Abstract
PrP106–126, a peptide corresponding to residues 107–127 of the human prion protein, induces neuronal cell death by apoptosis and causes proliferation and hypertrophy of glia, reproducing the main neuropathological features of prion-related transmissible spongiform encephalopathies, such as bovine spongiform encephalopathy and Creutzfeldt–Jakob disease. Although PrP106–126 has been shown to form amyloid-like fibrils in vitro, their structural properties have not been elucidated. Here, we investigate the conformational characteristics of a fibril-forming fragment of the mouse prion protein, MoPrP106–126, by using electron microscopy, CD spectroscopy, NMR-detected hydrogen–deuterium exchange measurements, and molecular dynamics simulations. The fibrils contain ≈50% β-sheet structure, and strong amide exchange protection is limited to the central portion of the peptide spanning the palindromic sequence VAGAAAAGAV. Molecular dynamics simulations indicate that MoPrP106–126 in water assumes a stable structure consisting of two four-stranded parallel β-sheets that are tightly packed against each other by methyl–methyl interactions. Fibril formation involving polyalanine stacking is consistent with the experimental observations.
Prions are infectious particles that cause transmissible spongiform encephalopathies in animals and humans. Prions are composed of PrPSc, a conformationally altered form of a host-encoded glycoprotein, PrPC (1). Although the two isoforms are chemically identical, they possess very different physicochemical properties. In particular, PrPC is mostly helical, whereas the scrapie form PrPSc contains ≈40% β-sheet (2). A synthetic peptide, PrP106–126, was shown to aggregate into protease-resistant amyloid fibrils and induce neuronal cell death by apoptosis, causing proliferation and hypertrophy of cultured glia (3, 4). This segment corresponds to an unstructured region just outside of the globular C-terminal domain of PrPC (5, 6). Analysis of deletion mutants of human prion protein (PrP) showed that a large N-terminal fragment (residues 23–88) and a segment within the structured domain of PrPC (residues 141–176) could be deleted without affecting its conversion into the protease-resistant PrPSc, whereas deletion of segments 95–107, 108–121, or 122–140 abolished the conformational transition (7). PrP106–126 is located within this critical region (residues 95–140), has been shown to adopt different secondary structures under different solution conditions (8, 9), and is thus a relevant model for investigating the mechanism of fibril formation and PrPSc-mediated cell death. Recent solid-state NMR results showed that fibrils of the mouse prion peptide 89–143 are composed predominantly of β-structure, and they suggested that its pathogenicity is related to the specific β-sheet conformation (10, 11). However, little is known presently about the detailed structure of PrPSc or any fibril-forming fragments of the PrP. In fact, there are very few direct experimental observations available for any amyloid fibril. A detailed structural model for Alzheimer's disease β-amyloid fibrils was proposed recently on the basis of solid-state NMR data (12). Another promising approach for characterizing amyloid fibrils makes use of hydrogen–deuterium (H/D) exchange coupled with NMR (13) or mass spectrometry (14). In this study, we combined NMR-based H/D exchange and other physicochemical techniques with molecular dynamics (MD) simulations to obtain an atomic-resolution model of the mouse PrP fragment MoPrP106–126, which contains an unusual palindromic sequence motif (VAGAAAAGAV).
Materials and Methods
Amyloid Fibrils from MoPrP106–126. MoPrP106–126 peptide was synthesized from amino acid residues protected by 9-fluorenylmethoxycarbonyl (Fmoc) on a Plus PepSynthesizer (model 9050; Millipore). Ethanedithiol/thioanisole/thiophenol/95% trifluoroacetic acid (1:2:2:35, vol) was used for cleavage. The N terminus of the peptide was free, and the C terminus was amidated. The peptide was purified by reverse-phase HPLC. To remove residual trifluoroacetic acid, a 3-fold excess of hydrochloric acid was added, and the fraction was lyophilized, suspended in 50% acetonitrile/50% water (vol/vol), and relyophilized. The peptide was converted to fibrils by rehydration in 100 mM sodium acetate, containing 150 mM NaCl (pH 5.5) and 50% (vol/vol) acetonitrile.
Electron Microscopy. Electron micrographs of fibrils were taken with a JEM-1200EX electron microscope (JEOL), operating at 100 kV at a magnification of ×40,000. The samples were applied to carbon grids and stained with 2% uranyl acetate. The images were recorded on FG electron microscope film (Fuji), developed in a D-19 developer (Kodak) for 7 min, and digitized to 2,048 × 2,048 pixels with a SCA1 precision scanner (Zeiss) at a 7-μm step size.
CD Spectroscopy. CD spectra were recorded in 0.1-cm pathlength quartz cells on a CD spectrometer (model 62A DS; Aviv Associates, Lakewood, NJ). Peptide concentrations were 25–35 μM. Spectra were recorded at 25°C over a wavelength of 200–250 nm. Extension of the range to lower wavelengths was not possible because of excessive background absorbance due to acetonitrile and salts.
NMR Spectroscopy. To assign the backbone NH resonances of MoPrP106–126, homonuclear total correlation spectroscopy (TOCSY) spectra with the DIPSI2 sequence (15) were recorded on a DMX 600-MHz spectrometer (Bruker, Billerica, MA). Typically, 16 scans consisting of 4,096 data points at a spectral width of 7,000 Hz were recorded for each of 512 increments. The water resonance was selectively suppressed by excitation sculpting with gradients (15).
H/D Exchange Measurements. After lyophilizing the amyloid fibril solution, the same volume of deuterated amyloid-forming buffer [100 mM sodium acetate, containing 150 mM NaCl (pH 5.5) and 50% (vol/vol) acetonitrile] was added to the fibrils, mixed vigorously, and incubated for H/D exchange at 25°C. After a variable exchange period, the solution was lyophilized immediately and kept at –80°C. For NMR analysis of H/D exchange, samples were dissolved in 95% DMSO-d6/4.5% D2O/0.5% dichloroacetate-d2 (vol/vol/vol), pH 5.0, uncorrected pH meter reading. After 30 s, the sample solution was loaded immediately into an NMR tube and TOCSY spectra were recorded at 25°C. Intrinsic exchange rates (16) were calculated by using hxpro (available as sphere at www.fccc.edu/research/labs/roder).
MD Simulations. MD simulations were performed in the canonical NPT (number of particles–pressure–temperature) ensemble at 25°C by using the program discover, Version 2.98 (Accelrys, San Diego). All atoms of the system were considered explicitly, and their interactions were computed by using the CFF91 force field with periodic boundary conditions. The potential energy functions include bond stretching, bending angles, torsion, and out-of-plane angle deformation terms, and they contain cross-terms to describe bond–bond, angle–angle, bond–angle, bond–torsion, torsion–angle, and angle–angle–torsion couplings. A distance cutoff of 10 Å was used for van der Waals interactions and electrostatic interactions. The time step in the MD simulations was 1 fs (10–15 s). Our model included eight copies of the PrP106–126 peptide solvated with ≈5,000 water molecules in a 46 × 60 × 60-Å3 rectangular box. The effective water density in the solvation box was 1.006 g/cm3. The pH of the system was set to 5.5, and neutralizing Cl– ions were added. The starting conformations of the peptide complex were chosen to represent a β-sheet cluster. To generate β-strands, the dihedral angles were set to values corresponding to the β-sheet by using the biopolymer module in the insight ii molecular modeling package (Accelrys). We used ideal planar (parallel) four-strand β-sheets as a starting conformation. To explore different intersheet orientations, two four-strand β-sheets were placed on top of each other at angles of 90° (orthogonal), 0° (parallel), or 180° (antiparallel). The H-bonded chains were placed at ≈5-Å separation, and the distance between the sheets was set to ≈10 Å, which corresponds to the average distance in a cross-β structure (17). The complex was then solvated with water molecules and subjected to 500 steps of energy minimization to relax local forces. Subsequently, the system was heated to the desired temperature in 10,000 steps. Unless specified, simulations were performed at 298 K. Most simulations were run for ≈1 ns, with the stable conformations being tested for up to 2 ns. Simulations were repeated five times. All computations were run in parallel by using eight processors of the Origin 2000 machine (Silicon Graphics, Mountain View, CA).
Results
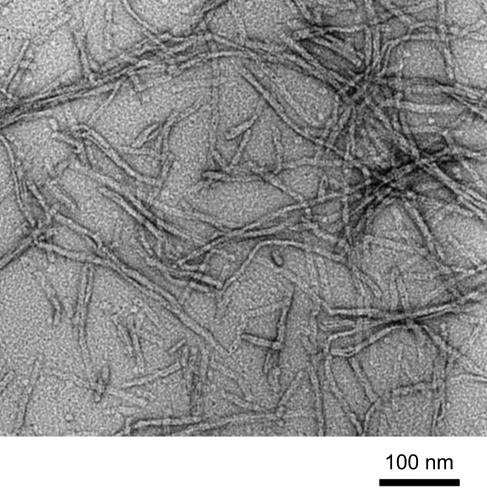
Fibril Morphology. The MoPrP106–126 peptide (TNVKHVAGAAAAGAVVGGLGG, palindrome underlined) studied here is homologous to residues 107–127 of human PrP (TNMKHMAGAAAAGAVVGGLGG). The fibril-forming tendencies of PrP106–126 depend critically on the solvent environment (8, 9). The conditions chosen for this work [100 mM sodium acetate/150 mM NaCl, pH 5.5/50% (vol/vol) acetonitrile] have been shown to facilitate fibril formation in a related prion peptide (9). An electron micrograph of MoPrP106–126 in this solvent confirmed formation of rods 6–8 nm in diameter and >100 nm in length (Fig. 1). The rods are bent in many places, suggesting that the fibrils are quite flexible, in contrast to other fibrils such as Alzheimer's disease β-amyloid fibrils, which have a more rigid appearance (18). However, compared with another fibril-forming prion fragment (residues 127–147), the PrP106–126 fibrils are less bent and more uniform in diameter (4).
Fig. 1.
Electron micrograph of a negatively stained preparation of the peptide MoPrP106–126 in 100 mM sodium acetate/150 mM NaCl, pH 5.5/50% (vol/vol) acetonitrile. The average length and width of the fibrils is 0.2 μm and 80 Å, respectively.
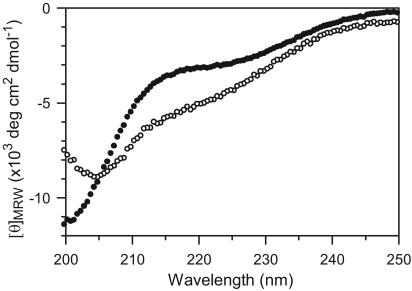
Characterization of Secondary Structure. The CD spectrum of MoPrP106–126 in a 1:1 mixture of acetonitrile and buffer shows a strong negative band near 200 nm and a less pronounced band in the 220- to 225-nm range (Fig. 2, •), which is consistent with a mixture of β-sheet and random coil (wavelengths <200 nm are obscured by the strong absorption of acetonitrile). Fibrils were formed within 1 min after dissolving the peptide in the solvent mixture, and the CD spectra remained unchanged even after 1 month. By using the program k2d (19), it was estimated that the fibrils contain ≈50% β-sheet and very low amounts of α-helix. Addition of 75% (vol/vol) trifluoroethanol to the fibril-containing solution resulted in a shift in the main band to higher wavelengths and enhanced the negative band near 220 nm (Fig. 2, ○), suggesting an increase in α-helix content at the expense of β-structure and random coil. The CD spectrum was unaffected by further addition of >50% (vol/vol) acetonitrile, indicating that fibril formation had reached a stable plateau (8). Although the rate of fibril formation for MoPrP106–126 appears to be faster than the rate reported for other peptides, such as Alzheimer's disease β-peptides (20), an exact comparison would require a more detailed analysis, including studies of the effects of seeding.
Fig. 2.
CD spectra of MoPrP106–126 in 100 mM sodium acetate/150 mM NaCl, pH 5.5/50% (vol/vol) acetonitrile, recorded at 25°C in the absence (•) and presence (○) of 75% (vol/vol) trifluoroethanol. MRW, mean residue weight.
Determination of the Protection Factors. Quenched hydrogen-exchange measurements coupled with NMR have been used widely for obtaining residue-specific structural information on protein folding intermediates, denatured states, and complexes that would be difficult to characterize by direct NMR or crystallographic methods (21–24). The method relies on the fact that stably folded proteins contain a large number of slowly exchanging amide protons that can be used as structural probes. However, the application of quenched H/D exchange methods for characterizing fibrous aggregates of peptides is hampered by the fact that, after dissolving the fibrils, the rate of back-exchange is too fast for 2D NMR analysis, even under optimal quench conditions (e.g., pH 3 and temperature ≈0°C). In a recent study of a fragment of the Alzheimer's disease β-peptide, Ippel et al. (25) were able to measure the level of exchange in the fibrous state by following the time course of exchange during and after dissolution of the fibrils in the presence of trifluoroethanol and extrapolating back in time. Hoshino et al. (13) used a similar approach to characterize the β2-microglobulin fibril. Their method is based on earlier work by Zhang et al. (26), who developed a quenched H/D exchange protocol involving mixtures of DMSO, water, and an organic acid to slow down the rate of back-exchange under quench conditions. For example, in 95% DMSO-d6/4.5% D2O/0.5% dichloroacetic-d2 acid (vol/vol/vol), pH 5.5, uncorrected, H/D exchange rates for model peptides are ≈100-fold slower than H/D exchange rates in 100% D2O (26), thus providing ample time to record a 2D NMR spectrum with minimal back-exchange. At the same time, DMSO is highly efficient for dissolving amyloid fibrils, including MoPrP106–126, resulting in monomeric peptide with a well resolved NMR spectrum (Fig. 3). For NMR analysis of H/D exchange, samples were dissolved in 95% DMSO-d6/4.5% D2O/0.5% dichloroacetic-d2 acid (vol/vol/vol), pH 5.0, uncorrected. The sample solution was immediately loaded into an NMR tube and a 1D 1H NMR spectrum was acquired every 5 min for 30 min. The spectral changes observed indicate that, in the quench solvent, the fibrils are disassembled into monomers at a rate of ≈0.1 min–1 (data not shown). For the measurement of H/D exchange rates, 1D 1H-NMR spectra were measured for 15 min to confirm complete disassembly of the fibrils, followed by consecutive acquisition of three 2D TOCSY spectra with an acquisition time of 6 h per spectrum. The first TOCSY spectrum recorded for each exchange time point was used to determine exchange rates.
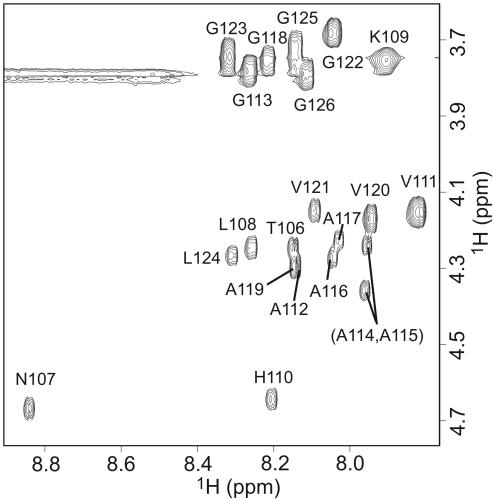
Fig. 3.
NH–CαH region of a TOCSY spectrum of MoPrP106–126 in 95% DMSO-d6/4.5% D2O/0.5% dichloroacetic-d2 acid (vol/vol/vol), pH 5.0, uncorrected.
Fig. 3 shows the fingerprint region of the TOCSY spectrum of a sample of MoPrP106–126 (lyophilized from H2O), which was freshly dissolved in 95% DMSO-d6/4.5% D2O/0.5% dichloroacetic-d2 acid (vol/vol/vol), pH 5.0. The NH–CαH cross peaks are well dispersed and could be readily assigned. The line widths are consistent with a monomeric peptide. Because the H/D exchange reaction is effectively quenched in this solvent (time constant >10 h; ref. 26), the integrated peak intensity for a resolved NH–CαH cross peak at a given exchange time in the fibril state, relative to that of a fully protonated control sample (Fig. 3), provides a direct measure of the degree of exchange (H/D ratio) of an individual amide group in the NH–CαH state. We repeated this procedure for a series of carefully chosen incubation times of MoPrP106–126 under conditions favoring fibril formation, i.e., a 1:1 mixture of D2O buffer (0.1 M sodium acetate/0.15 M sodium chloride, pH 5.5, uncorrected) and acetonitrile. The intensities of resolved cross peaks were found to decay exponentially with exchange time, yielding the rate of exchange of individual amide protons in the fibril state.
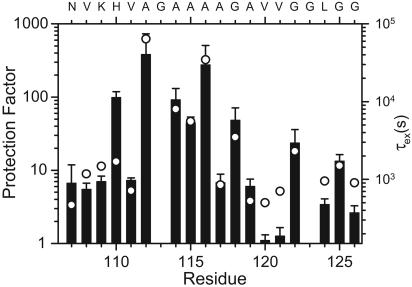
In contrast to previous work on other amyloid fibrils (13, 25), we were thus able to measure quantitative exchange rates, which allowed us to determine protection factors for the majority of residues in MoPrP106–126. Fig. 4 shows a logarithmic plot of the time constants of exchange vs. residue number along with protection factors (Fig. 4, bars), calculated as the ratio of the intrinsic exchange rate (predicted on the basis of model peptide data; ref. 16) to the observed exchange rate. Protection factors >30 indicative of amide protons involved in stable H bonds are observed for five of the eight Ala and Gly residues in the middle of the peptide, as well as for His-110. Two residues near the C terminus (Gly-122 and Gly-125) show intermediate protection factors in the range of 10–30. The protection factors for the remaining amide groups range between 1 and 10, indicating that these amides are exposed or that they form marginally stable H bonds in the fibrous form of the peptide. Jarvis et al. (27) measured the H/D exchange reaction of a transthyretin fragment (YTIAALLSPYS) in the presence of 30% acetonitrile/70% D2O (vol/vol). The rates observed are consistent with the rates of unstructured peptides in D2O (25), indicating that the presence of acetonitrile does not affect the intrinsic exchange rates significantly. Therefore, protection factors in the range of ≈5–10 (residues 106–109, 117, and 119) probably reflect weak structural protection rather than a solvent-induced slowing effect.
Fig. 4.
H/D exchange protection factors of individual amide protons in MoPrP106–126 amyloid fibrils. Bars indicate the protection factors, and ○ indicate H/D exchange time constants. Reliable exchange rates could not be measured for the Gly-113 and Gly-123 NH peaks because of their proximity to the water resonance.
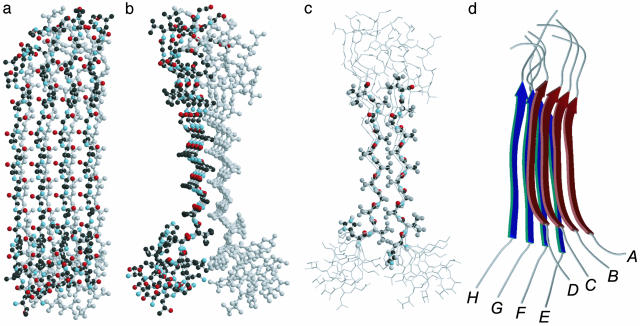
Modeling of PrP106–126 Fibrils by Using MD Simulation. MD simulations have been used extensively to model amyloid fibril formation for various peptides, including fragments of PrP (17, 28). Together with our experimental constraints, this approach promises to provide further insight into the structural basis of fibril formation. MD simulations were performed on an octamer of the peptide TNVKHVAGAAAAGAVVGGLGG solvated with ≈5,000 water molecules. The solvated oligomer was subjected to 500 steps of initial minimization to relax local forces. Subsequently, the system was heated from 0 to 298 K in 10,000 steps. Given the size of the system, only a limited number of simulations with different starting conformations were feasible. In their recent MD simulations on oligomers of alanine-rich peptides, including the conserved PrP fragment AGAAAAGA, Ma and Nussinov (17) found that two layers of four antiparallel β-strands, stacked in parallel orientation, formed the most stable complex. However, when we performed MD simulations of the longer MoPrP106–126 by using either parallel or antiparallel interstrand orientations as a starting structure, we found that two layers of parallel β-sheets (stacked in different orientations; see below) formed stable octameric complexes, whereas two layers of antiparallel β-sheet collapsed quickly into irregular globular structures. This finding is supported by recent electron microscopy data on 2D crystals of a truncated form of PrP (PrP27–30), which are consistent with a core of parallel β-helices (29). Thus, longer PrP fragments apparently show a preference to form parallel β-sheets, whereas both orientations can occur for shorter peptides (12).
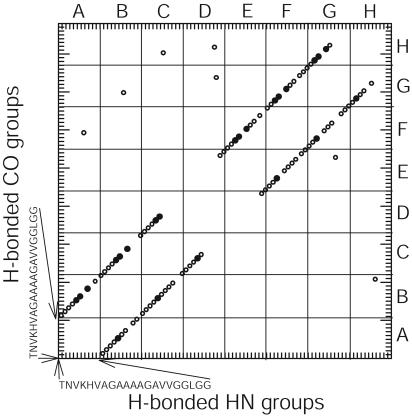
For further simulations, we chose two parallel β-sheets and explored different starting configurations with orthogonal and parallel/antiparallel stacking arrangements, respectively. The initial structure with an antiparallel stacking arrangement failed to converge into a stable structure. The simulation with an orthogonal starting configuration moved during the simulation toward the antiparallel orientation and converged to a stable structure (total energy –16,000 kcal/mol) with an angle of ≈120° between strands in opposite layers. The most stable complex was obtained in a simulation starting from two ideal parallel β-sheets that were stacked in parallel intersheet orientation. As illustrated in Fig. 5, the simulation assumed a very stable structure (average energy –44,000 kcal/mol) with highly regular parallel β-sheets and persistent H bonds for residues His-110 through Val-120 in all eight strands. The two layers of β-sheet form intimate van der Waals contacts by intercalation of methyl groups in the central hydrophobic region, spanning Val-111 to Val-121 (Fig. 5c). The side views (Fig. 5 b and c) show that the strands in opposite layers are parallel in the central Ala/Gly region (average backbone–backbone distance 5.8 Å), but they begin to diverge at the valine positions at each end. All of the four histidine side chains from one layer (Fig. 5c) show regular ring stacking along the N-terminal opening of the sandwich. Fig. 6 shows the distribution of amide H bonds (<2.2 Å, measured between hydrogen and oxygen) in the model. Backbone H bonds formed more frequently in the central palindromic region (•) compared with the peripheral regions (○). The large majority of H bonds are regular backbone–backbone H bonds between adjacent strands (intrasheet), especially for the two central strands of each layer. One interesting exception is a set of intersheet H bonds involving the NH of Ala-119 in strands A, B, and C of one layer (Fig. 5 b and c, left) and the CO of Gly-118 in strands F, G, and H of the opposite layer (Fig. 5 b and c, right). This break in the regular H-bonding structure may be responsible for the relatively low protection factors observed for Ala-117 and Ala-119 (Fig. 4).
Fig. 5.
Representative structure of the octameric complex of PrP106–126 calculated by using MD simulations. All peptides are oriented with the N terminus at the bottom. (a) Front view, using functional coloring for the top layer (strands A–D) and gray for the bottom layer (strands E–H). (b) Side view, using the same coloring scheme as in a. (c) Side view, emphasizing the methyl packing interactions in the core of the complex. Residues (His-110 to Val-120) on two opposite strands (B and F) are shown as a ball-and-stick representation (including H), and other residues are shown as a wire diagram. (d) Schematic ribbon diagram of the predicted PrP106–126 fibril structure.
Fig. 6.
Distribution of H bonds (<2.2 Å) that were formed during the simulation of octameric PrP106–126 in the palindromic (•) and nonpalindromic (○) regions of the sequence. Eight 21-residue peptides (A–H; see Fig. 5d) are aligned along the horizontal and vertical axes.
Overall, our simulations predict a parallel β-sheet arrangement with the most persistent H bonds and close-packed side-chain interactions in a core region spanning the central hydrophobic sequence VAGAAAAGAV, whereas the flanking regions appear more dynamic and partially frayed, judging from the structural variation among the different strands (Fig. 5) and the degree of mobility seen in the MD simulations (data not shown). These predictions are consistent with our hydrogen exchange results (Fig. 4), which show that the majority of well protected amide groups (five of a total of six residues with protection factors >30) are located within the central AGAAAAGA octapeptide. The observation that the distribution of protection factors is skewed toward the N terminus (Fig. 4) is also in line with simulated structure, which shows larger variation among individual strands in the Gly-rich C-terminal region compared with the N-terminal region (Fig. 5).
Discussion
Our findings are consistent with results on polyalanine-based peptides by Blondelle et al. (30), who found that peptides with the sequence Ac-KAnK-NH2 (i) are monomeric if n < 10, (ii) begin to show formation of β-structure for n = 10–14, and (iii) form soluble aggregates with high β-sheet content if n > 14, indicating that a minimum of ≈12 apolar residues is required to provide the hydrophobic surface for stacking β-strands into stable fibril structures. Their host–guest analysis of β-sheet formation for a series of peptides of the form Ac-KYA7XA5K-NH2 indicates that the Gly and Val residues found in the middle portion of MoPrP106–126 are also compatible with β-structure formation, whereas a single Lys, His, and Leu at the guest position abolished formation of the β-sheet complex. Thus, the MoPrP106–126 peptide (TNVKHVAGAAAAGAVVGGLGG) contains an uninterrupted stretch of 13 amino acids that favor β-sheet complex formation (underlined), flanked by residues that tend to disrupt β-structure (bold). Our hydrogen exchange results (Fig. 4) further support the conclusion that residues 111–123, which contain the majority of the stably H-bonded NH groups, comprise the core region of the β-sheet complex.
Two possible models for fibril extension are that the fibrils grow perpendicular to the β-sheet plane by stacking peptides or that they grow by extending the β-sheet by addition of monomers on each side. The hydrogen exchange data support the second possibility that in an extended fibril formed by stacking of four-strand sheets, alternating NH groups on the edge strands are not H bonded, which might give rise to heterogeneous (double-exponential) exchange kinetics. We observe single-exponential kinetics for all amides and slow rates for three Ala and one Gly of the central octapeptide, favoring a model in which the fiber grows by extending the β-sheet along the edge (i.e., the fiber axis is approximately parallel to the H bonds). This conclusion is further supported by our MD simulations, which show highly favorable packing interactions between two parallel β-sheets stacked in parallel, which allows further extension of the sheet by adding strands on each side. In fact, the strands at the edges of the β-sheet are structurally very similar to the interior ones (Fig. 5a), indicating that the regular H-bonding interactions in the core of the β-sheet can be readily extended by adding strands along the edge of the octamer consistent with fibril growth in a cross-β configuration. The β-sheets exhibit a small propeller twist of ≈2.3°, indicating that ≈150 strands are required to complete a full rotation along the fibril axis (with an average interstrand distance of 5 Å; this twist gives rise to a helical pitch of ≈75 nm). Finally, the sheets curl away from the central plane of the sandwich at each end (Fig. 5 b–d), which makes it sterically unfavorable to interact with additional sheets. Thus, our results argue against a growth mechanism involving stacking of β-sheets in the direction normal to the plane of the sheets. Our model is topologically similar to that of the Alzheimer's disease β-amyloid fibrils proposed by Petkova et al. (12) in that both models exhibit a double-layered parallel β-structure, stabilized by a tightly packed hydrophobic core and interstrand H bonding.
The end-to-end distance of the peptides in the model is ≈60 Å, which is comparable with the 6- to 8-nm diameter of the rods seen in the electron micrographs of the MoPrP106–126 fibrils (Fig. 1). The well protected NH groups are all between residues 110 and 121, indicating that this largely hydrophobic stretch of amino acids corresponds to the β-sheet core of the fibril. Thus, 12 of 21 residues (57%) are expected to assume regular β-structure, which is consistent with the β-sheet content of ≈50% that was estimated from the CD spectrum (Fig. 2). All of these findings are well supported by the predicted structure (Fig. 5), which shows regular β-structure for the central 12 residues of MoPrP106–126 and increased fraying toward each end. Other independent experimental results are consistent with this model also. For example, 13C isotope-edited IR experiments indicated that the peptide AGAAAAGA assumes a β-sheet conformation in the fibril (11), and a model containing parallel β-helical structure has been proposed on the basis of electron crystallographic data on truncated forms of PrP (29). Intriguingly, the spider silk protein contains similar sequence motifs, such as GAAAAG, and also has a tendency to form superfibers (31) and amyloid fibrils (32). Moreover, the structure obtained for the fibril resembles the nanostructure formed by polyalanine films (33).
Under the relatively acidic conditions that favor fibril formation, MoPrP106–126 contains two positively charged residues, Lys-109 and His-110, in addition to the N-terminal amino group. Because of this concentration of positive charge near the N terminus, one might expect that electrostatic interactions would favor antiparallel over parallel strand orientations. However, the MD simulations showed a preference for parallel interactions, both within each β-sheet and between the two layers (Fig. 5d). Apparently, the parallel configuration optimizes the packing interactions between the methyl groups of the central Ala and Val residues, outweighing any repulsive electrostatic interactions. This arrangement may be further stabilized by favorable interactions of histidine rings, which are neatly stacked along the crevice of the β-sandwich (Fig. 5c). Interestingly, replacement of this conserved histidine residue with a d-amino acid (d-His) was found to disrupt fibril formation of the human PrP106–126 peptide (8), confirming the importance of this residue.
Although the occurrence of a palindromic sequence within PrP (VAGAAAAGAV) may be fortuitous (note that the first Val is a Met in the human and Syrian hamster PrP sequences), an approximately symmetric arrangement of small apolar amino acids can form a β-sheet with relatively smooth surfaces that can pack favorably with a second layer of β-sheet. Symmetric sequences may also favor fibril formation because parallel and antiparallel conformations are energetically more similar compared with nonsymmetric sequences (34). Although the MD simulations suggest a preference for parallel orientations, the symmetric sequence motif may allow more heterogeneous structures containing both parallel and antiparallel orientations (depending on other regions of the protein), resulting in increased entropy for amyloid formation. Given the high stability and favorable structural properties of our computational model of the PrP106–126 fibrils, which is fully consistent with all experimental observations reported here (amide protection, CD, and electron microscopy) and with the results of previous biophysical and mutational studies on related peptides (7–11, 29), it appears likely that this unusual sequence motif plays an important role in converting the monomeric PrPC into its neurotoxic and infectious scrapie form.
Acknowledgments
We thank R. F. Latypov for his help with computer graphics, R. L. Dunbrack for his valuable advice on MD calculations and the manuscript, and K. Maki and G. D. Makham for their careful reading of the manuscript. The work was supported by National Institutes of Health Grants R01 GM056250 (to H.R.); National Cancer Institute Core Grant CA06927; an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center; Ministry of Education, Science, Culture, and Sports of Japan Grants-in-Aid for Scientific Research 14380314 and 14037224 (to K.K.); and the National Project on Protein Structural and Functional Analyses. The NMR Facility of the Fox Chase Cancer Center is supported by a grant from the Kresge Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PrP, prion protein; PrPSc, scrapie form of PrP; PrPC, cellular PrP; MoPrP106–126, mouse PrP fragment; H/D exchange, hydrogen–deuterium exchange; MD, molecular dynamics; TOCSY, total correlation spectroscopy.
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan, K.-M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., Mehlhorn, I., Huang, Z., Fletterick, R. J., Cohen, F. J. & Prusiner, S. B. (1993) Proc. Natl. Acad. Sci. USA 90, 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forloni, G., Angeretti, N., Chiesa, R., Monzani, E., Salmona, M., Bugiani, O. & Tagliavini, F. (1993) Nature 362, 543–546. [DOI] [PubMed] [Google Scholar]
- 4.Tagliavini, F., Prelli, F., Verga, L., Giaccone, G., Sarma, R., Gorevic, P., Ghetti, B., Passerini, F., Ghibaudi, E., Forloni, G., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 9678–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riek, R., Hornemann, S., Wider, G., Billeter, M., Glockshuber, R. & Wüthrich, K. (1996) Nature 382, 180–182. [DOI] [PubMed] [Google Scholar]
- 6.James, T. L., Liu, H., Ulyanov, N. B., Farr-Jones, S., Zhang, H., Donne, D. G., Kaneko, K., Groth, D., Mehlhorn, I., Prusiner, S. B. & Cohen, F. E. (1997) Proc. Natl. Acad. Sci. USA 94, 10086–10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muramoto, T., Scott, M., Cohen, F. E. & Prusiner, S. B. (1996) Proc. Natl. Acad. Sci. USA 93, 15457–15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salmona, M., Malesani, P., De Gioia, L., Gorla, S., Bruschi, M., Molinari, A., Vedova, F. D., Pedrotti, B., Marrari, M. A., Awan, T., et al. (1999) Biochem. J. 342, 207–214. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, H., Kaneko, K., Nguyen, J. T., Livshits, T. L., Baldwin, M. A., Cohen, F. E., James, T. L. & Prusiner, S. B. (1995) J. Mol. Biol. 250, 514–526. [DOI] [PubMed] [Google Scholar]
- 10.Laws, D. D., Bitter, H.-M. L., Liu, K., Ball, H. L., Kaneko, K., Wille, H., Cohen, F. E., Prusiner, S. B., Pines, A. & Wemmer, D. E. (2001) Proc. Natl. Acad. Sci. USA 98, 11686–10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldwin, M. A. (1999) Methods Enzymol. 309, 576–591. [DOI] [PubMed] [Google Scholar]
- 12.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshino, M., Katou, H., Hagihara, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Nat. Struct. Biol. 9, 332–336. [DOI] [PubMed] [Google Scholar]
- 14.Kheterpal, I., Zhou, S., Cook, K. D. & Wetzel, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13597–13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang, T. L. & Shaka, A. J. (1998) J. Magn. Reson. 135, 280–287. [DOI] [PubMed] [Google Scholar]
- 16.Bai, Y., Milne, J. S. & Englander, S. W. (1993) Proteins Struct. Funct. Genet. 17, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, B. & Nussinov, R. (2002) Protein Sci. 11, 2335–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh, D. M., Hartley, D. M., Kusumoto, Y., Fezoui, Y., Condron, M. M., Lomakin, A., Benedek, G. B., Selkoe, D. J. & Teplow, D. B. (1999) J. Biol. Chem. 274, 25945–25952. [DOI] [PubMed] [Google Scholar]
- 19.Andrade, M. A., Chacón, P., Merelo, J. J. & Morán, F. (1993) Protein Eng. 6, 383–390. [DOI] [PubMed] [Google Scholar]
- 20.Kirkitadze, M. D., Condron, M. M. & Teplow, D. B. (2001) J. Mol. Biol. 312, 1103–1119. [DOI] [PubMed] [Google Scholar]
- 21.Roder, H., Elöve, G. A. & Englander, S. W. (1988) Nature 335, 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udgaonkar, J. B. & Baldwin, R. L. (1988) Nature 335, 694–699. [DOI] [PubMed] [Google Scholar]
- 23.Roder, H. (1989) Methods Enzymol. 176, 446–473. [DOI] [PubMed] [Google Scholar]
- 24.Paterson, Y., Englander, S. W. & Roder, H. (1990) Science 249, 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ippel, J. H., Olofsson, A., Schleucher, J., Lundgren, E. & Wijmenga, S. S. (2002) Proc. Natl. Acad. Sci. USA 99, 8648–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Y.-Z., Paterson, Y. & Roder, H. (1995) Protein Sci. 4, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvis, J. A., Kirkpatrick, A. & Craik, D. J. (1994) Int. J. Pept. Protein Res. 44, 388–398. [DOI] [PubMed] [Google Scholar]
- 28.Alonso, D. O. & Daggett, V. (2001) Adv. Protein Chem. 57, 107–137. [DOI] [PubMed] [Google Scholar]
- 29.Wille, H., Michelitsch, M. D., Guénebaut V., Supattapone, S., Serben, A., Cohen, F. E., Agard, D. A. & Prusiner S. B. (2002) Proc. Natl. Acad. Sci. USA 99, 3563–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blondelle, S. B., Forood, B., Houghten, R. A. & Pérez-Payá, E. (1997) Biochemistry 36, 8393–8400. [DOI] [PubMed] [Google Scholar]
- 31.Xu, M. & Lewis, R. V. (1990) Proc. Natl. Acad. Sci. USA 87, 7120–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenney, J. M., Knight, D., Wise, M. J. & Vollrath, F. (2002) Eur. J. Biochem. 269, 4159–4163. [DOI] [PubMed] [Google Scholar]
- 33.Bensing, J. L. & Pysh, E. S. (1971) Biopolymers 10, 2645–2648. [DOI] [PubMed] [Google Scholar]
- 34.Steward, R. E. & Thornton, J. M. (2002) Proteins 48, 178–191. [DOI] [PubMed] [Google Scholar]