Abstract
Metabolites of vitamin D3 (D3) (cholecalciferol) are recognized as enzymatically formed chemicals in humans that can influence a wide variety of reactions that regulate a large number of cellular functions. The metabolism of D3 has been extensively studied, and a role for three different mitochondrial cytochrome P450s (CYP24A, CYP27A, and CYP27B1) has been described that catalyze the formation of the 24(OH), 25(OH), and 1(OH) metabolites of D3, respectively. The hormone 1,25-dihydroxyvitamin D3 has been most extensively studied and is widely recognized as a regulator of calcium and phosphorous metabolism. Hydroxylated metabolites of D3 interact with the nuclear receptor and thereby influence growth, cellular differentiation, and proliferation. In this article, we describe in vitro experiments using purified mitochondrial cytochrome P450scc (CYP11A1) reconstituted with the iron-sulfer protein, adrenodoxin, and the flavoprotein, adrenodoxin reductase, and show the NADPH and time-dependent formation of two major metabolites of D3 (i.e., 20-hydroxyvitamin D3 and 20,22-dihydroxyvitamin D3) plus two unknown minor metabolites. In addition, we describe the metabolism of 7-dehydrocholesterol by CYP11A1 to a single product identified as 7-dehydropregnenolone. Although the physiological importance of these hydroxylated metabolites of D3 and their in vivo formation and mode of action remain to be determined, the rate with which they are formed by CYP11A1 in vitro suggests an important role.
Cytochrome P450scc (CYP11A1) is a mitochondrial hemeprotein oxygenase of great interest because of its identification as a key enzyme in the metabolism of cholesterol to pregnenolone (1). This reaction is the initial step in the pathway leading to the synthesis of a number of metabolically important steroid hormones. Like other mitochondrial P450s this enzyme requires a minielectron transport chain consisting of the iron-sulfur protein, adrenodoxin (Adr), and the FAD-containing NADPH-Adr reductase (AdrR) for the transfer of reducing equivalents from NADPH to the P450. To date, CYP11A1 has no known natural substrates other then cholesterol. The conversion of cholesterol to pregnenolone (P5) is reported to occur by forming in sequence the monohydroxylated product [22R-(OH)cholesterol] followed by the formation of the dihydroxylated intermediate [20α,22R-(OH)2cholesterol] with subsequent carbon–carbon bond cleavage at the C20–C22 position to form the C19 steroid pregnenolone (1–5). It has been difficult to identify these hydroxylated intermediates of cholesterol metabolism during catalysis by CYP11A1 because they are not released from the active site of CYP11A1 during metabolism (1–5). Of interest is the recent report that the oxysterol 22R-(OH)cholesterol plays an important role as a ligand for the liver X receptor, a transcription factor (6).
In mammalian tissues 7-dehydrocholesterol (7-DHC) is the direct precursor of cholesterol in the Kandutsch–Russell cholesterol biosynthetic pathway. A deficiency of Δ7-DHC reductase, the enzyme responsible for catalyzing the final step of cholesterol formation, results in low plasma levels of cholesterol and increased concentrations of 7-DHC [the autosomal recessive malformation syndrome, Smith–Lemli–Opitz syndrome (SLOS)]. The presence of several C19 and C21 steroids with an unsaturated B ring has been shown recently in humans with SLOS (7), and it has long been known that these unsaturated B-ring steroids are normal constituents of equine urine in the forms of equilin and equilinin (8–10).
Interestingly, 7-DHC is not only the precursor of cholesterol but also gives rise to vitamin D3 (D3). Exposure of 7-DHC to sunlight, i.e., high-energy UV β photons (290–315 nm), photolyzes cutaneous stores of 7-DHC to form the pre-D3 (for review see ref. 11). Once formed, pre-D3 undergoes a thermal isomerization that results in the formation of D3.
D3 is biologically inert and requires successive hydroxylations by mitochondrial P450s present in the liver and kidney to form the biologically active hormone 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] (12). A major physiologic function of 1,25(OH)2D3 is to regulate the absorption of calcium and phosphorus from the intestine and maintain plasma concentrations of these ions. There are a significant number of metabolic disorders that are related to defects in the synthesis and metabolism of D3. Besides its calcemic activity, 1,25(OH)2D3 is a potent antiproliferative factor for cells and tissues that possess the vitamin D receptor. D3 (13) and its metabolites have clinical utility in that 1,25(OH)2D3 and analogs are successfully used for the treatment of the hyperproliferative skin disease and psoriasis and the prevention of osteoporosis. The inactivation of 25-hydroxyvitamin D3 [25(OH)D3] or 1α,25(OH)2D3 is catalyzed by a mitochondrial P450 that 24-hydroxylates these chemicals to form 24,25(OH)2D3, or 1α,24,25(OH)3D3. It must be emphasized that mitochondrial P450s are responsible for the hydroxylation reactions at the C1 (CYP27B1), C25 (CYP27A), and C24 (CYP24A) positions of D3 and for the 27-hydroxylation of other sterols (for review see ref. 14). Recently, the microsomal P450 CYP2D6 in the pig (15) and CYP2R1 in the mouse and human (16) have been reported to metabolize D3.
The present study was undertaken to identify natural substrates of CYP11A1 (P450scc) other than cholesterol. We show that CYP11A1 catalyzes in vitro the side-chain cleavage of 7-DHC to form 7-dehydropregnenolone (7-DHP5) at a rate comparable to that of cholesterol. Further, we discovered that D3 is metabolized by CYP11A1 to form two hydroxylated products, which unlike the proposed hydroxylated intermediates of cholesterol are released from the substrate-binding site of the P450 enzyme. Surprisingly, cytochrome b5 (b5), a microsomal hemeprotein, stimulates the in vitro reactions of CYP11A1 by using either cholesterol, 7-DHC, or D3 as substrates. The enzymatic formation of additional unique metabolites of D3 offers the opportunity to seek new functions for these D3 metabolic products.
Materials and Methods
Materials. Cholesterol, 7-DHC, D3, 2-hydroxypropyl-β-cyclodextrine, glucose-6-phosphate (G6P), G6P dehydrogenase, and NADPH were purchased from Sigma. Radioactive [3H]cholesterol was purchased from NEN. Deuterated chloroform [CDCl3 (99.9%)] used as the solvent for NMR studies was obtained from Cambridge Isotope Laboratories (Andover, MA). Samples of 1α(OH)D3, 25(OH)D3, and 1α,25(OH)2D3 were provided by J. Omdahl (University of New Mexico, Albuquerque).
Purified cytochrome P450scc (CYP11A1) and purified Adr were isolated from bovine adrenal cortical mitochondria as described (17, 18). A preparation of pure recombinant bovine AdrR was a generous gift of I. Pikuleva (University of Texas Medical Branch, Galveston).
Recombinant rat b5 was expressed in Escherichia coli and purified as described (19).
Analytical Methods. The concentration of CYP11A1 was determined from the difference spectrum of the reduced plus CO minus the reduced hemeprotein by using a molar extinction coefficient at 450 minus 490 nm of 91 mM–1·cm–1 (20). Concentrations of AdrR, Adr, and b5 were determined by using molar extinction coefficients (mM–1·cm–1) at 450 nm [11.3], 414 nm [9.8] (18), and 413 nm [117] (21). The concentrations of 7-DHC and D3 were determined by using published molar extinction coefficients (mM–1·cm–1) at 262 nm [9.8] (22) and 265 nm [18.0] (23), respectively.
Determination of Enzymatic Activities Using Reconstituted CYP11A1. Measurements of the rate of steroid hydroxylation using a CYP11A1 reconstituted assay system were carried out at 37°Cby using 50 mM K-phosphate buffer, pH 7.4. Reconstitution was established by mixing an aliquot of concentrated CYP11A1 with aliquots of concentrated AdrR and Adr followed by incubation for 5 min at 37°C. The concentrated mix was then diluted with buffer solution to give a final concentration of 1.0 μM CYP11A1, 2.0 μM AdrR, and 10 μM Adr. Steroids [dissolved in 45% 2-hydroxypropyl-β-cyclodextrine (24)] were added to the concentrations indicated. The reaction was started by addition of NADPH (final concentration 0.5 mM) and an NADPH-regenerating system. Samples of 0.5 ml were removed from the reaction mixture at the times indicated and rapidly mixed with 5 ml of dichloromethane, and the layers were separated by centrifugation. The dichloromethane layer was carefully removed and dried under a stream of nitrogen. The residue was dissolved in 100 μl of methanol, and aliquots were injected into a computerized Waters 840 HPLC instrument equipped with a 10-μmC18 Bondapak column (3.9 × 300 mm), a Spectraflow 757 absorbance detector, and a β-RAM radioactive flow detector (IN/US, Tampa, FL). Steroids were separated by using a methanol/water (vol/vol) gradient elution program starting initially at 64% methanol/36% water and ending with 100% methanol.
Spectrophotometric measurements were made by using an Aminco DW2 spectrophotometer modernized by Olis (Bogart, GA).
Mass Spectroscopic Analyses. The HPLC fractions containing products of the reaction were collected, extracted with dichloromethane, and evaporated to near dryness under a stream of nitrogen. The steroids were dissolved in dichloromethane and treated with N,O-Bis(trimethylsilyl)acetamide. Samples were analyzed by using a Shimadzu QP 5000 Gas Chromatograph Mass Spectrometer.
NMR Spectroscopy. Samples of D3, D3 hydroxylated-metabolites, cholesterol, and 20α(OH)- and 22R(OH)-cholesterol were dissolved in 150 μl of 99.9% CDCl3. 1H NMR spectra were acquired by using a 600-MHz Varian INOVA spectrometer equiped with a 3-mm inverse detection probe (Nalorac, Martinez, CA). An interpulse delay of 15 s was used to ensure full relaxation of each of the 1H resonances. 1H-1H COSY spectra were acquired by using a standard d1–90°-t1-90°-acquisition pulse sequence (25, 26). Each COSY spectrum consisted of 1,024 (t2, complex) by 512 (t1, real) data points covering a 6,300-Hz sweep width. Standard sine apodisation functions and zero filling were used in both dimensions before Fourier transformation.
Results
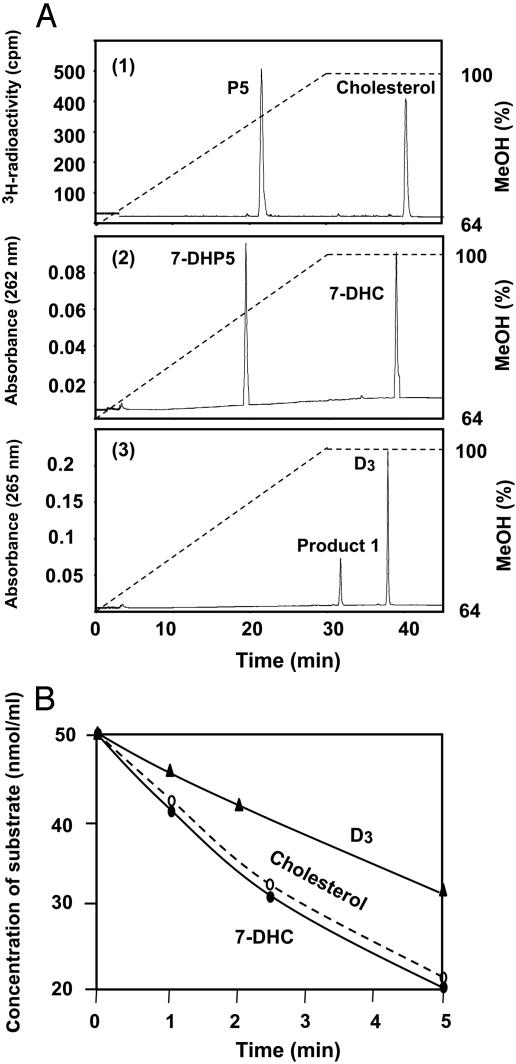
Reactions of CYP11A1 with Cholesterol, 7-DHC, or D3 and Detection of the Products of These Reactions. When radioactive cholesterol was incubated with the CYP11A1-reconstituted system containing Adr, AdrR, NADPH, and an NADPH-regeneration system, the time-dependent formation of pregnenolone was detected (retention time 21.6 min) (Fig. 1A, tracing 1). When 7-DHC was used as substrate in a parallel experiment, a single reaction product was also observed (retention time 19.5 min) (Fig. 1 A, tracing 2). When D3 was incubated in a similar way with the CYP11A1-reconstituted system a single product (product 1) was initially detected (retention time 31.7 min) (Fig. 1 A, tracing 3). However, as described below, longer incubation times with D3 revealed the formation of a second metabolite (product 2) (retention time 27.8 min). The retention times for the metabolites of cholesterol and 7-DHC significantly differ from the retention times of the metabolites of D3.
Fig. 1.
(A) Formation of metabolites during the short time reaction of reconstituted CYP11A1 with cholesterol (no. 1), 7-DHC (no. 2), and D3 (no. 3). The reaction mixtures contained 1 μM CYP11A1, 2 μM AdrR, 10 μM Adr, and 50 μM substrate diluted in 50 mM potassium phosphate buffer, pH 7.4, as described in Materials and Methods. The reactions were initiated by addition of 0.5 mM NADPH (final concentration) containing an NADPH-regeneration system and were incubated for 5 min at 37°C before terminating the reaction by addition of dichloromethane. (B) The time-dependent metabolism of cholesterol, 7-DHC, and D3 by the reconstituted CYP11A1 system. Reaction conditions are those described above. Samples were removed at the times indicated for analysis by HPLC.
Fig. 1B shows the time-dependent metabolism of cholesterol, 7-DHC, and D3 by the NADPH-dependent reconstituted CYP11A1 system. The rate of 7-DHC metabolism is essentially equal to the rate of cholesterol metabolism, whereas the rate of D3 metabolism is approximately half that of the other two substrates.
Determination of Km and Vmax Values for the Reaction of CYP11A1 with Different Substrates. When the concentrations of cholesterol, 7-DHC, or D3 were varied and the initial rate of metabolism for each substrate was determined, the results appeared to follow saturation-type kinetics. The Km and Vmax values were estimated graphically, allowing an estimate of the Vmax values for cholesterol and 7-DHC of 7.5 and 7.8 nmol product/min per nmol of CYP11A1, respectively. However, a significantly smaller value for the Vmax was estimated for D3, i.e., a Vmax = 3.2 nmol product/min per nmol of CYP11A1. Similarly, Km values (the concentration of substrate required for half-maximal rates of metabolism) were determined to be 1.6, 1.2, and 1.1 μM, for cholesterol, 7-DHC, and D3, respectively.
Identification of the Product Formed During the Metabolism of 7-DHC by CYP11A1. The structure of the product formed during the metabolism of 7-DHC by the CYP11A1 reconstituted system was determined by collecting fractions from several HPLC cycles and then subjecting the isolated steroid to GC–MS analysis with trimethylsilyl (TMS) derivatization (Fig. 2). The mass spectrum of this product was identified as 7-DHP5. As shown in Fig. 2, a molecular ion was detected at m/z 386. The ion at m/z 296 (M-90) is caused by elimination of TMS at C3, whereas the ion at m/z 281 (M-90–15) is caused by elimination TMS and the loss of one methyl group. Important ions at m/z 253 (M-90–43) and 211 (M-90–85) were formed by cleavage of the side chain and breakage of the D ring with loss of carbons 15, 16, and 17 as shown in Fig. 2.
Fig. 2.
Mass spectral analysis of the trimethylsilyl ether of the product formed from 7-DHC during incubation with the reconstituted CYP11A1 system. This product was identified as 7-DHP5. See the text for details.
Effect of b5 on the Reaction of CYP11A1 with Different Substrates. As illustrated in Fig. 1 A, trace 3, the rate of formation of product 1 during the metabolism of D3 by CYP11A1 is considerably slower than the rate of metabolism of cholesterol and 7-DHC. Unfortunately, NMR identification of the structure of a metabolite requires comparatively large amounts of the chemical for analysis. This problem prompted us to test ways of increasing the yield of D3 products. It has been reported that the addition of purified rat liver microsomal b5 to a reaction mixture containing the reconstituted CYP11A1 system increases the rate of cholesterol metabolism (27).
Fig. 3A shows the HPLC profile of metabolites formed when D3 is incubated for 30 min with the CYP11A1 system. It was noted that added recombinant b5 increased the rate of formation of the D3 metabolite with a retention time at ≈50 min (product 1). Moreover, in these experiments a second product was formed with a retention time of ≈43 min. We call this second metabolite product 2. Note the time of elution of products shown in Fig. 3 differs from that shown in Fig. 1 because a different time gradient was used for increasing the concentration of methanol in Fig. 3A to obtain better separation of the products.
Fig. 3.
(A) HPLC profile of the products formed during extended incubation of the CYP11A1 reconstitution system with D3 in the presence of b5. The reaction mixture (20 ml final volume) of 50 mM potassium phosphate buffer, pH 7.4, contained 1 μM CYP11A1, 2 μM AdrR, 10 μM Adr, 5 μM b5, 100 μM D3, 0.5 mM NADPH plus an NADPH-regeneration system. The reaction mixture was incubated for 30 min at 37°C. Note the difference in the gradient of methanol used for this experiment compared with Fig. 1. (B) Effect of b5 on the CYP11A1-dependent hydroxylation of D3. The reaction mixtures contained 1 μM CYP11A1, 2 μM AdrR, 10 μM Adr, 100 μM D3, 0.5 mM NADPH, and an NADPH-regeneration system in 50 mM potassium phosphate buffer, pH 7.4, as described. Aliquots were removed for HPLC analyses at the times indicated. Incubation temperature was 37°C. Reactions in the presence of 2 μM b5 are shown.
The Conversion of Large Amounts of D3 to Hydroxylated Products Suitable for NMR Analysis. To increase the yield of D3 products, we used longer incubation times of D3 with the CYP11A1-reconstituted system in the presence of b5. After an extended incubation, four main product peaks were observed (Fig. 3A). Peaks corresponding to product 1 and product 2 were collected manually from several HPLC cycles and subjected to 1H-NMR analyses (see below). Substances from the two other peaks, called product 3 and product 4, were subjected to electrospray ionization MS analysis. It was determined that product 3 has a molecular mass of 416, and product 4 has a molecular mass of 432. These masses correspond to unknown dihydroxy- and trihydroxy-D3 metabolites, respectively (data not shown). The rates of formation of the two major metabolites (product 1 and product 2) in the presence and absence of b5 are shown in Fig. 3B. In agreement with earlier reports (27), the presence of b5 results in an ≈2-fold stimulation of CYP11A1.
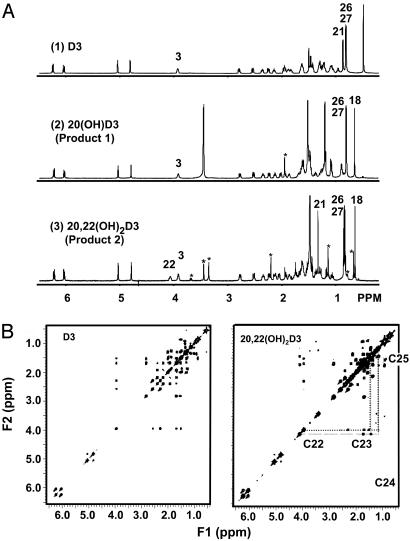
Identification of the Structure of Product 1 by NMR Analyses as 20-Hydroxyvitamin D3 [20(OH)D3]. Identification of product 1 was accomplished by analysis of the 1D 1H-NMR spectrum of this compound and comparison (25, 26, 28) with the 1D 1H NMR spectra of D3 (Fig. 4A), cholesterol, 20-hydroxycholesterol, and 22-hydroxy cholesterol. Product 1 was assigned the structure of 20(OH)D3. Upon substitution of the proton in position at C20 of product 1 with a hydroxyl group the doublet 21-methyl resonance (3JHH = 6.5 Hz) is converted into a singlet and is shifted to lower field [0.92 ppm in D3 to 1.23 ppm in 20(OH)D3]. This substitution is also observed when comparing the 1H NMR spectra of cholesterol with 20(OH)-cholesterol, i.e., the doublet component of the 20-hydroxy derivative disappears concomitant with the shift of the methyl resonance to lower field. One interpretation of this modification in the orientation of the methyl group at C20 is a change from a planar to a tilted up position for the C21 methyl group, suggesting an epi-type structural conformation.
Fig. 4.
(A) The 600-MHz 1H-NMR spectra of the products formed from D3 as described in Fig. 3A. (1) D3; (2) product 1 identified as 20(OH)D3; (3) product 2 identified as 20,22(OH)2D3. Numbers refer to the proton positions according to conventional numbering of D3 (28). The peaks marked by * are caused by unidentified impurities. (B) The 600-MHz COSY spectra of D3 (Left) and product 2 (Right) identified as 20,22(OH)2D3. Numbers refer to the proton positions according to convention (28).
Identification of Product 2 as 20,22-Dihydroxyvitamin D3 [20,22(OH)2D3]. The 1H-NMR spectrum of product 2 has a 1H resonance at 4.10 ppm, which is not present in the NMR spectrum of D3 or in the NMR spectrum of 20(OH)D3. This resonance appears at a chemical shift similar to the proton of position 3 (3.97 ppm), which is a proton characteristic of a vicinal hydroxyl group. The C21-methyl resonance of product 2 is a singlet, in accordance with a hydroxyl substitution at position C20. To identify the location of the second hydroxyl group, a complete analysis of the 1H-NMR spectrum of the dihydroxy derivative was necessary. This process consisted of first attributing the 1H resonances of D3 by analysis of the scalar–scalar connectivities on the COSY spectrum (Fig. 4B Left) and then performing a similar analysis of the COSY spectrum of the dihydroxy derivative, product 2 (Fig. 4B Right). The extra resonance at 4.10 ppm (at C22) has a strong correlation with protons at position C23 and a weaker correlation with the protons at position C24, demonstrating that it represents the proton at position C22 as a vicinal hydroxyl group.
Discussion
In recent years, the cytochrome P450s have taken on a new role in mammalian metabolism. Initially, this superfamily of enzymes was thought to serve only as hydroxylases (oxygenases) for reactions of steroid hormone synthesis or for the detoxification of xenobiotics, such as drugs and environmental pollutants. Today, we recognize the role of P450s in the formation of metabolites that serve as ligands that bind to receptors in the nucleus, thereby participating in a lipid-based metabolic signaling cascade (29, 30). Examples of P450-generated metabolites that act as ligands for transcription factors include metabolites of retinoic acid, eicosanoids, sterols, bile acids, ecdysone, and D3, to name but a few.
D3 and its metabolites occupy a central role in our understanding of ligand–receptor interactions because of the extensive contributions of DeLuca and his students (12, 31). Numerous studies have described a role for the hormone 1,25(OH)2D3 in the regulation of fetal development, cellular growth and differentiation, absorption and mobilization of calcium, and as agonists for receptor-mediated gene expression. In addition, a large number of synthetic chemicals mimic the actions of 1,25(OH)2D3, such as 20-epi-22-oxa-24α,26α,27α-trihomo-1α 25(OH)vitamin D3, which is reported to have 500- to 1,000-fold greater vitamin D receptor-binding affinity than the parent hormone (32). Of interest is the possibility that the 20(OH)D3 has an epi-like structure as a result of the hydroxyl-group at C20. Thus, there is great interest in the possible biological roles for the D3 metabolites that are generated enzymatically with CYP11A1.
Cytochrome P450s play a major role in the synthesis of 1,25(OH)2D3. Two mitochondrial P450s catalyze the sequential hydroxylation of D3, forming first the 25(OH)D3 in the liver (CYP27A1) followed by a 1α-hydroxylation reaction catalyzed by CYP27B1 in kidney mitochondria. We report here the role for a third mitochondrial P450 (CYP11A1) in the metabolism of D3. This P450 catalyzes the side-chain cleavage reaction for the conversion of cholesterol to pregnenolone and is of great interest because it represents a rate-limiting step in steroid hormone biosynthesis (1–5). During experiments designed to develop a simpler assay for measuring cholesterol metabolism we turned to the use of 7-DHC because one can monitor absorbance changes with the latter substrate rather than radioactive cholesterol. Also, 7-DHC might obviate the problem of endogenous cholesterol pools in membranes. As shown in this article, 7-DHC is metabolized in vitro by a reconstituted CYP11A1 system to a product at a rate comparable to that of cholesterol. We have identified the product of this reaction as 7-DHP5. In the current studies, we confirm and extend earlier observations that the mono- and dihydroxy metabolites of cholesterol (or 7-DHC) do not leave the active site of CYP11A1 during metabolism to the C19 steroid products.
Because 7-DHC is the precursor of D3 by irradiation our attention turned to the question of D3 metabolism by the reconstituted CYP11A1 system. As described here, such a reaction does occur when studied by in vitro techniques. The two primary metabolites are formed that have mobilities on HPLC analysis that indicate they are C27 compounds and had not undergone a C–C bond cleavage (desmolase reaction) of the side chain in a manner similar to that observed when cholesterol or 7-DHC are substrates. Further analysis by MS and NMR showed that these metabolites were 20(OH)D3 and 20,22(OH)2D3. Incubation of 20(OH)D3 with the reconstituted CYP11A1 system revealed the formation of 20,22(OH)2D3, albeit at a slow rate (0.3 nmol/min per nmole of CYP11A1). In a similar way 1α(OH)D3 is also metabolized slowly by the CYP11A1-reconstituted reaction system, but 25(OH)D3 and 1α,25(OH)2D3 are not metabolized.
Of major interest in the future is the question of the ligand-binding properties of these metabolites of D3.
The present study has expanded knowledge of the role of CYP11A1 in sterol metabolism. As summarized by the scheme presented in Fig. 5, we propose that CYP11A1 has at least three roles in the metabolism of endogenous sterols. First is the well-known pathway of conversion of cholesterol to pregnenolone. As noted in Fig. 1, no accumulation of intermediates during this three-step reaction is observed, confirming the conclusion that the intermediates formed during cholesterol metabolism do not leave the active site of CYP11A1 during the reaction. Likewise, a similar study of the metabolism of 7-DHC does not show the presence of intermediates. These are not designated in the scheme shown in Fig. 5 because we have not as yet measured the stoichiometry of NADPH utilization or oxygen incorporation into product, 7-DHP5. In unpublished experiments, we have demonstrated the further metabolism of 7-DHP5 by CYP17A to unknown products.
Fig. 5.
Pathways of sterol metabolism catalyzed by CYP11A1.
In contrast, the secosteroid metabolites, 20(OH)D3 and 20,22(OH)2D3, leave the active site of CYP11A1 unlike the cholesterol intermediates. Further, we are intrigued by the observation that 20(OH)D3 is formed first in the sequence of D3 metabolism in contrast to the 22(OH)-cholesterol intermediate observed for cholesterol metabolism.
Acknowledgments
We thank Professors Dean Sherry and Craig R. Malloy (University of Texas, Dallas, and University of Texas Southwestern Medical Center, Dallas) for providing availability to the NMR spectrometer required for this study and for their assistance with interpreting NMR spectra and Bikash Pramanik for assistance with collecting and interpreting mass spectral results. Dr. Irina Pikuleva (University of Texas Medical Branch, Galveston) kindly provided a sample of NADPH-AdrR. We are indebted to Dr. David Russell for his helpful comments and review of this manuscript. This work was supported in part by National Institute of General Medical Sciences Grant GM16488 (to R.W.E. and O.G.), the Chilton Foundation (S.U. and A.G.), and Foundation for Science and Technology Grant FCT-POCTI/CBO/38611/01 (to R.A.C.).
Abbreviations: Adr, adrenodoxin; AdrR, Adr reductase; D3, vitamin D3; 25(OH)D3, 25-hydroxyvitamin D3; 1,25(OH)2D3, 1,25-dihydroxyvitamin D3; 20(OH)D3, 20-hydroxyvitamin D3; 20,22(OH)2D3, 20,22-dihydroxyvitamin D3; 7-DHC, 7-dehydrocholesterol; 7-DHP5, 7-dehydropregnenolone; b5, cytochrome b5.
References
- 1.Solomon, S., Levitan, P. & Lieberman, S. (1956) Rev. Can. Biol. 15, 282–283. [Google Scholar]
- 2.Shimizu, K., Gut, M. & Dorfman, R. I. (1962) J. Biol. Chem. 237, 699–702. [PubMed] [Google Scholar]
- 3.Constantopoulos, G. & Tchen, T. T. (1961) Biochem. Biophys. Res. Commun. 4, 460–463. [DOI] [PubMed] [Google Scholar]
- 4.Hume, R., Kelly, R. W., Taylor, P. L. & Boyd, G. S. (1984) Eur. J. Biochem. 140, 583–591. [DOI] [PubMed] [Google Scholar]
- 5.Burstein, S. & Gut, M. (1976) Steroids 28, 115–131. [DOI] [PubMed] [Google Scholar]
- 6.Janowski, B. A., Willy, P. J., Devi, T. R., Falck, J. R. & Mangelsdorf, D. J. (1996) Nature 383, 728–731. [DOI] [PubMed] [Google Scholar]
- 7.Shackleton, C. H. L., Roitman, E. & Kelley, R. (1999) Steroids 64, 481–490. [DOI] [PubMed] [Google Scholar]
- 8.Bhavnani, B. R. (1988) Endocr. Rev. 9, 396–416. [DOI] [PubMed] [Google Scholar]
- 9.Tait, A. D., Hodge, L. C. & Allen, W. R. (1985) FEBS Lett. 182, 107–110. [DOI] [PubMed] [Google Scholar]
- 10.Tait, A. D., Santikarn, S. & Allen, W. R. (1983) J. Endocrinol. 99, 87–92. [DOI] [PubMed] [Google Scholar]
- 11.Kato, S. (2000) J. Biochem. 127, 717–722. [DOI] [PubMed] [Google Scholar]
- 12.Beckman, M. J. & DeLuca, H. F. (1998) Prog. Med. Chem. 35, 1–56. [DOI] [PubMed] [Google Scholar]
- 13.Norman, A. W., Ishizuka, S. & Okamura, W. H. (2001) J. Steroid Biochem. Mol. Biol. 76, 49–59. [DOI] [PubMed] [Google Scholar]
- 14.Pikuleva, I. & Waterman, M. (1999) Mol. Aspects Med. 20, 33–42. [PubMed] [Google Scholar]
- 15.Hosseinpour, F., Hidestrand, M., Ingelman-Sundberg, M. & Wikvall, K. (2001) Biochem. Biophys. Res. Commun. 288, 1059–1063. [DOI] [PubMed] [Google Scholar]
- 16.Cheng, J. B., Motola, D. L., Mangelsdorf, D. J. & Russell, D. W. (2003) J. Biol. Chem. 278, 38084–38093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilevich, S. N., Guryev, O. L., Shkumatov, V. M., Chashchin, V. L. & Akhrem, A. A. (1987) Biochemistry (Moscow) 52, 198–213. [PubMed] [Google Scholar]
- 18.Akhrem, A. A., Lapko, V. N., Lapko, A. G., Shkumatov, V. M. & Chashchin, V. L. (1979) Acta Biol. Med. Ger. 38, 257–273. [PubMed] [Google Scholar]
- 19.Holmans, P. L., Shet, M. S., Martin-Wixtrom, C. A., Fisher, C. W. & Estabrook, R. W. (1994) Arch. Biochem. Biophys. 312, 554–565. [DOI] [PubMed] [Google Scholar]
- 20.Omura, T. & Sato, R. (1964) J. Biol. Chem. 239, 2370–2379. [PubMed] [Google Scholar]
- 21.Strittmatter, P. & Velick, S. P. (1956) J. Biol. Chem. 221, 253–264. [PubMed] [Google Scholar]
- 22.Weast, R.C., ed. (1975) Handbook of Chemistry and Physics (CRC, Boca Raton, FL), 56th Ed.
- 23.Hiwatashi, A., Nishii, Y. & Ichikawa, Y. (1982) Biochem. Biophys. Res. Commun. 105, 320–327. [DOI] [PubMed] [Google Scholar]
- 24.Petrack, B. & Latario, B. J. (1993) J. Lipid Res. 34, 643–649. [PubMed] [Google Scholar]
- 25.Aue, W. P., Bartholdi, E. & Ernst, R. R. (1976) J. Chem. Phys. 64, 2229–2246. [Google Scholar]
- 26.Bax, A. & Freeman, R. (1981) J. Magn. Reson. 44, 542–561. [Google Scholar]
- 27.Usanov, S. A. & Chashchin, V. L. (1991) FEBS Lett. 278, 279–282. [DOI] [PubMed] [Google Scholar]
- 28.Nomenclature Committee of IUB (NC-IUB) and IUB-IUPAC Joint Commission on Biochemical Nomenclature (JCBN) (1983) Arch. Biochem. Biophys. 220, 321–324. [DOI] [PubMed] [Google Scholar]
- 29.Chawla, A., Repa, J. J., Evans, R. M. & Mangelsdorf, D. J. (2001) Science 294, 1866–1870. [DOI] [PubMed] [Google Scholar]
- 30.Nebert, D. W. & Russell, D. W. (2002) Lancet 360, 1155–1162. [DOI] [PubMed] [Google Scholar]
- 31.Jones, G., Strugnell, S. A. & DeLuca, H. F. (1998) Physiol. Rev. 78, 1193–1231. [DOI] [PubMed] [Google Scholar]
- 32.van den Bend, G. C., Dilworth, F. J., Makin, H. L., Prahl, J. M., DeLuca, H. F., Jones, G., Pols, H. A. & van Leeuwen, J. P. (2000) Biochem. Pharmacol. 59, 621–627. [DOI] [PubMed] [Google Scholar]