Summary
Biomarkers in human body fluids have great potential for use in screening for diseases such as cancer and diabetes, diagnosis, determining the effectiveness of treatments, and detecting recurrence. Present 96-well immunoassay technology effectively analyzes large numbers of samples; however, this approach is more expensive and less time effective on single or a few samples. In contrast, microfluidic systems are well suited for assaying small numbers of specimens in a point-of-care setting, provided suitable procedures are developed to work within peak capacity constraints when analyzing complex mixtures like human blood serum. Here, we developed integrated microdevices with an affinity column and capillary electrophoresis channels to isolate and quantitate a panel of proteins in complex matrices. To form an affinity column, a thin film of a reactive polymer was photopolymerized in a microchannel, and four antibodies were covalently immobilized to it. The retained protein amounts were consistent from chip to chip, demonstrating reproducibility. Furthermore, the signals from four fluorescently labeled proteins captured on-column were in the same range after rinsing, indicating the column has little bias toward any of the four antibodies or their antigens. These affinity columns have been integrated with capillary electrophoresis separation, enabling us to simultaneously quantify four protein biomarkers in human blood serum in the low ng/mL range using either a calibration curve or standard addition. Our systems provide a fast, integrated and automated platform for multiple biomarker quantitation in complex media such as human blood serum.
Introduction
Due to earlier stage diagnosis and advances in cancer treatment, the five-year relative survival rate (of patients compared with controls) for all cancers has improved from 50% in 1975-1977 to 66% in 1996-2004.1 Presently, cancer diagnosis is based mainly on morphological examination of a tumor biopsy, which is expensive, time consuming, and hence low in throughput.2 As an earlier stage tool, biomarkers can play an important role in cancer screening, diagnosis, and recurrence detection.3, 4 For instance, prostate-specific antigen (PSA) is a widely used analyte for prostate cancer screening.5 However, an abnormal level of a single biomarker alone is not generally sufficient to diagnose cancer.6 Thus, many men with PSA levels less than the 4.0 ng/mL action threshold had prostate cancer detected by biopsy (i.e., false-negatives).7 Furthermore, PSA levels above 4 ng/mL are associated with other conditions such as prostatitis, reducing the specificity (i.e., false-positives).5 To overcome these shortcomings, the simultaneous detection of multiple markers8 would enable more sensitive and accurate cancer screening with higher throughput. For instance, Yang et al.9 evaluated 12 biomarkers for gastrointestinal cancer diagnosis, and a combination of five markers significantly improved the diagnostic rate to ~40% relative to the ~27% rate achieved with just carcinoembryonic antigen (CEA).
Currently, most biomarkers are detected via immunoassays such as enzyme linked immunosorbent assay (ELISA).10 Recently, Ladd et al.11 developed a label-free detection protocol for cancer biomarker candidates using surface plasmon resonance imaging, with a limit of detection as low as a few ng/mL. Unfortunately, significant nonspecific adsorption was observed in diluted serum analysis, which led to a high background and much poorer detection limit. A recent review summarizes the advances and challenges of multiplexed immunoassay platforms.12 However, these multimarker systems need further validation and quality control. Transferring these approaches to a microfluidic format could provide higher speed and lower reagent consumption.13 Yet, analyzing real samples in complex matrices using microdevices is challenging, and the small microchip platform reduces resolving power and peak capacity relative to full-size instruments.14 Furthermore, due to small injected sample volumes and a short optical path, the concentration detection limit in microchips is often higher than in conventional techniques.15 To overcome these shortcomings of microfluidic systems, multiple analysis functions can be integrated on a single device, enabling sample purification and preconcentration.16 Many processing steps including sample desalting,17 labeling,18 and extraction19 have been successfully performed in microchip systems. Because extraction can purify target components from complex matrices, it is an especially attractive technique for the pretreatment of real samples.
Solid phase extraction (SPE) is used heavily in sample purification. The principle of SPE is as follows: the targeted component (or components) is retained on a solid medium to separate it from the matrix, and retained materials can then be eluted for analysis. SPE has been applied successfully in a microfluidic format;20, 21 however, nonspecific interactions like hydrophobic absorption alone do not provide high selectivity. To circumvent this shortcoming, enzymes or antibodies can be immobilized on the solid surface.21, 22 For instance, pisum sativum agglutinin has been immobilized on monolithic substrates to retain glycoproteins, which can be eluted in several fractions based on their affinities.23 A recent review summarizes the application of immunoaffinity capillary electrophoresis (CE) for biomarker, drug and metabolite analysis in biological samples.24 These studies indicate a promising future for immunoaffinity extraction as a pretreatment method for biological specimens in microdevices. We recently demonstrated an integrated microfluidic system that coupled immunoaffinity extraction with rapid microchip CE separation for quantitation of alpha-fetoprotein (AFP) in human blood serum, using either standard addition or a calibration curve for determining concentrations.19 Although this method was effective at quantifying AFP, the affinity column properties were not fully characterized, and only one biomarker was detected.
Here, we demonstrate an integrated microfluidic system that can simultaneously quantify multiple cancer biomarkers in human blood serum. We selected four commercially available biomarkers as test proteins (Table 1).25-28 Antibodies were attached to microchip columns, and the amounts of immobilized antibodies were characterized. We used our integrated microdevices to quantify these four proteins at low ng/mL levels, which are in the range of their action thresholds in human blood serum. These results demonstrate that our platform is generalizable and applicable for the simultaneous quantification of multiple biomarkers in complex samples.
Table 1.
Properties of the cancer biomarkers detected in this study
| Biomarker | Clinical use | Normal level (ng/mL) |
Action threshold (ng/mL) |
|---|---|---|---|
| AFP25 | liver cancer marker | <10 | 20 |
| CEA26 | colorectal cancer marker | <5 | 20 |
| Cytochrome C (CytC)27 |
prognostic marker during cancer therapy |
<0.5 | 25 |
| Heat shock protein 90 (HSP90)28 |
many oncogenic proteins are HSP90 clients |
n/a | Overexpression (no action threshold) |
Results and Discussion
Characterization of affinity columns
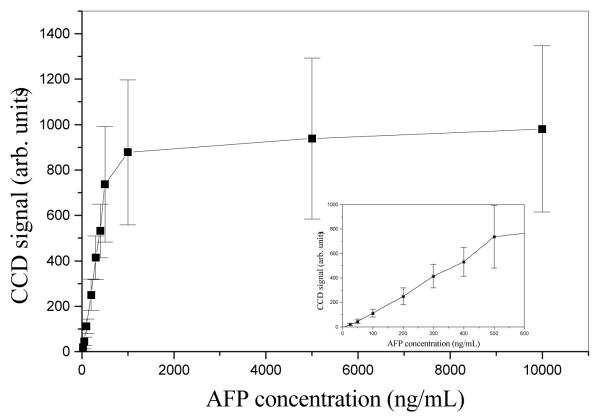
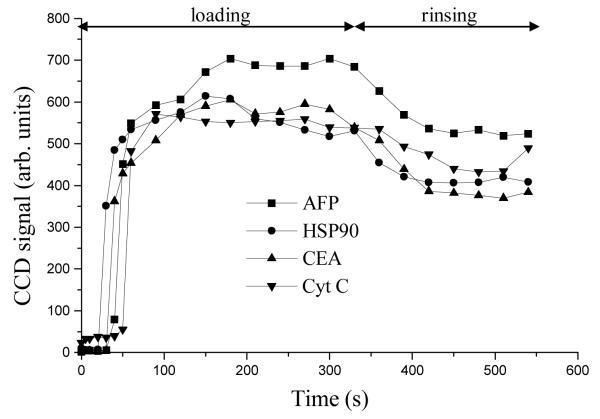
The fluorescence signal on affinity columns in our microdevices (Figure 1), as a function of AFP concentration is shown in Figure 2. The relationship between CCD signal and AFP concentration was linear up to ~500 ng/mL, and the signal approached a plateau at 1 μg/mL. Above ~1 μg/mL AFP, the antibody sites were all occupied with fluorescently labeled AFP (column saturation), such that the fluorescence signal did not change with further AFP concentration increases. Thus, after loading ~1 μg/mL of a target protein on the affinity column and washing off unbound material, the maximum amount of retained antigen can be monitored, as shown in Figure 3. During the rinsing step, the fluorescence signal decreased by ~15% due to the removal of some unbound protein. Importantly, the signal remained stable after this initial decline during rinsing, indicating strong interaction between antigens and antibodies. In addition, the fluorescence signals of all four proteins were in the same range after rinsing, indicating that the derivatization reaction had little bias toward any of the four antibodies we used.
Figure 1.
Layout of an integrated microdevice. (a) Schematic diagram and (b) photograph of a typical microchip with integrated affinity column. See the text for reservoir numbering.
Figure 2.
Background-subtracted fluorescence signal on a typical affinity column after washing, for multiple AFP concentrations. The lower concentration points are expanded in the inset.
Figure 3.
Fluorescence signal from the affinity column during loading and rinsing steps. All points are average values from CCD images, and standard deviations (not shown, ~200 units) were calculated from ~32,000 pixels in the CCD images. The relative standard deviation values reflect some heterogeneity in the density of immobilized antibodies on the column, as well as minor imperfections on the PMMA surfaces from device bonding.
Calibration curves relating fluorescence signal and standard protein concentration were generated in Figure 4 to convert the CCD signal into the effective concentration of fluorescently labeled protein attached to the column at saturation. For all four proteins, the CCD signal had a linear relationship with protein concentration (R2 > 0.95). The difference in mass per volume sensitivity for the various analytes is due to molecular differences in terms of number of available fluorescent labeling sites and molecular weight. Based on the CCD signal during the rinsing step (Figure 3) and the 6-nL column volume, the amounts of retained proteins on the affinity column were determined (Figure 5). The retained protein amounts were all in the range of 2 to 7 pg, and were also consistent from chip to chip, indicating that immunoaffinity extraction is not affected adversely by multiplexing antibodies on the column. There was a 10–30% between-device variability in the amount of retained proteins, due to small differences in column surfaces and operation conditions. Importantly, since samples and standards are both analyzed on the same column, any minor between-device differences have no effect on quantitation. Assuming the antigen-antibody interaction occurs with a 1:1 molar ratio, the average amounts of immobilized anti-AFP, anti-CEA, anti-CytC, and anti-HSP90 were 60, 30, 190, and 45 amol, respectively (~0.1 nmol/m2). Our channel wall coated affinity columns have a lower density of immobilized antibodies than high surface area, porous beads (2–35 nmol/m2).29 Since submicroliter volumes of sample are loaded on our affinity columns, the present binding capacity is not a serious issue for trace (<μg/mL) biomarker analysis. In addition, the density of binding sites in our devices can be easily increased by using a porous material as the solid support. These results demonstrate that affinity columns with four antibodies can be integrated reproducibly in our microdevices with good functionality.
Figure 4.
The relationship between background subtracted CCD signal and the concentration of fluorescently labeled proteins. Error bars indicate standard deviations (n = 3).
Figure 5.

The amounts of retained proteins on the affinity columns in three different microdevices. Standard deviations were calculated from the regression data in Figure 4.
Separation of a model protein mixture
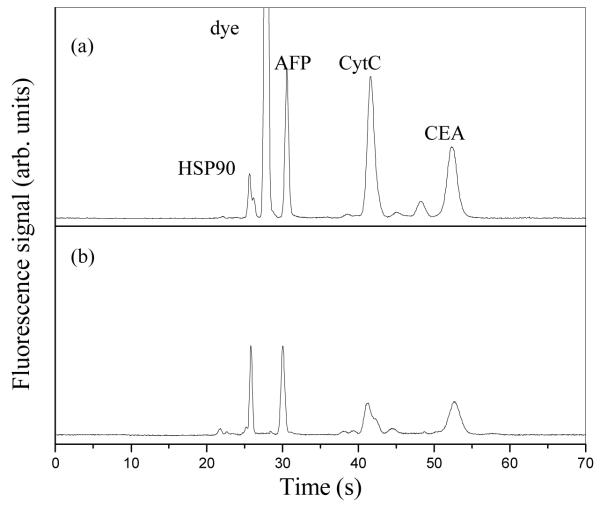
To demonstrate the feasibility of integrated microchip immunoaffinity extraction and CE for multiple biomarker analysis, a mixture of Alexa Fluor 488-labeled AFP, CytC, HSP90 and CEA at 1 μg/mL each in carbonate buffer was analyzed. Five baseline-resolved peaks, including a significant fluorescent dye peak, were observed when this mixture was analyzed by standard microchip CE (without affinity extraction), as shown in Fig. 6a. On the other hand, Figure 6b shows the electropherogram after this mixture was loaded on an affinity column having the requisite antibodies and then separated by microchip CE after rinsing and elution/injection. With on-chip affinity purification, the dye peak was essentially eliminated (over 10,000-fold reduction), while the four biomarker peaks remained. In addition, the HSP peak was sharpened after extraction because of the removal of a co-eluting impurity from the sample. These results indicate that our integrated microdevices can selectively retain and analyze targeted compounds in samples.
Figure 6.
Alexa Flour 488-labeled biomarker mixture (1 μg/mL for each protein), run by microchip electrophoresis (a) before and (b) after integrated affinity column extraction.
Multiplexed biomarker quantitation in human serum
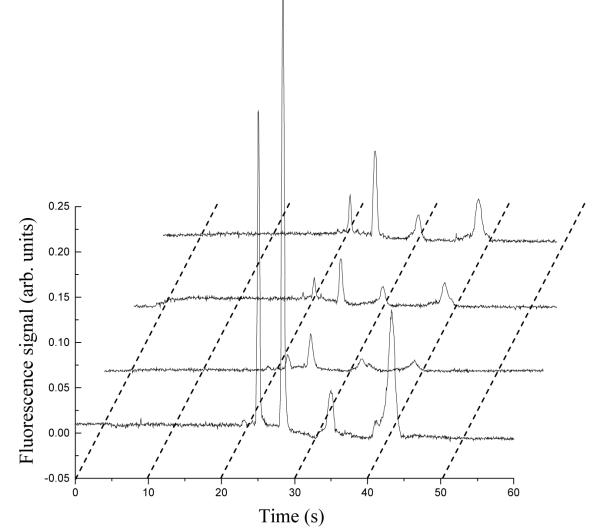
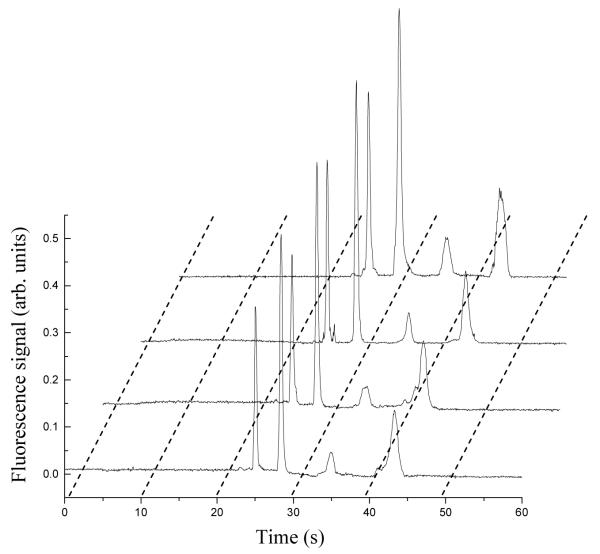
To assess the ability of our approach to quantify biomarkers in real samples, we analyzed a series of human blood serum specimens that had been spiked with four proteins and fluorescently tagged with Alexa Fluor 488 TFP Ester. Spiked biomarker concentrations in human serum were determined in the integrated affinity extraction and microchip CE devices using either a linear calibration curve (Fig. 7) or the standard addition method (Fig. 8). In Figure 7, the peak heights of standards increased proportionally going from 5 ng/mL to 20 ng/mL, and the peak heights increased with spiked protein concentration in Figure 8. In all electropherograms after on-chip affinity purification, only four clean baseline-resolved protein peaks were observed, indicating the efficacy of the multiplexed immunoaffinity extraction column. We tested four spiked human blood serum samples, and the calibration curve and standard addition results overall matched the known spiked concentrations well (Table 2). In general, the standard deviations for the calibration curve were smaller than those for standard addition; quantitation by standard addition involves extrapolation, which may partially explain the higher standard deviations. Because we eliminated the serum matrix in the affinity purification step, the results were similar for the calibration curve compared to standard addition, which is most effective in complex mixtures. The total analysis time for these samples (including labeling) was <60 min; therefore, our integrated devices are well suited for point-of-care (POC) applications. To verify the ability of these microchips to quantify biomarkers at native levels, we also analyzed an unspiked serum sample (Table 3). The biomarker concentrations are all <6 ng/mL, indicating that this system works effectively with naturally produced biomarkers at trace levels. The calibration curve method provided more reliable and precise results than standard addition in these analyses.
Figure 7.
Microchip CE of Alexa Fluor 488-labeled human serum and of standard solutions after affinity column extraction. Curves from bottom to top are: unknown spiked human serum sample, 5 ng/mL standard mixture, 10 ng/mL standard mixture, and 20 ng/mL standard mixture, respectively. Analyte elution order is the same as in Figure 6.
Figure 8.
Microchip electrophoresis of Alexa Fluor 488-labeled human serum after standard addition and affinity column extraction. Curves from bottom to top are: unknown spiked human serum sample, serum sample + 5 ng/mL standard mixture, serum sample + 10 ng/mL standard mixture, and serum sample + 20 ng/mL standard mixture, respectively. Analyte elution order is the same as in Figure 6.
Table 2.
Results from a blinded study with the integrated microfluidic biomarkers assay chip for spiked human serum samples (all concentrations are ng/mL)
| Analyte | Sample number |
Concentration | Standard deviation | |||
|---|---|---|---|---|---|---|
| Spiked | Calibration curve |
Standard addition |
Calibration curve |
Standard addition |
||
| HSP90 | 1 | 110 | 116 | 87 | 7 | 7 |
| 2 | 183 | 200 | 140 | 13 | 94 | |
| 3 | 219 | 206 | 201 | 13 | 31 | |
| 4 | 58 | 73 | 60 | 4 | 12 | |
| AFP | 1 | 116 | 106 | 128 | 7 | 5 |
| 2 | 140 | 136 | 166 | 10 | 35 | |
| 3 | 37 | 27 | 50 | 2 | 13 | |
| 4 | 70 | 63 | 92 | 4 | 12 | |
| CytC | 1 | 200 | 152 | 156 | 25 | 37 |
| 2 | 53 | 38 | 22 | 5 | 3 | |
| 3 | 106 | 104 | 142 | 16 | 42 | |
| 4 | 160 | 118 | 128 | 19 | 27 | |
| CEA | 1 | 27 | 38 | 42 | 2 | 8 |
| 2 | 50 | 60 | 50 | 4 | 7 | |
| 3 | 83 | 95 | 131 | 6 | 21 | |
| 4 | 100 | 118 | 136 | 8 | 8 | |
Table 3.
Results for an unspiked human serum sample with the integrated microfluidic biomarkers assay chip (all concentrations are ng/mL)
| Analyte | Concentration | Standard deviation | ||
|---|---|---|---|---|
| Calibration curve |
Standard addition |
Calibration curve |
Standard addition |
|
| HSP90 | 5.6 | 0.4 | 0.9 | 2.0 |
| AFP | 3.5 | 1.9 | 0.1 | 1.0 |
| CytC | 3.5 | 0.9 | 1.9 | 1.3 |
| CEA | 3.6 | 4.9 | 0.1 | 0.2 |
Our approach could be easily extended up to ~10 biomarker detection by simply immobilizing more antibodies on the affinity column. The surface area of our open channel affinity column (i.e., column saturation) could be an obstacle to scaling to tens of biomarkers, although we note that the column saturation level is a factor of at least 25 above the diagnostic threshold for our markers. Furthermore, the binding capacity could be raised by increasing the surface area of columns (e.g., using a monolith material as the solid support). For more than ~10 components, the peak capacity in our present device design could be an issue, but a longer folded separation channel30 (e.g. 8-cm length) could increase peak capacity to ~30. Peak capacity could also be raised through spectral multiplexing, wherein several distinct fluorescent labels are used on different proteins. Thus, higher-level multiplexing should be able to significantly increase the number of biomarkers that can be quantified.
To make a real POC assay, the laser-induced fluorescence (LIF) system and power supplies would need to be miniaturized. A shoebox-size LIF package has been successfully demonstrated for microchip CE analysis of DNA, indicating strong potential to miniaturize the platform for POC applications.31 In addition, post-column labeling could be used to decrease the labeling time and reduce operator intervention.18 We further note that device throughput could be increased by performing separations in parallel,15 with multiple extraction and separation units on a single chip. Such integrated capillary array devices would enable either replicate sample analysis or higher-level multiplexing.
Experimental
Reagents and materials
CytC (from bovine heart), CEA (from human fluids), monoclonal anti-AFP antibody (produced in mouse), monoclonal anti-CEA antibody (produced in mouse), anti-CytC antibody (produced in sheep), glycidyl methacrylate (GMA, 97%), poly(ethylene glycol) diacrylate (PEGDA, 575 Da average molecular weight), and 2,2-dimethoxy-2-phenylacetophenone (DMPA, 99%) were purchased from Sigma-Aldrich (St. Louis, MO). HSP90 and monoclonal anti-HSP90 antibody (produced in mouse) were obtained from Stressgen (Ann Arbor, MI). AFP was from Lee Biosolutions (St. Louis, MO). Human blood serum from a healthy male (Sigma-Aldrich) was spiked with different concentrations of AFP, CytC, CEA, and HSP90 in the range of 20 to 250 ng/mL (all above normal clinical levels). These unknown serum samples were then labeled with Alexa Fluor 488 TFP Ester (Invitrogen, Eugene, OR) following an Invitrogen protocol (MP 00143). Briefly, 0.1 mg fluorescent dye was dissolved in 10 μL dimethyl sulfoxide (DMSO), and a 2-μL aliquot of DMSO solution was mixed with 98 μL of spiked human serum. The mixture was left to react in the dark at room temperature for 15 min. For protein standards, a 5-μL aliquot of the DMSO solution containing the fluorescent label was mixed with 0.2 mL of 1 mg/mL protein in 10 mM carbonate buffer (pH 9.0). All solutions were prepared with deionized water (18.3 MΩ-cm) purified by a Barnstead EASYpure UV/UF system (Dubuque, IA). Poly(methyl methacrylate) (PMMA, Acrylite FF) was purchased from Cyro Industries (Rockaway, NJ) and was cut into 4.0 × 5.5 cm2 blanks using a CO2 laser cutter (VLS2.30, Universal Laser Systems, Scottsdale, AZ) before device fabrication.
Layout and fabrication of microfluidic devices
The device layout (Figure 1) and fabrication protocol were adapted from our previous work.19, 32 Briefly, the microchips contained a sample reservoir (1), two SPE processing reservoirs (2-3) for wash buffer and elution solution, respectively; three reservoirs (4-6) having different standard concentrations for quantification; a waste reservoir (8) for the immunoaffinity extraction step; a reservoir (7) for basic solution (5 mM NaOH) to neutralize the acidic elution solution; and three reservoirs (9, 10 and 12) for standard microchip CE separation. The additional reservoir 11 was originally designed to facilitate the integration of a semi-permeable membrane near the injection intersection, but this capability was not utilized in the present experiments. The microchip pattern was transferred to silicon template wafers using photolithography and wet etching.32 PMMA substrates (1.5-mm thick) were imprinted by hot embossing against the etched Si templates.21 The patterned PMMA was thermally bonded to an unimprinted PMMA substrate (3.0-mm thick, to provide ~10 μL reservoir volume capacities) with laser-cut holes (2.0-mm diameter). Channel widths were ~50 μm, except the affinity column which was 100-μm wide, and channel depths were ~20 μm.
Since PMMA is inert to many chemical reactions, the microchannel surface was coated to form affinity columns. Briefly, a prepolymer mixture containing GMA (~60%), PEGDA (~40%), and DMPA (0.5%) was sonicated and then purged with nitrogen for 3 min to remove dissolved oxygen. The degassed mixture was introduced into the affinity microchannel region via reservoir 7, and a ~3 μm coating of the prepolymer mixture remained on the channel walls after applying vacuum to reservoir 7 and flowing nitrogen (~50 psi) from reservoir 1. The microchip was covered with an aluminum photomask, placed on a copper plate in an icebath, and exposed to UV light (320–390 nm, 200 mW/cm2) for 5 min. Finally, unpolymerized material was removed via flushing of 2-propanol through the microchip using a syringe pump.
For immobilization on the patterned affinity channel surface, the four antibodies (anti-AFP, anti-CEA, anti-CytC and anti-HSP90) were mixed at 0.5 mg/mL each in 50 mM borate buffer (pH 8.6). The antibody mixture was pipetted into reservoir 8 and the affinity column filled via capillary action. Borate buffer was placed into all other microchip reservoirs to avoid evaporation during reaction. The entire chip was sealed with 3M Scotch tape (St. Paul, MN), and the mixture was left to react at 37 °C for 24 h in the dark.33 After reaction, the device was flushed using 100 mM Tris buffer (pH 8.3) for 0.5 h. This process also blocked any remaining epoxy groups on the column. Finally, the entire chip was rinsed with carbonate buffer (pH 9.1) before use.
LIF detection setup
LIF detection was performed on a Nikon Eclipse TE300 inverted optical microscope equipped with a photomultiplier tube (PMT) detector (Hamamatsu, Bridgewater, NJ) and CCD camera (Coolsnap HQ, Roper Scientific, Sarasota, FL). The LIF detection system and data collection setup have been described previously.21, 32 CCD images were collected at 10 Hz and analyzed using V++ Precision Digital Imaging software (Auckland, New Zealand). The sampling rate for PMT detection was 20 Hz.
Characterization of affinity columns
To estimate the saturation point of affinity columns, different concentrations of fluorescently labeled AFP were loaded for 5 min by applying 400 V at reservoir 8 and 0 V at reservoir 1. Then, unbound AFP was rinsed off the affinity column with PBS buffer for 3 min with 400 V applied to reservoir 8 while grounding reservoir 2. The fluorescence signal on the affinity column was monitored via CCD during the loading and rinsing processes (Fig. 3).
For each analyte, standards of different concentrations were loaded into a microchannel, and fluorescence signal versus protein concentration plots were generated. These calibration curves provided the relationship between CCD signal and the concentration of fluorescently labeled protein in the column in Figure 4. To determine the amount of immobilized antibodies on the affinity column, 1 μg/mL biomarker standards were loaded on the column with 400 V between reservoirs 1 and 8 for 330 s to saturate all active antibody sites, and the column was washed with Tris buffer using 400 V between reservoirs 2 and 8 for 210 s to remove unbound material. CCD images of the affinity column were recorded at 10 s intervals for the first 60 s, and then 30 s intervals for the remaining time during the loading and washing processes. The CCD signal after washing corresponded to column saturation with antigen; this signal was converted into the equivalent antigen concentration in the column based on the obtained calibration curves. From the column volume of 6 nL (length: 3 mm), we determined the mass of each protein bound on-chip at saturation. Then, assuming the antigen-antibody interaction occurred with a 1:1 molar ratio, the quantity of immobilized antibodies on the column was determined.
Immunoaffinity extraction and electrophoretic separation
The operation of our integrated microchips and the data analysis were adapted from previous work.19 To demonstrate proof-of-principle of multiplexed operation, a mixture of fluorescently labeled AFP, CytC, CEA and HSP90 in buffer was compared before and after microchip immunoaffinity extraction. A double-T microchip layout32 was used to directly separate the mixture (without immunoaffinity extraction). The mixture was then pipetted onto an integrated microdevice, loaded for 5 min on the affinity column, rinsed for 5 min, eluted through the injection intersection for 45 s, and then separated by microchip CE.
For calibration curve quantitation, each standard solution containing all four proteins was loaded on the affinity column (5 min), rinsed with PBS buffer (5 min), eluted through the injection intersection for 1 min with phosphate buffer (pH 2.1), and separated by microchip CE, by applying a sequence of potentials to the various reservoirs for all steps.19 The sample was analyzed by loading it on the affinity column, rinsing, eluting/injecting and separating the same as for the standards. The peak heights from each standard electropherogram were plotted against the series of known protein concentrations, and linear regression was used to fit a line to the data. The concentration of each component in the sample was calculated from its peak height in the electropherogram and the linear fit equation.
For standard addition quantification, sample was first analyzed the same way as for the calibration curve. Next, sample was loaded on the affinity column for 5 min, followed by loading of the first standard mixture for 5 min; the rinsing, elution/injection and microchip CE separation steps were then carried out as before. This same set of processes was repeated to spike the other two standards into the sample and analyze them.19 A linear fit was generated from the peak heights in the electropherograms of the unknown sample and of the sample spiked with standards, plotted against the standard concentrations spiked into the sample. The concentration of each protein was calculated from the intercept and slope of this line.
Conclusion
Sample pretreatment, cleanup, and quantitation are essential in biomarker analysis in complex media. In this study, affinity purification columns with four different antibodies were prepared in polymer microfluidic devices. The amounts of antibodies immobilized on our columns were consistent from chip to chip, and comparable, low femtomole amounts of each of the four antibodies were attached to the columns. Analysis of four proteins in buffer solution demonstrated that multiplexed immunoaffinity columns could selectively extract the desired species for subsequent CE analysis. With spiked human blood serum samples, four proteins in the ng/mL range were simultaneously quantified using both calibration curves and standard addition. In general, the calibration curve and standard addition results were close to the known spiked concentrations. These microdevices provide an excellent platform for fast, integrated and automated biomarker quantitation. Furthermore, our system could be expanded to ~30 biomarker quantitation by immobilizing additional different antibodies on the affinity column, in conjunction with using porous materials for the solid support to improve binding capacity, and longer separation channels as well as spectral multiplexing to raise peak capacity. Importantly, with improvements in engineering and miniaturization, a straightforward POC instrument for multiple biomarker quantitation could result.
Acknowledgements
This work was supported by a Presidential Early Career Award for Scientists and Engineers through the National Institutes of Health (R01 EB006124).
References
- 1.American Cancer Society [Access date: 3/23/2010];Cancer Facts and Figures. 2009 http://www.cancer.org/downloads/STT/500809web.pdf.
- 2.Nass SJ, Moses HL, editors. Cancer biomarkers: the promises and challenges of improving detection and treatment. National Academies Press; Washington, D.C.: 2007. [Google Scholar]
- 3.Kiviat NB, Critchlow CW. Dis. Markers. 2002;18:73–81. doi: 10.1155/2002/589075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma M, Seminara D, Arena FJ, John C, Iwamoto K, Hartmuller V. Mol. Diagn. Ther. 2006;10:1–15. doi: 10.1007/BF03256438. [DOI] [PubMed] [Google Scholar]
- 5.Makarov DV, Loeb S, Getzenberg RH, Partin AW. Annu. Rev. Med. 2009;60:139–151. doi: 10.1146/annurev.med.60.042307.110714. [DOI] [PubMed] [Google Scholar]
- 6.Sinise GA, editor. Tumor markers research perspectives. Nova Science Publishers; New York: 2007. [Google Scholar]
- 7.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford ED, Crowley JJ, Coltman CA., Jr. N. Eng. J. Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 8.Sunami E, Shinozaki M, Higano CS, Wollman R, Dorff TB, Tucker SJ, Martinez SR, Singer FR, Hoon DSB. Clin. Chem. 2009;55:559–567. doi: 10.1373/clinchem.2008.108498. [DOI] [PubMed] [Google Scholar]
- 9.Yang X-Q, Yan L, Chen C, Hou J-X, Li Y. Hepatogastroenterology. 2009;56:1388–1394. [Google Scholar]
- 10.Bronchud MH, Foote M, Giaccone G, Olopade O, Workman P, editors. Principles of molecular oncology. 2nd Ed. Humana Press; Totowa, N.J.: 2004. [Google Scholar]
- 11.Ladd J, Taylor AD, Piliarik M, Homola J, Jiang S. Anal. Bioanal. Chem. 2009;393:1157–1163. doi: 10.1007/s00216-008-2448-3. [DOI] [PubMed] [Google Scholar]
- 12.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Clin. Chem. 2010;56:186–193. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittrich PS, Tachikawa K, Manz A. Anal. Chem. 2006;78:3887–3907. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 14.Bharadwaj R, Santiago JG, Mohammadi B. Electrophoresis. 2002;23:2729–2744. doi: 10.1002/1522-2683(200208)23:16<2729::AID-ELPS2729>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 15.Yang W, Woolley AT. J. Assoc. Lab. Autom. 2010;15:198–209. doi: 10.1016/j.jala.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagally ET, Emrich CA, Mathies RA. Lab Chip. 2001;1:102–107. doi: 10.1039/b109031n. [DOI] [PubMed] [Google Scholar]
- 17.Lion N, Gobry V, Jensen H, Rossier JS, Girault H. Electrophoresis. 2002;23:3583–3588. doi: 10.1002/1522-2683(200210)23:20<3583::AID-ELPS3583>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Yu M, Wang H-Y, Woolley AT. Electrophoresis. 2009;30:4230–4236. doi: 10.1002/elps.200900349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, Sun X, Wang H-Y, Woolley AT. Anal. Chem. 2009;81:8230–8235. doi: 10.1021/ac901566s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen J, Guillo C, Ferrance JP, Landers JP. Anal. Chem. 2007;79:6135–6142. doi: 10.1021/ac0703698. [DOI] [PubMed] [Google Scholar]
- 21.Yang W, Sun X, Pan T, Woolley AT. Electrophoresis. 2008;29:3429–3435. doi: 10.1002/elps.200700704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips TM, Wellner EF. Electrophoresis. 2009;30:2307–2312. doi: 10.1002/elps.200900095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao X, Luo Y, Dai Z, Wang K, Du Y, Lin B. Anal. Chem. 2004;76:6941–6947. doi: 10.1021/ac049270g. [DOI] [PubMed] [Google Scholar]
- 24.Guzman NA, Blanc T, Phillips TM. Electrophoresis. 2008;29:3259–3278. doi: 10.1002/elps.200800058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright LM, Kreikemeier JT, Fimmel CJ. Cancer Detect. Prev. 2007;31:35–44. doi: 10.1016/j.cdp.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Park Y-A, Lee KY, Kim NK, Baik SH, Sohn SK, Cho CW. Ann. Surg. Oncol. 2006;13:645–650. doi: 10.1245/ASO.2006.03.090. [DOI] [PubMed] [Google Scholar]
- 27.Barczyk K, Kreuter M, Pryjma J, Booy EP, Maddika S, Ghavami S, Berdel WE, Roth J, Los M. Int. J. Cancer. 2005;116:167–173. doi: 10.1002/ijc.21037. [DOI] [PubMed] [Google Scholar]
- 28.Mahalingam D, Swords R, Carew JS, Nawrocki ST, Bhalla K, Giles FJ. Br. J. Cancer. 2009;100:1523–1529. doi: 10.1038/sj.bjc.6605066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke W, Beckwith JD, Jackson A, Reynolds B, Karle EM, Hage DS. J. Chromatogr. A. 2000;888:13–22. doi: 10.1016/s0021-9673(00)00548-3. [DOI] [PubMed] [Google Scholar]
- 30.Paegel BM, Hutt LD, Simpson PC, Mathies RA. Anal. Chem. 2000;72:3030–3037. doi: 10.1021/ac000054r. [DOI] [PubMed] [Google Scholar]
- 31.Lagally ET, Scherer JR, Blazej RG, Toriello NM, Diep BA, Ramchandani M, Sensabaugh GF, Riley LW, Mathies RA. Anal. Chem. 2004;76:3162–3170. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- 32.Kelly RT, Woolley AT. Anal. Chem. 2003;75:1941–1945. doi: 10.1021/ac0262964. [DOI] [PubMed] [Google Scholar]
- 33.Sun X, Yang W, Pan T, Woolley AT. Anal. Chem. 2008;80:5126–5130. doi: 10.1021/ac800322f. [DOI] [PMC free article] [PubMed] [Google Scholar]