Abstract
Fibronectin (FN) forms fibrillar networks coupling cells to the extracellular matrix. The formation of FN fibrils, fibrillogenesis, is a tightly regulated process involving the exposure of cryptic binding sites in individual FN type III (FN-III) repeats presumably exposed by mechanical tension. The FN-III1 module has been previously proposed to contain such cryptic sites that promote the assembly of extracellular matrix FN fibrils. We have combined NMR and steered molecular dynamics simulations to study the structure and mechanical unfolding pathway of FN-III1. This study finds that FN-III1 consists of a β-sandwich structure that unfolds to a mechanically stable intermediate about four times the length of the native folded state. Considering previous experimental findings, our studies provide a structural model by which mechanical stretching of FN-III1 may induce fibrillogenesis through this partially unfolded intermediate.
Fibronectin (FN) is a key extracellular matrix protein that not only provides a substrate for cell anchorage but also serves as a regulatory protein in processes such as cell adhesion, motility, differentiation, and proliferation (1). Structurally, human FN is an ≈500-kDa multimodular protein existing both as a soluble dimer and as an insoluble fibrillar component that incorporates into the extracellular matrix (ECM). Cells bind FN through transmembrane proteins of the integrin family, which mechanically couple the actin cytoskeleton to the ECM. The mechanical responses of FN are defined by its multimodular structure, composed predominantly of three different repeats termed FN-I, -II, and -III. Individual FN-III modules have been proposed to unfold on mechanical stretching of FN, providing for the elasticity of FN fibrils (2). Consistent with this hypothesis, cells have been observed to stretch FN up to 4-fold their relaxed length (3–5).
In addition to providing the necessary elasticity for accommodating cell movements, stretching of FN-III modules is believed to expose buried binding sites that, for example, serve as nucleation sites for the assembly of FN into its fibrillar form. These buried binding sites, termed cryptic sites, presumably exist either within the FN-III core or buried between the hinge regions of two neighboring FN-III modules. However, the role of mechanical force in this process of transforming FN to a fibrillar form, fibrillogenesis, is only partially understood. It is known that cell-derived mechanical force is a prerequisite for FN polymerization in vitro. Fibrillogenesis is initiated by both artificial (6) and cell-derived tensile forces and by addition of partially or completely denatured FN-III modules (see review in ref. 1). In contrast, fibrillogenesis does not occur if the mechanical linkage between cells and FN has been disrupted by Rho inhibitors, myosin inhibitors, or actin-disrupting agents (7–9).
Cryptic sites for fibrillogenesis have been proposed to exist on FN-III1 (10, 11), -III2 (12), -III7 (13), -III9 (14), -III10 (15), and -III13–15 (13, 16). Thermal or chemical unfolding of these modules is associated with increased binding by either FN or a 70-kDa NH2-terminal FN fragment. In particular, partially folded FN-III1 has been found to induce fibrillogenesis. Studies have shown that heat-denatured FN-III1 was able to bind FN and its 70-kDa N-terminal fragment (10). Furthermore, a 76-aa fragment from FN-III1, termed anastellin and obtained by cutting the A and B strands off the N terminus, is structurally stable and promotes assembly of so-called super-FN macromolecular structure (10, 13, 17–19), which was found to inhibit the growth of malignant cells (20).
Although FN-III1 and other FN-III modules have been probed for biologically relevant cryptic sites, these experiments were typically conducted by using thermal and chemical denaturing or proteolytic cleavage. Consequently, it remained unclear whether partial denaturing can occur under physiological conditions, and whether these denatured fragments are thus physiologically relevant. Recently, single-molecule experiments using atomic force microscopy (AFM) have revealed that the mechanical unfolding pathway of FN-III1 is markedly different from that of other FN-III modules such as FN-III10, -III12, and -III13 (21). FN-III1 exhibits pronounced intermediate states during forced unfolding. Because no structural information of the FN-III1 module or its forced-unfolding intermediate was available, it was not known whether these intermediates were functionally relevant or correlated to fragments promoting fibrillogenesis. To address this issue, we determined the structure of FN-III1 by using NMR spectroscopy. We then used steered molecular dynamics (SMD) (22), which has been successfully applied to identify the mechanical unfolding intermediate of titin Ig domains previously (23, 24), to simulate the forced unfolding of FN-III1 and probe its mechanical intermediates. We find that the structure of one stable mechanical intermediate is closely related to the adhesive FN-III1 fragment previously implicated to initiate formation of FN. Taken together with known experimental findings, these results suggest how mechanical stretching induces partial unfolding and exposure of cryptic sites.
Materials and Methods
Sample Preparation. Human FN cDNA encoding FN-III1 repeat (residues S609–S701 in Swiss-Prot accession no. P02751) was subcloned into a pGEX-6P-2 expression vector (Pharmacia). The recombinant module was expressed in Escherichia coli BL21 strain as a GST fusion protein. Uniformly 15N-labeled FN-III1 was expressed in M9 minimal media by using 0.1% (wt/vol) 15NH4Cl, as the sole nitrogen source. Recombinant FN-III1 was purified by using the protocol previously used for FN-III13 preparation (25). The identity and purity of the protein were confirmed by electrospray mass spectrometry. NMR samples typically comprised 1–2 mM protein in either 90% H2O/10% D2O or 100% D2O containing 50 mM sodium phosphate buffer at pH 6. The sample used for residual dipolar coupling measurement contained 5% (wt/vol) of a 30/10/1 molar ratio of ditridecyl-phosphatidylcholine/dihexyl-phosphatidylcholine/cetyltrimethylammonium bromide.
NMR Spectroscopy. NMR experiments were acquired at 1H frequencies of 500.1, 600.1, and 750.1 MHz on spectrometers built in-house at the Oxford Centre for Molecular Sciences, incorporating Oxford Instruments magnets, and on a Bruker (Billerica, MA) DMX500 spectrometer. 3D 1H-15N experiments were recorded at 500.1 MHz at 20°C and 40°C. 1H and 15N resonance assignments were obtained by using 3D NOESY-heteronuclear sequential quantum correlation (HSQC) (75- and 125-ms mixing times) and 3D total correlation spectroscopy (TOCSY)-HSQC (34- and 61-ms isotropic mixing times). Vicinal 3JHNHα coupling constants were measured from a 3D HNHA (correlation of 1HN, 15N, 1Hα resonances) experiment. Homonuclear 2D NOESY and TOCSY spectra were recorded at 750 MHz and 40°C with mixing times of 125 and 55 ms, respectively. 2D NOESY, TOCSY, and double quantum filtered-COSY spectra were also recorded at 500 MHz with a sample dissolved in D2O. Slow-exchanging amide protons were identified on a series of 2D 1H-15N HSQC spectra recorded after dissolving the sample in D2O. Residual dipolar couplings were measured from in-phase antiphase 1H-15N HSQC spectra (26). Data were processed by using the felix 2.3 software package (Accelrys, San Diego) and the Bruker xwinnmr program, running on Sun and Silicon Graphics (Mountain View, CA) workstations. Spectra were analyzed with the aid of the xeasy program (27).
Structure Calculation. Structures were calculated by torsion angle dynamics using dyana (28). A set of 100 structures was calculated, starting with randomized conformers and using a standard annealing protocol. The input for the structure calculation consisted of 1,113 distance restraints and 81 dihedral angle restraints. Twenty-three hydrogen bonds were constrained, corresponding to slow-exchanging amide protons. The 25 conformers with the lowest residual target function (1.33 ± 0.31 Å2) were subsequently refined by using the x-plor-nih program (29) and topallhdg force field. The structures were submitted to a simulated annealing protocol, during which 57 dipolar coupling restraints were introduced. Finally the structures were energy-minimized by using the CHARMm22 force field (30). One structure was eliminated from the final set because of large residual violations.
MD Simulations. The MD simulations of FN-III1 were carried out by following the method described previously (23, 31). The program namd (32) was used with the CHARMm22 force field and TIP3 water parameters (33). Simulations were performed with a time step of 1 fs, a uniform dielectric constant of 1, and a cutoff of nonbonded forces with a switching function starting at 10 Å and reaching zero at 13 Å. FN-III1 was solvated in a 64-Å diameter water sphere, resulting in a system of 12,532 atoms. The whole system was minimized for 2,000 steps, followed by gradually heating from 0 to 300 K in 10 ps. The temperature was maintained at 300 K for another 10 ps, and the system was then equilibrated for 1.8 ns. Starting configurations for SMD simulations were obtained every 100 ps after the protein had been equilibrated for 1.5 ns. During SMD simulations, external harmonic force was applied to the C terminus of the protein while fixing the N terminus, with the stretching direction chosen along the vector pointing from N to C terminus and a spring constant of 11.7 kBT/Å2. Trajectories were saved every picosecond. The end-to-end distance RNC is defined as the distance between the Cα atoms of the two termini. Force and RNC presented in Fig. 3 were averaged every 4 and 20 ps for simulations at pulling velocities 0.05 Å/ps and 0.01 Å/ps, respectively. The hydrogen bond energy presented in Fig. 4 was calculated by using explicit hydrogen bond parameters in the CHARMm22 force field. The total simulation time was 82 ns.
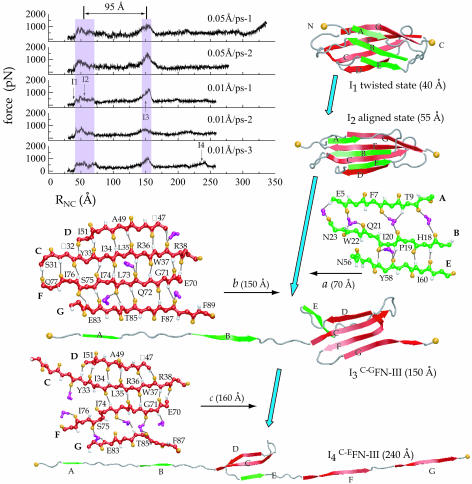
Fig. 3.
Mechanical unfolding intermediates of FN-III1. Curves shown (Top Left) are force vs. distance RNC profiles from constant velocity SMD unfolding simulations of FN-III1 at speeds of 0.05 and 0.01 Å/ps. RNC is defined as the end-to-end distance between two termini. Multiple runs at the same velocity are differentiated by a numeric suffix. Two force peak regions separated ≈95 Å are highlighted. Four unfolding intermediates (I1-I4) characterizing the unfolding pathway of FN-III1 are shown as snapshots in cartoon representation. Transition from aligned state I2 to I3 and from I3 to I4 requires disrupting certain interstrand hydrogen bonds between two β-sheets, ABE (a) and GFCD (b and c). The disruptions need assistance from surrounding water molecules (purple) that attack hydrogen bonds. Formed hydrogen bonds between backbone oxygen (orange) and hydrogen (white) atoms are represented as thick black lines.
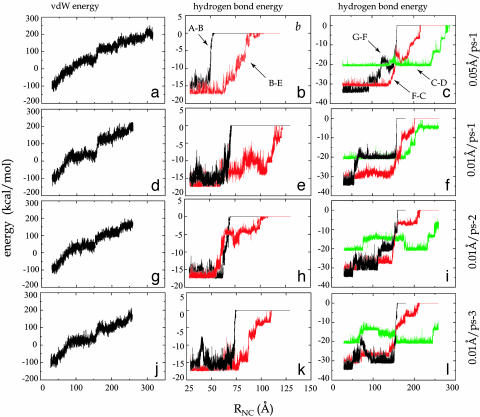
Fig. 4.
Energy of unfolding FN-III1. Profiles of energy vs. end-to-end distance RNC for four SMD unfolding simulations: (a–c) 0.05 Å/ps-1, (d–f) 0.01 Å/ps-1, (g–i) 0.01 Å/ps-2, (j–l) 0.01 Å/ps-3. Curves shown in the three rows are van der Waals energy of FN-III1, energy of interstrand hydrogen bonds between A and B strands (denoted as A-B, black); between B and E strands (B-E, red); and energy of interstrand hydrogen bonds between G and F strands (G-F, black); between F and C strands (F-C, red); and between C and D strands (C-D, green).
Results
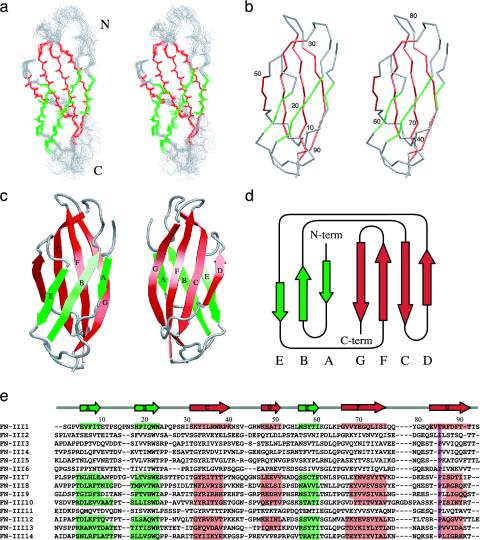
Structure of FN-III1. The structure of the recombinant FN-III1 module was determined by 1H, 15N NMR spectroscopy. The NMR structures exhibit few residual violations and low energies, indicating good agreement with experimental restraints and good geometry (Table 1). The structure of the FN-III1 module consists of two antiparallel β-sheets arranged as a β-sandwich with a long axis of ≈35 Å (Fig. 1). One β-sheet is formed by three strands (A, B, E), and the other β-sheet contains four strands (G, F, C, D) as in other FN-III modules resolved previously (34–36). The β-strands are well defined, with a backbone rms deviation (rmsd) of 0.37 Å (Table 1). Loops DE and EF show good precision, whereas loops AB, BC, CD, and FG are more disordered, especially loop AB (backbone rmsd of 1.95 Å), which is longer than in other structurally known FN-III modules.
Table 1. Structure statistics.
| Residual distance constraint violations* | |
| Number >0.1 Å | 2.4 ± 1.4 |
| rmsd, Å | 0.014 ± 0.001 |
| Residual dihedral constraint violations | |
| Number >5° | 3.0 ± 1.0 |
| rmsd, ° | 2.1 ± 0.3 |
| rmsd of dipolar couplings, Hz | 0.11 ± 0.003 |
| rmsd from idealized covalent geometry | |
| Bond lengths, pm | 0.94 ± 0.01 |
| Bond angles, ° | 1.97 ± 0.02 |
| Improper torsion angles, ° | 2.35 ± 0.11 |
| van der Waals energy, kcal·mol-1 | -427 ± 8 |
| Ramachandran diagram (% residues) | |
| Most favored regions | 80.7 |
| Additional allowed regions | 17.6 |
| Generous allowed regions | 1.6 |
| Disallowed regions | 0.2 |
| rmsd to mean structure,† Å | |
| N, Cα, C′ (1-93) | 0.88 ± 0.19 |
| N, Cα, C′ of β-strands | 0.37 ± 0.08 |
| Heavy atoms of β-strands | 0.88 ± 0.09 |
| N, Cα, C′ in loops‡ | 1.26 ± 0.28 |
The final input for the structure calculation consisted of 1, 113 distance restraints (206 intraresidual, 362 sequential, 93 medium-range, and 452 long-range); 81 dihedral angle restraints (71 ϕ, 10χ1); and 57 1H-15N dipolar coupling restraints.
Average coordinates for the 24 conformers after superposition for best fit of N, Cα, and C′ atoms of residues in β-strands (residues 5-8, 17-23, 30-39, 47-51, 57-61, 67-77, and 82-91).
rmsd of N, Cα, C′ atoms for individual loops are (in Å): loop AB, 1.95 ± 0.73; loop BC, 1.00 ± 0.30; loop CD, 1.08 ± 0.39; loop DE, 0.27 ± 0.13; loop EF, 0.54 ± 0.22; and loop FG, 1.10 ± 0.38.
Fig. 1.
FN-III1 structures and FN-III module sequence alignment. (a) Stereo views of the backbone heavy atom superposition of 24 minimized NMR structures. The two β-sheets and loops connecting β-strands are colored green, red, and gray, respectively. The same color code was used for all other representations of FN-III1. Stereo views of the Cα atom trace (b) and cartoon drawings of a single FN-III1 structure (c) in two different orientations are shown. (d) Schematic diagram of FN-III1. (e) Sequence alignment of 14 FN-III modules. The secondary structures of solved modules are highlighted. A key conserved proline among all other modules, but not in FN-III1 and -III2, is highlighted in purple. Protein snapshots were generated with vmd (40).
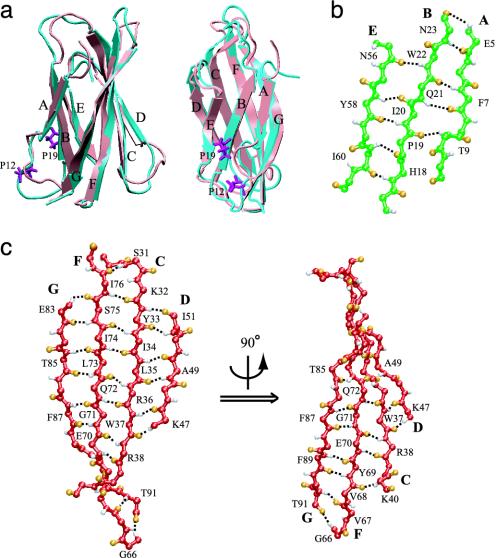
Equilibration of the explicitly solvated protein for 1.8 ns yields a stabilized structure with backbone rmsd from the NMR structure of <2.5 Å (Fig. 2a). The A strand in the triple-stranded β-sheet ABE is unusually short, with only four backbone hydrogen bonds between Glu-5, Phe-7 and Asn-23, Gln-21 of the B strand in the NMR structure. This hydrogen-bonding network was marginally extended during the equilibration through the formation of an additional hydrogen bond between Thr-9 and Pro-19 (Fig. 2b). Furthermore, two prolines, Pro-12 and -19, twist the A and B strands away from each other and act as β-sheet breakers, disrupting further hydrogen bonding between these two strands and resulting in an unusually weak β-sheet compared to other FN-III modules (Fig. 2a). In contrast, in the four-stranded β-sheet GFCD, the G and F strands form 10 interstrand hydrogen bonds (Fig. 2c), more than the F···G hydrogen bonds of any other structurally resolved FN-III modules. This structural difference, i.e., strengthening of the GFCD sheet, can be attributed to the mutation of a proline located in the FG loop, well conserved among other FN-III modules (Fig. 1e).
Fig. 2.
Structures of equilibrated FN-III1. (a) Alignment of NMR (cyan) and equilibrated (pink) structures shown in two different orientations. Two prolines that prevent A and B strands forming more interstrand hydrogen bonds are colored purple. Interstrand hydrogen bond networks of the ABE (b) and of the GFCD β-sheets (c), the latter in different orientations. Hydrogen and oxygen atoms are colored white and orange. Hydrogen bonds are represented by black dashed lines.
Forced-Unfolding Intermediates. Because interstrand hydrogen bonds have been shown to play important roles in the mechanical stability of proteins (24, 37), the unusual hydrogen-bonding network of FN-III1 suggests a unique mechanical design. Indeed, mechanical unfolding experiments using AFM have shown that FN-III1 has distinct intermediate states (21), which are not typical for other FN-III modules such as FN-III12 and -III13. To further examine the mechanical response of FN-III1, we conducted five SMD simulations at constant stretching velocities of 0.05 Å/ps and 0.01 Å/ps for a total time of 82 ns.
Two pronounced peaks were observed at end-to-end distances RNC of ≈55 and ≈150 Å in all five unfolding simulations, and four intermediates have been identified along the mechanical unfolding pathway (Fig. 3). On stretching, the module first elongates the coiled termini to ≈40 Å with no obvious disruption of the tertiary structure, reaching state I1. This state, also called “twisted state,” is similar to the first transitional state observed previously in studies of FN-III7–10 modules (37). After I1, the two β-sheets rotate to align themselves in the direction of the force vector, leading the protein to the second transitional state I2 at ≈55 Å, also termed aligned state (37). The aligned state occurs before separation of the A strand and corresponds to disrupting the hydrophobic core of the module, requiring a force of ≈500 pN in 0.01 Å/ps stretches. Entering the first peak area the force subsequently remains high, ≈500–800 pN, until the A strand separates from the fold.
Although the unfolding pathway is similar to that of other FN-III modules before the first β-strand detaches from the remaining fold (37), a significant divergence in the unfolding pathway for FN-III1 was observed after the detachment. The A and B strands gradually unravel first under a lower force until they are completely separated from the folding core, extending the protein to ≈150 Å. At this point, the protein reaches its most stable intermediate, termed I3, or C-GFN-III1, because it contains C to G strands in the remaining folded core. Transiting further from I3 requires a second peak force of up to ≈1,400 pN in 0.01 Å/ps stretches to disrupt the remaining GFCD β-sheet. After this most prominent force peak, the module gradually extends under a much lower force. Occasionally a fourth intermediate, I4, was seen before a third force peak; the intermediate, also termed C-EFN-III1, contains only C, D, and E strands folded. Finally, the protein fully extended at ≈330 Å.
Transition from one intermediate to the next requires forces to overcome energy barriers contributed not only from interstrand hydrogen bonds but also from van der Waals (vdW) and electrostatic packing interactions of the protein (Fig. 4). Transition from I2 is connected to the first force peak between 40 and 70 Å. The force peak is correlated with the breaking of five intrastrand hydrogen bonds between A and B strands, the breaking of two to four hydrogen bonds between G and F strands near the C terminus, and a severe perturbation of the hydrophobic core. Reaching I3 and stretching the protein away from the intermediate, the second force peak at ≈150 Å arises, correlating with the burst of six to eight F···C hydrogen bonds and two to four G···F hydrogen bonds, as well as a sudden increase of the protein's vdW energy. Breaking interstrand hydrogen bonds other than those between A and B strands and between G and F strands generally requires a relatively small force, because the respective hydrogen bonds rupture one by one in a “zipper-like” fashion. Rarely, a third peak force arises corresponding to destabilizing I4 by breaking the remaining hydrogen bonds, mainly between C and D strands.
Discussion
Comparison with AFM Experiments. Unfolding experiments of FN-III1 repeats using AFM has revealed several mechanically stable conformations, separated by 90 or 200 Å and characterized through rupture forces of 90 and 120 pN, respectively (21). This suggests the existence of intermediates that are consistent with our SMD results, despite the fact that rupture forces measured in SMD are about one order of magnitude higher than in AFM experiments due to the different pulling velocity regimes (38). According to SMD simulations, the extensions of 90 and 200 Å observed in AFM experiments are caused by the transition from I2 to I3 and by completely unraveling I3. The transition from I2 to I3 is characterized by breaking the A···B interstrand hydrogen bonds at distance RNC of 40–70 Å (55 ± 15 Å), and unraveling I3 is characterized by breaking the G···F hydrogen bonds at RNC of ≈150 Å. Accordingly, our simulations give an extension of ≈95 Å from I2 to I3, and an extension of ≈180 Å for unraveling I3, in close agreement with AFM experiments. Furthermore, the SMD rupture force corresponding to disrupting I3 is higher than the force needed to extend I2, in qualitative agreement with AFM experiments.
Comparison with Anastellin NMR Structure. Ruoslahti and colleagues (17) have found that anastellin, a 76-residue FN-III1 fragment beginning with residues NAPQ of the BC loop and spanning the remaining C to G strands, is able to stimulate fibrillogenesis of FN to form so-called super-FN (17). This fragment has shown antimetastatic effects on tumors grown in mice (20). Unless the 76-residue fragment assumes a new conformation, structurally it would correspond to the folded core of the I3 intermediate C-GFN-III1 found in our SMD studies, containing only the C to G strands. Indeed, the predicted structure of intermediate I3 is in excellent agreement with a recently published anastellin NMR structure (19), which exhibits a similar single antiparallel β-sheet consisting of C, D, F, and G strands, as well as a flexible E strand covering partially the hydrophobic core. Formed interstrand hydrogen bonds shown in Fig. 3b match well with those observed in the NMR structure (figure 3 in ref. 19). A difference is found at the G···F interstrand hydrogen bonding. Two bonds at residues Glu-83 and Thr-85 are missing in case of anastellin but are present in the simulated I3, whereas two bonds of Phe-89 near the C terminus were observed in the NMR structure but are broken in the simulated I3.
Tuning Mechanical Stability of Intermediates. Assuming the prominent unfolding intermediate I3 has functional relevance, it is of interest to understand the origin of this intermediate, based on the NMR structure reported here. In this respect, we note that the NMR structure shows that FN-III1 contains four more hydrogen bonds between the F and G strands than FN-III10, which exhibits an intermediate similar to I3, but only rarely (39). In fact, the intermediate is found only in the seldom case that stretching of FN-III10 unravels the A strand first. However, as a consequence of the strong F···G interstrand hydrogen bonding, unfolding of FN-III1 always begins with unraveling the A strand and, accordingly, always leads to the I3 intermediate. The structural reason for the strong F···G hydrogen bonding is the absence of a proline residue that is otherwise highly conserved and found at the beginning of the G strand of other FN-III modules (Fig. 1e). Based on other FN-III modules with known structure (FN-III7–10,12–14), this proline creates a β-bulge, preventing formation of more backbone hydrogen bonds and increasing solvent accessibility to existing backbone hydrogen bonds between the F and G strands. In contrast, FN-III1 and FN-III2 belong to the few FN-III modules that do not include this proline and, consequently, do not contain the β-bulge. The absence of this proline thus enhances interactions between the F and G strands and stabilizes FN-III1 and -III2 over other FN-III modules. This hypothesis is consistent with AFM experiments in which FN-III1 and -III2 modules were found to be the mechanically most stable FN-III modules (21). The absence of the stated proline in FN-III2 implies that FN-III2 is capable of forming an intermediate similar to I3 described above. Interestingly, FN-III2 has also been proposed to contain buried cryptic sites necessary for fibrillogenesis (12). However, further research is needed to determine under what physiological conditions the I3 intermediate may be exposed by mechanical stretching.
Significance of the Intermediates. The intermediates of FN-III1 can have profound physiological relevance. The states I1 and I2 may expose cryptic sites buried between domains or doubling the length of modules without inducing the unraveling of the entire FN-III tertiary structure. The presence of two late-stage intermediates after the unraveling of the first β-strands, I3 and I4, can expose further cryptic sites buried deep in the hydrophobic core. It was previously suggested that fibrillogenesis involves the swapping of β-strands with other partially unfolded FN-III modules (14). Our findings of a stable intermediate I3 in the mechanical unfolding pathway support this suggestion and provide a structural model revealing how cells exploit mechanical forces to expose cryptic sites on the FN-III1 module. Once an FN-III1 module is stretched into this intermediate, the straightened A and B strands can potentially be swapped with adjacent partially unfolded FN-III modules, thereby promoting binding among FN molecules.
Acknowledgments
We thank colleagues at the University of Oxford, Sachchidanand and David Staunton for recombinant FN-III1 production, and Jen Potts and Jorn M. Werner for residual dipolar coupling refinement. This work was supported by National Institutes of Health Grants PHS5P41RR05969 (to K.S.), 1R01GM60946 (to K.S.), R01EB00249 (to V.V.), and 5T32GM08268 (to D.C.); the Federation of European Biochemical Societies (to O.L.); the Wellcome Trust (to I.D.C.); a University of Washington University Initiative Fund grant (to D.C.); and National Science Foundation Grant NRAC MCA93S028 (to K.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FN, fibronectin; FN-III, FN type III; MD, molecular dynamics; SMD, steered MD; AFM, atomic force microscopy; rmsd, rms deviation.
Data deposition: The atomic coordinates for the 24 NMR structures of FN-III1 have been deposited in the Protein Data Bank, www.rcsb.org (PCB ID code 1OWW).
References
- 1.Geiger, B., Bershadsky, A., Pankov, R. & Yamada, K. M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 793–805. [DOI] [PubMed] [Google Scholar]
- 2.Erickson, H. P. (1994) Proc. Natl. Acad. Sci. USA 91, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohashi, T., Kiehart, D. P. & Erickson, H. P. (1999) Proc. Natl. Acad. Sci. USA 96, 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baneyx, G., Baugh, L. & Vogel, V. (2002) Proc. Natl. Acad. Sci. USA 99, 5139–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baneyx, G., Baugh, L. & Vogel, V. (2001) Proc. Natl. Acad. Sci. USA 98, 14464–14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baneyx, G. & Vogel, V. (1999) Proc. Natl. Acad. Sci. USA 96, 12518–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu, C., Keivens, V. M., O'Toole, T. E., McDonald, J. A. & Ginsberg, M. H. (1995) Cell 83, 715–724. [DOI] [PubMed] [Google Scholar]
- 8.Halliday, N. L. & Tomasek, J. J. (1995) Exp. Cell Res. 217, 109–117. [DOI] [PubMed] [Google Scholar]
- 9.Zhong, C. L., Chrzanowska Wodnicka, M., Brown, J., Shaub, A., Belkin, A. M. & Burridge, K. (1998) J. Cell Biol. 141, 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hocking, D. C., Sottile, J. & McKeown-Longo, P. J. (1994) J. Biol. Chem. 269, 19183–19187. [PubMed] [Google Scholar]
- 11.Hocking, D. C. & Kowalski, K. (2002) J. Cell Biol. 158, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sechler, J. L., Rao, H., Cumiskey, A. M., Vega-Colon, I., Smith, M. S., Murata, T. & Schwarzbauer, J. E. (2001) J. Cell Biol. 154, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingham, K. C., Brew, S. A., Huff, S. & Litvinovich, S. V. (1997) J. Biol. Chem. 272, 1718–1724. [DOI] [PubMed] [Google Scholar]
- 14.Litvinovich, S. V., Brew, S. A., Aota, S., Akiyama, S. K., Haudenschild, C. & Ingham, K. C. (1998) J. Mol. Biol. 280, 245–258. [DOI] [PubMed] [Google Scholar]
- 15.Hocking, D. C., Smith, R. K. & McKeown-Longo, P. J. (1996) J. Cell Biol. 133, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bultmann, H., Santas, A. J. & Peters, D. M. (1998) J. Biol. Chem. 273, 2601–2609. [DOI] [PubMed] [Google Scholar]
- 17.Morla, A., Zhang, Z. & Ruoslahti, E. (1994) Nature 367, 193–196. [DOI] [PubMed] [Google Scholar]
- 18.Morla, A. & Ruoslahti, E. (1992) J. Cell. Biol. 118, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briknarova, K., Akerman, M. E., Hoyt, D. W., Ruoslahti, E. & Ely, K. R. (2003) J. Mol. Biol. 332, 205–215. [DOI] [PubMed] [Google Scholar]
- 20.Yi, M. & Ruoslahti, E. (2001) Proc. Natl. Acad. Sci. USA 98, 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberhauser, A. F., Badilla-Fernandez, C., Carrion-Vazquez, M. & Fernandez, J. M. (2002) J. Mol. Biol. 319, 433–447. [DOI] [PubMed] [Google Scholar]
- 22.Isralewitz, B., Gao, M. & Schulten, K. (2001) Curr. Opin. Struct. Biol. 11, 224–230. [DOI] [PubMed] [Google Scholar]
- 23.Lu, H., Isralewitz, B., Krammer, A., Vogel, V. & Schulten, K. (1998) Biophys. J. 75, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marszalek, P. E., Lu, H., Li, H., Carrion-Vazquez, M., Oberhauser, A. F., Schulten, K. & Fernandez, J. M. (1999) Nature 402, 100–103. [DOI] [PubMed] [Google Scholar]
- 25.Sachchidanand, Lequin, O., Staunton, D., Mulloy, B., Forster, M. J., Yoshida, K. & Campbell, I. D. (2002) J. Biol. Chem. 277, 50629–50635. [DOI] [PubMed] [Google Scholar]
- 26.Ottiger, M., Delaglio, F. & Bax, A. (1998) J. Magn. Reson. 131, 373–378. [DOI] [PubMed] [Google Scholar]
- 27.Bartels, C., Xia, T. H., Billeter, M., Guntert, P. & Wuthrich, K. (1995) J. Biomol. NMR 5, 1–10. [DOI] [PubMed] [Google Scholar]
- 28.Guntert, P., Mumenthaler, C. & Wuthrich, K. (1997) J. Mol. Biol. 273, 283–298. [DOI] [PubMed] [Google Scholar]
- 29.Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Clore, G. M. (2003) J. Magn. Reson. 160, 65–73. [DOI] [PubMed] [Google Scholar]
- 30.MacKerell, A. D., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., et al. (1998) J. Phys. Chem. B 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- 31.Krammer, A., Lu, H., Isralewitz, B., Schulten, K. & Vogel, V. (1999) Proc. Natl. Acad. Sci. USA 96, 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kale, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K. & Schulten, K. (1999) J. Comput. Phys. 151, 283–312. [Google Scholar]
- 33.Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, J. W. & Klein, M. L. (1983) J. Chem. Phys. 79, 926–935. [Google Scholar]
- 34.Main, A. L., Harvey, T. S., Baron, M., Boyd, J. & Campbell, I. D. (1992) Cell 31, 671–678. [DOI] [PubMed] [Google Scholar]
- 35.Leahy, D. J., Aukhil, I. & Erickson, H. P. (1996) Cell 84, 155–164. [DOI] [PubMed] [Google Scholar]
- 36.Sharma, A., Askari, J. A., Humphries, M. J., Jones, E. Y. & Stuart, D. I. (1999) EMBO J. 18, 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Craig, D., Krammer, A., Schulten, K. & Vogel, V. (2001) Proc. Natl. Acad. Sci. USA 98, 5590–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craig, D., Gao, M., Schulten, K. & Vogel, V. (2004) Structure (Cambridge, U.K.), in press.
- 39.Gao, M., Craig, D. W., Vogel, V. & Schulten, K. (2002) J. Mol. Biol. 323, 939–950. [DOI] [PubMed] [Google Scholar]
- 40.Humphrey, W., Dalke, A. & Schulten, K. (1996) J. Mol. Graphics 14, 33–38. [DOI] [PubMed] [Google Scholar]