Abstract
Aims: Efficient genotyping methods for many biologically significant repeat genetic polymorphisms, particularly in GC-rich regions of the genome, are limited. In particular, a short tandem repeat polymorphism [GGCGGG] in the promoter region of ALOX5 has been implicated as an important marker for inflammatory diseases. We developed a pyrosequencing assay to genotype the ALOX5 short tandem repeat polymorphism using pyrosequencing technology that will make assessing this important genetic marker in large, diverse populations more accessible than using current methods. Materials and Methods: We used a nested polymerase chain reaction approach to amplify DNA for pyrosequencing. Population allele frequencies were assessed in two cohorts of previously collected human DNA samples with 188 and 1032 samples, respectively. Sixteen genetic samples with known genotypes were used to confirm the accuracy of the method. Results and Discussion: Genotypes were 100% concordant with samples of known genotype. Genotype frequencies in European American, Hispanic, and African American agreed with previously published results (wild-type homozygotes 66%, 64%, and 19%, respectively). The method presented here will facilitate both genetic association and pharmacogenomic research on this polymorphism in large samples that are ethnically and/or racially admixed.
Introduction
Among methods for genotyping polymorphisms in large samples, pyrosequencing is one of the most versatile. This technology is based on sequencing by synthesis that efficiently determines genotypes for different types of genetic variations. This method can be used to determine unknown DNA sequence and is amenable to genotyping multiple allele systems or tandem repeat polymorphisms. The read length (up to 250 base pairs depending on the application) makes it particularly relevant in short tandem repeat polymorphism (STRPs) genotyping. However, the pyrosequencing method is not generally conducive to genotyping long stretches of the same nucleotide (homopolymers), and users encounter problems when genotyping GC-rich regions similar to traditional sequencing methods (Hube et al., 2005). Further, beyond traditional sequencing, there are few other genotyping methods that can be used for polymorphisms with these properties. This poses a considerable research obstacle because many important and interesting genetic polymorphisms in human disease and in pharmacogenomic studies are located in GC-rich regions of the genome and/or are composed of repeat elements.
One such important genetic polymorphism is located in the promoter region of the ALOX5 gene. ALOX5 encodes the enzyme arachidonic acid 5-lipoxygenase, which is the rate-limiting enzyme in the arachidonate 5-lipoxygenase pathway. Specifically, the repeat polymorphism is thought to modify the number of binding sites for both Sp1 and Egr-1 zinc-finger transcription factors in the promoter region of ALOX5 (Silverman et al., 1998). Recent evidence indicates that this pathway plays a role in the pathogenesis of inflammatory conditions such as asthma and atherosclerotic heart disease (Dwyer et al., 2004; Helgadottir et al., 2004, 2005, 2006; Cipollone et al., 2005; Iovannisci et al., 2007). The polymorphism in the promoter region has been studied in both in vitro and clinical studies that implicate it as an important marker for inflammatory diseases, including cardiovascular disease and asthma (Drazen et al., 1999; Dwyer et al., 2004; Kim et al., 2005; Kalayci et al., 2006). This association is thought to be important across race populations, although limited numbers of subjects from various race or ethnicity groups in previous studies limit the power in these studies to draw conclusions (Dwyer et al., 2004; Lima et al., 2006).
Genotyping this STRP in large clinical studies to further assess its role in disease or in drug response poses particular challenges. The polymorphism is a series of tandem repeats of the nucleotide sequence 5′-GGGCGG-3′. The variant alleles are formed by varying numbers of these repeats that can range from two to eight in humans. The most common allele contains five repeats, and the three most common variants have a 6-base-pair (one repeat) deletion, a 12-base-pair (two repeats) deletion, or a 6-base-pair addition, respectively. Because of the GC-rich nature of this genetic region (about 78%), there is presumably considerable secondary structure. As such, this region is difficult to sequence using traditional methods. Further, this region is not amenable to common methods of medium to high-throughput genotyping such as allelic discrimination because it is a series of repeat elements. In previous studies, this polymorphism has only been genotyped by either direct sequencing technology or traditional STRP size fractionation (In et al., 1997; Lima et al., 2006; Poole et al., 2006). Pyrosequencing technology has known difficulties with genotyping repeat sequences that include long homopolymers, particularly when this is compounded by a long read-length polymorphism (Moore et al., 2006). There may also be other factors that cause this particular polymorphism to be so challenging that are not identified. The primary problem is amplifying the region sufficiently with biotinylated primers for pyrosequencing, Currently, published procedures for amplification do not amplify the region sufficiently for pyrosequencing based on our experience.
Recently, a single-nucleotide polymorphism in the ALOX5 promoter region has been identified that may be in sufficient linkage disequilibrium to serve as a marker SNP for the wild-type allele of the promoter polymorphism as compared to the next most frequent allele. Although this result promises to improve the ability to study the role of genetic polymorphism in this gene, these researchers found that this SNP does not sufficiently correlate with the STRP in African Americans. Further, the correlation between this SNP and the STRP in the Hispanic population in this study is based on a very small sample size (Helgadottir et al., 2006). As such, it is not clear whether this SNP is a suitable marker in highly admixed Hispanic (or other) populations (Assimes et al., 2008). Therefore, an alternative method to genotype this STRP directly is still needed.
Materials and Methods
Genotyping
We optimized the polymerase chain reaction (PCR) protocol for amplification of the genetic polymorphism using DNA samples at a concentration of 10 ng/mL. A gene amplicon surrounding the promoter repeats was prepared using nested PCR. Our PCR reactions were conducted using approximately 5 ng of genomic DNA derived from buccal cells. PCR reagents and PCR conditions for both T1c nested reactions are given in Table 1. Primers were purchased from Operon® (Operon Biotechnologies, Huntsville, AL), and MJ® Thermal cyclers (MJ Research, Waltham, MA) were used. Primer sequences are listed in Table 1. Both nested PCR reactions were conducted using Platinum® Taq polymerase (Life Technologies, Foster City, CA), GeneAmp® 10 × Gold Buffer (Life Technologies), and 7-deaza-deoxyguanine (Roche, Nutley, NJ).
Table 1.
Methods Used in PCR and Pyrosequencing Assay
|
PCR reagents and concentrations for both nested reactions | |
|---|---|
| Reagents | Concentration |
| Platinum Taq polymerase | 1.25 U |
| 10 × ABI Gold Buffer | 1X |
| Dimethyl sulfoxide | 2% |
| Nucleotide mixture | 0.1 mM |
| MgCL | 0.75 mM |
| Genomic DNA | 5 ng |
| Primers (5′ to 3′) | |
| PCR 1 Forward: GGGTGGCAGCCGAGGT | 10 pmol |
| PCR 1 Reverse: TGTCCAGCAGGTGCTTCTCGC | 10 pmol |
| PCR 2 Forward: Biotin-AGGAACAGACACCTCGCTGAGGAGAGAC | 10 pmol |
| PCR 2 Reverse: GAGCAGCGAGCGCCGGGAGCCTCGGC | 10 pmol |
| Sequencing: CAGGCTCCCGGCTGCCC | 10 pmol |
|
Nested PCR Conditions | ||
|---|---|---|
| 1st Reaction | Number of cycles | |
| Initiate | 94/2 min | 1 |
| Denature | 94/30 s | |
| Annealing | 60/30 s | 40 |
| Extension | 72/30 s | |
| Extension | 72/5 min | 1 |
| 2nd Reaction | Number of cycles | |
|---|---|---|
| Initiate | 95/12 min | 1 |
| Annealing | 94/1 min | 10 |
| Extension | 68/2 min | |
| Denature | 94/30 s | |
| Annealing | 60/30 s | 25 |
| Extension | 72/45 s | |
| Extension | 72/5 min | 1 |
7-deaza-deoxyguanine was added to deoxyguanosine triphosphate in a ratio of 3:1, and this mixture was added in a 1:1:1:1 ratio with 2′-deoxyadenosine-5′-triphosphate, 2′-deoxycytidine-5′-triphosphate, and 2′-deoxythymidine-5′-triphosphate to form the mixture of nucleotides. The second PCR was conducted using biotinylated primers appropriate for pyrosequencing, as previously described (Langaee and Ronaghi, 2005). Ten microliters of this reaction mix was used for pyrosequencing.
Genotyping by pyrosequencing was conducted using the standard manufacturer protocol (Biotage AB, Uppsala, Sweden). Briefly, 10 μL of biotinylated PCR product was immobilized on streptavidin-coated Sepharose beads in binding buffer (10 mM Trizma base, 2 M NaCl, 1 mM EDTA, and 0.1% Tween-20, pH 7.6) to a volume of 82 μL at room temperature for 10 min. After incubation, beads with bound DNA were isolated by hand-held vacuum probe, and then treated for 5 s successively with 70% ethanol, 0.1 M NaOH, and washing buffer (10 mM Tris-acetate). This is released into a solution of annealing buffer (20 mM Tris acetate and 5 mM magnesium acetate, pH 7.6) and 10 pmol of sequencing primer of the sequence 5′-CAGGCTCCCGGCTGCCC-3′. This was heated to 80°C for 3 min and then cooled to room temperature.
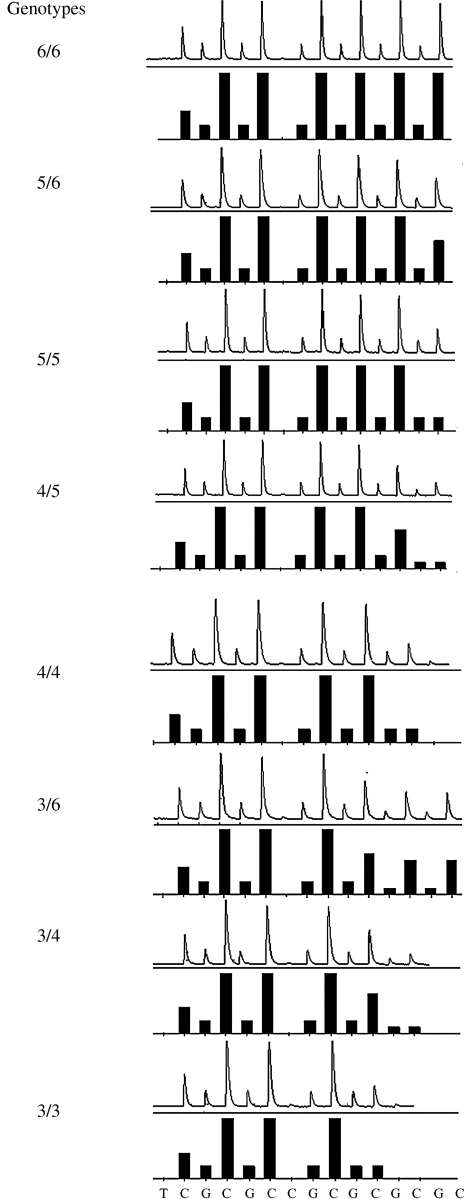
Pyrosequencing was performed using a Biotage PSQ HS 96 System. The sequence to analyze is CCGCCCCCGCCC[CCGCCC][CCGCCC][CCGCCC][CCGCCC]CCGCA. The pyrosequencing method was capable of detecting two to six repeats on each allele. During the development of this method, it was observed that the pyrosequencing signal deteriorated some after sequencing six repeats (>40 base pairs). This led to some ambiguity at this number of repeats once the number was greater than six; therefore, it was decided to design the assay to detect up to six repeats. Any allele with more than six repeats appeared to the observer as the presence of six repeats, and therefore particular genotypes with alleles having greater than six repeats cannot be determined using this method. Because the frequency of subjects with more than seven repeats is expected to be very low in all populations (<1%) (Lima et al., 2006), we found this caveat acceptable. The resulting pyrograms were visually assessed and verified by a second observer. We confirmed our pyrosequencing results by comparing them with 16 DNA samples with known STRP genotypes (In et al., 1997; Pemberton et al., 2008) determined by previous methods as positive control samples.
Allele frequency determination
Genotype and allele frequencies were studied in two populations. The first was in 188 genetic samples from the St. James Women Take Heart (WTH) study (Gulati et al., 2003). In WTH, race was collected as a baseline characteristic by subject self-report. Genomic DNA was isolated from blood samples using commercially available kits. DNA was normalized to a concentration of 10 ng/μL. In addition, genotype frequencies were studied in 834 samples from INVEST-GENES (International VErapamil SR/Trandolapril STudy–GENEtic Substudy) for which ancestry information was available (Pepine et al., 2003; Langaee and Ronaghi, 2005). In INVEST-GENES, race was also collected by self-report as part of the baseline information. Further, an ancestral informative marker analysis using 87 markers was conducted (Gerhard et al., 2008), and was used to assess the degree of admixture in the INVEST-GENES population. In this population, African Americans had 74 ± 18% African ancestry (n = 106). The European Americans had 92 ± 9% European ancestry (n = 503). The Hispanic population was primarily recruited from Puerto Rico and were of 65 ± 16% European ancestry, 19 ± 15% African ancestry, and 16 ± 10% Native American ancestry (n = 225). Genomic DNA was isolated from buccal cells obtained from mouthwash samples using commercially available kits (PureGene; Gentra Systems, Minneapolis, MN). DNA was normalized to a concentration of 20 ng/μL. The above protocol was followed for determining the genotypes of these samples. Allele frequencies were collected and reported as “#repeats in allele 1/# repeats in allele 2” in the WTH study, and as five repeat-carriers, homozygous for five repeats, and carriers of two variant alleles in the INVEST-GENES study.
Results and Discussion
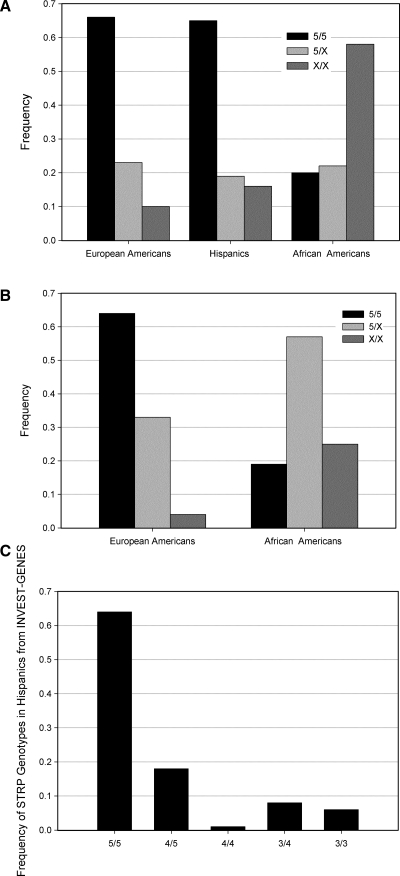
The predicted histograms and resulting pyrograms from pyrosequencing are shown in Figure 1. Comparison of our analysis with known genotypes resulted in 100% concordance. ALOX5 STRP genotype frequencies for European Americans, Hispanics, and African Americans from the 834 INVEST-GENES samples and in the 188 WTH samples (European Americans and African Americans) are given in Figure 2A and B. In addition, genotype frequencies for each common genotype in Hispanics from INVEST-GENES are given in Figure 2C.
FIG. 1.
For each genotype listed (notation is # ALOX5 STRP repeats in allele1/# ALOX5 STRP repeats in allele 2), example pyrograms (top half for each genotype) are given from actual pyrosequencing assays for each ALOX5 genotype observed and confirmed by direct sequencing. Predicted histograms (bottom half for each genotype) are given for each genotype generated by Pyrosequencing software for ALOX5 STRP genotypes observed in the analysis.
FIG. 2.
(A) Genotype frequencies of the ALOX5 STRP in European Americans, Hispanics, and African Americans in INVEST-GENES and (B) in European Americans and African Americans from WTH (Hispanic ethnicity is not largely represented in WTH). Genotypes are given as wild-type (5/5), one wild-type allele and one variant allele (5/X), and two variant alleles (X/X) in both figures. (C) Genotype frequencies for each common genotype in Hispanics INVEST-GENES. Genotypes are given as # ALOX5 STRP repeats in allele1/# ALOX5 STRP repeats in allele 2.
In this study we describe a method for analyzing this STRP in the promoter region of the ALOX5 gene that will facilitate further analysis of this genetic polymorphism in large, racially and ethnically diverse populations. We have successfully implemented an alternative method for genotyping the ALOX5 promoter tandem repeat polymorphism using a nested PCR method and pyrosequencing technology. The relative differences between the genotype frequencies between European American and African American subjects that have been previously identified remained true in our sample, which supports the accuracy of this method for genotyping the ALOX5 promoter polymorphism. Although the frequency of homozygote variants in African American subjects was significantly higher in our samples as compared to previous studies, we are confident that our genotyping is correct based on both the comparison to samples of known genotypes and also because the DNA of subjects from all race groups were positioned randomly on the 96-well plates. Of note, because the DNA samples used in this study come from patients with CAD or those with risk factor for CAD, and the ALOX5 gene purportedly confers risk for CAD, we would expect that the frequency of variant carrier alleles in our population may differ from previous reports in healthy individuals or those without CAD or significant risk factors for CAD.
Several studies have reported that the ALOX5 STRP plays a role in coronary artery disease (Dwyer et al., 2004) and asthma (Kim et al., 2005; Kalayci et al., 2006), and influences response to leukotriene modifiers used in the treatment of asthma (Lima et al., 2006). Further, ALOX5 promoter variants appear to have functional consequences in terms of gene expression (In et al., 1997; Kalayci et al., 2006), although these differences are not consistent across cell-types (Silverman and Drazen, 2000). Importantly, because the allele frequencies of ALOX5 promoter variants vary considerably between race groups and heretofore identified marker SNPs are not in sufficient linkage disequilibrium with the repeat polymorphism alleles to appropriately serve as surrogate markers, the ability to characterize the promoter repeat polymorphism directly will be required to validate its importance across diverse populations.
In conclusion, pyrosequencing is a time-efficient, cost-effective, and accurate method for many types of genotyping assays (Aquilante et al., 2004), and, in particular, the ALOX5 promoter tandem repeat polymorphism up to six repeats per allele. Therefore, the method presented here will facilitate both genetic association and pharmacogenomic research on this polymorphism in large samples that are ethnically and/or racially diverse.
Acknowledgments
We would like to acknowledge the work of Stephanie Roberts in contributing many hours to perfecting the PCR protocols. We would also like to thank the American Heart Association for supporting Dr. Schentrup's work with a Predoctoral Fellowship.
Disclosure Statement
No competing financial interests exist.
References
- Aquilante CL. Lobmeyer MT. Langaee TY. Johnson JA. Comparison of cytochrome P450 2C9 genotyping methods and implications for the clinical laboratory. Pharmacotherapy. 2004;24:720–726. doi: 10.1592/phco.24.8.720.36074. [DOI] [PubMed] [Google Scholar]
- Assimes TL. Knowles JW. Priest JR, et al. Common polymorphisms of ALOX5 and ALOX5AP and risk of coronary artery disease. Hum Genet. 2008;123:399–408. doi: 10.1007/s00439-008-0489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollone F. Mezzetti A. Fazia ML, et al. Association between 5-lipoxygenase expression and plaque instability in humans. Arterioscler Thromb Vasc Biol. 2005;25:1665–1670. doi: 10.1161/01.ATV.0000172632.96987.2d. [DOI] [PubMed] [Google Scholar]
- Drazen JM. Yandava CN. Dube L, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet. 1999;22:168–170. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- Dwyer JH. Allayee H. Dwyer KM, et al. Arachidonate 5-lipoxygenase promoter genotype, dietary arachidonic acid, and atherosclerosis. N Engl J Med. 2004;350:29–37. doi: 10.1056/NEJMoa025079. [DOI] [PubMed] [Google Scholar]
- Gerhard T. Gong Y. Beitelshees AL, et al. Alpha-adducin polymorphism associated with increased risk of adverse cardiovascular outcomes: results from GENEtic Substudy of the INternational VErapamil SR-trandolapril STudy (INVEST-GENES) Am Heart J. 2008;156:397–404. doi: 10.1016/j.ahj.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati M. Pandey DK. Arnsdorf MF, et al. Exercise capacity and the risk of death in women: the St James women take heart project. Circulation. 2003;108:1554–1559. doi: 10.1161/01.CIR.0000091080.57509.E9. [DOI] [PubMed] [Google Scholar]
- Helgadottir A. Gretarsdottir S. St Clair D, et al. Association between the gene encoding 5-lipoxygenase-activating protein and stroke replicated in a Scottish population. Am J Hum Genet. 2005;76:505–509. doi: 10.1086/428066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A. Manolescu A. Helgason A, et al. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet. 2006;38:68–74. doi: 10.1038/ng1692. [DOI] [PubMed] [Google Scholar]
- Helgadottir A. Manolescu A. Thorleifsson G, et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36:233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- Hube F. Reverdiau P. Iochmann S. Gruel Y. Improved PCR method for amplification of GC-rich DNA sequences. Mol Biotechnol. 2005;31:81–84. doi: 10.1385/MB:31:1:081. [DOI] [PubMed] [Google Scholar]
- In KH. Asano K. Beier D, et al. Naturally occurring mutations in the human 5-lipoxygenase gene promoter that modify transcription factor binding and reporter gene transcription. J Clin Invest. 1997;99:1130–1137. doi: 10.1172/JCI119241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovannisci DM. Lammer EJ. Steiner L, et al. Association between a leukotriene C4 synthase gene promoter polymorphism and coronary artery calcium in young women: the muscatine study. Arterioscler Thromb Vasc Biol. 2007;27:394–399. doi: 10.1161/01.ATV.0000252680.72734.10. [DOI] [PubMed] [Google Scholar]
- Kalayci O. Birben E. Sackesen C, et al. ALOX5 promoter genotype, asthma severity and LTC4 production by eosinophils. Allergy. 2006;61:97–103. doi: 10.1111/j.1398-9995.2006.00979.x. [DOI] [PubMed] [Google Scholar]
- Kim SH. Bae JS. Suh CH, et al. Polymorphism of tandem repeat in promoter of 5-lipoxygenase in ASA-intolerant asthma: a positive association with airway hyperresponsiveness. Allergy. 2005;60:760–765. doi: 10.1111/j.1398-9995.2005.00780.x. [DOI] [PubMed] [Google Scholar]
- Langaee T. Ronaghi M. Genetic variation analyses by pyrosequencing. Mutat Res. 2005;573:96–102. doi: 10.1016/j.mrfmmm.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Lima JJ. Zhang S. Grant A, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006;173:379–385. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. Dhingra A. Soltis PS, et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ. Mehta NU. Witonsky D, et al. Prevalence of common disease-associated variants in Asian Indians. BMC Genet. 2008;9:13. doi: 10.1186/1471-2156-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepine CJ. Handberg EM. Cooper-DeHoff RM, et al. A calcium antagonist vs a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease: The International Verapamil-Trandolapril Study (INVEST): A Randomized Controlled Trial. JAMA. 2003;290:2805–2816. doi: 10.1001/jama.290.21.2805. [DOI] [PubMed] [Google Scholar]
- Poole EM. Bigler J. Whitton J, et al. Prostacyclin synthase and arachidonate 5-lipoxygenase polymorphisms and risk of colorectal polyps. Cancer Epidemiol Biomarkers Prev. 2006;15:502–508. doi: 10.1158/1055-9965.EPI-05-0804. [DOI] [PubMed] [Google Scholar]
- Silverman ES. Drazen JM. Genetic variations in the 5-lipoxygenase core promoter. Description and functional implications. Am J Respir Crit Care Med. 2000;161:S77–S80. doi: 10.1164/ajrccm.161.supplement_1.ltta-16. [DOI] [PubMed] [Google Scholar]
- Silverman ES. Du J. De Sanctis GT, et al. Egr-1 and Sp1 interact functionally with the 5-lipoxygenase promoter and its naturally occurring mutants. Am J Respir Cell Mol Biol. 1998;19:316–323. doi: 10.1165/ajrcmb.19.2.3154. [DOI] [PubMed] [Google Scholar]