Abstract
Here we report the demonstration of an integrated microfluidic chip that performs helicase dependent amplification (HDA) on samples containing live bacteria. Combined chip-based sample preparation and isothermal amplification are attractive for world health applications, since the need for instrumentation to control flow rate and temperature changes are reduced or eliminated. Bacteria lysis, nucleic acid extraction, and DNA amplification with a fluorescent reporter are incorporated into a disposable polymer cartridge format. Smart passive fluidic control using a flap valve and a hydrophobic vent (with a nanoporous PTFE membrane) with a simple on-chip mixer eliminates multiple user operations. The device is able to detect as few as ten colony forming units (CFU) of E. coli in growth medium.
Keywords: DNA, Bacteria, Helicase dependent amplification, Isothermal amplification, Microfluidic chip, Plastic, Solid phase extraction
1 Introduction
Although the polymerase chain reaction (PCR) is the predominant technique of nucleic acid amplification for DNA (or RNA) diagnostics, it has proven difficult to implement in devices meant for use in global health environments. The possiblility of reducing or eliminating the requirement for accurate temperature control and cycling makes amplification methods that do not require thermo-cycling attractive alternatives for these applications.
Several isothermal amplification methods have been developed as alternatives to PCR that reach sensitivities similar or equivalent to PCR. The most common isothermal amplification methods are reviewed (Gill and Ghaemi 2008), including loop-mediated amplification (LAMP), transcription-mediated amplification (TMA), rolling-circle amplification (RCA), strand displacement amplification (SDA), and helicase-dependent amplification (HDA). In this study, we focus on HDA in a disposable microfluidic chip. Similar to PCR, the steps in HDA comprise numerous cycles of DNA denaturation, primer annealing, and primer extension. However, instead of heat denaturation as in PCR, the separation of the two complementary DNA strands is achieved enzymatically by employing a heat-stable DNA helicase (Vincent et al. 2004). The main advantage of HDA over other isothermal methods is that the reaction can be directly performed on extracted DNA (or RNA) without intermediate steps, and requires only one primer set (Gill and Ghaemi 2008). The HDA method has a simple reaction scheme with a pair of primers, reaction buffer, and enzyme mix similar to PCR. The reaction is unlike other isothermal methods that have more complicated priming and enzymatic requirements that actually require two temperatures (Gill and Ghaemi 2008; Vincent et al. 2004). The method is well developed. Standardized reagents are available commercially and there have been multiple reports of the use of HDA for detection of bacterial and viral target DNA and RNA, using both SYBR green and TaqMan chemistries in standard microtube protocols (Chow et al. 2008; Goldmeyer et al. 2007, 2008; Tong et al. 2008; Vincent et al. 2004).
Several integrated devices have been presented that start with whole cells and accomplish both nucleic acid extraction and amplification on-chip (Chen et al. 2007; Dimov et al. 2008; Easley et al. 2006; Lien et al. 2009; Liu et al. 2009; Sauer-Budge et al. 2009; Yobas et al. 2009). There are many fewer reports of HDA reactions on-chip (Andresen et al. 2009; Ramalingam et al. 2009), and none of these include integration with on-chip sample preparation. HDA was demonstrated on a glass-PDMS hybrid chip with previously purified high copy number plasmid DNA (Ramalingam et al. 2009), and HDA was performed on glass slides with separately purified bacterial genomic DNA (Andresen et al. 2009). Our approach combines a micro solid phase extraction (μSPE) column for DNA isolation from whole bacteria and isothermal HDA in a disposable plastic cartridge. We believe that this is the first report of a disposable microfluidic chip that performs both sample preparation HDA starting from whole cells.
2 Materials and methods
2.1 Chip design
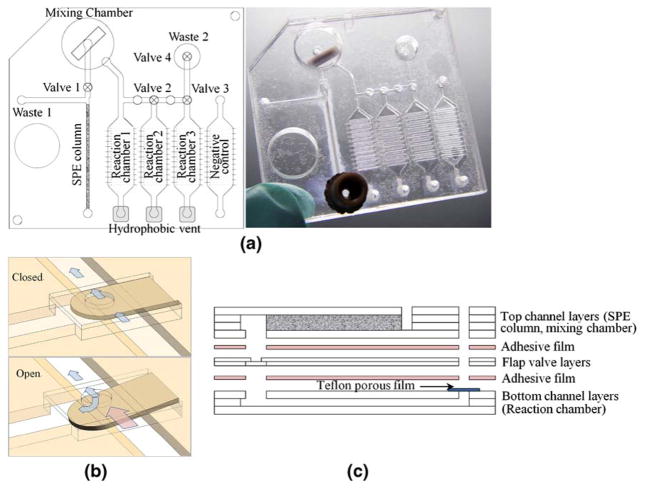
For the initial HDA experiments, we used DNA extracted from E. coli bacteria using μSPE columns formed in cyclic olefin polymer (COP) microchannels. Detailed methods on the manufacture of these channels and the μSPE columns are found in our previous reports of DNA extraction from bacteria in urine, blood and stool (Gillers et al. 2009; Kulinski et al. 2009; Mahalanabis et al. 2009). The HDA was first performed in microtubes to establish that μSPE-extracted DNA was suitable for HDA and to determine the optimal concentrations and conditions for the reactions. The integrated chip is built up from layers of COP and porous Teflon films. A schematic layout and a picture (Fig. 1) of the integrated μSPE-HDA chip are shown. The chip includes a μSPE column, flap valves (Fig. 1(b)), hydrophobic vents, micro channels, triplicate HDA reaction chambers for experimental replicates, and a fourth HDA chamber to contain the negative control reaction. The μSPE column (0.55×0.55×20 mm) is used to lyse and extract DNA from whole bacteria in a liquid sample. During the sample preparation step, flap valve one in front of the μSPE column keeps the sample preparation solutions from being injected into the mixing chamber. First, the sample is mixed with lysis buffer and pushed over the μSPE. The DNA in the sample will bind to the μSPE during this step. Waste buffer from this step and the subsequent wash steps flow through the μSPE and into the waste resevoir. After the last wash step, the waste chamber is filled and the flap valve opens, directing the elution buffer (water) into the mixing chamber, which contains the HDA reagents. Mixing of the eluted DNA and HDA reagents occurs by a miniature magnetic stir bar in the chamber (ø10×2.5 mm). After mixing is complete, the solution is pushed into the three reaction chambers, which fill sequentially.
Fig. 1.
(a) Schematic layout of the integrated disposable microfluidic device; a μSPE (solid-phase extraction) column, a flap valve, a hydrophobic vent, and microchannels are integrated on a disposable cartridge format, and photograph of the fabricated device. (b) Illustration showing operation of a flap valve; a flap valve opens with the pressure over 3.5 psi inside. (c) Fabrication process of the device; a top channel layer, a flap valve layer and a bottom reactor layer were cut and thermally bonded each other and then assembled using double sided adhesive film. The μSPE column was made in the final step
The three 25 μL reaction volumes allow for replicate experiments on the same loci. The bottoms of the reaction chambers have a shallow scratched pattern to prevent bubble trapping, which allows for uniform filling of the low aspect ratio chamber by providing even surface tension. A flap valve (valves 2–4) and a hydrophobic vent are located at each end of each reaction chamber. Once the previous chamber is completely filled, the micro valve opens and the HDA reaction mixture fills the next reactor in series. Any trapped air can exit via the hydrophobic vent, but the liquid remains trapped in the reaction chamber. A waste chamber is located after the last reaction chamber to collect any excess HDA reaction mix and to prevent cross-contamination. After the waste chamber fills, air can be pushed into the cross channel to isolate the individual reaction chambers. The vents and valves are sealed with PCR tape to prevent evaporation and the integrated device is heated to 65°C on a dry heat block for up to 30 min for the HDA step.
2.2 Chip fabrication
Figure 1(c) schematically illustrates the fabrication process of the integrated disposable microfluidic device. The top microfluidic channels were prepared by cutting 188 μm thick COP (Zeon Chemicals, Louisville, KY) films using a cutter plotter (Bartholomeusz et al. 2005). Three sheets of cut film formed the μSPE channel, and 12 sheets formed the mixing chamber. These were stacked and thermally bonded with the magnetic stir bar inside the chamber. The flap valve layer in the middle was prepared by cutting a 100 μm thick COP film and thermally bonding it to the top channels. For the bottom reaction chamber layer, the scratch pattern was made on a 1 mm thick COP plaque using a cutter plotter to enable uniform filling (Do et al. 2009). Two cut films (188 μm thick) patterned with the reaction chamber design were also prepared. A hydrophobic porous film (PTFE unlaminated membrane filter, Sterlitech, Kent, WA) was inserted between the 1 mm plaque and the last cut sheet of 188 μm film and then thermally bonded. All thermally bonded layers (the top channel layer, the flap valve layer, and the bottom reaction chamber layer) were assembled using double sided adhesive film (GRAFIX, Cleveland, OH). All fabrication steps were completed after fabrication of the μSPE column inside a channel in the top layer. All thermal bonding described above was done at 125°C for 5 min at 0.5 tons followed by 5 min of annealing at 137°C.
2.3 Bacterial culture and DNA extraction for off-chip HDA assay development
The protocol for the E. coli cultures and genomic DNA extraction on-chip has been previously described in detail (Kulinski et al. 2009; Mahalanabis et al. 2009). Briefly, E. coli DH5α were grown in shaking LB liquid culture overnight (16 h) at 37°C. Dilutions were prepared in sterile LB broth from the overnight culture and the cells were pelleted at 8,000 RPM for 8 min. Positive control DNA extractions were performed using a commercial Qiagen DNEasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. To create the serial dilutoins of bacteria in LB broth, samples were prepared by removing the supernatant and resuspending the cell pellet in the appropriate amount of broth. Then 50 μL of the LB broth was mixed 1:1 with 50 μL 3 M GuSCN buffer containing 0.01% Triton X-100 and 0.8 mg/mL proteinase K and was immediately loaded onto the μSPE column. After the sample was loaded, the bound sample was washed sequentially with aliquots of 70% and 100% ethanol, and eluted into two sequential fractions of 50 μL of PCR grade water. All steps were performed with a KDS100 syringe pump (KD Scientific Inc., Holliston, MA) at a flow rate of 450 μL/h. The second elution was used for all off-chip HDA reactions.
2.4 PCR and HDA amplification and detection
In order to compare the performance of the HDA reaction to PCR using chip-extracted DNA, an assay for the single copy dxs gene of E. coli present in genomic DNA was used as the target sequence for both amplification schemes. PCR of the dxs gene was performed as previously described (Mahalanabis et al. 2009). For isothermal HDA amplification, we used the IsoAmp II Universal tHDA kit from Biohelix (Beverly, MA, USA). The HDA primers used to amplify a 70 bp product were forward primer 5′-AGAAAAAGACCCGATCACTTTCCACG-3′ and reverse primer 5′-AGATTTTTGAATAGCTCGGCAAACCG-3′. The HDA reaction was prepared as per the manufacturer’s directions except the reagent volumes were halved since total reaction volumes were reduced to 25 μL from 50 μL. For our target sequence-primer choices, 50 or 100 ng DNA amplified well; therefore we used 50 ng of DNA per reaction after quantification by a Nanodrop 1000 (Thermo Fisher Scientific, Wilmington, DE). The reaction was run either in a thermocycler (AB 7300 Real-time (RT) system, Applied Biosystems, Foster City, CA) or on-chip using a dry heat block as a heat source at 65°C for 1 h. To run HDA reactions in the RT machine (which is not generally set up to run isothermal reactions), cycles were entered as 66°C for 5 s and 65°C for 1 min 55 s. Thus one cycle represented 2 min. EvaGreen and ROX dyes (Biotium Inc., Hayward, CA, USA) were used for detection in the thermocycler and to visualize amplified products on-chip by UV illumination in a Versadoc Gel Imaging System (Bio-Rad, Hercules, CA). The presence of amplicons and the yield (ng/μL) was determined by gel capillary electrophoresis in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
3 Results
3.1 Off-chip amplification from on-chip bacterial DNA extraction
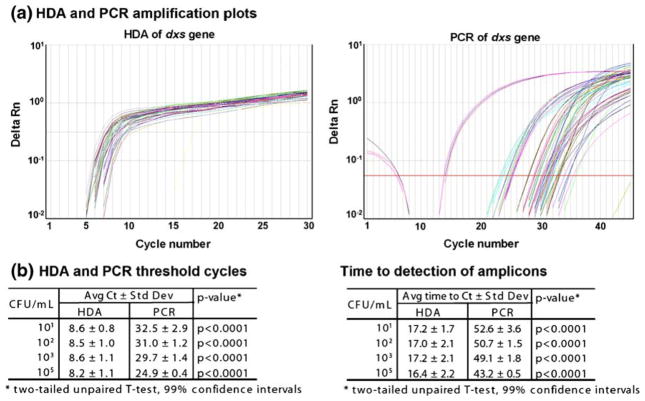
E. coli cells ranging from 105 to 101 colony forming units (CFU, or number of viable cells) total (resuspended and diluted in LB broth as described above; a CFU represents one living organism) were extracted using μSPE-only chips (chips containing only μSPE channels and not the mixing or reaction chambers). The eluted genomic DNA was amplified in a real-time (RT) PCR machine using both HDA and standard RT-PCR protocols for comparison. Each dilution was extracted in triplicate. We have previously reported successful RT-PCR from μSPE column extracted E. coli DNA from as little as 10 CFU in human urine and blood (Kulinski et al. 2009; Mahalanabis et al. 2009). Using the same μSPE-only chips, we replicated those results here for E. coli in LB broth as expected (Fig. 2(b)). In addition, the data indicate that the μSPE extraction on-chip yielded DNA of sufficient quantity and quality for amplification by HDA from each of the 105–101CFU dilutions (Fig. 2). The delta Rn over time is shown in Fig. 2(a) and the average Ct and time to detection of the amplicons is plotted in Fig. 2(b) for all E. coli dilutions extracted. The HDA reactions reached the maximum amplicon yield (Rn) faster, as measured by fluorescence in the RT thermocycler. The HDA consistently detected the E. coli DNA at significantly lower threshold cycles (Ct) than the PCR (p<0.0001, two-tailed unpaired t-test, 99% CI) with an average Ct around 8.5 as compared to average Ct values of 25–32 by PCR (Fig. 2(b)). Recall, however that the “cycles” for the HDA reaction are only measures of time, since the reaction is kept at the same temperature throughout. Therefore the average time to detection of the amplicon was significantly faster by HDA (p<0.0001, two-tailed unpaired t-test, 99% CI) at 17 min by HDA as compared to 43–53 min by PCR (Fig. 2(b)). As expected, the Ct values increased with fewer E. coli CFU in the quantitative PCR reactions. However, for the HDA reactions we did not observe this dilution effect, and the Ct value remained constant. As shown in other publications showing the kinetics of HDA, this technique reaches the maximum Rn faster than PCR (steeper slope) and does not distinguish well between amount of input DNA unless the input DNA concentrations vary by orders of magnitude (Goldmeyer et al. 2007; Tong et al. 2008). The literature shows that microtube HDA assays have Rn curves that cluster together, shifting only two cycles with 1–3 log differences in bacterial CFU or DNA and RNA input concentrations from nucleic acid isolated with commercial kits (Chow et al. 2008; Goldmeyer et al. 2007, 2008; Tong et al. 2008). We speculate that our channels all retrieve similar concentrations of DNA at these lower bacterial CFU (as compared to the undiluted overnight culture containing 109CFU/ml); thereby further reducing the bacterial dilution effect. It is likely that the amount of μSPE channel extracted DNA does not differ by orders of magnitude within this range of input CFU and may be in the picogram range for all of the input dilutions of E. coli extracted (data not shown). This is supported by the RT-PCR data in this report and in our previous work (Mahalanabis et al. 2009), as the Ct values converge around 30 cycles for the lower bacterial CFU dilutions. Once the μSPE extracted DNA was determined to be suitable for HDA, we investigated amplification by HDA in the integrated thermoplastic microfluidic chip.
Fig. 2.
Comparison of quantitative HDA and PCR methods from μSPE channel extracted DNA. E. coli DNA from 105 to 101CFU total was extracted by μSPE channels in triplicate. (a) Real time quantitative HDA and PCR results are shown for amplification of the dxs gene. Reactions were performed in an Applied Biosystems 7300 PCR thermocycler. PCR cycling conditions: 50°C, 2 min; 95°C, 10 min; 40 cycles of 95°C, 15 s; 55°C, 1 min. HDA cycling conditions: 40 cycles of 66°C, 5 s; 65°C 1 min 55 s. (b) The average Ct and time to detection of the HDA versus PCR products was determined
3.2 Detection of bacteria on the integrated μSPE-HDA chip
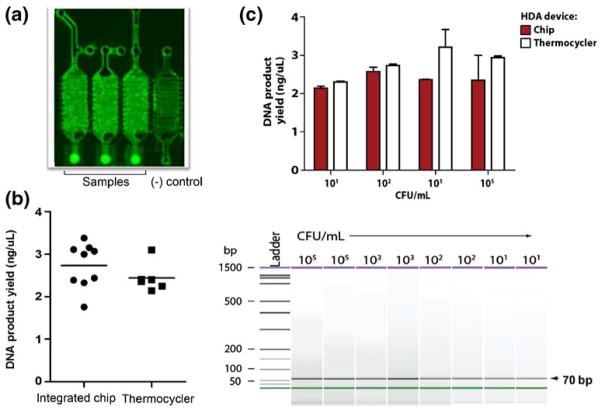
The integrated chip (Fig. 1) was initially tested using a high concentration of E. coli at 109CFU/ml. Three combined μSPE-HDA chips with three reaction wells each and a negative control well were run. E. coli cells were mixed with lysis buffer as they were introduced into the chip (as described above) and the DNA was extracted, washed and eluted from the μSPE channel on the chips. The DNA was then directly eluted into the HDA mixing chamber with HDA reaction mix. A small aliquot was also removed from each chip (using a pipettor and an extra access port) after mixing with the HDA reagents for use in HDA reactions off the chip in the RT thermocycler run in parallel. These reactions were positive controls. Qiagen kit extracted E. coli DNA was used as a positive control for the HDA reaction in the RT thermocycler. The HDA reactions in all three chips were positive for the HDA product as visualized by fluorescence (Fig. 3(a)). The negative control well, containing water instead of eluted DNA, was negative for any amplified product by both fluorescence visualization (Fig. 3(a)) and by slab gel electrophoresis (not shown). The resulting on-chip and off-chip HDA product yields from the μSPE channel extracted DNA were compared (Fig. 3(b)). Amplification on-chip worked as well as in the RT thermocycler on average with yields around 2.5 ng/μl (p= 0.252, two tailed t-test, 95%CI) as compared to the positive control DNA, which had a slightly greater average yield of 3.4 ng/μL±0.4 ng/μL (n=6).
Fig. 3.
On-chip detection of whole bacterial cells. (a) Fluorescence imaging of a representative integrated μSPE-HDA chip showing positive HDA reactions done in triplicate and a negative control water reaction. Reactions were positive for HDA of E. coli dxs from broth culture containing 109CFU loaded into the integrated chip. DNA was extracted by the μSPE column, mixed with HDA reaction mix in the HDA chamber, and flowed into the separate reaction wells. (b) The HDA products from three integrated chips (imaged in a) were analyzed for yield concentrations and compared to products from reactions done in an RT thermocycler from integrated chip μSPE DNA extractions. (c) Dilutions of E. coli were prepared and 105–101 CFU were run through the μSPE channels and then HDA was performed on-chip and in an RT thermocycler. The product yields were compared using an Agilent 2100 Bioanalyzer and were shown to be positive for the correct size 70 bp product
Finally, we determined the sensitivity of the on-chip assay by testing lower concentrations of E. coli from 105 to 101 total CFU. These experiments were again performed in the same manner described above. The DNA extracted from each dilution of E. coli after the μSPE step was positive for the amplified product both on-chip and in the RT thermocycler with comparable product yields ranging from 2 to 3 ng/μl (Fig. 3(c)). Thus, we are able to detect as little as 10 CFU of E. coli by HDA from DNA extraction and amplification on-chip.
4 Discussion
The data presented in this proof-of-concept study highlights the novel use of a single disposable thermoplastic device that combines isothermal amplification by HDA and μSPE DNA extraction to detect the presence of whole bacterial cells in a liquid sample. The low cost of materials, isothermal amplification and integrated nature of this device make it a good candidate for further development for point-of-care testing; it is our goal to simplify chip design and fabrication to allow for high-throughput manufacturing and eventual use in resource-limited settings. The existing chip design allows for multiplexing via parallel reactions. Chips with more reaction chambers can also be envisioned.
The molecular assay can also be further optimized to potentially provide lower limits of detection and greater amplicon yields. Of course, the primer design is also highly flexible and can be aimed at any genomic target. The amplicon yield obtained on-chip and off-chip may be improved in the future by altering the primer set, using template targets that are present in high numbers in bacteria such as 16S rDNA or multi-copy internal transcribed sequences (ITS), optimizing the HDA reaction concentrations of primer and MgSO4, and adding blocking agents to on-chip reactions to prevent potential loss of enzyme to the COP surface. Other improvements to consider will include on-board storage of primers and HDA buffer components, multiplexing to detect multiple target pathogens or strains and decreasing HDA reaction volumes on-chip. Although we used E. coli in broth medium for our proof-of-concept study, the same method of extraction with our μSPE chips has been shown previously to work in the background of complex biological samples of blood, urine, and stool for gram-negative bacteria such as E. coli and gram-positive bacteria such as Enterococcus faecalis, Bacillus subtilis and Clostridium difficile, and for RNA isolation from viruses and mammalian cells (Bhattacharyya and Klapperich 2006, 2008; Gillers et al. 2009; Kulinski et al. 2009; Mahalanabis et al. 2009). Thus we anticipate that this integrated device may be applied for a wide variety of point-of-care diagnostic applications.
5 Conclusion
In this study, we deomstrate detection of as little as 10 CFU of E. coli in a broth medium. We were able to detect bacterial DNA by extraction using μSPE channels coupled with isothermal amplification by HDA on an integrated thermoplastic chip. We chose HDA as our amplification method, since it is an isothermal method for which the processes of strand denaturation, primer annealing, and extension occur at a single temperature. This is highly amenable for use in point-of-care microfluidic devices meant to be deployed in low resource settings. When used as an endpoint assay, it can also allow for faster target detection (Vincent et al. 2004). By following the kinetics of the HDA reaction with our target-primer set in the real-time machine, we show that the product may be detected in as little as 50 min including all sample preparation steps.
Contributor Information
Madhumita Mahalanabis, Department of Biomedical Engineering, Boston University, Boston, MA 02215, USA.
Jaephil Do, Department of Biomedical Engineering, Boston University, Boston, MA 02215, USA.
Hussam ALMuayad, Department of Biomedical Engineering, Boston University, Boston, MA 02215, USA.
Jane Y. Zhang, Department of Biomedical Engineering, Boston University, Boston, MA 02215, USA
Catherine M. Klapperich, Email: catherin@bu.edu, Department of Biomedical Engineering, Boston University, Boston, MA 02215, USA, Department of Mechanical Engineering, Boston University, Boston, MA 02215, USA
References
- Andresen D, von Nickisch-Rosenegk M, Bier FF. Helicase dependent onchip-amplification and its use in multiplex pathogen detection. Clin Chim Acta. 2009;403(1–2):244–248. doi: 10.1016/j.cca.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Bartholomeusz DA, Boutte RW, Andrade JD. Xurography: rapid prototyping of microstructures using a cutting plotter. J Micro-electromech S. 2005;14(6):1364–1374. [Google Scholar]
- Bhattacharyya A, Klapperich CM. Thermoplastic microfluidic device for on-chip purification of nucleic acids for disposable diagnostics. Anal Chem. 2006;78(3):788–792. doi: 10.1021/ac051449j. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Klapperich CM. Microfluidics-based extraction of viral rna from infected mammalian cells for disposable molecular diagnostics. Sens Actuators B Chem. 2008;129(2):693–698. [Google Scholar]
- Chen L, Manz A, Day PJR. Total nucleic acid analysis integrated on microfluidic devices. Lab Chip. 2007;7(11):1413–1423. doi: 10.1039/b708362a. [DOI] [PubMed] [Google Scholar]
- Chow WH, McCloskey C, Tong Y, Hu L, You Q, Kelly CP, Kong H, Tang YW, Tang W. Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic clostridium difficile. J Mol Diagnostics. 2008;10(5):452–458. doi: 10.2353/jmoldx.2008.080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimov IK, Garcia-Cordero JL, O’Grady J, Poulsen CR, Viguier C, Kent L, Daly P, Lincoln B, Maher M, O’Kennedy R, Smith TJ, Ricco AJ, Lee LP. Integrated microfluidic tmrna purification and real-time nasba device for molecular diagnostics. Lab Chip. 2008;8(12):2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- Do J, Zhang JY, Klapperich CM. The 13th International Conference on Micro Total Analysis Systems; Jeju, Korea. 2009. [Google Scholar]
- Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci U S A. 2006;103(51):19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleosides Nucleotides Nucleic Acids. 2008;27(3):224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- Gillers S, Atkinson CD, Bartoo AC, Mahalanabis M, Boylan MO, Schwartz JH, Klapperich C, Singh SK. Microscale sample preparation for pcr of c. Difficile infected stool. J Microbiol Methods. 2009;78(2):203–207. doi: 10.1016/j.mimet.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmeyer J, Kong H, Tang W. Development of a novel one-tube isothermal reverse transcription thermophilic helicase-dependent amplification platform for rapid rna detection. J Mol Diagnostics. 2007;9(5):639–644. doi: 10.2353/jmoldx.2007.070012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmeyer J, Li H, McCormac M, Cook S, Stratton C, Lemieux B, Kong H, Tang W, Tang YW. Identification of staphylococcus aureus and determination of methicillin resistance directly from positive blood cultures by isothermal amplification and a disposable detection device. J Clin Microbiol. 2008;46(4):1534–1536. doi: 10.1128/JCM.02234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulinski MD, Mahalanabis M, Gillers S, Zhang JY, Singh S, Klapperich CM. Sample preparation module for bacterial lysis and isolation of DNA from human urine. Biomed Microdevices. 2009;11(3):671–678. doi: 10.1007/s10544-008-9277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien KY, Liu CJ, Kuo PL, Lee GB. Microfluidic system for detection of α-thalassemia-1 deletion using saliva samples. Anal Chem. 2009;81(11):4502–4509. doi: 10.1021/ac900453d. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Lien KY, Weng CY, Shin JW, Chang TY, Lee GB. Magnetic-bead-based microfluidic system for ribonucleic acid extraction and reverse transcription processes. Biomed Micro-devices. 2009;11(2):339–350. doi: 10.1007/s10544-008-9240-1. [DOI] [PubMed] [Google Scholar]
- Mahalanabis M, ALMuayad H, Kulinski MD, Altman D, Klapperich CM. Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip. 2009;9:2811–2817. doi: 10.1039/b905065p. [DOI] [PubMed] [Google Scholar]
- Ramalingam N, San TC, Kai TJ, Mak MYM, Gong H-Q. Micro-fluidic devices harboring unsealed reactors for rel-time isothermal helicase-dependent amplification. Microfluid Nanofluid. 2009 doi: 10.1007/s10404-008-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer-Budge AF, Mirer P, Chatterjee A, Klapperich CM, Chargin D, Sharon A. Low cost and manufacturable complete microtas for detecting bacteria. Lab Chip. 2009;9(19):2803–2810. doi: 10.1039/b904854e. [DOI] [PubMed] [Google Scholar]
- Tong Y, Tang W, Kim HJ, Pan X, Ranalli T, Kong H. Development of isothermal taqman assays for detection of biothreat organisms. Biotechniques. 2008;45(5):543–557. doi: 10.2144/000112959. [DOI] [PubMed] [Google Scholar]
- Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5(8):795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yobas L, Feng Cheow L, Tang K-C, Yong S-E, Kye-Zheng Ong E, Wong L, Cheng-Yong Teo W, Ji H, Rafeah S, Yu C. A self-contained fully-enclosed microfluidic cartridge for lab on a chip. Biomed Microdevices. 2009;11(6):1279–1288. doi: 10.1007/s10544-009-9347-z. [DOI] [PubMed] [Google Scholar]