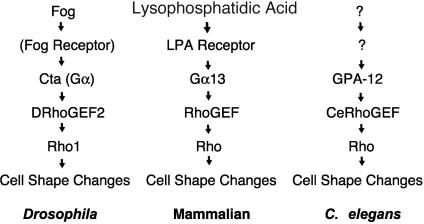
The GPA12–CeRhoGEF pathway in C. elegans shows conservation of the G12/13 pathway with previously described Fog-induced Rho1 activation in Drosophila and lysophosphatidic acid (LPA)-induced Rho activation in mammals.
The Gα12–RhoGEF pathway has been reported to be critically important in embryonic development. In Drosophila, defects in gastrulation associated with the invagination of the mesoderm and midgut were observed by genetically deleting the components of this pathway (15, 16). In mice, Gα13 deficiency resulted in embryonic lethality at embryonic day (E) 10, which was apparently caused by impaired angiogenesis in the yolk sac and embryo. Although, Gα12 simple knockout mice were viable and did not show any obvious abnormal phenotypes, double knockout mice of Gα12 and Gα13 died between E8 and E8.5. Embryos carrying one Gα12 allele with Gα13 null survived until E9, whereas those that had one Gα12 and one Gα13 alleles were viable. These genetic studies revealed that both an overlapping and a distinct function of Gα12- and Gα13-mediated signaling exists in mouse embryonic development (8, 29). The Gα12–RhoGEF–Rho pathway in C. elegans is also critical in embryonic development. From our RNAi studies, loss of function of CeRhoGEF or gpa-12 resulted in embryonic arrest with a significantly reduced brood size, as compared with the controls. Further genetic analysis of CeRhoGEF and gpa-12 in embryos will define the critical role of this pathway in early development.
In adult animals, coexpression of CeRhoGEF and gpa-12 was observed in motor neurons in the ventral cord and several neurons in the head. In agreement with this finding, the expression of gpa-12 in precursor cells of motor neurons in the ventral cord was indicated in young larva (22). The locomotive defects induced by RNAi of gpa-12 or CeRhoGEF may be the result of impaired signaling of the GPA-12–CeRhoGEF pathway in ventral cord motor neurons. The uncoordinated movement and abnormal touch response was also observed for RNAi of both genes, suggesting an overlapping sensory function. Thus, in adult animals, the GPA-12–CeRhoGEF pathway seems to function mainly in neuronal cells. Because Gα12–RhoGEF signaling will likely regulate cytoskeletal organization of cells, this pathway may be responsible for the establishment of neuronal circuits in C. elegans (30). The involvement of Rho protein in neuronal function in C. elegans has previously been indicated. Expression of RhoA, the only Rho gene in C. elegans, was enriched in the nerve ring and in the chemosensory and mechanosensory neurons in the head during larval development (31). This expression pattern of RhoA coincides with that of its upstream regulator CeRhoGEF or gpa-12. More precise identification of gpa-12 or CeRhoGEF expressing neurons and the ligands for this pathway will be crucial for understanding the physiological roles of the GPA-12–CeRhoGEF pathway in the nervous system.
Expression analysis using the promoter-GFP fusion construct revealed that gpa-12 is highly expressed in the hypodermis, pharynx, and muscle cells. The result agrees well with recent observations (21, 22). Because CeRhoGEF is not expressed in these cells, the existence of another effector molecule for GPA-12 is suggested. In mammalian cells, multiple target molecules for Gα12 or Gα13 different from RGS-RhoGEFs, such as radixin, AKAP110, or cadherin were isolated (32–34). It is possible that a homologue of one of these target molecules or other forms of RGS-RhoGEF in C. elegans may function downstream of GPA-12 in the hypodermis or muscle cells.
TPA-1, a protein kinase C homologue in C. elegans, was recently identified as a possible downstream component of gpa-12-mediated signaling in pharyngeal muscle cells (22). However, whether GPA-12 directly interacts with TPA-1 is unknown. It will be important to examine the functional interaction between the GPA-12–CeRhoGEF pathway and TPA-1 activation. Because TPA-1 can be activated by the Gαq–PLC pathway, the data also suggest a possible connection between the Gα12 and Gαq pathways in vivo. The network formation between different G protein-mediated pathways in vivo, such as the Gq and Go pathways, has previously been reported in C. elegans (35, 36).
Several lines of evidence have been presented to indicate that RGS-RhoGEFs in mammalian cells receive multiple signals from other pathways in addition to the input from Gα12/13. For example, nonreceptor tyrosine kinases, such as Tec, Btk, or Pyk2, have been shown to participate in Gα12/13-mediated signaling (14, 37–39). Indeed, LARG or PDZ-RhoGEF could serve as substrates for tyrosine phosphorylation by FAK or Tec kinase. It was further demonstrated that tyrosine phosphorylation of LARG by Tec was required for Gα12 to activate RhoGEF activity of LARG in vitro (14). Thus, Gα12/13-mediated signaling and tyrosine kinase signaling can be integrated at the level of RGS-RhoGEFs. It has also been reported recently that the interaction of the cytoplasmic region of plexin B with the PDZ domain of LARG or PDZRhoGEF could mediate Rho activation induced by semaphorin 4D, which is a ligand for plexin B (40, 41). The GPA-12–CeRhoGEF pathway in C. elegans identified in this study will become a valuable model system for analyzing the in vivo effect of this signaling network on RGS-RhoGEF.
The Gα12/13–RhoGEF pathway was recently identified as a unique G protein-mediated signaling pathway. Unlike other G protein-mediated pathways, this signaling mechanism does not change the concentration of soluble second messengers or ions. In contrast, it directly regulates the activity of low molecular weight GTPase, Rho, resulting in actin-cytoskeletal changes. The involvement of this pathway in cellular processes such as cell growth, cell shape changes, or cell migration has been demonstrated. The elucidation of the physiological roles of this G protein pathway in vivo by using the C. elegans system will provide critical information for understanding these fundamental cellular processes.
Supplementary Material
Acknowledgments
We thank Drs. S. Mitani, P. Okkema, N. Suzuki, and Shin Takagi for plasmids and technical assistance. We thank T. Stiernagle and the Caenorhabditis Genetics Center for providing nematode strains. This work was supported in part by the National Institutes of Health (T.K.) and the American Heart Association (T.K.). T.K. is an Established Investigator of the American Heart Association. S.S.S. thanks Dr. A. B. Malik for support. D.M.Y. was supported by a training grant from the National Institutes of Health.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RGS, regulator of G protein signaling; GEF, guanine nucleotide exchange factor; PDZ, PSD-95/Dlg/ZO-1; DH, Dbl homology; PH, pleckstrin homology; LARG, leukemia-associated RhoGEF; RNAi, RNA-mediated genetic interference; SRE, serum response element; SRF, serum response factor; NLS, nuclear localization signal; dsRNA, double-stranded RNA; DiI, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine; EGFP, enhanced GFP.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY436362).
References
- 1.Hall, A. (1998) Science 279, 509–514. [DOI] [PubMed] [Google Scholar]
- 2.Whitehead, I. P., Campbell, S., Rossman, K. L. & Der, C. J. (1997) Biochim. Biophys. Acta 1332, F1–F23. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt, A. & Hall, A. (2002) Genes Dev. 16, 1587–1609. [DOI] [PubMed] [Google Scholar]
- 4.Gohla, A., Harhammer, R. & Schultz, G. (1998) J. Biol. Chem. 273, 4653–4659. [DOI] [PubMed] [Google Scholar]
- 5.Aragay, A. M., Collins, L. R., Post, G. R., Watson, A. J., Feramisco, J. R., Brown, J. H. & Simon, M. I. (1995) J. Biol. Chem. 270, 20073–20077. [DOI] [PubMed] [Google Scholar]
- 6.Kranenburg, O., Poland, M., van Horck, F. P. G., Drechsel, D., Hall, A. & Moolenaar, W. H. (1999) Mol. Biol. Cell 10, 1851–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu, N., Voyno-Yasenetskaya, T. & Gutkind, J. S. (1994) Biochem. Biophys. Res. Commun. 201, 603–609. [DOI] [PubMed] [Google Scholar]
- 8.Offermans, S., Mancino, V., Revel, J. P. & Simon, M. I. (1997) Science 275, 533–556. [DOI] [PubMed] [Google Scholar]
- 9.Katoh, H., Aoki, J., Yamaguchi, Y., Kitano, Y., Ichikawa, A. & Negishi, M. (1998) J. Biol. Chem. 273, 28700–28707. [DOI] [PubMed] [Google Scholar]
- 10.Kozasa, T., Jiang, X., Hart, M. J., Sternweis, P. M., Singer, W. D., Gilman, A. G., Bollag, G. & Sternweis, P. C. (1998) Science 280, 2109–2111. [DOI] [PubMed] [Google Scholar]
- 11.Hart, M. J., Jiang, S., Kozasa, T., Roscoe, W., Singer, W. D., Gilman, A. G., Sternweis, P. C. & Bollag, G. (1998) Science 280, 2112–2114. [DOI] [PubMed] [Google Scholar]
- 12.Fukuhara, S., Murga, C., Zohar, M., Igishi, T. & Gutkind, J. S. (1999) J. Biol. Chem. 274, 5868–5879. [DOI] [PubMed] [Google Scholar]
- 13.Fukuhara, S., Chikumi, H. & Gutkind, J. S. (2000) FEBS Lett. 485, 183–188. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki, N., Nakamura, S., Mano, H. & Kozasa, T. (2003) Proc. Natl. Acad. Sci. USA 100, 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett, K., Leptin, M. & Settleman, J. (1997) Cell 91, 905–915. [DOI] [PubMed] [Google Scholar]
- 16.Hacker, U. & Perrimon, N. (1998) Genes Dev. 12, 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa, M., Wilson, E. T. & Wieschaus, E. (1994) Cell 76, 1075–1089. [DOI] [PubMed] [Google Scholar]
- 18.Parks, S. & Weischaus, E. (1991) Cell 64, 447–458. [DOI] [PubMed] [Google Scholar]
- 19.Mendel, J. E., Korswagen, H. C., Liu, K. S., Haidu-Cronin, Y. M., Simon, M. I., Plasterk, R. H. & Sternberg, P. W. (1995) Science 267, 1652–1655. [DOI] [PubMed] [Google Scholar]
- 20.Brundage, L., Avery, L., Katz, A., Kim, U. J., Mendel, J. E., Sterberg, P. W. & Simon, M. I. (1996) Neuron 16, 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen, G., Thijssen, K. L., Werner, P., van der Horst, M., Hazendonk, E. & Plasterk, R. A. (1999) Nat. Genet. 21, 414–419. [DOI] [PubMed] [Google Scholar]
- 22.van der Linden, A. M., Moorman, C., Cuppen, E., Korswagen, H. C. & Plasterk, R. H. (2003) Curr. Biol. 13, 516–521. [DOI] [PubMed] [Google Scholar]
- 23.Simmer, F., Tijsterman, M., Parrish, S., Koushika, S. P., Nonet, M. L., Fire, A., Ahringer, J. & Plasterk, R. H. (2002) Curr. Biol. 12, 1317–1319. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, J. A. & Fleming, J. T. (1995) Methods Cell Biol. 48, 4–27. [PubMed] [Google Scholar]
- 25.Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. (1991) EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedgecock, E. M., Culotti, J. G., Thomson, J. N. & Perkins, L. A. (1985) Dev. Biol. 111, 158–170. [DOI] [PubMed] [Google Scholar]
- 27.Fromm, C., Coso, O. A., Montaner, S., Xu, N. & Gutkind, J. S. (1997) Proc. Natl. Acad. Sci. USA 94, 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 29.Gu, J. L., Muller, S., Mancino, V., Offermanns, S. & Simon, M. I. (2002) Proc. Natl. Acad. Sci. USA 99, 352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddiqui, S. S. (1990) Neurosci. Res. Suppl., 13, S171–S190. [DOI] [PubMed] [Google Scholar]
- 31.Chen, W. & Lim, L. (1994) J. Biol. Chem. 269, 32394–32404. [PubMed] [Google Scholar]
- 32.Vaiskunaite, R., Adarichev, V., Furthmayr, H., Kozasa, T., Gudkov, A. & Voyno-Yasenetskaya, T. A. (2000) J. Biol. Chem. 275, 26206–26212. [DOI] [PubMed] [Google Scholar]
- 33.Niu, J., Vaiskunaite, R., Suzuki, N., Kozasa, T., Carr, D. W., Dulin, N. & Voyno-Yasenetskaya, T. A. (2001) Curr. Biol. 11, 1686–1690. [DOI] [PubMed] [Google Scholar]
- 34.Meigs, T. E., Fedor-Chaiken, M., Kaplan, D. D., Brackenbury, R. & Casey, P. J. (2002) J. Biol. Chem. 277, 24594–24600. [DOI] [PubMed] [Google Scholar]
- 35.Hajdu-Cronin, Y. M., Chen, W. J., Patikoglou, G., Koelle, M. R. & Sternberg, P. W. (1999) Genes Dev. 13, 1780–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chase, D. L., Patikoglou, G. A. & Koelle, M. R. (2001) Curr. Biol. 11, 222–231. [DOI] [PubMed] [Google Scholar]
- 37.Mao, J., Xie, W., Yuan, H., Simon, M. I., Mano, H. & Wu, D. (1998) EMBO J. 17, 5638–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang, Y., Ma, W., Kozasa, T., Hattori, S. & Huang, X. Y. (1998) Nature 395, 808–813. [DOI] [PubMed] [Google Scholar]
- 39.Shi, C. S., Sinnarajah, S., Cho, H., Kozasa, T. & Kehrl, J. H. (2000) J. Biol. Chem. 275, 24470–24476. [DOI] [PubMed] [Google Scholar]
- 40.Aurandt, J., Vikis, H. G., Gutkind, J. S., Ahn, N. & Guan, K. L. (2002) Proc. Natl. Acad. Sci. USA 99, 12085–12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swiercz, J. M., Kuner, R., Behrens, J. & Offermanns, S. (2002) Neuron 35, 51–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.