Abstract
Organic cation/carnitine transporter (OCTN2; SLC22A5) is an important transporter for L-carnitine homeostasis, but can be inhibited by drugs, which may cause L-carnitine deficiency and possibly other OCTN2-mediated drug-drug interactions. One objective was to develop a quantitative structure–activity relationship (QSAR) of OCTN2 inhibitors, in order to predict and identify other potential OCTN2 inhibitors and infer potential clinical interactions. A second objective was to assess two high renal clearance drugs that interact with OCTN2 in vitro (cetirizine and cephaloridine) for possible OCTN2-mediated drug-drug interactions. Using previously generated in vitro data of 22 drugs, a 3D quantitative pharmacophore model and a Bayesian machine learning model were developed. The four pharmacophore features include two hydrophobic groups, one hydrogen-bond acceptor, and one positive ionizable center. The Bayesian machine learning model was developed using simple interpretable descriptors and function class fingerprints of maximum diameter 6 (FCFP_6). An external test set of 27 molecules, including 15 newly identified OCTN2 inhibitors, and a literature test set of 22 molecules were used to validate both models. The computational models afforded good capability to identify structurally diverse OCTN2 inhibitors, providing a valuable tool to predict new inhibitors efficiently. Inhibition results confirmed our previously observed association between rhabdomyolysis and Cmax/Ki ratio. The two high renal clearance drugs cetirizine and cephaloridine were found not to be OCTN2 substrates and their diminished elimination by other drugs is concluded not to be mediated by OCTN2.
Keywords: Bayesian model, pharmacophore, transporters
INTRODUCTION
The Organic Cation/Carnitine Transporter (OCTN2) is a high affinity cation/carnitine transporter widely expressed in human tissues, including skeletal muscle, kidney, brain, heart, and placenta. OCTN2 is essential for L-carnitine transport in humans.1 Mutations of the OCTN2 gene in humans cause primary systemic carnitine deficiency, with clinical significant symptoms that include hypoketotic hypoglycemia, cardiomyopathy, and skeletal myopathy.2, 3 In addition to L-carnitine, some drugs are also transported by OCTN2,4-7 implying a potentially important role of OCTN2 in the disposition of such drugs in humans.
The role of OCTN2 in L-carnitine transport provides for potential drug-L-carnitine and drug-drug interactions. Long-term treatment with structurally and therapeutically diverse drugs has been reported to cause secondary L-carnitine deficiency. In the kidney, OCTN2 is localized on the apical membrane of renal proximal tubular cells and ensures minimal loss of L-carnitine by re-absorption. One possible mechanism for drug induced secondary L-carnitine deficiency is inhibition of OCTN2-mediated L-carnitine renal re-absorption. For example, the use of cephaloridine is associated with L-carnitine deficiency and enhanced renal excretion of L-carnitine8, consistent with the in vitro studies that cephaloridine competitively inhibited OCTN2-mediated L-carnitine transport.
No crystal structure or three dimensional (3D) protein model of OCTN2 exists. Hence, in a previous study, we generated an in silico common features (HipHop) pharmacophore model that consisted of three hydrophobic features and a positive ionizable feature, derived from initial screening data using an in vitro cell culture.9 The HipHop pharmacophore was used to predict the molecular requirements of OCTN2 inhibition and identify more potent inhibitors of OCTN2.9 Among the 33 tested drugs that were predicted to map to the pharmacophore, 27 inhibited OCTN2 in vitro. The pharmacophore was shown to accurately prioritize compounds for testing. We identified novel low micromolar inhibitors that belonged to diverse therapeutic classes of drugs, such as vinblastine, carvedilol, raloxifene, thioridazine and clozapine. We also identified an association that compounds were more likely to cause rhabdomyolysis if the Cmax/Ki ratio was higher than 0.0025.9 This preliminary work illustrated that a combined computational pharmacophore and in vitro approach could suggest new, structurally diverse inhibitors for OCTN2 that may possibly cause clinical significant toxicity.
One objective in the present work was to extend the earlier HipHop pharmacophore by developing a quantitative structure–activity relationship (QSAR) of OCTN2 inhibitors, in order to predict additional molecules and identify potential OCTN2 inhibitors. To date, no homology model or QSAR model for OCTN2 has been generated. In the current study, the first 3D-QSAR model for OCTN2 inhibition is described, as well as a Bayesian model, both of which were subjected to validation with external molecules. Inhibition results are discussed in terms of possible relationship to rhabdomyolysis or carnitine deficiency.
A second objective was to assess two high renal clearance drugs that interact with OCTN2 in vitro (cetirizine and cephaloridine) for possible OCTN2-mediated drug-drug interactions. Cetirizine and cephaloridine exhibit reduced renal clearance with coadministration of ritonavir and furosemide, respectively, yielding the hypothesis that cetirizine and cephaloridine tubular secretion via OCTN2 can be inhibited by other drugs to reduce their renal clearance. However, neither cetirizine nor cephaloridine was an OCTN2 substrate, such that their diminished elimination upon co-administration of other drugs is concluded not to be mediated by OCTN2.
EXPERIMENTAL SECTION
Materials
Fetal bovine serum, trypsin-EDTA, and Dulbecco’s modified Eagle medium (DMEM) were purchased from Invitrogen Corporation (Carlsbad, CA). L-[3H]carnitine was purchased from American Radiolabeled Chemicals (St. Louis, MO). All other drugs were purchased from Spectrum Chemicals & Laboratory Products (Gardena, CA), Sigma Chemical (St. Louis, MO), AK Scientific (Mountain View, CA), or TCI America (Portland, OR).
Cell Culture
Stably transfected hOCTN2-MDCK cells and MDCK cells were cultured at 37 °C, 90% relative humidity, and 5% CO2 atmosphere and fed every 2 days. Media was composed of DMEM supplemented with 10% FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin. Cells were passaged after reaching 80% confluence.
Inhibition Study
Inhibition studies of L-carnitine were conducted as previously described.9 Briefly, after reaching 90% confluence, cells were seeded in 12 well cluster plates at a density of 1.5 million cells/well and cultured for four days. The culture medium was changed every 48 hr. Uptake studies were performed on the fourth day and were conducted in presence of Hank’s balance salts solution (HBSS). Cells were exposed to donor solution containing 2.5 μM L-carnitine (spiked with L-[3H]-carnitine) in the presence or absence of drug at 37 °C and 50 rpm orbital shaking for 10 min. The donor solution was removed and the cells were washed thrice with ice-cold sodium-free buffer (SFB). Subsequently, cells were lysed using 0.25 mL of 1 M NaOH for 2 hr at room temperature and neutralized with 0.25 mL of 1 M HCl. Cell lysate was then counted for associated radioactivity using a liquid scintillation counter. Jmax of L-carnitine was measure on each inhibition study occasion. Unless otherwise noted, data are summarized as mean (±SEM) of three measurements.
Kinetic Analysis
To measure Ki, inhibition studies were performed as described above, where a range of drug concentrations were applied to inhibit L-carnitine uptake. The following competitive inhibition model was applied:
| (1) |
where Ki is the competitive inhibition coefficient, I is the concentration of inhibitor, and S is the 2.5 μM concentration of L-carnitine. In applying eqn 1, only Ki was estimated using nonlinear regression fitting performed by WinNonlin 4.1 (Pharsight, Mountain View, CA). The other three parameters (i.e. Jmax, Kt, and Pp) were estimated from L-carnitine uptake studies without inhibitor.
Cetirizine and cephaloridine studies
Cetirizine and cephaloridine are drugs that are mainly renally eliminated. Uptake studies of cetirizine and cephaloridine into OCTN2-MDCK cells were conducted, along with inhibition studies. Inhibition studies were performed to study the possible effect of ritonavir on OCTN2-mediated transport of cetirizine, as well as the possible effect of furosemide on OCTN2-mediated transport of cephaloridine. Cetirizine and cephaloridine samples were quantified by LC/MS/MS.
Analytical methods
L-[3H]-carnitine was quantified by scintillation counting. Cetirizine and cephaloridine samples were quantified by LC/MS/MS. The LC/MS/MS instrumental system consisted of Finnigan Surveyor® Plus Autosampler, Finnigan Surveyor® LC Pump Plus and Finnigan TSQ® Quantum Discovery MAX™ mass spectrometer with an electrospray ionization source and triple-quadrupole mass analyzer.
The column used was Synergi™ Polar-RP column (4 μm, 50 × 2.00 mm; Phenomenex, Torrance, CA). Gradient of acetonitrile with 0.1% formic acid and 50 mM ammonia formate (pH 3.2) was used as the mobile phase, with organic phase rising from 50% to 80% in 3 mins. Cetirizine and cephaloridine were monitored by selective reaction monitoring (SRM) at m/z transition 389.3 → 200.9 and 416.1 → 151.9, respectively. The flow rate was 0.4 mL/min. The assay was linear (r2>0.999) over 10–1000 nM for both cetirizine and cephaloridine.
Data analysis
Data were expressed as mean ± SEM derived from three independent wells. Statistical significance was evaluated using Graphpad Prism (Graphpad Software, Inc.; La Jolla, CA).
Quantitative pharmacophore development
We generated a 3D-QSAR using HypoGen in Discovery Studio version 2.1 or 2.5.5. (San Diego, CA) that has been previously applied to P-glycoprotein10. The pharmacophore model training set included 22 compounds, including phenothiazine antipsychotics, atypical antipsychotics, selective estrogen receptor modulators, calcium channel blockers, anti-cancer compounds, and tricyclic antidepressants drug classes. Previously reported OCTN2 Ki values were used as the biological activity.9 In the HypoGen approach, ten hypotheses were generated, using hydrophobic, hydrogen bond acceptor, hydrogen bond donor, and the positive and negative ionizable features. After assessing all ten generated hypotheses, the hypothesis with lowest energy cost was selected for further analysis, as this model possessed features representative of all the hypotheses and had the lowest total cost.
The total energy cost of the generated pharmacophore was calculated from the deviation between the estimated activity and the observed activity, combined with the complexity of the hypothesis (i.e. the number of pharmacophore features). A null hypothesis, which presumed no relationship between chemical features and normally distributed biological activities, was also calculated. The greater the difference between the energy cost of the generated and null hypotheses, the less likely the generated hypothesis reflected a chance correlation. Also, the quality of the structure-activity correlation between the predicted and observed activity values was estimated via correlation coefficient.
For testing the quantitative pharmacophore, two validation test sets of molecules were used. The first test set included additional 15 retrieved compounds from the SCUT 2008 database (796 molecules), US Drug Microsource database (1034 molecules) and Natural products Microsource database (787 molecules), and 12 drugs from our previous published study (i.e. 11 non-inhibitors and the renally-eliminated compound cephaloridine). The second test set consisted of molecules collated from the literature. These structures were retrieved from the ChemSpider database (www.chemspider.com) as .mol files and imported in Discovery Studio.
In all cases, the databases or test sets were created from sdf files and up to 100 conformers/molecule were created with the Build 3D database protocol using the FAST conformation method. The BEST method was also used for the test set molecules; results employing the BEST method note its use. Each dataset was then searched using the search 3D database protocol. The ligand pharmacophore mapping protocol was also used to search these databases and allowed one or more feature misses.
Model assessment via test sets was pursued by treating observed and predicted data as binary using 1000 μM as the Ki upper limit for an inhibitor. Hence, each prediction was assessed as being a true positive, false negative, false positive, or true negative. This binary approach was taken since the goal was to assess model predictability simply in terms of inhibitors versus non-inhibitors.
Bayesian machine learning model development
Laplacian-corrected Bayesian classifier models were generated using Discovery Studio 2.1 or 2.5.5. This approach employs a machine learning method with 2D descriptors. Molecular function class fingerprints of maximum diameter 6 (FCFP_6), AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular polar surface area were calculated from input sdf files using the “calculate molecular properties” protocol. The “create Bayesian model” protocol was used for model generation. A custom protocol for validation, involving leave 50% out 100 times, was also used.
Comparison of training and test sets
Principal Component Analysis (PCA) available in Discovery Studio version 2.5.5 was used to compare the molecular descriptor space for the test and training sets (using descriptors ALogP, molecular weight, number hydrogen bond donors, number of hydrogen bond acceptors, number of rotatable bonds, number of rings, number of aromatic rings, and molecular fractional polar surface area). In each case, the respective test set and the training set compounds were combined and used to generate the PCA analysis.
Additionally, each molecule from the two test sets were evaluated for their similarity to training set molecules using MDL keys and the Tanimoto similarity in Discovery Studio version 2.5.5. MDL keys are a set of 960, mostly substructural features, developed for rapid substructural searching. The presence or absence of features was denoted with bits (e.g. 1 or 0). The Tanimoto is:
| (2) |
where SA represents the number of bits present in both the target and the reference molecules, SB is the number of bits in the target but not the reference molecule, and SC is the number of bits in the reference but not the target molecule.
RESULTS
Quantitative Pharmacophore
A 3D-QSAR pharmacophore model was developed using 22 compounds from diverse drug classes in the previous study.9 Of the 22 compounds, 14 were OCTN2 inhibitors, while 8 were non-inhibitors. The resulting quantitative pharmacophore is illustrated in Figure 1, composed of one hydrogen bond acceptor, two hydrophobic features, and a positive ionizable feature. Table 1 lists the predicted and observed Ki values of the training set. Energy cost values for the generated hypothesis were total cost = 101.67, fixed cost = 90.98, and null cost = 122.93. Ideally, the magnitude of the difference between the total and null cost should be larger. The correlation coefficient r for the training set was 0.89, indicating that the model was acceptable.
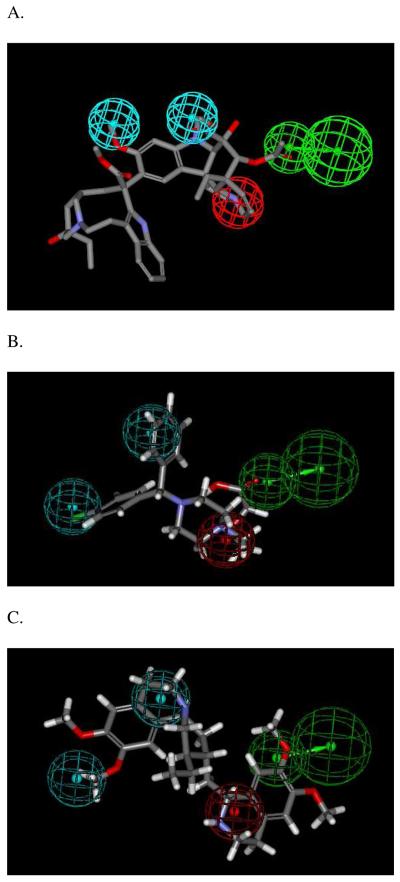
Figure 1.
Quantitative pharmacophore for OCTN2 inhibition. Panel A shows the most active drug in the training set, vinblastine, mapped to the quantitative pharmacophore. Panel B shows cetirizine mapped to the quantitative pharmacophore. Panel C shows emetine mapped to the quantitative pharmacophore. Quantitative pharmacophore features represent: cyan = hydrophobes, red = positive ionizable which maps to the basic nitrogen, green = hydrogen bond acceptor.
Table 1.
Training set of 22 compounds for the quantitative pharmacophore and the Bayesian model. Compounds are listed in decreasing order of fit value
| Drug | Observed Ki (μM)a (±SEM) |
Quantitative pharmacophore fit valueb |
Predicted Ki (μM)b |
Bayesian scorec |
|---|---|---|---|---|
| Vinblastine | 4.85±0.71 | 5.82 | 3.5 | 15.095 |
| Verapamil | 17.6±3.1 | 5.00 | 23 | 7.212 |
| Carvedilol | 10.7±1.6 | 4.89 | 30 | 4.550 |
| Amlodipine | 96.0±15.2 | 4.86 | 32 | −0.250 |
| Bromocriptine | 16.6±2.0 | 4.79 | 37 | 11.148 |
| Thioridazine | 23.0±4.1 | 4.74 | 42 | 9.155 |
| Raloxifine | 13.8±2.4 | 4.73 | 44 | 7.882 |
| Duloxetine | 118±13 | 4.64 | 53 | 4.684 |
| Propantheline | 20.4±4.1 | 4.62 | 56 | 5.448 |
| Trifluoperazine | 67.3±10.1 | 4.62 | 55 | 10.320 |
| Prochlorperazine | 51.3±10.5 | 4.54 | 67 | 10.375 |
| Cerivastatin | 425±12 | 4.52 | 70 | 3.770 |
| Clozapine | 47.3±8.6 | 4.43 | 87 | 6.082 |
| Desloratidine | 53.3±6.7 | 4.33 | 110 | 4.369 |
| Atenolol | 3000* | 3.38 | 970 | −14.701 |
| Atropine | 3000* | 3.27 | 1200 | −15.079 |
| Lomefloxacin | 3000* | 3.20 | 1500 | −26.875 |
| Acyclovir | 3000* | 3.20 | 1500 | −30.617 |
| Pramipexole | 3000* | 3.20 | 1500 | −20.237 |
| Guanosine | 3000* | 3.20 | 1500 | −35.675 |
| Hydrochlorothiazide | 3000* | 3.05 | 2100 | −18.578 |
| Sulfanilamide | 3000* | 1.60 | 58000 | −12.923 |
Observed Ki values were from our previous study.9
Fit values and predicted Ki values are derived from the pharmacophore using the BEST conformer method.
Bayesian score was predicted from the Bayesian model, using the best split value of −3.512. A score higher than −3.512 predicts the compound to be an inhibitor; a score lower than −3.512 predicts the compound to be a non-inhibitor.
Ki value was estimated from a single concentration and indicates drug was not a potent inhibitor. A Ki value of 3000 μM was assigned.
Bayesian Model
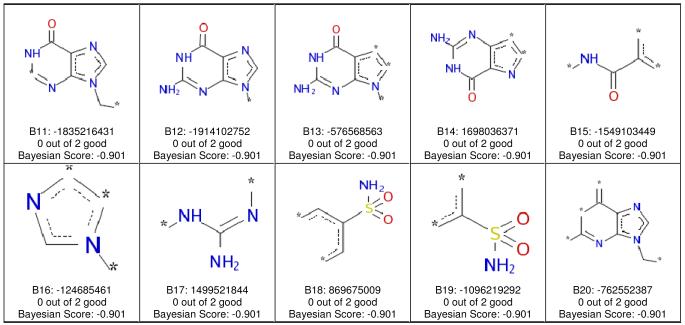
The same training set of 22 compounds was also applied to develop a Bayesian model11 with molecular function class fingerprints of maximum diameter 6 (FCFP_6) and eight interpretable descriptors. The model had a leave-one-out cross-validation receiver operator curve (ROC) statistic of 0.93 (Supplemental Table S1) and enrichments (Supplemental Table S2 and S3) that suggested that OCTN2 inhibitors (Ki < 1000 μM) were well separated from non-inhibitors (Supplemental Table S4). After leaving 50% out 100 times, three models failed. When these three were removed, the mean values for the 97 remaining models were calculated. The external ROC [mean (±SD)] was 0.90 (±0.09), while internal ROC was 0.79 (±0.18%); concordance was 73.4% (±14.5); specificity was 88.2% (±21.2); and sensitivity was 64.2% (±26.17). The Bayesian method showed favorable cross validation data. However, these results may be impacted by the small training set (N= 22), especially when 50% are left out. Use of the FCFP_6 descriptors allowed the identification of molecular features that favored inhibition (Figure 2), as well as features that did not promote inhibition (Figure 3).
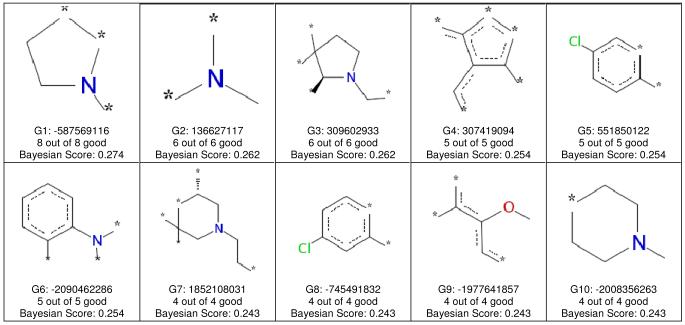
Figure 2.
FCFP_6 features associated with OCTN2 inhibitors. Each of the 20 panels shows the naming convention for one fragment, the numbers of inhibitors containing the fragment, and the Bayesian score for the fragment. In all cases, a compound containing any of these 20 fragments was an OCTN2 inhibitor.
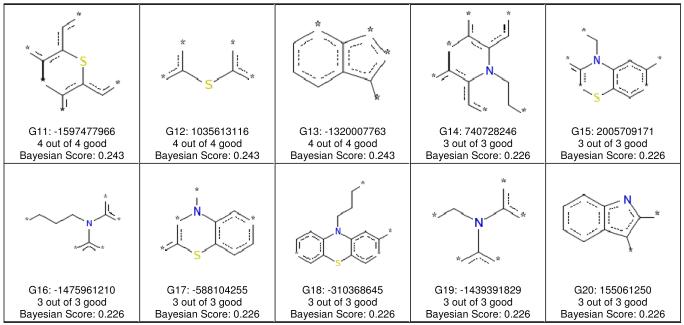
Figure 3.
FCFP_6 features associated with OCTN2 non-inhibitors. Each of the 20 panels shows the naming convention for one fragment, the numbers of inhibitors containing the fragment, and the Bayesian score for the fragment. In all cases, a compound containing any of these 20 fragments was not an OCTN2 inhibitor.
Table 1 lists the observed Ki values, as well as the Bayesian score of the training set. The best split value was −3.512 (Supplemental Table S1) and demarcated inhibitors from non-inhibitors (Table 1). The best split value was calculated by minimizing the number of compounds that were incorrectly predicted as either inhibitors or non-inhibitors, using the cross-validated score for each sample.
The Bayesian model correctly ranked the most active compounds, producing only 4.5% false negatives and 4.5% false positives. Model evaluations using test sets are described below and represent a better evaluation of the model. This Bayesian model with 2D fingerprints also represents a classification approach to building models that can be used for rapid screening of compound libraries. From the molecular fingerprints, descriptors identified regions in the training set molecules (e.g., the tertiary amine) that were likely important for OCTN2 inhibition (Figure 2), as well as substructures that were associated with non-inhibitors (Figure 3).
Test Set Evaluations
To validate the quantitative pharmacophore and the Bayesian model, 27 additional compounds were used as the first test set. This test set included an additional 15 retrieved compounds from SCUT 2008 database, US Drug Microsource database and Natural products Microsource database, and tested here, as well as 12 drugs (11 non-inhibitors and the weak inhibitor cephaloridine) from our previous published study 9. Figure 1B shows cetirizine, a representative molecule from the literature test set, mapped to the quantitative pharmacophore.
Table 2 lists the predicted Ki values using the quantitative model and experimentally observed counterparts, as well as the Bayesian scores from the Bayesian model. For the quantitative pharmacophore model using FAST conformer method, the majority were correctly predicted (over 70%). There were 29.6% true positives plus 40.7% true negatives; 29.6% of the predictions were false negatives and zero percent were false positives. Use of the BEST conformer methods provided similar results (Table 3). For the Bayesian model, the majority were also correctly predicted (over 80%). There were 48.1% true positives plus 33.3% true negatives; 11.1% of the predictions were false negatives and 7.4% were false positives.
Table 2.
Test set of 27 drugs with Ki data generated in our laboratory. Predictions from the quantitative pharmacophore model and Bayesian model are listed. Compounds are listed in decreasing order of pharmacophore fit value (FAST conformer method)
| Drug | Observed Ki (μM) (±SEM) |
Quantitative pharmacophore fit valuea |
Predicted Ki (μM)a |
Quantitative pharmacophore fit valueb |
Predicted Ki (μM)b |
Bayesian score |
Maximal Tanimoto similarity to training set compound |
|---|---|---|---|---|---|---|---|
| Roxithromycin | 333±25 | 16.5 | 5.14 | 5.34 | 10.5 | −2.693 | 0.60 (vinblastine) |
| Reserpine | 32.2±4.8 | 5.67 | 4.89 | 5.24 | 13.1 | 5.787 | 0.75 (vinblastine) |
| Cisapride | 66.7±9.8 | 5.52 | 6.94 | 3.85 | 327 | −2.431 | 0.66 (vinblastine) |
| Penfluridol | 26.5±4.1 | 5.46 | 8.01 | 4.27 | 124 | 2.729 | 0.57 (raloxifene) |
| Vincristine | 15.9±1.4 | 4.79 | 37.1 | 5.88 | 3.03 | 14.316 | 0.97 (vinblastine) |
| Propafenone | 74.2±6.1 | 4.78 | 38.5 | 4.30 | 115 | −5.110 | 0.72 (carvedilol) |
| Cetirizine | 79.8±8.0 | 4.25 | 130 | 5.53 | 6.71 | 0.175 | 0.60 (raloxifene) |
| Nizatidine | 183±17 | 3.49 | 741 | 4.42 | 86.7 | −5.003 | 0.49 (pramipexole) |
| Daunorubicin | 502±41 | 2.89 | 2930 | 2.87 | 3110 | −1.630 | 0.63 (vinblastine) |
| Cephaloridine | 379* | 1.56 | 63700 | 2.21 | 14300 | −1.229 | 0.57 (bromocriptine) |
| Vinorelbine | 26.8±1.5 | 1.55 | 644000 | 5.90 | 2.91 | 11.739 | 0.92 (vinblastine) |
| Irinotecan | 219±18 | 0.946 | 260000 | 3.20 | 15500 | 3.182 | 0.75 (bromocriptine and vinblastine) |
| Risperidone | 144±17 | 0.566 | 623000 | 2.81 | 3590 | 3.427 | 0.49 (lomefloxacin and bromocriptine) |
| Erythromycin | 2870* | NM | NM | 4.76 | 39.8 | −2.334 | 0.64 (vinblastine) |
| Ritonavir | 7.73±0.67 | NM | NM | NM | NM | −4.684 | 0.61 (bromocriptine) |
| Ezetimibe | 29.3±5.0 | NM | NM | NM | NM | 1.952 | 0.48 (lomefloxacin) |
| Simvastatin | 12.4±1.2 | NM | NM | NM | NM | −1.172 | 0.48 (lomefloxacin) |
| Furosemide | 1350* | NM | NM | NM | NM | −8.403 | 0.69 (hydrochlorothiazide) |
| Procainamide | 1400* | NM | NM | NM | NM | −9.243 | 0.49 (atenolol and lomefloxacin) |
| Metformin | 2050* | NM | NM | NM | NM | −3.945 | 0.32 (clozapine) |
| Zidovudine | 2140* | NM | NM | NM | NM | −7.485 | 0.63 (guanosine) |
| Lamivudine | 2590* | NM | NM | NM | NM | −6.327 | 0.70 (guanosine) |
| Ketorolac | 1530* | NM | NM | NM | NM | −4.922 | 0.53 (atropine, vinblastine and carvedilol) |
| Gabapentin | 1700* | NM | NM | NM | NM | −4.666 | 0.40 (carvedilol) |
| Levofloxacin | 1890* | NM | NM | NM | NM | −10.388 | 0.81 (lomefloxacin) |
| Succinylcholine | 2700* | NM | NM | NM | NM | 1.263 | 0.52 (verapamil) |
| Procarbazine | 2520* | NM | NM | NM | NM | −7.436 | 0.53 (atenolol) |
Fit values and predicted Ki values are derived from the pharmacophore using the FAST conformer method.
Fit values and predicted Ki values are derived from the pharmacophore using the BEST conformer method.
For each of these 12 compounds, Ki value was estimated from a single concentration, as reported previously.9
NM denotes compound did not map to the pharmacophore.
Table 3.
Validation analysis for the training and test sets when treating the data as binary using 1000μM as the Ki upper limit for an inhibitor.a
| true positivesb | false negativesc | false positivesd | true negativese | |
|---|---|---|---|---|
| quantitative model training set (n=22) | 14 (63.6%) | 0 (0%) | 1 (4.55%) | 7 (31.8%) |
| quantitative model test set (n=27, FAST conformer) | 8 (29.6%) | 8 (29.6%) | 0 (0%) | 11(40.7%) |
| quantitative model test set (n=27, BEST conformer) | 9 (33.3%) | 6 (22.2%) | 1 (3.70%) | 11(40.7%) |
| quantitative model literature test set (n=32, FAST conformer) | 2 (6.25%) | 12 (37.5%) | 0 (0%) | 18 (56.25%) |
| quantitative model literature test set (n=32, BEST conformer) | 2 (6.25%) | 12 (37.5%) | 2 (6.25%) | 16 (50%) |
| Bayesian model training set (n=22)f | 13 (59.1%) | 1 (4.55%) | 1 (4.55%) | 7 (31.8%) |
| Bayesian model test set (n=27) | 13 (48.1%) | 3 (11.1%) | 2 (7.41%) | 9 (33.3%) |
| Bayesian model literature test set (n=32) | 9 (28.1%) | 5 (15.6%) | 6 (18.8%) | 12 (37.5%) |
Values in table are numbers of compounds that were true positives, false negatives, false positives, or true negatives. Values in parenthesis are simply the percent of compounds that were true positives, false posi tives, true negatives, or false negatives.
True positives were both predicted and observed to be inhibitors (Ki < 1000μM).
False negatives were predicted to non- inhibitors but were inhibitors.
False positives were predicted to be inhibitors but were non-inhibitors.
True negatives were both predicted and observed to be non-inhibitors (Ki > 1000μM).
The calculation was based on the leave-one-out cross-validation approach.
Furthermore, a second test set (Table 4) was used employing 32 chemically-diverse OCTN2 inhibitors from the literature. Figure 1C shows emetine, a representative molecule from the literature test set, mapped to the quantitative pharmacophore. Table 4 lists the predicted Ki values, the reported Ki or IC50 values, and the Bayesian score for the literature test set. The experimental literature data is derived from different expression systems and different laboratories, which can be interpreted as a strength or weakness in the QSAR validation approach here. We view this approach as a significant challenge of the developed QSAR models, since biological activity can depend upon assay conditions (e.g. different cell cultures). Previous studies have also used additional external test sets derived from the literature as a means to further test models derived and this is acceptable as long as the limitations of such data are fully understood this should not be an impediment to their use.12,13
Table 4.
Literature test set predictions from the quantitative pharmacophore model and Bayesian model. Compounds are listed in decreasing order of fit value (FAST conformer method)
| Drug | Ki or IC50 (μM) |
Quantitative pharmacophore fit valuea |
Predicted Ki (μM)a |
Quantitative pharmacophore fit valueb |
Predicted Ki (μM)b |
Bayesian score |
Maximal Tanimoto similarity to training set compound |
Experimental system |
|---|---|---|---|---|---|---|---|---|
| Cetirizine28 | 27 | 4.24 | 133 | 5.34 | 10.8 | 0.175 | 0.60 (raloxifene) | Caki-1 (human proximal tubule cell line) |
| Emetine29 | 4.2 | 4.29 | 118 | 4.15 | 162 | 2.92 | 0.70 (verapamil) | Xenopus laevis oocytes |
| Acebutolol6 | Not inhibitor |
2.59 | 5860 | 2.46 | 7960 | −11.3 | 0.79 (atenolol) | hOCTN2-Transfected MDCKII Cells |
| Cefoselis7 | 6400 | 0.134 | 0.134 | 3.48 | 754 | −5.78 | 0.65 (acyclovir) | Hela with human OCTN2 |
| Famotidine28 | 1920 | 0.0259 | 2.16E+06 | 0.376 | 963000 | −6.35 | 0.56 (hydrochlorthiazide) |
Caki-1 |
| Zidovudine30 | 6 | NM | NM | NM | NM | −7.49 | 0.63 (guanosine) | Mouse myoblastic C2C12 cells |
| Omeprazole31 | 14.6 | NM | NM | NM | NM | −0.37 | 0.52 (vinblastine) | Rat kidney OCTN2 reconstituted in liposomes |
| Mildronate6 | 26 | NM | NM | NM | NM | −0.967 | 0.50 (propantheline) | hOCTN2-Transfected MDCKII Cells |
| Spironolactone6 | 26 | NM | NM | NM | NM | −0.589 | 0.39 (atropine) | hOCTN2-Transfected MDCKII Cells |
| Ciclotropium28 | 30 | NM | NM | NM | NM | −2.93 | 0.72 (atropine) | Caki-1 |
| Ipratropium4, 28 | 95 | NM | NM | NM | NM | −9.62 | 0.89 (atropine) | HEK stable cell line with human OCTN2 |
| Valproic acid28 | 139 | NM | NM | NM | NM | −0.767 | 0.23 (atropine, lomefloxacin, verapamil and vinblastine) |
Caki-1 |
| Cephaloridine7, 28 | 230/248 | NM | NM | NM | NM | −1.23 | 0.57 (bromocriptine) | Hela/ Caki-1 (human proximal tubule cell line) |
| Grepafloxacin32 | 300 | NM | NM | NM | NM | −16.3 | 0.96 (lomefloxacin) | BeWo (human placental cell line) |
| Cimetidine28 | 336 | NM | NM | NM | NM | −6.00 | 0.56 (pramipexole) | Caki-1 |
| Tacrine33 | 500 | NM | NM | NM | NM | −3.13 | 0.54 (desloratidine) | HEK stable cell line with rat OCTN2 |
| (−)-N- butylscopolamine28 |
1007 | NM | NM | NM | NM | −9.31 | 0.73 (atropine) | Caki-1 |
| Cefepime7 | 1700 | NM | NM | 3.80 | 363 | −5.42 | 0.63 (bromocriptine) | Hela with human OCTN2 |
| Levofloxacin32 | 3000 | NM | NM | NM | NM | −10.4 | 0.81 (lomefloxacin) | BeWo |
| Metformin28 | 4963 | NM | NM | NM | NM | −4.03 | 0.32 (clozapine) | Caki-1 |
| Captopril6 | Not inhibitor |
NM | NM | NM | NM | −4.32 | 0.53 (atropine) | hOCTN2-Transfected MDCKII Cells |
| Cisplatin34 | Not inhibitor |
NM | NM | NM | NM | −2.98 | 0.28 (sulfanilamide) | L6 stable cell line with rat OCTN2 |
| Cefazolin35 | 6740 | NM | NM | NM | NM | −3.88 | 0.57 (guanosine) | L6 cells overexpressing hOCTN2 |
| Cefuroxime35 | 525 | NM | NM | NM | NM | −5.25 | 0.57 (bromocriptine) | L6 cells overexpressing hOCTN2 |
| Cephalexin35 | 3037 | NM | NM | NM | NM | −6.35 | 0.53 (lomefloxacin) | L6 cells overexpressing hOCTN2 |
| Digoxin6 | Not inhibitor |
NM | NM | NM | NM | −1.62 | 0.56 (vinblastine) | hOCTN2-Transfected MDCKII Cells |
| Diphenhydramine36 | Not inhibitor |
NM | NM | NM | NM | −0.0798 | 0.63 (propantheline) | Mouse enterocyte |
| Lidocaine36 | Not inhibitor |
NM | NM | NM | NM | −4.13 | 0.48 (atenolol) | Mouse enterocyte |
| Memantine6 | Not inhibitor |
NM | NM | NM | NM | −1.73 | 0.29 (pramipexole) | hOCTN2-Transfected MDCKII Cells |
| Metoprolol6 | Not inhibitor |
NM | NM | NM | NM | −10.9 | 0.76 (atenolol) | hOCTN2-Transfected MDCKII Cells |
| Probenecid36 | Not inhibitor |
NM | NM | NM | NM | −3.22 | 0.54 (sulfanilamide) | Mouse enterocyte |
| Oxaliplatin34 | Not inhibitor |
NM | NM | NM | NM | −2.34 | 0.50 (guanosine) | L6 stable cell line with rat OCTN2 |
Fit values and predicted Ki values are derived from the pharmacophore using the FAST conformer method.
Fit values and predicted Ki values are derived from the pharmacophore using the BEST conformer method.
NM denotes compound did not map to the pharmacophore.
In Table 3, the quantitative pharmacophore model miss-predicted compounds that were inhibitors in the literature test set, yielding a false negative rate of 37.5%, but a favorable zero false positive rate, using the FAST method. The BEST method provided similar results. The Bayesian model was more balanced, with lower false negative rate of 15.6% and a false positive rate of 18.8%, however it did not perform as well on the literature test set compared with the test set generated in our laboratory for 27 compounds.
DISCUSSION
Quantitative Pharmacophore and Bayesian Modeling
The quantitative pharmacophore, derived from the HypoGen approach, was developed by using a training set of 22 compounds (Figure 1). This quantitative pharmacophore differs slightly from our previously developed common features (HipHop) pharmacophore, which was devised using chlorpheniramine, imipramine, and diltiazem as actives, and physostigmine and guanfacine as ‘inactive’ compounds. This common features pharmacophore consisted of three hydrophobic features and a positive ionizable feature. The current quantitative pharmacophore model replaced a hydrophobic feature with a hydrogen bond acceptor and is acceptable based on models statistics and, more importantly, external test set performance. These results indicate the 3D-QSAR model is useful for identifying additional hOCTN2 inhibitors from compound databases.
The Bayesian model performed well in correctly ranking the most active compounds and represents a machine learning classification approach to building models that can be used for rapid screening of compound libraries. Molecular fingerprint descriptors identified molecular regions (e.g., the tertiary amine, aromatic hydrophobic groups) that were likely important for hOCTN2 inhibition (Figure 2), as well as substructures (e.g. purine, imidazole, and sulfonamides) that were associated with non-inhibitors (Figure 3).
Validation Results
Both the quantitative pharmacophore and the Bayesian classification model performed better on the test set generated in this study, compared with the literature test set. Although there were some compounds in common in both datasets (e.g. cetirizine, cephaloridine, metformin, zidovudine and levofloxacin), these findings suggest that the training set and test set should (ideally) be generated under identical conditions in the same laboratory. In both cases, the test and training sets overlapped in simple descriptor molecular space upon PCA analysis (Supplemental Figures S1 and S2). However, when each of the training and test set molecules were analyzed using the Tanimoto similarity using the MDL keys (Table 2 and 4), very few of the test set molecules had a similarity > 0.75 to any one training set molecule (i.e. only four molecules in our test set and only three molecules in the literature test), which is considered an acceptable arbitrary cutoff for making reliable predictions.14,15 The mean maximal Tanimoto similarity for the test set in Table 2 is 0.61 (± 0.15), while the mean maximal Tanimoto similarity for the literature test set shown in Table 4 is 0.58 (± 0.17). These mean values are not statistically different when analyzed for all molecules in this study using the Students t-test (JMP, SAS Institute Cary, NC), so data variability rather than molecular similarity was the more likely reason for the poor predictions for the literature test set. The correlation between the maximal Tanimoto similarity and Bayesian score was very low, but statistically significant, for the test set in Table 2 (r2 = 0.19, p = 0.02, data not shown) and Table 4 (r2 = 0.42, p = 0.0001, data not shown). Considering the low level of similarity to any training set member, it is remarkable that the pharmacophore and Bayesian models performed as well as observed.
This observation suggests that prediction would likely improve dramatically if the maximal Tanimoto similarity of test sets to training set members was > 0.75. It is also interesting to note that ritonavir, ezetimide, and simvastatin (maximal Tanimoto similarity to training set 0.48-0.61) were not predicted by the pharmacophore, but have Ki values less than 30 μM. In cases in which the molecules did not map to the pharmacophore (database searching requires hits to map all features in Discovery Studio), the ‘ligand mapping protocol’ in the software was subsequently applied to enable at least one feature miss. In these cases, some molecules mapped three out of four features (data not shown). For example, omeprazole did not map to the positive ionizable feature. Such molecules would be expected to have a lower Fit score. Interestingly, in cases in which molecules miss a feature, results suggest that OCTN2 may enable some flexibility in the number of pharmacophore features mapped. Omeprazole was however predicted by the Bayesian model to be an inhibitor.
Overall, there was little difference in the binary validation analysis (Table 3) between using the FAST and BEST conformer generation for the test sets. The FAST conformers provided fewer false positives. The FAST conformation method also quickly produces diverse low energy conformations whereas BEST conformer covers greater conformation space which requires more computational time. The results suggest that the FAST method adequately covers the conformational space.
hOCTN2 Inhibition and Rhabdomyolysis or Carnitine Deficiency
The main objective was to develop a QSAR for OCTN2 inhibition, in order to predict and identify potential OCTN2 inhibitors. This objective was motivated by OCTN2 being an important transporter for L-carnitine homeostasis (e.g. energy utilization in muscle tissue) and the basis for OCTN2-mediated drug-drug interactions (e.g. rhabdomyolysis). We previously observed that compounds were more likely to cause rhabdomyolysis if the Cmax/Ki ratio was higher than 0.0025.9
Drugs from the test set in Table 2 and literature test set in Table 4 are collated in Table 5 to further evaluate the possible association between hOCTN2 inhibition and clinical rhabdomyolysis or carnitine deficiency. Twenty-three drugs exhibited a Cmax/Ki ratio higher than 0.0025, while the remaining 23 drugs were lower than 0.0025. Ki of 18 drugs were measured here, while Ki of the remaining 28 other drugs were obtained from the literature. Among the 21 drugs that were associated with rhabdomyolysis or carnitine deficiency, 14 (66.7%) provide a Cmax/Ki ratio higher than 0.0025. In contrast, among 25 drugs that were not associated with rhabdomyolysis or carnitine deficiency, only 9 (36.0%) show a Cmax/Ki ratio higher than 0.0025. Therefore, consistent with the previous observation9, clinical rhabdomyolysis or carnitine deficiency was associated with a Cmax/Ki value above 0.0025 (Pearson’s chi-square test p=0.0382). The limitations of Cmax/Ki serving as a predictor for rhabdomyolysis have been previously discussed9. For example, Cmax/Ki does not consider the effects of drug tissue distribution or plasma protein binding.
Table 5.
Evaluation of possible association between clinical rhabdomyolysis and/or carnitine deficiency and hOCTN2 inhibition. Cmax/Ki was computed for compounds in Table 2 and 4.a Compounds in bold text employ Ki (or IC50) values from literature (i.e. Table 4). Compounds listed in descending order of Cmax/Ki value. See Supplemental Table S6 for references 1-65 in this table
| Compound name | Ki (μM) | Documented to cause rhabdomyolysis and/or carnitine deficiency in the literature |
Cmax (μM) b | Cmax/Ki |
|---|---|---|---|---|
| Mildronate | 26 | No | 174 | 6.71 |
| Valproic acid | 139 | Yes1 | 159 | 1.15 |
| Cefazolin | 6740 | Yes2 | 1640 | 0.252 |
| Cefepime | 1700 | Yes3 | 378 | 0.222 |
| Cephaloridine | 230 | Yes3 | 41.7 | 0.181 |
| Cephaloxin | 3037 | Yes4 | 74.0 | 0.135 |
| Gabapentin | 1700 | No | 172 | 0.101 |
| Ritonavir | 7.73 | Yes with statin5 | 0.610 | 0.079 |
| Omeprazole | 14.6 | Yes6 | 0.909 | 0.0622 |
| Cefoselis | 6400 | Yes3 | 174 | 0.0271 |
| Irinotecan | 219 | Yes7 | 5.01 | 0.0229 |
| Cetirizine | 79.8 | No | 1.73 | 0.022 |
| Cimetidine | 336 | No | 6.97 | 0.0208 |
| Vincristine | 15.9 | Yes8 | 0.325 | 0.0204 |
| Cefuroxime | 525 | Yes9 | 10.6 | 0.0201 |
| Grepafloxacin | 300 | No | 5.51 | 0.0184 |
| Roxithromycin | 333 | Yes with statin10 | 6.06 | 0.0182 |
| Propafenone | 74.2 | No | 0.812 | 0.0109 |
| Nizatidine | 183 | No | 1.67 | 0.00911 |
| Vinorelbine | 26.8 | No | 0.133 | 0.00497 |
| Spironolactone | 26 | No | 0.114 | 0.00438 |
| Emetine | 4.2 | Yes11 | 0.0173 | 0.00413 |
| Simvastatin | 12.4 | Yes12 | 0.0410 | 0.00331 |
| Cisapride | 66.7 | Yes13 | 0.159 | 0.00239 |
| Metformin | 4963 | Yes14 | 10.1 | 0.00203 |
| Furosemide | 1350 | No | 2.70 | 0.00200 |
| Ezetimibe | 29.3 | Yes with statin15 | 0.0559 | 0.00191 |
| Ipratropium | 30 | No | 0.139 | 0.00146 |
| Procainamide | 1400 | Yes16 | 1.55 | 0.00111 |
| Ciclotropium | 95 | No | 0.0206 | 6.88×10−4 |
| Penfluridol | 26.5 | No | 0.0104 | 3.93×10−4 |
| Tacrine | 500 | No | 0.134 | 2.68×10−4 |
| Risperidone | 144 | Yes with statin17 | 0.0188 | 1.31×10−4 |
| Famotidine | 1920 | Yes18 | 0.240 | 1.25×10−4 |
| Daunorubicin | 502 | No | 0.0290 | 5.78×10−5 |
| Reserpine | 32.2 | No | 0.00181 | 5.61×10−5 |
| Acebutolol | Not inhibitor | No | 2.59 | 0 |
| Captopril | Not inhibitor | No | 3.70 | 0 |
| Cisplatin | Not inhibitor | Yes19 | 8.13 | 0 |
| Digoxin | Not inhibitor | No | 0.00282 | 0 |
| Diphenhydramine | Not inhibitor | No | 0.343 | 0 |
| Lidocaine | Not inhibitor | No | 1.21 | 0 |
| Memantine | Not inhibitor | No | 0.26 | 0 |
| Metoprolol | Not inhibitor | No | 0.357 | 0 |
| Oxaliplatin | Not inhibitor | No | 5.91 | 0 |
| Probenecid | Not inhibitor | No | 701 | 0 |
The following compounds from Table 2 or 4 are not listed below, since they were previously considered9 in evaluating a possible association to hOCTN2 inhibition: zidovudine, lamivudine, ketorolac, levofloxacin, succinylcholine, and procarbazine. Although listed in Table 4, (−)-N-butylscopolamine is not included in Table 5 since its Cmax is unknown.
Values for Cmax in units of μM were computed using compound molecular weight and Cmax from the literature. References 20-65 provide literature Cmax values for listed compounds in Table 5, respectively. See Supplemental Table S6 for references 1-65 in this table.
Drug-drug interaction
Among the newly discovered OCTN2 inhibitors in this study, some are mainly renally excreted and involved in drug-drug interactions. These observations motivated a second objective, which was to assess two high renal clearance drugs that interact with OCTN2 in vitro (cetirizine and cephaloridine) for possible OCTN2-mediated drug-drug interactions. This objective emanated from the hypothesis that competition for tubular excretion via OCTN2 expressed on the apical side of proximal tubular cells reduced cetirizine or cephaloridine renal clearance.
Coadministration of racemic cetirizine and ritonavir increases the plasma AUC and half-life of cetirizine by approximately 42% and 53%, respectively, and resulted in a 29% reduction in cetirizine clearance.16 In vitro experiments were conducted here to examine if OCTN2 was the possible transporter involved in the drug-drug interaction. Cetirizine (10 – 500 μM) uptake into OCTN2-MDCK cells in HBSS was not inhibited by L-carnitine (100 μM). Additionally, cetirizine (100 μM) uptake into OCTN2-MDCK cells in HBSS was not affected by ritonavir (1-50 μM). Uptake and inhibition results are provided in Supplemental Figure S3. Therefore, cetirizine was likely not a substrate for OCTN2; other transporters such as P-glycoprotein (P-gp) might mediate the interaction between cetirizine and ritonavir.17
Another drug-drug interaction is cephaloridine and furosemide. Concurrent administration of furosemide and cephaloridine results in a decrease in the renal clearance and a corresponding increase in the plasma concentrations of cephaloridine.18-19 Nephrotoxicity, reversible encephalopathy, and acute renal failure have been reported during high-dose therapy with cephaloridine, and the concomitant use of furosemide may increase the risk of developing cephaloridine toxicity.20-22 In vitro experiments were thus conducted to examine if OCTN2 was the possible transporter involved in the drug-drug interaction. No significant difference was observed between cephaloridine (0.5 mM) uptake into OCTN2-MDCK cells and MDCK cells (Supplemental Figure S4A). Furthermore, cephaloridine (0.5 mM) uptake into OCTN2-MDCK cells was not affected by furosemide from 0.5 to 2.5 mM (Supplemental Figure S4B). Basolateral uptake is the first step of tubular secretion. Both furosemide and cephaloridine were substrates for organic anion transporters (OATs) expressed on the basolateral side of proximial tubular cells.23-27 Therefore, the competition for tubular excretion between furosemide and cephaloridine might occur at the basolateral side, perhaps mediated by OATs.
In summary, for the first time, a 3D-QSAR pharmacophore model and a Bayesian model of OCTN2 inhibition were developed and validated using external test sets with data generated in a single laboratory. When predictions from both modeling approaches are treated as binary (i.e. inhibitor or non-inhibitor), we can directly compare the approaches in terms of the number of false positives and false negatives. Using the test set from our laboratory, the Bayesian method correctly identified over 80% of the compounds (i.e. 48.1% true positives plus 33.3% true negatives), whereas the pharmacophore could classify over 70% of the compounds (i.e. 29.6% true positives plus 40.7% true negatives). There was minimal difference between using the FAST and BEST conformer generation for the test sets using binary validation analysis (Table 3), although the FAST conformers yielded fewer false positives. Results show the benefit of obtaining data from the same laboratory for training and testing the models; that the training and test sets covered similar chemical space using PCA and simple descriptors (Supplemental Figure S1 and S2); and that the test set compounds did not possess a high degree of structural similarity to the training set based on MDL keys, yet predictions were still useful. In the absence of a crystal structure of OCTN2, we believe that our pharmacophore model coordinates (Supplemental Table S5 and Supplemental Figure S5) and the Bayesian model may provide novel insights into the molecular interaction of inhibitors with OCTN2 and identify important molecular features for OCTN2 inhibition, which might be useful for predicting potential drug induced secondary L-carnitine deficiency and OCTN2 mediated drug-drug interactions. A second objective was to assess cetirizine and cephaloridine for possible OCTN2-mediated drug-drug interactions. However, neither was an OCTN2 substrate, such that their reduced clearance by other drugs is concluded to not be mediated by OCTN2. Finally, our results here continue to support the previous observation that clinical rhabdomyolysis or carnitine deficiency may be associated with a high Cmax/Ki ratio higher than 0.0025.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health Grant DK67530.
S.E. gratefully acknowledges Dr. Matthew D. Krasowski for his assistance in creating the SCUT 2008 database supplemented with metabolites and drugs of abuse. S.E. also thanks Accelrys (San Diego, CA) for making Discovery Studio available.
ABBREVIATIONS
- 3D-QSAR
three-dimensional quantitative structure-activity relationship
- HBSS
Hanks balanced salt solution
- MDCK
Madin-Darby canine kidney
Footnotes
SUPPORTING INFORMATION
Supporting information includes model performance results and drug-drug interaction results. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Koepsell H, Lips K, Volk C. Polyspecific Organic Cation Transporters: Structure, Function, Physiological Roles, and Biopharmaceutical Implications. Pharm. Res. 2007;24:1227–1251. doi: 10.1007/s11095-007-9254-z. [DOI] [PubMed] [Google Scholar]
- 2.Nezu JI, Tamai I, Oku A, Ohashi R, Yabuuchi H, Hashimoto N, Nikaido H, Sai Y, Koizumi A, Shoji Y, Takada G, Matsuishi T, Yoshino M, Kato H, Ohura T, Tsujimoto G, Hayakawa J. i., Shimane M, Tsuji A. Primary systemic carnitine deficiency is caused by mutations in a gene encoding sodium ion-dependent carnitine transporter. Nat. Genet. 1999;21:91–94. doi: 10.1038/5030. [DOI] [PubMed] [Google Scholar]
- 3.Tein I. Carnitine transport: Pathophysiology and metabolism of known molecular defects. J. Inherited Metab. Dis. 2003;26:147–169. doi: 10.1023/a:1024481016187. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura T, Nakanishi T, Haruta T, Shirasaka Y, Keogh JP, Tamai I. Transport of Ipratropium, an anti-chronic obstructive pulmonary disease drug, is mediated by Organic Cation/Carnitine Transporters in human bronchial epithelial cells: implications for carrier-mediated pulmonary absorption. Mol. Pharmaceutics. 2009;7:187–195. doi: 10.1021/mp900206j. [DOI] [PubMed] [Google Scholar]
- 5.Kano T, Kato Y, Ito K, Ogihara T, Kubo Y, Tsuji A. Carnitine/Organic Cation Transporter OCTN2 (Slc22a5) is responsible for renal secretion of cephaloridine in mice. Drug Metab. Dispos. 2009;37:1009–1016. doi: 10.1124/dmd.108.025015. [DOI] [PubMed] [Google Scholar]
- 6.Grube M, Meyer zu Schwabedissen HEU, Prager D, Haney J, Moritz KU, Meissner K, Rosskopf D, Eckel L, Bohm M, Jedlitschky G, Kroemer HK. Uptake of cardiovascular drugs into the human heart: expression, regulation, and function of the carnitine transporter OCTN2 (SLC22A5) Circulation. 2006;113:1114–1122. doi: 10.1161/CIRCULATIONAHA.105.586107. [DOI] [PubMed] [Google Scholar]
- 7.Ganapathy ME, Huang W, Rajan DP, Carter AL, Sugawara M, Iseki K, Leibach FH, Ganapathy V. Beta-lactam antibiotics as substrates for OCTN2, an Organic Cation/Carnitine Transporter. J. Biol. Chem. 2000;275:1699–1707. doi: 10.1074/jbc.275.3.1699. [DOI] [PubMed] [Google Scholar]
- 8.Tune BM. Effects of L-carnitine on the renal tubular transport of cephaloridine. Biochem. Pharmacol. 1995;50:562–564. doi: 10.1016/0006-2952(95)00169-z. [DOI] [PubMed] [Google Scholar]
- 9.Diao L, Ekins S, Polli J. Novel inhibitors of human Organic Cation/Carnitine Transporter (hOCTN2) via computational modeling and in vitro testing. Pharm. Res. 2009;26:1890–1900. doi: 10.1007/s11095-009-9905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. Rapid identification of P-glycoprotein substrates and inhibitors. Drug Metab. Dispos. 34:1976–1984. doi: 10.1124/dmd.106.012351. [DOI] [PubMed] [Google Scholar]
- 11.Hassan M, Brown R, Varma-O’Brien S, Rogers D. Cheminformatics analysis and learning in a data pipelining environment. Mol. Diversity. 2006;10:283–299. doi: 10.1007/s11030-006-9041-5. [DOI] [PubMed] [Google Scholar]
- 12.Zientek M, Stoner C, Ayscue R, Klug-McLeod J, Jiang Y, West M, Collins C, Ekins S. Integrated in silico-in vitro strategy for addressing cytochrome P450 3A4 time-dependent inhibition. Chem. Res. Toxicol. 2010;23:664–76. doi: 10.1021/tx900417f. [DOI] [PubMed] [Google Scholar]
- 13.Chang C, Pang KS, Swaan PW, Ekins S. Comparative pharmacophore modeling of organic anion transporting polypeptides: a meta-analysis of rat Oatp1a1 and human OATP1B1. J. Pharmacol. Exp. Ther. 2005;314:533–41. doi: 10.1124/jpet.104.082370. [DOI] [PubMed] [Google Scholar]
- 14.Arimoto R, Prasad MA, Gifford EM. Development of CYP3A4 inhibition models: Comparisons of machine-learning techniques and molecular descriptors. J. Biomol. Screening. 2005;10:197–205. doi: 10.1177/1087057104274091. [DOI] [PubMed] [Google Scholar]
- 15.Ekins S, Andreyev S, Ryabov A, Kirillov E, Rakhmatulin EA, Sorokina S, Bugrim A, Nikolskaya T. A combined approach to drug metabolism and toxicity assessment. Drug Metab. Dispos. 2006;34:495–503. doi: 10.1124/dmd.105.008458. [DOI] [PubMed] [Google Scholar]
- 16.Product Information: XYZAL(R) oral tablets, levocetirizine dihydrochloride oral tablets. UCB Pharma,Inc; May 1, 2007. [Google Scholar]
- 17.Polli JW, Baughman TM, Humphreys JE, Jordan KH, Mote AL, Salisbury JA, Tippin TK, Serabjit-Singh CJ. P-glycoprotein influences the brain concentrations of cetirizine (Zyrtec (R)), a second-generation non-sedating antihistamine. J. Pharm. Sci. 2003;92:2082–2089. doi: 10.1002/jps.10453. [DOI] [PubMed] [Google Scholar]
- 18.Tilstone WJ, Semple PF, Lawson DH, Boyle JA. Effects of furosemide on glomerular filtration rate and clearance of practolol, digoxin, cephaloridine, and gentamicin. Clin. Pharmacol. Ther. 1977;22:389–94. doi: 10.1002/cpt1977224389. [DOI] [PubMed] [Google Scholar]
- 19.Norrby R, Stenqvist K, Elgefors B. Interaction between cephaloridine and furosemide in man. Scand. J. Infect. Dis. 1976;8:209–212. doi: 10.3109/inf.1976.8.issue-3.17. [DOI] [PubMed] [Google Scholar]
- 20.Simpson IJ. Nephrotoxicity and acute renal failure associated with cephalothin and cephaloridine. N. Z. Med. J. 1971;74:312–315. [PubMed] [Google Scholar]
- 21.Gabriel R, Foord RD, Joekes AM. Reversible encephalopathy and acute renal failure after cephaloridine. Br. Med. J. 1970;4:283–284. doi: 10.1136/bmj.4.5730.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodds MG, Foord RD. Enhancement by potent diuretics of renal tubular necrosis induced by cephaloridine. Br. J. Pharmacol. 1970;40:227–236. doi: 10.1111/j.1476-5381.1970.tb09916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasannejad H, Takeda M, Taki K, Shin HJ, Babu E, Jutabha P, Khamdang S, Aleboyeh M, Onozato ML, Tojo A, Enomoto A, Anzai N, Narikawa S, Huang XL, Niwa T, Endou H. Interactions of human organic anion transporters with diuretics. J. Pharmacol. Exp. Ther. 2004;308:1021–9. doi: 10.1124/jpet.103.059139. [DOI] [PubMed] [Google Scholar]
- 24.Eraly SA, Vallon V, Vaughn DA, Gangoiti JA, Richter K, Nagle M, Monte JC, Rieg T, Truong DM, Long JM, Barshop BA, Kaler G, Nigam SK. Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J. Biol. Chem. 2006;281:5072–83. doi: 10.1074/jbc.M508050200. [DOI] [PubMed] [Google Scholar]
- 25.Ueo H, Motohashi H, Katsura T, Inui K. Human organic anion transporter hOAT3 is a potent transporter of cephalosporin antibiotics, in comparison with hOAT1. Biochem. Pharmacol. 2005;70:1104–13. doi: 10.1016/j.bcp.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 26.Jung KY, Takeda M, Shimoda M, Narikawa S, Tojo A, Kim DK, Chairoungdua A, Choi BK, Kusuhara H, Sugiyama Y, Sekine T, Endou H. Involvement of rat organic anion transporter 3 (rOAT3) in cephaloridine-induced nephrotoxicity: in comparison with rOAT1. Life Sci. 2002;70:1861–74. doi: 10.1016/s0024-3205(02)01500-x. [DOI] [PubMed] [Google Scholar]
- 27.Takeda M, Babu E, Narikawa S, Endou H. Interaction of human organic anion transporters with various cephalosporin antibiotics. Eur. J. Pharmacol. 2002;438:137–42. doi: 10.1016/s0014-2999(02)01306-7. [DOI] [PubMed] [Google Scholar]
- 28.Glube N, Closs E, Langguth P. OCTN2-mediated carnitine uptake in a newly discovered human proximal tubule cell line (Caki-1) Mol. Pharmaceutics. 2006;4:160–168. doi: 10.1021/mp060073a. [DOI] [PubMed] [Google Scholar]
- 29.Wagner CA, Lukewille U, Kaltenbach S, Moschen I, Broer A, Risler T, Broer S, Lang F. Functional and pharmacological characterization of human Na+-carnitine cotransporter hOCTN2. Am. J. Physiol.-Renal Physiol. 2000;279:F584–F591. doi: 10.1152/ajprenal.2000.279.3.F584. [DOI] [PubMed] [Google Scholar]
- 30.Georges B, Galland S, Rigault C, Borgne FL, Demarquoy J. Beneficial effects of L-carnitine in myoblastic C2C12 cells: Interaction with zidovudine. Biochem. Pharmacol. 2003;65:1483–1488. doi: 10.1016/s0006-2952(03)00110-2. [DOI] [PubMed] [Google Scholar]
- 31.Pochini L, Scalise M, Indiveri C. Inactivation by omeprazole of the carnitine transporter (OCTN2) reconstituted in liposomes. Chem. Biol. Interact. 2009;179:394–401. doi: 10.1016/j.cbi.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 32.Hirano T, Yasuda S, Osaka Y, Asari M, Kobayashi M, Itagaki S, Iseki K. The inhibitory effects of fluoroquinolones on L-carnitine transport in placental cell line BeWo. Int. J. Pharm. 2008;351:113–118. doi: 10.1016/j.ijpharm.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 33.Sung JH, Yu KH, Park JS, Tsuruo T, Kim DD, Shim CK, Chung SJ. Saturable distribution of tacrine into the striatal extracellular fluid of the rat: evidence of involvement of multiple organic cation transporters in the transporter. Drug Metab. Dispos. 2005;33:440–448. doi: 10.1124/dmd.104.002220. [DOI] [PubMed] [Google Scholar]
- 34.Haschke M, Vitins T, Lude S, Todesco L, Novakova K, Herrmann R, Krahenbuhl S. Urinary excretion of carnitine as a marker of proximal tubular damage associated with platin-based antineoplastic drugs. Nephrol. Dial. Transplant. 2010;25:426–433. doi: 10.1093/ndt/gfp456. [DOI] [PubMed] [Google Scholar]
- 35.Todesco L, Bur D, Brooks H, Török M, Landmann L, Stieger B, Krähenbühl S. Pharmacological manipulation of L-carnitine transport into L6 cells with stable overexpression of human OCTN2. Cell Mol. Life Sci. 2008;65:1596–608. doi: 10.1007/s00018-008-8065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato Y, Sugiura M, Sugiura T, Wakayama T, Kubo Y, Kobayashi D, Sai Y, Tamai I, Iseki S, Tsuji A. Organic cation/carnitine transporter OCTN2 (Slc22a5) is responsible for carnitine transport across apical membranes of small intestinal epithelial cells in mouse. Mol. Pharmacol. 2006;70:829–37. doi: 10.1124/mol.106.024158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.