SUMMARY
Traditional proteomics methodology allows global analysis of protein abundance but does not provide information about the regulation of protein activity. Proteases in particular are known for their multilayered posttranslational activity regulation which can lead to a significant difference between protease abundance levels and their enzyme activity. To address those issues, the field of activity based proteomics has been established in order to characterize protein activity and monitor the functional regulation of enzymes in complex proteomes. In this review, we present structural features of activity based probes for proteases and discuss their applications in proteomic profiling of various catalytic classes of proteases.
Keywords: Activity based probes, proteomics, proteolysis, proteases
INTRODUCTION
All organisms express a large variety of structurally and catalytically diverse proteases. Genome sequencing projects have revealed the presence of at least 569 human proteases that can be divided into several distinct catalytical classes: metalloproteinases, serine, cysteine, threonine and aspartic proteases [1]. Interestingly, the mouse and rat genomes show even higher protease variety with 644 and 629 members respectively [2]. Proteases have the unique ability to hydrolyze peptide bonds and therefore irreversibly modify the function of target proteins. They play crucial roles in diverse physiological processes such as protein turnover, blood coagulation, apoptosis, hormone processing and bone remodeling. Because proteolytic processing is an irreversible event, it must be tightly regulated to avoid catastrophic consequences. The most common way that proteases are regulated is by expression as an inactive proenzyme that requires activation by proteolytic cleavage. The activation process can be regulated by cellular signaling as well as by changes in the microenvironment. If this regulation fails, endogenous extracellular and intracellular protease inhibitors (such as serpins and cystatins) prevent any unwanted proteolysis. This multilayered post-translational regulation can lead to significant difference between overall abundance levels and activity of proteases. Thus, methods that rely on detection of protease abundance rather than activity do not always provide sufficient information to allow functional assignments of specific proteases.
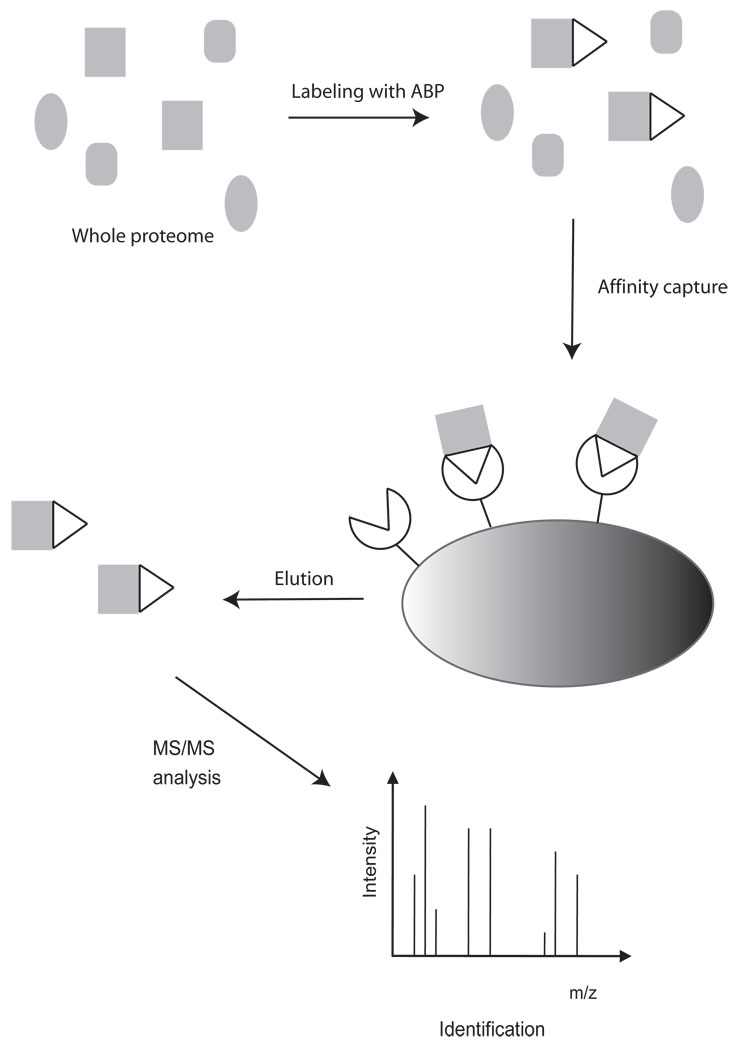
Traditional proteomics methodology based on 2D gel chromatograhpy coupled with mass spectrometry has recently been complemented with non-gel based methods such as MudPIT and SELDI [3, 4]. Chromatography separations employed in these methods significantly increase resolving power and proteome coverage but they still lack the ability to resolve active and inactive populations of enzymes. Activity based proteomics is a relatively new sub-division of proteomics that has been developed to characterize protein activity and directly monitor the functional regulation of enzymes in complex proteomes. This method utilizes small molecule activity based probes (APBs) which covalently modify the enzyme active site and enable detection and affinity purification of a target enzyme population (Fig. 1). Activity based probes are highly selective and can be used for analysis of complex samples such as cell lysates, intact cells and even whole organisms. In this review, we will outline advances in the development of activity based probes and highlight the numerous applications of these reagents for the study of various protease classes and their roles in human disease.
Figure 1. Workflow of activity based proteomics.
Target proteins in the complex proteome are labeled with activity based probes. Labeled proteins are enriched by affinity purification and identified by mass spectrometry.
STRUCTURE OF ACTIVITY BASED PROBES
All activity based probes share a similar basic design, which incorporates elements required for targeting, modification and detection of labeled proteins. Structurally, those elements can be divided into the reactive group, linker and tag (Fig 2A).
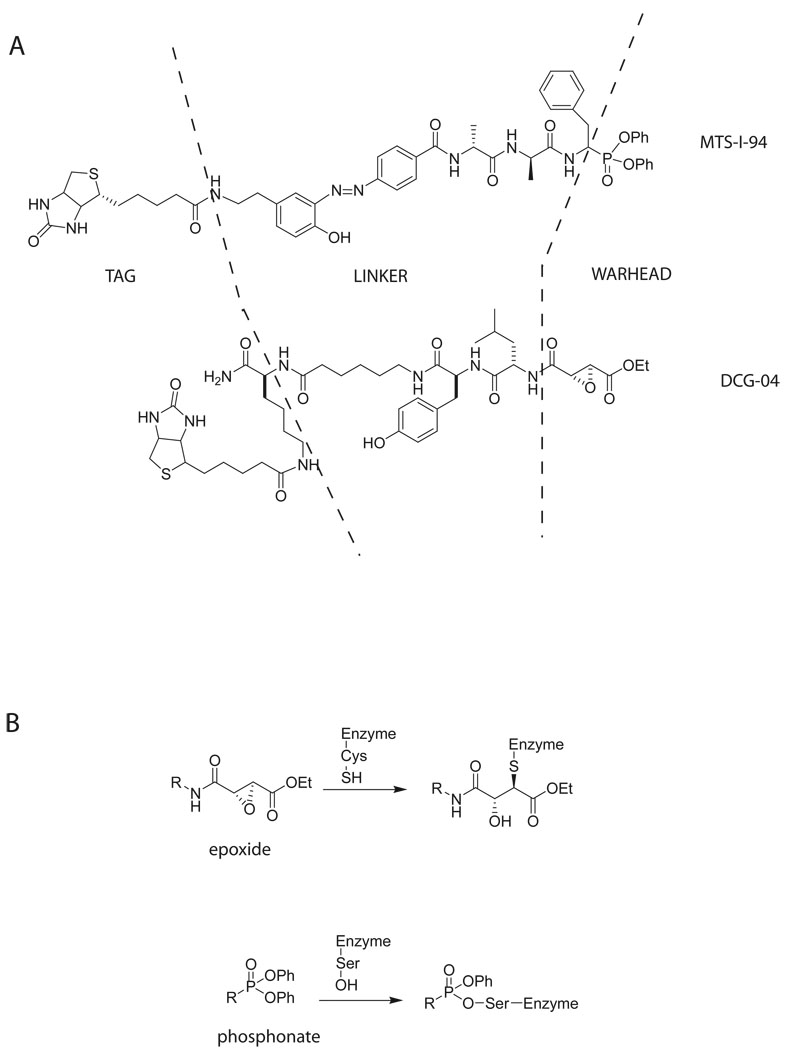
Figure 2. Structure and binding of activity based probes.
A: Structural features of two types of activity based probes are shown. Probe MTS-I-94 targets serine proteases. It is composed of phosphonate warhead, a cleavable diazo linker and a biotin tag. DCG-04 is a cysteine protease probe with epoxide warhead.
B: Probes that target serine and cysteine proteases irreversibly and covalently bind to the active site nucleophile (serine or cysteine).
Reactive group
The reactive functional group (also termed as ‘’the warhead”) provides a covalent attachment of the probe to the catalytic residue of the protease active site. The majority of protease families have well-established catalytic mechanisms that have been defined using abundant structural and kinetic data. This data has enabled the synthesis of a large variety of specific, irreversible small molecule inhibitors, that can be transformed into activity based probes by the addition of an appropriate reporter tag [5]. Probes for serine, cysteine and threonine proteases utilize electrophile groups that form a covalent bond with their active site nucleophile (serine, cysteine or threonine residue). The electrophile has to be strong enough to react with the active site nucleophile but not with other free nucleophiles in the proteome (Fig 2B). Various reactive groups have been successfully applied to protease probe design (Table 1). The phosphonate group is highly reactive towards the hydroxyl nucleophile of serine proteases and has therefore found use a number of probes for both trypsin and chymotrypsin fold serine proteases [6]. The proteasome, on the other hand, has been targeted with probes containing a vinyl sulfone reactive group even though this class of compounds was originally designed to react with cysteine proteases [7]. Interestingly, a,b-epoxy ketones are also known to form highly selective bounds with the N-terminal threonine of the proteasome [8]. Cysteine proteases are the family most extensively studied using ABPs and probes containg a number of different reactive functional groups including epoxides, diazomethyl ketones, acyloxymethyl ketones and vinyl sulfones have been designed [9–11]. Unfortunately, the same electrophile-based approach does not work well for metalloproteinases and aspartic proteinases because they do not form covalent intermediates with their substrates. In this case, probes are designed using reversible inhibitor scaffolds coupled to a photoreactive group that provides covalent attachment upon irradiation with UV light [12, 13].
Table 1.
Types of reactive groups used for targeting of various protease classes.
| Reactive group | Targeted protease class | References |
|---|---|---|
| phosphonate | serine proteases | 6, 67–70 |
| sulfonate | serine proteases | 68 |
| vinyl sulphone | cysteine proteases threonine proteases (proteasome) | 7, 11, 30, 79 |
| epoxide | cysteine proteases | 10, 41–47 |
| diazomethyl ketone | cysteine proteases | 5 |
| acyloxymethyl ketone | cysteine proteases | 9, 89 |
| hydroxamate | metalloproteases | 13, 72–77 |
Linker
The most basic function of the linker is to separate the reactive functional group from the tag. This improves probe accessibility and reduces steric hindrance between both groups. For this purpose, the linker can be as simple as an extended alkyl or polyethylene glycol spacer. Alternatively, the linker can be used to increase probe specificity by addition of structural elements (i.e. peptides) that enhance the specificity of the probe. For most protease probes, the linker is a short peptide sequence that mimics a true protein substrate. This peptide linker can be optimized to target distinct subfamilies of proteases by changing its aminoacid sequence [14]. A more recent improvement in linker design has been the development of a region that can be enzymatically or chemically cleaved to release probe labeled proteins. This simplifies the elution process and decreases background protein contamination during an affinity purification. Various cleavable linker elements have recently been reported. Cravatt and coworkers developed a cleavable linker that has a peptide recognition site for tobacco etch virus protease (TEV) and applied it to the identification of proteins that had been labeled with a sulfonate ester probe [15, 16].
One of the first chemically cleavable linkers described makes use of a disulfide bond that is readily cleaved in mild reductive conditions [17–19] (Fig. 3A). Disulfide linkers work well in some applications, such as profiling of serine hydrolases, but they are not compatible with reductive buffers which are needed for profiling of cysteine proteases. Disulfides are also quite labile and prone to disulfide exchange, which can also lead to a premature release and nonspecific labeling of proteins containing free thiol groups [20]. These problems have been addressed with development of a novel diazo cleavable linker, which enables selective elution under mild reductive conditions [21] (Fig. 3A). The diazo linker was successfully incorporated into ABPs that were used for labeling and proteomic identification of cysteine and serine proteases [22]. As an alternative to cleavage by chemical reduction, acid cleavable linkers have been developed and are also commercially available for ICAT applications [23–25]. However, the strong acids required for cleavage (TFA) also release nonspecifically bound proteins. Finally, the linker region can be used for isotopic labeling in ICAT-like applications. Incorporation of light and heavy linkers have been demonstrated for the general papain family protease probe DCG-04 which was used for quantitative profiling of cysteine proteases by direct mass spectrometry based methods [26].
Figure 3. Cleavable and ‘’click” chemistry linkers.
A: Reduction cleavage of disulfide and diazo cleavable linkers.
B: Mechanism of ‘’click” chemistry. Cycloaddition of an azide and alkyne functional group is used for the tag attachment after protein labeling.
Tag
Incorporation of a tag group into an activity based probe enables the visualization and/or purification of labeled proteins. Radioisotopes are commonly used in biological applications and were also the first to be used as tags for activity based proteomics [27]. The gamma ray emitter 125I can be introduced by iodination of any covalent inhibitor that has a phenol group. Labeled proteins can then be analyzed by 1D or 2D electrophoresis and visualized by autoradiography. This approach was used in activity profiling of cysteine proteases such as cathepsins [28], caspases [29] and the proteasome [30, 31]. Protease probes that incorporate 3H as a tag have also been reported [32]. Tritium replaces hydrogen and this modification does not affect the probe structure. However, its use is limited by low specific activity that requires long exposure times to obtain labeling profiles. Although radiolabeled probes are easy to prepare, their storage time is limited by the half-life of the radiolabel. They also require special handling and laboratory safety procedures and cannot be used to directly identify target proteins.
The application of fluorescent tags was a big step forward in the development of activity based probes. Fluorophores are safe to use and are commercially available in a variety of excitation and emission spectral ranges. The first fluorescent APBs were demonstrated for cysteine proteases and serine hydrolases [33, 34]. Various types of dyes can be used as fluorophores but their chemical properties can significantly influence their range of applications. Fluorescein and rodamine are inexpensive, but susceptible to photobleaching which limits their imaging applications. The fluorophosphonate warhead containing a PEG linker labeled with rhoadamine tag has been demonstrated to be an effective probe for profiling of serine hydrolases in cell homogenates [33]. Fluorescein and rodamine are not cell permeable and therefore cannot be used for labeling of intracellular targets in vivo. Dansyl and NBD (nitrobenz-2-oxa-1,3-diazole) are inexpensive fluorophores that have been used for in vitro activity based profiling [34, 35]. BODIPY (dipyrromethene boron difluoride) and cyanine (Cy) dyes are photostable with high absorption coefficients, high quantum yields and narrow absorption peaks. They are also cell permeable making them useful in a variety of biological applications. BODIPY labeled epoxide probes have been used for labeling of cysteine proteases in lysates and intact cells [36]. Caspase probes containing the Cy fluorophore have been reported [37]. Because of their superior chemical properties, cyanine dyes have a great potential for use in imaging applications. Unfortunately, they are also extremely expensive which limits their use for large scale synthesis.
Fluorophores are perfect for rapid determination of labeling patterns after SDS-PAGE analysis, since gels can be directly scanned using a laser scanner and there is no need for time consuming blotting procedures required for biotinylated probes. However, for the purpose of isolation and identification of proteins targeted by APBs, biotin still remains the affinity tag of choice. Strong, diffusion limited interaction between biotin and immobilized avidin provides efficient enrichment of even low abundant labeled targets. Avidin based enrichment also reduces complexity of biological samples which is extremely beneficial for proteomic applications. Unfortunately, the strong avidin-biotin interaction requires harsh elution conditions which are not directly compatible with mass spectrometry analysis. Eluted samples must first be analyzed by SDS-PAGE and proteins cut from a gel for identification. Alternatively, enriched proteins can be prepared for mass spectrometry by ‘’on bead” digestion. In this approach, proteins are reduced, alkylated and digested while they are still bound to immobilized avidin. In both approaches the final sample is contaminated with natively biotinylated proteins and abundant, nonspecifically bound proteins. This contamination can be minimized by usage of probes with cleavable linkers, which enable specific elution [22]. The biggest disadvantage of the biotin tag is its overall poor cell permeability which limits its use for in situ and in vivo applications. Since protease activity is often regulated by factors within its intracellular microenvironment, labeling of cell lysates often does not provide an accurate picture of protease activity profiles. To resolve this problem, a tandem labeling strategy which utilizes ‘’click chemistry” was developed. Çlick chemistry is based on a [2+3] cycloaddition of an azide and alkyne functional group in the presence of copper catalyst (Fig. 3B). This method was originally developed by Sharpless and coworkers but in the modified form was applied to activity based proteomics [38, 39]. In this approach, intact cells are labeled with a cell permeable probe, which instead of a biotin tag carries an alkyne or azido group. After labeling, cells are homogenized and the biotin tag (which carries a complementary alkyne or azido functional group) is added by cycloaddition. Tagged proteins can than be enriched and identified by mass spectrometry [40].
APPLICATION OF PROBES FOR THE PROFILING OF PROTEASES
Cysteine, serine and threonine proteases all have an active site nucleophile that can be covalently modified by electrophiles and it is of no surprise that majority of reported activity based probes have been designed against these protease classes. Activity based probes for numerous cysteine and serine proteases are available but, so far, the proteasome is the only threonine protease to be profiled by ABPs. Probe design for proteases that do not form a covalent intermediate with its substrate have proven to be much more challenging. Probes that target metalloproteses were reported just few years ago but ABPs that target aspartic proteases still remain elusive. In this part of the review we will outline the development and application of activity based proteomics for each of the various protease classes.
CYSTEINE PROTEASES
The main catalytical feature of cysteine proteases is the use of a Cys residue that is activated for nucleophilic attack by a nearby His residue. Cyteine proteases are further divided into six clans based on the structure of their active site. The two most abundant and intensively studied clans are the CA and CD clans. Clan CA includes papain and calpain families of cysteine proteases as well as various families of the ubiquitin processing peptidases. E-64 is a well known natural product inhibitor of this family of cysteine proteases and its structure and reactive epoxide were a logical choice for the use in the design of cysteine protease activity based probes [41]. The general epoxide probe DCG-04 has a wide specificity toward calpains and cysteine cathepsins and was used to determine their roles in various physiological and pathological processes. DCG-04 was used to identify m-calpain and the related Lp82 as the main active cysteine proteases present in the eye lens that mediate γ-crystalin cleavage during cataract formation [42]. Activity based labeling and affinity purification with DCG-04 was also used for identification of the papain-like protease cathepsin L as the prohormone processing enzyme responsible for cleaving proenkephalin [43]. This probe was also successfully used to identify cysteine proteases involved in host cell invasion by the parasite Plasmodium falciparum and in the proteomic identification of papain-like proteases in plants [44, 45]. Papain-like cathepsins were also found to be involved in various stages of cancer progression. Fluorescent versions of DCG-04 were used to profile up-regulation of cathepsin activity in various stages of tumorigenesis in several mouse cancer models [46, 47]
Mammalian deubiquitinating enzymes (DUBs) are another important group of clan CA cysteine proteases. Many of these enzymes act as oncogenes, tumor suppressors or have some other connection to cancer and therefore represent important target for drug development. The first DUB-specific activity based probes were designed based on a full length ubiquitin (Ub) derivatized at the C-terminus with a reactive vinyl sulfone. Fluorogenic substrates for DUBs such as Ub-7-amido-4-methyl coumarin (UbAMC) as well as DUB inhibitors Ub-aldehyde and Ub-nitrile were also developed [48–51]. Development of the irreversible DUB inhibitor UbVS (ubiquitin vinyl sulfone) represented an important advancement in the study of DUB function. UbVS binding is SDS resistant and also enables radioiodination with subsequent autoradiographic visualization of labeled enzymes [52]. However, this probe still lacks the ability to enrich and isolate targets for the purpose of proteomic identification. To achieve this goal an intein-based chemical ligation method was introduced. Here, an N-terminally epitope tagged ubiquitin derivative with a C-terminal electrophile was prepared by combination of recombinant protein expression and synthetic chemistry [53]. This probe allowed activity based profiling and proteomic identification of previously uncharacterized DUBs in complex biological samples [54]. In a more complex study, DUB activity was monitored in tumor cell lines and was found to be increased as a direct consequence of cell transformation [55]. Activity based probes have also been used for proteomic identification of DUBs in various pathogenic organisms such as herpes simplex virus, human cytomegalovirus, chlamydia, Plasmodium falciparum and Escherichia coli [56–59].
Clan CD is the second most abundant clan of cysteine proteases. Its members include caspases, gingipains, legumains, clostripains and separases. Caspases are of particular interest due to their crucial role in apoptosis and it is not surprising that they were targets of the first generation of APBs for the CD clan. Caspase probes utilizing acyloxymethyl ketone (AOMK) and aldehyde reactive groups coupled to a specific peptide sequence and a biotin tag have been designed. In fact, the first caspase (caspase 1, β-interleukin converting enzyme- ICE) was identified using a biotinylated AOMK activity based probe [60]. Recently, a small molecule positional scanning library was used for the development of highly specific AOMK based probes for caspases 3, 7, 8 and 9. These probes could then be used for monitoring caspase activation kinetics upon stimulation of apoptosis in cell-free extracts and intact cells [61]. APBs were also designed for separases and legumain. Acyloxymethyl and chloromethyl ketone based probes were used to study the role of separase in cell division [62]. Legumain is thought to play a role in lysosomal protein degradation although its exact role still remains elusive. Legumain can participate in the processing of antigenic peptides and in generation of double chain forms of the papain-like cathepsins [63, 64]. A highly specific set of AOMK based probes for legumain was recently developed that could provide key insight into its physiological role [65].
SERINE PROTEASES
Serine proteases are part of the larger serine hydrolase family which represents approximately 1% of all genes in the human genome. Besides proteases, the serine hydrolase family also includes numerous lipases, esterases, amidases and transacylases. All of them share the same catalytical mechanism which involves a serine nucleophile, that is activated by a proton relay involving an acidic residue (aspartate or glutamate) and basic residue (usually histidine). The majority of serine hydrolases are irreversibly inhibited by the fluorophosphonate (PF) reactive group. Thus probes containing PEG or aliphatic linkers and various tags attached to this warhead have been developed as broad specificity serine hydrolyase ABPs [66]. These probes have been used in a range of profiling experiments, most notably in studies of serine hydrolase biomarkers in cancer. Cravatt and coworkers used fluorescent PF-Rhodamine probes to profile serine hydrolase activities in a series of cancer cell lines [67]. Samples were labeled, analyzed by SDS-PAGE and the labeled protein profile was visualized by laser scanning. In a parallel experiment, a biotinylated version of the probe was used to directly isolate targets by affinity chromatography for identification by mass spectrometry. The identified serine hydrolase profiles were used to classify cancer cell lines into functional subtypes based on tissue of origin and state of invasiveness, thus demonstrating the diagnostic power of the probes for disease classification.
A similar approach was also used to profile hydrolases found in different stages of breast cancer [68]. Serine hydrolase specific probes (fluorophosphonate and sulfonate ester reactive groups) were used to study the differences in activity profiles of MDA-MB-231 human breast cancer cells, when grown in cell culture and after tumor formation in mouse mammary fat pads. These studies showed that cancer cells exhibited distinct activity profiles when grown in vitro (in culture) and in vivo (xenograft tumors). More than seven types of enzyme activities with unique activity profiles were identified. These findings suggest that studies using human cancer cell lines grown in culture may not be predictive of the behaviour of these same cells in vivo. It was also noted that many dramatic alterations in enzyme activities occurred as a result of posttranscriptional events, again confirming the value of activity based profiling methods.
This cancer profiling methodology was soon enhanced by incorporation of non-gel methods for proteomic analysis of labeled samples. A two-phase functional proteomic platform was reported, where in the first phase serine hydrolases were labeled by fluorogenic FP-probe and an activity profile was determined by 1D SDS-PAGE analysis. This stage required a minimal amount of sample and could be applied to a wide range of primary human specimens. In the second phase, labeled targets were identified by multidimensional LC-MS/MS (MudPIT). Thus, the sensitivity and resolution of MudPIT was coupled to the high-content functional information obtained by activity based profiling. Using this approach, more than fifty enzyme activities were identified in breast tumor samples [69]. Comparison of activity based proteomics data with cDNA microarray analysis showed that for some enzymes, activity and mRNA levels were largely uncorrelated. This finding additionally emphasizes the importance of activity based proteomics, which can detect low abundance disease-associated enzymes, that might evade other molecular profiling methods. As an alternative to the FP probe, which shows broad reactivity with serine hydrolyases, the diphenylphosphonate warhead has been used to design serine protease specific probe. P1 basic amino acid probes were found to target only trypsin family serine proteases and proved valuable for the profiling of serine protease activity in the mast cells [70].
METALLOPROTEASES
Perhaps the most critical difference between metalloproteases and the serine and cysteine classes of proteases is that they do not use an aminoacid sidechain as a nucleophile for direct covalent attack on the substrate. Instead, they utilize a zinc ion in their active site to deprotonate a water molecule which then mediates hydrolysis of the peptide bond [71]. Since metalloproteases do not form acyl-enzyme intermediates they cannot be labeled using simple electrophiles fused to a peptide that mimics a protein substrate. To overcome this problem, probes containing a hydroxamate scaffold linked to a benzophenone crosslinking group have been developed [72]. The hydroxamate is a strong zinc chelating agent, which tightly binds in the protease active site. While this is a high affinity interaction, it is not irreversible and the benzophenone photocrosslinking group is required for the attachment of the probe to the protease active site. The main disadvantage of this type of ABPs is that they are not suitable for use in living cells and whole animals. Hydroxamate probes for metalloproteases were successfully applied in the search for diagnostic markers in an invasive melanoma cell line [72]. In this study, a trifunctional hydroxamate probe (with fluorescent and biotin tag) was used for detection, affinity isolation and identification of neprilysin. This metallo protease was also found to be highly upregulated in melanoma cell lines. Neprilysin is a membrane associated metalloprotease which is considered to be a negative regulator of tumorigenesis since it is known to degrade several mitogenic peptides [73]. The results reported with the metallo protease ABPs suggest that, in some tumor types, it may also promote cancer progression.
Actvity based probes usually only target a limited number of members within a specific enzyme class. This is problematic when trying to globally profile large and structurally diverse protease families such as metalloproteases. To overcome this problem, Cravatt and coworkers developed a profiling strategy using small libraries of structurally diverse photoreactive hydroxamate probes. By using libraries that were designed to have complementary metalloprotease selectivity it was possible to increase the overall coverage of the metalloproteases. The probes in the library were designed to be cell permeable by incorporating the alkyne group, so that a tag of choice (fluorophore or biotin) could be added after labeling using ‘’click” chemistry. Initial SDS-PAGE analysis enabled the selection of a ‘’cocktail” of optimal probes, that provided the greatest coverage of the family. This mixture of probes was used for more extensive ABPP-MudPIT analysis of cancer cell lines [74]. With this approach, the authors were able to identify over 20 metalloproteases in various human and mouse samples. Among the identified metalloproteases were members of all of the main branches of this enzyme family. Other examples of hydroxamate probes for matrix metalloproteases using ‘’click” chemistry have also been reported [75]. Metalloprotease probe libraries have also been created and used for the rapid determination of metalloprotease inhibitor specificity fingerprints using a microarray approach [76, 77].
THREONINE PROTEASES
So far the only threonine protease extensively studied using activity based proteomics is the proteasome. This multi-subunit complex is the central protease activity involved in ubiquitin mediated protein degradation. It degrades old and damaged proteins and fulfills several vital regulatory functions. The proteasome has six proteolytic sites in the central core particle that possess three distinct substrate specificities: chymotrypsin-like, trypsin-like and peptidyl-glutamyl peptide hydrolyzing [78]. The proteasome is a threonine protease since it uses the N-terminal threonine of the catalytic β-subunits as a nucleophile for the attack on the peptide bond. Although initially designed to target cysteine proteases, carboxy-terminal vinylsulfones have proven effective as irreversible inhibitors of active site N-terminal threonine and are now widely used as warheads for design of ABPs [30]. The first probes developed for characterization of catalytical proteasome β subunits made use of the vinylsulfone warheads coupled to a peptide that could be labeled with a radioisotope or biotin tag [79]. In addition, cell permeable proteasome probes have also been prepared by incorporation of azido linker (‘’click chemistry”) or hapten tag [80, 81]. Finally, cell permeable fluorescent probes were recently developed for visualization of labeled proteasomes in living cells and animals [82, 83]. The natural product epoxomicin was also found to be a highly selective, irreversible covalent inhibitor of the proteasome. This natural product contains a reactive a,b epoxyketone that forms a stable six member ring with the N-terminal threonine. A labeled version of this natural product was used as an ABP to identify the target as the proteasome [84]. Thus, this class of compounds is likely to find use as a highly specific ABP of the proteasome.
Expert Opinion and five-year view
The authors believe that the next five years will see a continued growth in the development and application of activity based probes for not only proteases but also for additional classes of important regulatory enzymes. The past five years has already seen a dramatic expansion in the diversity of enzyme families that can be studied using activity based proteomic methods. Creative work by chemists has lead to the development of probes for kinases, histone deacetylases, phosphatases and glycanases to name just a few [85–88]. The development of activity based probes that target diverse enzyme families has led to a range of applications for activity based proteomics. Probes can be applied to a large collection of complex biological samples and enable global profiling of enzyme activities. Comparison of enzyme activity profiles from healthy and disease samples can lead toward identification of novel biomarkers and drug targets. Important advantage of activity based probes is also in profiling of enzyme activities in vivo. Last year Blum and coworkers developed protease probes that enable monitoring of enzyme activities by whole body imaging. Furthermore, those probes also allow subsequent ex vivo identification and biochemical characterization of labeled targets [89]. Those results show that ABPs have a potential to become an important imaging tools used for disease diagnosis and for preclinical and clinical testing of therapeutic agents in vivo. ABP based in vivo assays significantly improve high throughput inhibitor screens needed for discovery of novel therapeutic agents. They enable screening of more than one target in a single experiment and because protein targets are screened in their native environment there is a better chance of detecting any unanticipated off-target inhibition or activation [90, 91].
We can expect that the number and types of applications for ABPs will continue to expand. This rapidly growing field is likely to have a significant impact on a number of disciplines of research with proteomics being perhaps the most directly impacted. Besides profiling of protease activities and target identification, chemical labeling has also become a powerful tool for proteomic identification of protease substrates. In this approach, complex proteomes are directly treated with specific proteases and newly formed N-terminal ends of cleaved substrates are chemically modified by biotinylated reagent. After affinity purification, substrates and their exact cleavage sites are identified by mass spectrometry [92]. Cleavage sites can also be quantitatively evaluated by incorporation of quantitative proteomic techniques such as iTRAQ [93, 94]. Determination of whole complement of cellular protease substrates (degradome) remains one of the main challenges that field of proteolysis faces today.
As the quality of instrumentation improves and we are able to analyze more and more complex, biologically relevant samples, we anticipate that APBs will help to siphon off the most relevant and important information from these large data sets. This will require a wide-spread integration of the probes and continued efforts from chemist to develop probes with enhanced functionality. We are confident that the early developments in this field have created the necessary positive momentum to see these goals realized.
KEY ISSUES
-
-
The main advantage of activity based proteomics is that it detects enzyme activity instead of abundance and therefore provides more accurate information about the biological roles of these enzymes in a given biological process.
-
-
Activity based proteomics makes use of small molecules probes that are designed to target specific enzyme classes.
-
-
Activity based probes are composed of a functional group (warhead), linker and tag, which enable specific labeling and enrichment of target proteins.
-
-
Activity based probes for all the main protease classes of proteases (serine, cysteine, threonine and metalloproteases) have been successfully developed and are in use for global functional studies of proteases.
REFERENCES
- 1.Rawlings ND, Morton FR, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puente XS, Lopez-Otin C. A genomic analysis of rat proteases and protease inhibitors. Genome Res. 2004;14:609–622. doi: 10.1101/gr.1946304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seibert V, Ebert MP, Buschmann T. Advances in clinical cancer proteomics: SELDI-ToF-mass spectrometry and biomarker discovery. Brief Funct Genomic Proteomic. 2005;4(1):16–26. doi: 10.1093/bfgp/4.1.16. [DOI] [PubMed] [Google Scholar]
- 4.Chen EI, Hewel J, Brunhilde Felding-Habermann B, Yates JR., 3rd Large Scale Protein Profiling by Combination of Protein Fractionation and Multidimensional Protein Identification Technology (MudPIT) Mol Cell Proteomics. 2006;5(1):53–56. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 5. Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine and threonine proteases. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v.. Extensive review of small molecule protease inhibitors and their inhibition mechanisms.
- 6.Oleksyszyn J, Powers JC. Irreversible inhibition of serine proteases by peptide derivatives of (alpha-aminoalkyl)phosphonate diphenyl esters. Biochemistry. 1991;30(2):485–493. doi: 10.1021/bi00216a026. [DOI] [PubMed] [Google Scholar]
- 7.Bogyo M, Gaczynska M, Ploegh HL. Proteasome inhibitors and antigen presentation. Biopolymers. 1997;43(4):269–280. doi: 10.1002/(SICI)1097-0282(1997)43:4<269::AID-BIP2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 8.Bo Kim K, Fonseca FN, Crews CM. Development and characterization of proteasome inhibitors. Methods Enzymol. 2005;399:585–609. doi: 10.1016/S0076-6879(05)99039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KAH, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nature Chemical Biology. 2005;1(1):33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 10.Sadaghiani AM, Verhelst SH, Gocheva V, Hill K, Majerova E, Stinson S, Joyce JA, Bogyo M. Design, synthesis, and evaluation of in vivo potency and selectivity of epoxysuccinyl-based inhibitors of papain-family cysteine proteases. Chem Biol. 2007;14(5):499–511. doi: 10.1016/j.chembiol.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Mahesh U, Chen GY, Yao SQ. Solid-phase synthesis of peptide vinyl sulfones as potential inhibitors and activity-based probes of cysteine proteases. Org Lett. 2003;5(5):737–740. doi: 10.1021/ol0275567. [DOI] [PubMed] [Google Scholar]
- 12.Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, Harrison T, Lellis C, Nadin A, Neduvelil JG, Register RB, Sardana MK, Shearman MS, Smith AL, Shi XP, Yin KC, Shafer JA, Gardell SJ. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405(6787):689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- 13.Hagenstein MC, Mussgnug JH, Lotte K, Plessow R, Brockhinke A, Kruse O, Sewald N. Affinity-based tagging of protein families with reversible inhibitors: a concept for functional proteomics. Angew Chem Int Ed Engl. 2003;42(45):5635–5638. doi: 10.1002/anie.200352084. [DOI] [PubMed] [Google Scholar]
- 14. Greenbaum DC, Arnold WD, Lu F, Hayrapetian L, Baruch A, Krumrine J, Toba S, Chehade K, Bromme D, Kuntz ID, Bogyo M. Small molecule affinity fingerprinting. A tool for enzyme family subclassification, target identification, and inhibitor design. Chem Biol. 2002;9(10):1085–1094. doi: 10.1016/s1074-5521(02)00238-7.. One of the crucial reports describing affinity fingerprinting method for functional characterisation of cysteine protease families. This approach significantly improved inhibitor design and target selection methodology.
- 15.Speers AE, Cravatt BF. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J Am Chem Soc. 2005;127(28):10018–10019. doi: 10.1021/ja0532842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2(6):1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 17.Shimkus M, Levy J, Herman T. A chemically cleavable biotinylated nucleotide: usefulness in the recovery of protein-DNA complexes from avidin affinity columns. Proc Natl Acad Sci U S A. 1985;82:2593–2597. doi: 10.1073/pnas.82.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hekmat O, Kim YW, Williams SJ, He S, Withers HG. Active site peptide ‘’fingerprinting” of glycosidases in complex mixtures by mass spectrometry. Discovery of a novel retaining beta-1,4-glycanase in Cellulomonas firmi. J Biol Chem. 2005;280:35126–35135. doi: 10.1074/jbc.M508434200. [DOI] [PubMed] [Google Scholar]
- 19.Everley PA, Gartner CA, Haas W, Saghatelian A, Elias JE, Cravatt BF, Zetter BR, Gygi SP. Assessing enzyme activities using stable isotope labeling and mass spectrometry. Mol Cell Proteomics. 2007;6(10):1771–1777. doi: 10.1074/mcp.M700057-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Ichikawa M, Ichikawa Y. A mechanism-based affinity-labeling agent for possible use in isolating N-acetylglucosaminidase. Bioorg Med Chem Lett. 2001;11(13):1769–1773. doi: 10.1016/s0960-894x(01)00300-6. [DOI] [PubMed] [Google Scholar]
- 21.Verhelst SH, Fonović M, Bogyo M. A mild chemically cleavable linker system for functional proteomic applications. Angew Chem Int Ed Engl. 2007;46(8):1284–1286. doi: 10.1002/anie.200603811. [DOI] [PubMed] [Google Scholar]
- 22.Fonović M, Verhelst SH, Sorum MT, Bogyo M. Proteomics Evaluation of Chemically Cleavable Activity-based Probes. Mol Cell Proteomics. 2007;6(10):1761–1770. doi: 10.1074/mcp.M700124-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.van der Veken P, Dirksen EH, Ruijter E, Elgersma RC, Heck AJ, Rijkers DT, Slijper M, Liskamp RM. Development of a novel chemical probe for the selective enrichment of phosphorylated serine- and threonine-containing peptides. Chembiochem. 2005;6(12):2271–2280. doi: 10.1002/cbic.200500209. [DOI] [PubMed] [Google Scholar]
- 24.Fauq AH, Kache R, Khan MA, Vega IE. Synthesis of acid-cleavable light isotope-coded affinity tags (ICAT-L) for potential use in proteomic expression profiling analysis. Bioconjug Chem. 2006;17(1):248–254. doi: 10.1021/bc0503059. [DOI] [PubMed] [Google Scholar]
- 25.An acid-cleavable ICAT reagent with undisclosed structure is available from Applied Biosystems.
- 26.van Swieten PF, Maehr R, van den Nieuwendijk AM, Kessler BM, Reich M, Wong CS, Kalbacher H, Leeuwenburgh MA, Driessen C, van der Marel GA, Ploegh HL, Overkleeft HS. Development of an isotope-coded activity-based probe for the quantitative profiling of cysteine proteases. Bioorg Med Chem Lett. 2004;14(12):3131–3134. doi: 10.1016/j.bmcl.2004.04.046. [DOI] [PubMed] [Google Scholar]
- 27.Mason RW, Bartholomew LT, Hardwick BS. The use of benzyloxycarbonyl[125I]iodotyrosylalanyldiazomethane as a probe for active cysteine proteinases in human tissues. Biochem J. 1989;263(3):945–949. doi: 10.1042/bj2630945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogyo M, Verhelst S, Bellingard-Dubouchaud V, Toba S, Greenbaum D. Selective targeting of lysosomal cysteine proteases with radiolabeled electrophilic substrate analogs. Chem Biol. 2000;7(1):27–38. doi: 10.1016/s1074-5521(00)00061-2. [DOI] [PubMed] [Google Scholar]
- 29.Methot N, Vaillancourt JP, Huang J, Colucci J, Han Y, Menard S, Zamboni R, Toulmond S, Nicholson DW, Roy S. A caspase active site probe reveals high fractional inhibition needed to block DNA fragmentation. J Biol Chem. 2004;279(27):27905–27914. doi: 10.1074/jbc.M400247200. [DOI] [PubMed] [Google Scholar]
- 30.Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh H. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc Natl Acad Sci U S A. 1997;94(13):6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessler BM, Tortorella D, Altun M, Kisselev AF, Fiebiger E, Hekking BG, Ploegh HL, Overkleeft HS. Extended peptide-based inhibitors efficiently target the proteasome and reveal overlapping specificities of the catalytic beta-subunits. Chem Biol. 2001;8(9):913–929. doi: 10.1016/s1074-5521(01)00069-2. [DOI] [PubMed] [Google Scholar]
- 32.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268(5211):726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 33.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1(9):1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Berkers CR, Verdoes M, Lichtman E, Fiebiger E, Kessler BM, Anderson KC, Ploegh HL, Ovaa H, Galardy PJ. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat Methods. 2005;2(5):357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- 35.Schmidinger H, Birner-Gruenberger R, Riesenhuber G, Saf R, Susani-Etzerodt H, Hermetter A. Novel fluorescent phosphonic acid esters for discrimination of lipases and esterases. Chembiochem. 2005;6(10):1776–1781. doi: 10.1002/cbic.200500013. [DOI] [PubMed] [Google Scholar]
- 36. Greenbaum D, Baruch A, Hayrapetian L, Darula Z, Burlingame A, Medzihradszky KF, Bogyo M. Chemical approaches for functionally probing the proteome. Mol Cell Proteomics. 2002;1(1):60–68. doi: 10.1074/mcp.t100003-mcp200.. One of the first reports describing functional proteomic approach for profiling of proteases in complex proteomes.
- 37.Liau ML, Panicker RC, Yao SQ. Design and synthesis of an affinity probe that targets caspases in proteomic experiments. Tetrahedron Lett. 2003;44:1043–1046. [Google Scholar]
- 38.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41(14):2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125(16):4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 40. Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11(4):535–546. doi: 10.1016/j.chembiol.2004.03.012.. Description of click chemistry application for proteomic analysis of proteases in complex samples.
- 41.Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol. 2000;7(8):569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 42.Baruch A, Greenbaum D, Levy ET, Nielsen PA, Gilula NB, Kumar NM, Bogyo M. Defining a link between gap junction communication, proteolysis, and cataract formation. J Biol Chem. 2001;276(31):28999–29006. doi: 10.1074/jbc.M103628200. [DOI] [PubMed] [Google Scholar]
- 43.Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, Schilling B, Petermann I, Dehnert J, Logvinova A, Goldsmith P, Neveu JM, Lane WS, Gibson B, Reinheckel T, Peters C, Bogyo M, Hook V. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc Natl Acad Sci U S A. 2003;100(16):9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenbaum DC, Baruch A, Grainger M, Bozdech Z, Medzihradszky KF, Engel J, DeRisi J, Holder AA, Bogyo M. A role for the protease falcipain 1 in host cell invasion by the human malaria parasite. Science. 2002;298(5600):2002–2006. doi: 10.1126/science.1077426. [DOI] [PubMed] [Google Scholar]
- 45.van der Hoorn RA, Leeuwenburgh MA, Bogyo M, Joosten MH, Peck SC. Activity profiling of papain-like cysteine proteases in plants. Plant Physiol. 2004;135(3):1170–1178. doi: 10.1104/pp.104.041467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5(5):443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 47.Vasiljeva O, Papazoglou A, Kr¸ger A, Brodoefel H, Korovin M, Deussing J, Augustin N, Nielsen BS, Almholt K, Bogyo M, Peters C, Reinheckel T. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66(10):5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- 48.Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37(7):1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 49.Hershko A, Rose IA. Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc Natl Acad Sci U S A. 1987;84(7):1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pickart CM, Rose IA. Mechanism of ubiquitin carboxyl-terminal hydrolase. Borohydride and hydroxylamine inactivate in the presence of ubiquitin. J Biol Chem. 1986;261(22):10210–10217. [PubMed] [Google Scholar]
- 51.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385(6618):737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 52.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20(18):5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3(6):429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 54.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9(10):1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 55.Ovaa H, Kessler BM, Rolén U, Galardy PJ, Ploegh HL, Masucci MG. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci U S A. 2004;101(8):2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell. 2005;19(4):547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Loveland AN, Kattenhorn LM, Ploegh HL, Gibson W. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J Virol. 2006;80(12):6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Misaghi S, Balsara ZR, Catic A, Spooner E, Ploegh HL, Starnbach MN. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol. 2006;61(1):142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 59.Artavanis-Tsakonas K, Misaghi S, Comeaux CA, Catic A, Spooner E, Duraisingh MT, Ploegh HL. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol Microbiol. 2006;61(5):1187–1195. doi: 10.1111/j.1365-2958.2006.05307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thornberry NA, Peterson EP, Zhao JJ, Howard AD, Griffin PR, Chapman KT. Inactivation of interleukin-1 beta converting enzyme by peptide (acyloxy)methyl ketones. Biochemistry. 1994;33(13):3934–3940. doi: 10.1021/bi00179a020. [DOI] [PubMed] [Google Scholar]
- 61.Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, Bogyo M. Identification of early intermediates of caspase activation using selective inhibitors and activity based probes. Mol Cell. 2006;23(4):509–521. doi: 10.1016/j.molcel.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 62.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103(3):375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 63.Manoury B, Mazzeo D, Li DN, Billson J, Loak K, Benaroch P, Watts C. Asparagine endopeptidase can initiate the removal of the MHC class II invariant chain chaperone. Immunity. 2003;18(4):489–498. doi: 10.1016/s1074-7613(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 64.Shirahama-Noda K, Yamamoto A, Sugihara K, Hashimoto N, Asano M, Nishimura M, Hara-Nishimura I. Biosynthetic processing of cathepsins and lysosomal degradation are abolished in asparaginyl endopeptidase-deficient mice. J Biol Chem. 2003;278(35):33194–33199. doi: 10.1074/jbc.M302742200. [DOI] [PubMed] [Google Scholar]
- 65.Sexton KB, Witte MD, Blum G, Bogyo M. Design of cell-permeable, fluorescent activity-based probes for the lysosomal cysteine protease asparaginyl endopeptidase (AEP)/legumain. Bioorg Med Chem Lett. 2007;17(3):649–653. doi: 10.1016/j.bmcl.2006.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40(13):4005–4015. doi: 10.1021/bi002579j.. First report on application of activity based proteomics for profiling of serine hydrolases.
- 67.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99(16):10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR, 3rd, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A. 2004;101(38):13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2(9):691–697. doi: 10.1038/nmeth778.. Functional proteomics strategy which combines activity based profiling with multidimensional protein analysis is presented. Authors showed the benefits of this approach on analysis of human samples and identification of previously unknown proteases involved in cancer progression.
- 70.Pan Z, Jeffery DA, Chehade K, Beltman J, Clark JM, Grothaus P, Bogyo M, Baruch A. Development of activity-based probes for trypsin-family serine proteases. Bioorg Med Chem Lett. 2006;16(11):2882–2885. doi: 10.1016/j.bmcl.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 71.Coleman JE. Zinc enzymes. Curr Opin Chem Biol. 1998;2(2):222–234. doi: 10.1016/s1367-5931(98)80064-1. [DOI] [PubMed] [Google Scholar]
- 72. Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci U S A. 2004;101(27):10000–10005. doi: 10.1073/pnas.0402784101.. Groundbreaking report on development of activity based probes for metalloproteases and their application in proteomic identification of metalloproteases involved in cancer progression.
- 73.Turner AJ, Isaac RE, Coates D. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays. 2001;23(3):261–269. doi: 10.1002/1521-1878(200103)23:3<261::AID-BIES1036>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 74. Sieber SA, Niessen S, Hoover HS, Cravatt BF. Proteomic profiling of metalloprotease activities with cocktails of active-site probes. Nat Chem Biol. 2006;2(5):274–281. doi: 10.1038/nchembio781.. An interesting functional proteomics approach where authors used mixture of activity based probes for targeting and identification of structurally diverse families of metalloproteases.
- 75.Wang J, Uttamchandani M, Li J, Hu M, Yao SQ. "Click" synthesis of small molecule probes for activity-based fingerprinting of matrix metalloproteases. Chem Commun (Camb) 2006;36:3783–3785. doi: 10.1039/b609446e. [DOI] [PubMed] [Google Scholar]
- 76.Lee WL, Li J, Uttamchandani M, Sun H, Yao SQ. Inhibitor fingerprinting of metalloproteases using microplate and microarray platforms: an enabling technology in Catalomics. Nat Protoc. 2007;2(9):2126–2138. doi: 10.1038/nprot.2007.305. [DOI] [PubMed] [Google Scholar]
- 77.Uttamchandani M, Lee WL, Wang J, Yao SQ. Quantitative inhibitor fingerprinting of metalloproteases using small molecule microarrays. J Am Chem Soc. 2007;129(43):13110–13117. doi: 10.1021/ja073914v. [DOI] [PubMed] [Google Scholar]
- 78.Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272(40):25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- 79.Bogyo M, Shin S, McMaster JS, Ploegh HL. Substrate binding and sequence preference of the proteasome revealed by active-site-directed affinity probes. Chem Biol. 1998;5(6):307–320. doi: 10.1016/s1074-5521(98)90169-7. [DOI] [PubMed] [Google Scholar]
- 80.Ovaa H, van Swieten PF, Kessler BM, Leeuwenburgh MA, Fiebiger E, van den Nieuwendijk AM, Galardy PJ, van der Marel GA, Ploegh HL, Overkleeft HS. Chemistry in living cells: detection of active proteasomes by a two-step labeling strategy. Angew Chem Int Ed Engl. 2003;42(31):3626–3629. doi: 10.1002/anie.200351314. [DOI] [PubMed] [Google Scholar]
- 81.van Swieten PF, Samuel E, Hernández RO, van den Nieuwendijk AM, Leeuwenburgh MA, van der Marel GA, Kessler BM, Overkleeft HS, Kisselev AF. A cell-permeable inhibitor and activity-based probe for the caspase-like activity of the proteasome. Bioorg Med Chem Lett. 2007;17(12):3402–3405. doi: 10.1016/j.bmcl.2007.03.092. [DOI] [PubMed] [Google Scholar]
- 82.Verdoes M, Florea BI, Menendez-Benito V, Maynard CJ, Witte MD, van der Linden WA, van den Nieuwendijk AM, Hofmann T, Berkers CR, van Leeuwen FW, Groothuis TA, Leeuwenburgh MA, Ovaa H, Neefjes JJ, Filippov DV, van der Marel GA, Dantuma NP, Overkleeft HS. A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem Biol. 2006;13(11):1217–1226. doi: 10.1016/j.chembiol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 83.Berkers CR, van Leeuwen FW, Groothuis TA, Peperzak V, van Tilburg EW, Borst J, Neefjes JJ, Ovaa H. Profiling proteasome activity in tissue with fluorescent probes. Mol Pharm. 2007;4(5):739–748. doi: 10.1021/mp0700256. [DOI] [PubMed] [Google Scholar]
- 84.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci U S A. 1999;96(18):10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blair JA, Rauh D, Kung C, Yun CH, Fan QW, Rode H, Zhang C, Eck MJ, Weiss WA, Shokat KM. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Biol. 2007;3(4):229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 86.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci U S A. 2007;104(4):1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lo LC, Pang TL, Kuo CH, Chiang YL, Wang HY, Lin JJ. Design and synthesis of class-selective activity probes for protein tyrosine phosphatases. J Proteome Res. 2002;1(1):35–40. doi: 10.1021/pr015506a. [DOI] [PubMed] [Google Scholar]
- 88.Witte MD, Descals CV, de Lavoir SV, Florea BI, van der Marel GA, Overkleeft HS. Bodipy-VAD-Fmk, a useful tool to study yeast peptide N-glycanase activity. Org Biomol Chem. 2007;5(22):3690–3697. doi: 10.1039/b711531h. [DOI] [PubMed] [Google Scholar]
- 89.Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol. 2007;3(10):668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 90.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21(6):687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 91.Gillet LC, Namoto K, Ruchti A, Hoving S, Boesch D, Inverardi B, Mueller D, Coulot M, Schindler P, Schweigler P, Bernardi A, Gil-Parrado S. In-cell selectivity profiling of serine protease inhibitors by activity-based proteomics. Mol Cell Proteomics. 2008;7(7):1241–1253. doi: 10.1074/mcp.M700505-MCP200. [DOI] [PubMed] [Google Scholar]
- 92.Timmer JC, Enoksson M, Wildfang E, Zhu W, Igarashi Y, Denault JB, Ma Y, Dummitt B, Chang YH, Mast AE, Eroshkin A, Smith JW, Tao WA, Salvesen GS. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407(1):41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Enoksson M, Li J, Ivancic MM, Timmer JC, Wildfang E, Eroshkin A, Salvesen GS, Tao WA. Identification of proteolytic cleavage sites by quantitative proteomics. J Proteome Res. 2007;6(7):2850–2858. doi: 10.1021/pr0701052. [DOI] [PubMed] [Google Scholar]
- 94.Dean RA, Overall CM. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol Cell Proteomics. 2007;6(4):611–623. doi: 10.1074/mcp.M600341-MCP200. [DOI] [PubMed] [Google Scholar]