Abstract
Most patients with Autosomal Dominant Polycystic Kidney Disease (ADPKD) harbor mutations in PKD1 the gene for Polycystin-1 (PC1), a transmembrane protein with a cytoplasmic C-terminus that interacts with numerous signaling molecules, including Gα12. The functions of PC1 and the mechanisms of cyst development leading to renal failure are complex. Recently, we reported that PC1 expression levels modulate activity of Gα12-stimulated apoptosis (Yu et al., J. Biol. Chem. 2010 285(14):10243-51). Herein, a mutational analysis of Gα12 and PC1 was undertaken to identify regions required for their interaction and ability to modulate apoptosis. A set of Gα12 mutations with systematic replacement of six amino acids with NAAIRS were tested for binding to the PC1 C-terminus in GST pulldowns. Additionally, a series of deletions within the PC1 C-terminus were examined for binding to Gα12. We identified 3 NAAIRS substitutions in Gα12 that completely abrogated binding, and identified a previously described 74 amino acid Gαi/o binding domain in the PC1 C-terminus as necessary for Gα12 interaction. The functional consequences of uncoupling PC1/Gα12 binding were studied in apoptosis assays utilizing HEK293 cells with inducible PC1 overexpression. Gα12 mutants deficient in PC1 binding were refractory to PC1 inhibition of Gα12-stimulated apoptosis. Likewise, deletion of the Gα12-interacting sequence from PC1 cytoplasmic domain abrogated its inhibition of Gα12-stimulated apoptosis. Based on the crystal structure of Gα12, the PC1 interaction sites are likely to reside on exposed regions within the G protein helical domain. These structural details should facilitate the design of reagents to uncouple PC1/Gα12 signaling in ADPKD.
1.1 INTRODUCTION
Autosomal dominant polycystic kidney disease (ADPKD1) develops as the result of mutations in the genes PKD1 (~70-85%) or PKD2 (~15-30%) that encode the protein products polycystin-1 (PC1) and polycystin-2 (PC2) respectively. PC1 is a ~460 kDa, eleven-transmembrane spanning protein containing an extensive extracellular domain and a relatively short (~225 amino acid) cytoplasmic domain that interacts with numerous signaling molecules including trimeric G proteins [1-3]. PC1 is localized in cilia and at sites of cell-matrix and cell-cell interactions [4-6]. Mutations in PKD1 lead to defects in cilia function and changes in epithelial cell growth/apoptosis, cell-cell and cell-matrix interactions. Many mutations have been identified in PKD1, and most are deletion, frameshift or nonsense mutations that lead to inactivation of one allele. However, disease development requires an additional insult (a somatic mutation or other injury) to promote cyst development and progression [7, 8]. PKD1 mRNA and protein are expressed throughout development and at moderate to low levels in collecting ducts and distal tubules in the adult. With development of ADPKD, PC1 protein levels are increased about two-fold [9, 10]. Homozygous loss of PKD1 is embryonic lethal with diffuse cystic disease [11] and reviewed [12]) and conditional knockouts of PKD1 reveal important roles during development and have yielded new insights into the mechanisms necessary for cyst formation and progression in vivo (see [12]). Although loss of PC1 leads to cyst development, there is also evidence that PC1 overexpression results in cystic disease [13] [14]. In patients with ADPKD, PC1 expression persists and is even enhanced in most but not all cysts [5] [15]. In addition, transgenic mice overexpressing PC1 develop PKD with renal failure suggesting that, in some cases, a gain of function may be a pathogenic mechanism.
Disregulated apoptosis is an important feature of ADPKD; for instance, increased apoptosis was detected in polycystic kidneys from patients with and without renal failure, but not in controls [16]. Animal models of PKD have also revealed important roles for apoptosis in cyst development in combination with changes in proliferation [17]. However, the focal nature of cyst development, the slow time course of progression, and changes in apoptosis/proliferation in specific nephron segments at different developmental time points has made identifying the role(s) of apoptosis in disease progression difficult to analyze. We recently demonstrated that PC1 expression levels determine activity of the Gα12/JNK apoptosis pathway in MDCK cells [18] suggesting a titration mechanism of regulation. Furthermore, we found that Gα12 but not the closely related G protein α-subunit Gα13 bound to the PC1 C-terminus.
In canonical G protein signaling, ligand binding to a G protein coupled receptor (GPCR) results in conformational changes in the Gα subunit that trigger dissociation of GDP and loss of affinity for the Gβγ dimer. GTP rapidly binds to Gα, and signaling through Gα and Gβγ subunits occurs until the intrinsic GTPase activity of Gα hydrolyzes GTP to GDP. G proteins also interact with numerous regulatory and scaffolding proteins. PC1 has been reported to function as an atypical GPCR, bind Gαo/i, and regulate calcium flux through PC2 (a member of the TRP family of calcium channels) by release of Gβγ subunits [19, 20]. Multiple G protein α-subunits [1-3] and at least one Regulator of G protein Signaling (RGS) protein [21] interact with PC1. Furthermore, we reported binding of wildtype and mutationally activated (GTPase deficient) Gα12 to the PC1 C-terminus [3] and recently extended this observation to show that thrombin-stimulated Gα12 preferentially bound to this PC1 domain [18]. Furthermore, in transient overexpression systems, Gα12 regulated AP1 transcription factor activity in a PC1 dependent manner [2]. Herein, we utilize mutant forms of Gα12 and PC1 to provide insights into the structural details of PC1/Gα12 binding, and demonstrate for the first time that apoptotic regulation can be uncoupled by disrupting the interaction between Gα12 and PC1. Based on the Gα12 crystal structure, the PC1 binding sites on Gα12 can be modeled, and implications for regulating the PC1/Gα12 interaction and its effects on apoptosis are discussed.
2. MATERIALS AND METHODS
2.1 Chemicals, Antibodies and cDNA Constructs
Anti-Gα12 (sc-409) and PC1 antibodies (7e12) were purchased from Santa Cruz Biotechnology, and alkaline phosphatase-conjugated anti-rabbit antibody was from Promega. Glutathione-sepharose was from Amersham Pharmacia Biotech. The PC1 C-terminal GST fusion protein was previously established and purified as described [3]. Substitution mutants within myc-tagged Gα12QL, in which regions of the cDNA encoding consecutive sextets of amino acids are replaced with a sequence encoding the sextet NAAIRS, were engineered using oligonucleotide-directed mutagenesis as described previously [22].
2.2 Creation of GST and CD16/CD7/ PC1 C-terminal Deletion Mutations
A GST-fusion construct of the C-terminal domain of PC1 (GST-PC1CT) was subjected to PCR-based mutagenesis to generate the deletion mutants used in this study. For mutants HQ128 and SH3, oligonucleotides were designed to amplify the PC1 cytoplasmic domain coding sequence, with a stop codon introduced immediately past the amino acids QATEDVYQ (for the HQ128 deletion mutant) or RGPSPGLR (for the SH3 deletion mutant). Each amplimer was then digested at its ends and ligated into the GST-PC1 plasmid in place of its original PC1 sequence. For deletion mutant CT-74, internal oligonucleotides 5′- tggcgctactcgcacccctccacctcctcc -3′ and 5′-ggggtgcgagtagcgccagcggagaataacag -3′ were designed to traverse the minimal G protein binding domain [1] of the PC1 cytoplasmic domain and thus remove the 74-amino acid sequence spanning from HALRGE to SAGSDA. Each internal oligonucleotide was next used in conjunction with an oligonucleotide at the end of the PC1 sequence, and the resulting two PCR amplimers (with 18 base overlap) were subjected to additional PCR using the end oligonucleotides to generate a PC1 C-terminus lacking sequence encoding the internal 74 amino acids. This amplimer was ligated into GST-PC1 in place of its original PC1 sequence. The construct FLM-PC1 (gift of Thomas Weimbs, Univ. of California Santa Barbara), containing the PC1 C-terminal sequence positioned downstream of the extracellular region of CD16 and a single CD7 transmembrane span, was used as a template for a second set of these PC1 deletion mutants. This construct was subjected to PCR-based mutagenesis in the same manner as GST-PC1, and amplimers were ligated into FLM-PC1 in place of its original PC1 C-terminal sequence. All constructs were verified by sequencing.
2.3 Preparation of Gα12 mutants from cell lysates
Human embryonic kidney (HEK293) cells were grown in DMEM (Mediatech) supplemented with 10% fetal bovine serum (Hyclone) and penicillin/streptomycin, and were maintained at 37°C in a 5.0% CO2 atmosphere. For each NAAIRS substitution mutant of myc-tagged QLα12, 7 μg of plasmid DNA were used to transfect a 10-cm dish of HEK293 cells at 80-90% confluence, using Lipofectamine 2000 reagent (Invitrogen) in accordance with the manufacturer’s instructions. At 36-42 h post-transfection, cells were rinsed twice with phosphate-buffered saline (PBS), scraped from the dish, pelleted at 800 × g, and then 0.5 mL of ice-cold Buffer A (50 mM HEPES pH 7.5, 1 mM EDTA, 3 mM dithiothreitol, 10 mM MgSO4) supplemented with 1% (w/v) polyoxyethylene-10-lauryl ether and protease inhibitors was used to solubilize each cell pellet. Samples were mixed by inversion at 4°C for 30 min, and then centrifuged at 100,000 × g, 4°C, for 1 h. Supernatants were snap-frozen in liquid N2 and stored at −80°C.
2.4 GST Pull-down of Gα12 Mutants
GST-PC1 C-terminus and GST were expressed in BL21- Gold(DE3) E. coli (Stratagene), with induction of protein expression by 0.5 mM isopropyl-β-D-thiogalactopyranoside 3 hours prior to cell harvesting, and were purified by immobilization onto glutathione-sepharose as described previously [23]. For each mutant of QLα12 tested, 60 μL of HEK293 cell supernatant (prepared as described above) was diluted to 600 μL using Buffer A lacking polyoxyethylene-10-lauryl ether (producing a final detergent concentration of 0.1%). For each sample, 5% of diluted lysate was set aside as “load” and the remainder was equally divided and incubated with glutathione-sepharose bound to either GST-PC1 C-terminus or GST. Samples were mixed by inversion for 2 h at 4°C, pelleted by centrifugation at 1,300 × g, and supernatants were discarded. Sepharose pellets were washed three times with 1 mL Buffer A containing 0.05% polyoxyethylene-10-lauryl ether, followed by resuspension of pellets in SDS sample buffer and heating to 72°C for 10 min. For each sample, 80% of the volume was subjected to SDS-PAGE and subsequent blot transfer. Immunoblots were blocked overnight at 4°C in TBST [50 mM Tris pH 7.7, 150 mM NaCl, 0.05% Tween-20] supplemented with 5% (w/v) powdered milk, and then were incubated with rabbit anti-Gα12 antibody (1:500 dilution) in TBST + 5% milk for 2 h. Following three 10-min washes using TBST, alkaline phosphatase-conjugated anti-rabbit antibody was diluted 1:7500 in TBST + 5% milk and applied for 1 h. Three additional washes were performed using TBST, and blots were developed using NBT/BCIP reagent (Promega) according to the manufacturer’s instructions. Results were documented using a Kodak Gel Logic 100 scanner and band intensities were quantified using Carestream Molecular Imaging software (New Haven, CT). To ensure equal distribution of the GST-fusion proteins in all pulldown assay samples, the remaining 20% of each sample volume was subjected to SDS-PAGE, and gels were stained overnight with Coomassie blue, destained, and photographed. For experiments examining the binding of PC1 C-terminal deletion mutants to myc-QLα12 expressed in HEK293 cells, identical conditions for bacterial protein expression, mammalian cell culture, and pulldown assays were employed as described above.
2.5 Tet-On PC1 Inducible HEK293 Cells
The Tet-regulated 293TRex (Invitrogen) cells overexpressing PC1 were established by transfecting PvuI linearized pcDNA4.1/YFP-Pkd1-Myc-HIS6 plasmid into 293TRex cells. Colonies resistant to Zeocin (400 μg/ml) were subcloned and expression of PC1 determined by Western blotting in the presence or absence of tetracycline (1μg/ml) using antibody 7e12 at 1:1000 dilution (Santa Cruz Biotechnology).
2.6 Transient Transfections for Determining Effects of Mutant Proteins on Apoptosis
To determine the effects of NAAIRS mutations on apoptosis in the presence and absence of PC1 overexpression, PC1-293TRex cells were transfected using Fugene 6 (Roche) according to the manufacturer’s protocol with 2 μg of pcDNA3.1 vector alone, myc-Gα12QL, or the NAAIRS mutants OO, TT, Z, O and VV in the absence of doxycycline (dox) and serum free DMEM. At 12 h after transfection, DMEM with 10% dox-free serum was added for an additional 24 h. Dox (5 μg/ml) was added to media 36 h after transfection. The analysis was performed in triplicate and a parallel set of transfections were established under identical conditions and maintained in dox-free media. Cells were serum starved overnight and analyzed in parallel by flow cytometry for determining the percentage of apoptotic cells. To characterize the effects of PC1 C-terminal mutations on QLα12-stimulated apoptosis, HEK293 cells were transiently transfected with 2 μg of pcDNA3.1, PC1-FLCT, HQ128, or SH3 with and without QLα12 using Fugene 6 and serum free DMEM. Approximately 12 h later, fresh DMEM with 10% serum was added to cells and incubated at 37°C for an additional 24 h. After serum starvation overnight, cells were then trypsinized and collected for apoptosis analysis by FACS. To analyze Gα12 effects on apoptosis +/− PC1 overexpression, PC1-293TRex cells were transfected with G228Aα12, QLα12, or wildtype Gα12, and apoptosis was measured by FACS +/− dox for 24 h and +/− thrombin (2 U/ml) for 24 h.
2.7 Apoptosis Determined by Flow Cytometry
Transfected cells were collected after trypsinization and pooled with floating cells by low speed centrifugation, washed with phosphate-buffered saline (PBS), and fixed with 70% ethanol in PBS at −20°C for 30 min. Subsequently, cells were incubated with 1 μg/ml RNase A and 50 μg/ml propidium iodide (Invitrogen) in PBS at 37°C for 30 min and analyzed by flow cytometry (propidium iodide/PE Texas Red channel).
2.8 Modeling of Crystal Structures
The Gα12 crystal structure 1ZCA [24] was viewed with FirstGlance in Jmol (http://molvis.sdsc.edu/fgij). Mutations were highlighted with yellow spheres and the image rotated to highlight specific orientations. The images were exported as jpeg files and assembled in Adobe Illustrator (Adobe, San Jose, CA).
2.9 Statistics and Quantification
Data was analyzed using t-test in Graphpad Prism (La Jolla, California) and significance reported at p <0.05.
3. RESULTS
3.1 Specific NAAIRS Mutations in Gα12 Uncouples Interaction with PC1 C-terminus
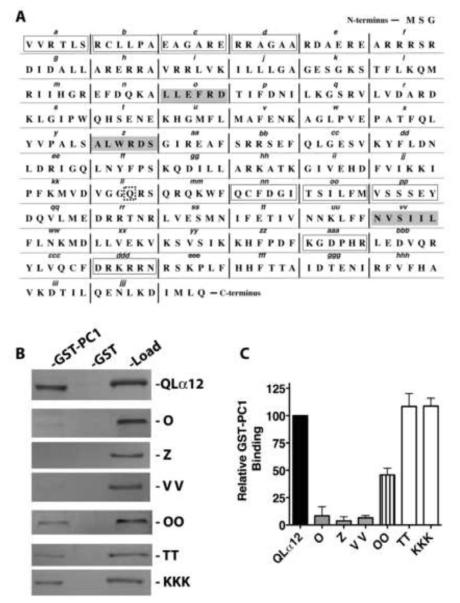
To identify regions of Gα12 important for interaction with PC1, we utilized a series of mutants that systematically replace six amino acids of the Gα12 sequence with the sequence asparagine-alanine-alanine-isoleucine-arginine-serine (NAAIRS). This series of mutations was created within the QLα12 coding sequence to facilitate the study of Gα12 binding proteins that would be predicted as downstream effectors in canonical G protein signaling. We have previously utilized this approach to map the binding domains of Gα12 and the scaffolding Aα subunit of PP2A [23], and this approach has been used successfully to identify unique Gα12 regions important for interaction with p115RhoGEF and LARG (leukemia-associated RhoGEF)[22]. The NAAIRS sequence is found naturally in both α-helical and β-sheet secondary structures and is believed to be a well-tolerated substitution [25]. Figure 1A shows the linear sequence of Gα12 with the designated NAAIRS mutations. Myc-tagged QLα12 was used as a starting template so that subsequently engineered NAAIRS mutants could be distinguished from endogenous Gα12 when expressed in cultured mammalian cells. These NAAIRS substitution mutants were given consecutive alphabetical designations: A to Z followed by AA to ZZ, ending with AAA to KKK (see Fig. 1A). We were able to express 61 of these 63 mutants in HEK293 cells, as determined by immunoblot analysis (data not shown). Mutant W was not engineered due to the positioning of the myc epitope tag within it, and mutant ZZ was poorly detected in cell lysates. Mutants were detergent-extracted from HEK293 lysates and then screened for binding to a GST fusion of the PC1 C-terminus (GST-PC1CT) as described in 2.4.
Figure 1. Identification of NAAIRS mutants that uncouple Gα12 binding to PC1.
(A) Native amino acid sequence of Gα12 is shown, with sextets replaced by the sequence NAAIRS flanked by vertical bars and alphabetical designations of mutants indicated in italic lowercase. The starting template for all mutants was myc-tagged QLα12 cDNA. The myc tag with flanking SGGGGS sequences was inserted between Pro139 and Val140, and the w sequence was not converted to a NAAIRS mutant. The site of the activating Q-to-L substitution is indicated by a dashed square. The three regions in which replacement by NAAIRS resulted in a >90% decrease in binding to PC1 are indicated by gray boxes. Mutants with 50-75% decrease in binding are indicated by open boxes. All remaining NAAIRS mutants displayed less than 50% decrease in binding compared to myc-tagged QLα12. (B) Representative Western blots of GST-PC1 pulldown experiments utilizing QLα12 variants. Lysates from HEK293 cells expressing either myc-tagged QLα12 or each indicated “NAAIRS” substitution mutant (e.g. O, Z) were pulled down with GST alone or GST-PC1CT as described in 2.4. For each individual panel, the left lane (GST-PC1) indicates Gα12 that was precipitated by GST-PC1CT, the middle lane (GST) indicates Gα12 precipitated by GST alone, and the right lane (load) indicates a fraction (5%) of the HEK293 cell lysate that was set aside prior to the pulldown steps. (C) Quantification of binding relative to QLα12. Western blot results for two independent experiments using the indicated Gα12 variants were quantified using Carestream Molecular Imaging software, and are presented as mean +/− range.
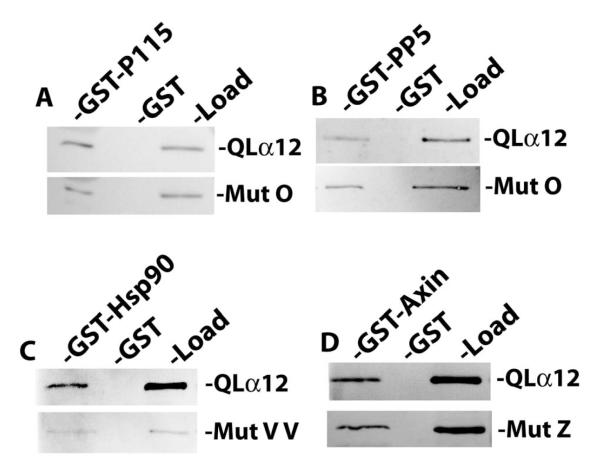
Of the NAAIRS mutants examined for interaction with GST-PC1CT, most displayed relative binding similar to the myc-QLα12 control protein (50-100% within the same experiment). There were nine mutants with a moderate reduction in binding (25-50% of control); these are highlighted with open boxes in Figure 1A, and the immunoblot results for one such mutant (OO) are shown in Fig. 1B. However, there were three additional mutants (O, Z, and VV) that consistently failed to interact with GST-PC1CT (<10% of control) and these are highlighted in gray boxes (Fig. 1A) with immunoblot results shown in Fig. 1B. Also shown in Figure 1B is a comparison of PC1 interaction for mutants O, Z, and VV with two additional mutants (TT, KKK) that displayed unimpaired binding. The relative affinity of each mutant (ratio of GST-PC1CT pulldown band intensity to load band intensity) is shown as a percent of the same ratio for myc-QLα12 (Fig. 1C). To exclude the possibility that mutants O, Z and VV are misfolded and incapable of productive interactions with any downstream targets, these variants were examined in binding experiments utilizing GST fusions of several other reported Gα12 effector proteins. Fig. 2 shows that these PC1 binding-impaired mutants are still capable of normal interactions with other known Gα12 binding partners [26-29]. Mutant O interacted with GST-p115RGS and GST-PP5 with an affinity indistinguishable from the QLα12 control (Fig. 2A, B). Mutant VV interacted with Hsp90 in a manner comparable to QLα12 (Fig. 2C), and mutant Z was similar to QLα12 in its ability to bind the RGS domain of axin (Fig. 2D). Taken together, this analysis identifies 3 specific NAAIRS substitutions that significantly diminish the affinity of Gα12 for the PC1 cytoplasmic tail, while preserving interaction with several other Gα12 binding partners (Fig. 2). Therefore, these regions of Gα12 are likely to contribute to the Gα12/PC1 binding surface. NAAIRS substitutions at these locations appear not to cause global disruptions in the conformation of Gα12. Furthermore, each of these mutants is capable of stimulating apoptosis (see Fig. 3).
Figure 2. Gα12 mutations uncoupled from PC1 binding interact normally with other Gα12 targets.
For all panels, the indicated NAAIRS mutants and QLα12 were expressed in HEK293 cells and examined in pulldown experiments as described in 2.4. (A, B) Mutant O was tested for in vitro interaction with GST fusions of p115RGS (GST-p115) and protein phosphatase-5 (GST-PP5). For each pulldown condition the binding of the mutant was compared to results for QLα12 analyzed in parallel. (C.) Mutant Z and QLα12 were tested for interaction with a GST fusion of the RGS domain of axin (GST-Axin). (D.) Mutant VV and QLα12 were tested for binding to GST-Hsp90. For all panels, the band intensities were quantified using Carestream Molecular Imaging software, and the ratio of pulldown-to-load for each mutant was compared to the pulldown-to-load ratio for QLα12 tested in parallel.
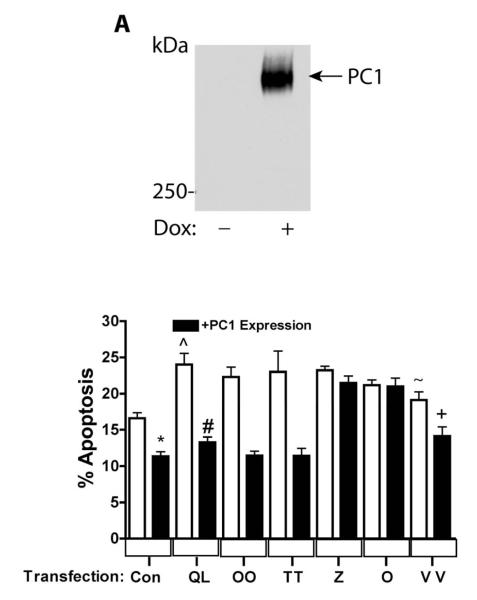
Figure 3. Gα12 mutations uncoupled from PC1 binding fail to regulate apoptosis.
(A) Characterization of Tet-On PC1 HEK293 Cells. (B) QLα12-stimulated apoptosis +/− PC1 over-expression. HEK293 cells in which PC1 over-expression was induced by dox (black bars) or parallel cultures maintained without dox (no PC1 expression; white bars) were transiently transfected with the vector pcDNA3.1 (Con), QLα12 (QL), and NAAIRS mutants OO, TT, Z, O, and VV. Apoptosis was determined by FACS 48 h after transfection as described in 2.7. (*) Control +/− PC1 expression, p=0.0004; (#) QLα12+/−dox, p<0.0001; (^) Control compared with QLα12 without PC1 expression, p=0.0017; (+) VV+/−PC1 expression, p=0.0147; Control versus VV without PC1 expression; n.s. p=0.08, (~) QLα12 versus VV p=0.029.
3.2 Gα12 Binding Mutants are Uncoupled From PC1/Gα12 Regulated Apoptosis
We recently demonstrated that PC1 overexpression in MDCK cells inhibits Gα12-stimulated apoptosis through JNK stimulation and Bcl-2 degradation [18]. Therefore, to examine the effects of uncoupling PC1/Gα12 interaction on apoptosis, we utilized HEK293 cells with dox-inducible PC1 overexpression, introduced Gα12 NAAIRS mutants by transient transfection, and then treated cells with +/−dox. Figure 3A shows a Western blot of dox-induced PC1 protein migrating on a 5% polyacrylamide gel significantly above the 250 kDa marker (estimated size of PC1 is ~460 kDa); this band was not seen in the absence of dox. Apoptosis was determined by FACS, as described in 2.7, in cells +/− PC1 overexpression after transient transfection of vector, QLα12, or the NAAIRS mutants OO, TT, Z, O, or VV (Fig. 3B). Consistent with the previously published finding that PC1 overexpression inhibits apoptosis [30], serum starvation of HEK293 cells overexpressing PC1 exhibited lower baseline apoptosis than the control (white and black columns at far left, Fig. 3B). QLα12 has been shown to stimulate apoptosis in MDCK cells [31], and in similar fashion, transfection of QLα12 into HEK293 cells without PC1 expression triggered an increase in apoptosis compared to vector-transfected cells (Fig. 3B; QL versus control; white bars). As expected, PC1 overexpression effectively blocked QLα12 stimulated apoptosis to levels similar to the vector controls. NAAIRS mutant OO showed no difference from QLα12 in its stimulation of apoptosis +/− PC1 overexpression, despite a partial reduction in PC1 binding (about 50%; see Fig. 1). Likewise, and as expected, mutant TT showed no significant difference from QLα12 in its ability to trigger apoptosis in the presence or absence of overexpressed PC1. However, two NAAIRS mutants (O, Z) that were incapable of PC1 binding were also uncoupled from PC1 inhibition of apoptosis. Of note, these variants of QLα12 effectively stimulated apoptosis, indicating that these mutations do not distort overall conformation or disrupt the ability to engage downstream signaling proteins required for apoptosis. Mutant VV displayed a partial phenotype with slightly less stimulation of apoptosis compared with QLα12 (24% versus 19%) and exhibited partial inhibition of apoptosis with PC1 overexpression. An explanation for these observations may be that the VV mutant, unlike the O and Z mutations, replaces native residues within the GTPase domain and may induce subtle differences in conformation or interactions with other components of the apoptosis signaling pathway.
3.3 The PC1 Minimal G Protein Binding Domain is Required for Gα12 Interaction and Regulation of Apoptosis
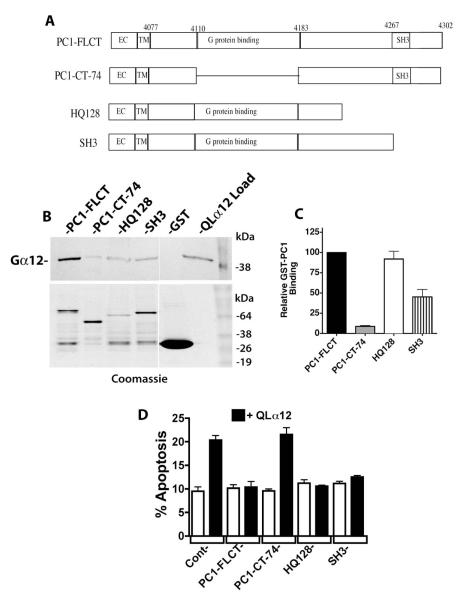
Trimeric G proteins of the Gi subfamily have been shown to interact with the PC1 C-terminus through a minimal 74 amino acid G protein binding sequence [1]. This domain also contains a putative 20 amino acid G protein activation sequence that, when synthesized as a short peptide, was able to accelerate GTP binding and hydrolysis on purified Gi subfamily proteins [1]. To identify regions of the PC1 C-terminus necessary for binding to Gα12, we utilized a GST fusion of the PC1 C-terminus (previously characterized [3]) and constructed several deletions in this region: PC1-CT-74, HQ128, and SH3 (defined in Fig. 4A and based on [1]). These were tested for binding to QLα12 in pulldown experiments, and Figure 4B shows the results for each of these GST-PC1 fusion proteins (Western blot for QLα12 in upper panel, Coomassie blue staining of immobilized proteins in lower panel). The band intensity of bound QLα12 for each PC1 variant was normalized to the amount of GST-fused protein present, and then calculated as a percentage of the same value for non-mutated GST-PC1CT (Fig. 4C). Deletion mutant HQ128 was previously shown to interact normally with Gαi proteins [1] and also interacted with Gα12 normally in our experiments (Fig. 4B, 4C). The extreme C-terminal region of PC1 contains an α-helix and SH3-binding motif, and when expressed as a 120 amino acid GST fusion protein occasionally bound Gα subunits [1]. We tested a shorter C-terminal deletion of 36 amino acids (termed SH3; see Fig. 4A) and observed ~50% of the binding seen with PC1-FLCT and HQ128 (Fig. 4B, 4C). As suggested from prior studies utilizing Gi subfamily proteins [1], deletion of the 74 amino acid minimal G protein binding domain almost completely abrogated binding to QLα12 (Fig. 4B, 4C). Unfortunately, we were unsuccessful in testing a deletion of only the 20 amino acid putative G protein activation sequence within PC1, because this GST fusion protein was poorly expressed in bacterial cells (data not shown).
Figure 4. PC1 C-terminus requires a minimal G protein binding domain for Gα12 interaction and regulation of apoptosis.
(A) Diagram of PC1 C-terminal deletions. Mutations were engineered as described in 2.2 within a GST fusion of PC1 (used for pulldown experiments; panels B and C) and a CD16/CD7/PC1 fusion protein (used for transient cellular expression and determination of apoptosis; panel D). (B) GST pulldowns of QLα12 using PC1 C-terminal deletion mutations. Top panel is a Western blot showing the intensity of QLα12 precipitated by the indicated GST-PC1 constructs, and lower panel shows abundance of each GST-PC1 fusion protein within the precipitated sample. (C) Quantification of pulldown results. Band intensity values for precipitated QLα12 were normalized to the band intensity of each GST-PC1 protein using Carestream Molecular Imaging software and presented as a percent of the value for QLα12 precipitated by full length PC1 C-terminus (PC1-FLCT). (D) Inhibition of QLα12-stimulated apoptosis +/− PC1 C-terminal Mutants. HEK293 cells were transfected with a CD16/CD7/PC1 fusion protein with no deletion (FLCT) or the deletions CT-74, HQ128 or SH3 (see panel A). Constructs were transfected +/− a plasmid encoding QLα12 (white vs. black bars). Cells were analyzed for apoptosis by FACS as described in 2.7.
We recently showed that a previously described C-terminal deletion (CTM-PC1; a membrane-bound PC1 cytoplasmic domain lacking amino acids 4191-4302 [32]) failed to inhibit thrombin/Gα12 stimulated apoptosis [18]. This construct lacks most of the 74 amino acids comprising the putative minimal G protein binding sequence [1]. To test the effects of the C-terminal deletions in Fig. 4A on PC1/Gα12 regulated apoptosis, we engineered these deletions within an isolated PC1 cytoplasmic domain fused to a single CD7 transmembrane domain and a CD16 extracellular domain [32], and then expressed these constructs in HEK293 cells +/− QLα12 and measured apoptosis as described in 2.7 (Fig. 4D). QLα12 transfection alone stimulated apoptosis in control cells, and there was no significant effect on apoptosis with any of the membrane-targeted PC1 C-terminal constructs when expressed without QLα12 (Fig. 4D). When QLα12 was co-expressed with the full-length PC1 C-terminal construct, QLα12-stimulated apoptosis was fully inhibited to baseline levels (Fig. 4D). However, the PC1-CT-74 construct (impaired in Gα12 binding; Fig 4B, 4C) had no effect on QLα12-stimulated apoptosis, indicating its functional uncoupling from Gα12. The remaining membrane-bound PC1 C-terminal constructs, with deletions termed HQ128 and SH3 were indistinguishable from the full length PC1 C-terminus in their inhibitory effect on QLα12-stimulated apoptosis (Fig 4D). Taken together, these results indicate the previously identified Gαi-binding 74 amino acid region within the PC1 C-terminus is required for interaction with Gα12 and for mediating inhibition of Gα12-stimulated apoptosis.
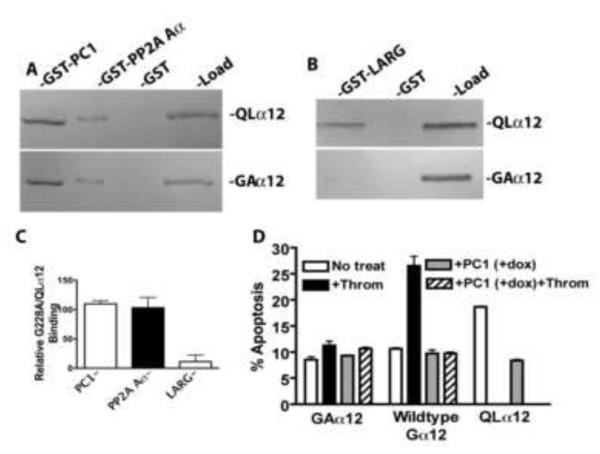
3.4 Dominant Negative and Constitutively Activated Gα12 Interact Similarly with PC1 C-Terminus
A novel aspect of PC1 regulation of Gα12 signaling is the consistent finding that both wildtype and active (QL) conformations of Gα12 interact with the PC1 C-terminus [3, 18]. In canonical G protein receptor signaling, the inactive conformation of Gα associates with a GPCR and Gβγ subunits, and upon activation dissociates from these proteins to bind downstream effectors. The observation that both constitutively active (QLα12) and wildtype Gα12 interact with the PC1 C-terminus suggests that this signaling mechanism may be more complex and raises the possibility that PC1 functions as a scaffold to organize multiple signaling complexes or potentially serves as a target (effector) for an activated Gα. In previous work, we observed that thrombin stimulation caused an increase in the amount of Gα12 pulled down from cell lysates by the PC1 C-terminus, but this experiment could not distinguish active from inactive Gα12 [18]. To more rigorously characterize the relative binding of the PC1 C-terminus to the active or inactive conformations of Gα12, we directly compared the binding of a constitutively GDP-bound Gα12 (Gly-Ala at position 228 in murine Gα12; based on [33]) with the GTPase-deficient QLα12 in pulldown experiments. Figure 5A shows in vitro interaction with GST fusions of PC1 and another Gα12 binding partner (PP2A Aα [34]) that also binds to Gα12 in both the active and inactive conformations. Somewhat surprisingly, there was no difference in the binding of G228Aα12 and QLα12 with the PC1 C-terminus or PP2A Aα (Fig. 5A, 5C). However, the Gα12-stimulated, Rho-specific guanine nucleotide exchange factor LARG preferentially bound to QLα12 in this assay (Fig. 5B, 5C), consistent with its known function [35]. We also examined the G228Aα12 mutant in apoptosis assays with and without PC1 overexpression. As expected for a dominant negative mutant, G228Aα12 was unable to increase apoptosis when cells were stimulated with thrombin (Fig. 5D). When G228Aα12 was expressed with PC1 there was no further reduction in apoptosis nor was there any effect of thrombin. Thrombin stimulation of HEK293 cells expressing wildtype Gα12, but without PC1 expression (-dox), led to a significant increase in apoptosis that was completely reversed with PC1 overexpression (+dox). As a positive control, QLα12 expression in the absence of PC1 (-dox) showed significant stimulation of apoptosis compared with wildtype Gα12 or G228Aα12. In cells with PC1 expression (+dox) and QLα12 expression, apoptosis was inhibited to control levels as shown in Figure 3. Taken together, the results with G228Aα12 and QLα12 are consistent with prior observations and suggest that at least in vitro, PC1 binds with similar affinity to both active and inactive Gα12 conformations. The observation that there are not additive effects on inhibition of apoptosis with G228Aα12 and PC1 overexpression is consistent with the notion that Gα12 and PC1 signal within the same pathway.
Figure 5. Constitutively GDP-bound and mutationally activated Gα12 bind PC1 in vitro with similar affinity.
For panels A-C, cellular expression of proteins and in vitro interaction assays were performed as described in 2.4. (A) Comparison of G228Aα12 (GAα12) with QLα12 in pulldowns using GST fusions of the PC1 C-terminus and the Aα subunit of PP2A. (B) Binding of G228Aα12 and QLα12 to GST-LARG. (C) Quantification of relative binding of G228Aα12 to each partner, compared with QLα12. (D) Apoptosis of HEK293 Cells +/− PC1 overexpression, transiently transfected with G228Aα12, wildtype Gα12 or QLα12. Cells were stimulated +/− thrombin (2 U/ml) for 24 h prior to determining apoptosis as described in Material and Methods.
4. DISCUSSION
ADPKD is a complex disorder, and the biologic functions of PC1 are only partially understood. PC1 has a large extracellular domain, eleven transmembrane domains, and a short cytoplasmic C-terminus that interacts with numerous signaling molecules. Understanding how this domain regulates intracellular signaling and interactions of PC1 with other proteins is essential to unraveling the cellular physiology of PC1 and the pathology of cyst development and progression. Several lines of evidence indicate important regulation of cellular functions via PC1 and G proteins. PC1 binds several Gα subunits and RGS proteins, and regulates PC2 function through release of Gβγ subunits [19, 20] suggesting that PC1 functions as an atypical GPCR. However, the specificity of these interactions leading to regulation of unique cellular effects remains to be determined. Although multiple Gα subunits can interact with the PC1 C-terminus, we recently demonstrated that Gα12 and not the related Gα13 subunit bound the PC1 C-terminus [18]. Furthermore, we have described an apoptosis pathway that is stimulated by Gα12 activation [31], and found that PC1 expression levels modulate apoptosis through a mechanism in which PC1 levels titrate the available pool of Gα12 [18]. Utilizing this phenotype of PC1 inhibition of Gα12-stimulated apoptosis, we have examined specific mutations in Gα12 and the PC1 C-terminus to identify binding regions essential to this protein interaction, and have demonstrated that these mutations also uncouple PC1/Gα12 regulation of apoptosis. To our knowledge, this is the first demonstration of uncoupling a PC1 signaling pathway.
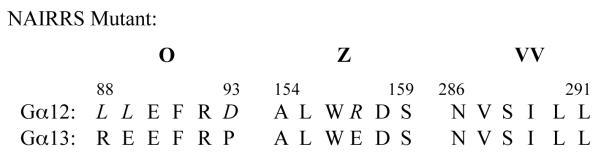
The use of NAAIRS amino acid sextet substitutions within a protein is a useful approach for identifying discrete amino acid regions necessary for protein interactions. We have utilized this approach to identify regions important for interactions with other Gα12 binding partners including the Aα subunit of PP2A, p115RhoGEF, and LARG [22, 23, 34]. Interestingly, there were only three NAAIRS mutations in Gα12 (O, Z, and VV) that displayed >90% impairment in binding the PC1 C-terminus, and nearby substitutions in Gα12 upstream and downstream of these mutations did not affect PC1 binding. This suggests that these substitutions are in discrete regions on the surface of the protein with access to neighboring structures. In addition, all three mutations had preserved interactions with other effectors and retained the ability to stimulate apoptosis, indicating that they are not significantly altered in overall conformation. Alignment of these Gα12 sequences with the PC1 non-binder Gα13[18] reveals that mutant O has the most differences, and that a charge reversal exists in Gα13 at the position corresponding to the Z mutant (Table 1). There is no significant similarity of the Gα12 O and Z regions with Gαi1 or Gαs, suggesting that these G protein subfamilies bind via a different mechanism to the PC1 C-terminus. Two mutants of Gα12 (O and Z) were completely uncoupled from PC1 mediated inhibition of apoptosis, indicating that these binding sites were not only essential for the interaction but also for PC1 mediated inhibition of Gα12-stimulated apoptosis. Mutant VV behaved differently than O and Z, displaying a slight impairment in stimulation of apoptosis and retaining sensitivity to PC1 inhibition of apoptosis.
Table 1.
Alignment of Mouse Gα12 and Gα13 Protein Sequences.
|
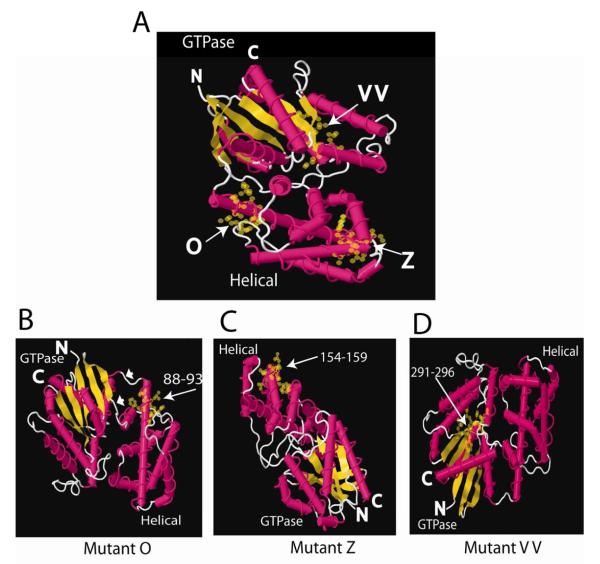
Crystal structures of Gα subunits reveal that a helical domain and a GTPase domain comprise the tertiary structure. The GTPase domain contains the amino and carboxyl termini, the guanine nucleotide binding site that is highly conserved among all Gα subunits and the conformationally sensitive regions that change in the active (GTP-liganded) and inactive (GDP-liganded) states (reviewed in [36]). The helical domain is attached to the GTPase domain through two linkers (see Fig. 6B). The helical domain lies over the guanine nucleotide binding pocket to sandwich the bound nucleotide. The helical domain may have GAP-like activity for Gα [37], but its function is not well understood. Three-dimensional modeling of the O, Z and VV mutations (Fig. 6) indicates that mutants O and Z replace amino acids located in α-helical regions that are exposed on the surface of the helical domain. Figure 6A shows the location of each mutation (O, Z, VV) in a single crystal structure. Rotating the structures reveals that both O and Z mutations are located on exposed surfaces (Fig. 6B,C). These regions are not conformationally sensitive, which is consistent with these sites mediating interaction between PC1 and Gα12 in both the active and inactive conformations. A loss-of-function screen in Dictyostelium, in which the helical domain was randomly mutated in Gα2, revealed seven single amino acid substitutions that resulted in defective aggregation [38]. One of these changes (R64G) is in the αA helical domain, and the equivalent site in QLα12 is L88N located within the O mutant. Although mutant VV is severely impaired in binding PC1 and capable of stimulating apoptosis (Fig. 3B), it retains partial sensitivity to inhibition of apoptosis by PC1, suggesting that the consequences of this NAAIRS substitution may be indirect. VV is within the GTPase domain (Fig. 6A, D) in a region that does not appear to be easily accessible to partnering proteins. Although it is possible that the PC1 terminus directly interacts with this region, it is more likely that this substitution indirectly affects PC1 binding through secondary changes in Gα12 structure.
Figure 6. Crystal Structure Models.
(A) Crystal structure model of Gα12 with mutants O, Z, and VV highlighted with yellow spheres and arrows. GTPase and helical domains are labeled and the N- and C-termini shown. (B-D) Mutants O, Z, and VV rotated to highlight the exposed amino acids in the NAAIRS-substituted regions of these mutants. Numbers refer to substituted amino acid positions and arrows highlight location of changes. White arrowheads in B mark linkers between the GTPase and helical domains. Gα12 crystal structure 1ZCA was viewed with FirstGlance in Jmol (http://molvis.sdsc.edu/fgij) and images were exported as jpeg files and assembled in Adobe Illustrator (Adobe, San Jose, CA).
The PC1 C-terminus contains a 20 amino acid G protein activating sequence that has been shown to stimulate GTPγP32 binding to purified brain Gαo/i protein and is contained within the 74 amino acid G protein binding region [1]. However, in some contexts, overexpression of the PC1 C-terminus functions as a dominant negative. Our approach was to test the NAAIRS mutants of Gα12 in an experimental system along with overexpressed full length PC1 and determine the effects on apoptosis. We found that QLα12-stimulated apoptosis was blocked by PC1 overexpression (Fig. 3B) and this is consistent with recent results in MDCK cells with stable PC1 overexpression [18]. Deletion of the previously identified 74 amino acid G protein binding region in the PC1 C-terminus also uncouples its ability to regulate Gα12-stimulated apoptosis. This finding is consistent with a requirement for direct binding of PC1 and Gα12 to regulate apoptosis. The minimal G protein binding sequence may be smaller than the 74 amino acids identified, but is likely to include the 20 amino acid G protein activating sequence [1]. Increased PC1 expression inhibits Gα12 signaling regardless of whether Gα12 is activated by Q-to-L mutation or through thrombin receptor stimulation. One possible model is that PC1 inhibits Gα12 signaling by binding to Gα12 and preventing it access to the thrombin receptor, as we recently suggested [18].
The finding that G228Aα12 binding to the PC1 C-terminus is indistinguishable from QLα12 suggests that Gα12 conformation and the identity of its bound nucleotide are not critical factors regulating the interaction. One possibility is that the PC1 C-terminus functions as a scaffold to organize a multi-protein signaling complex. In this model, Gα12 would not dissociate from PC1 as in canonical GPCR signaling, but rather would regulate release of Gβγ and interaction with other downstream molecules through nucleotide exchange without dissociation from PC1. Release of Gβγ from PC1 has been shown to modulate calcium channel activity of PC2 [19, 20], but the apoptosis pathway regulated by Gα12 is seen with QLα12 and therefore not dependent upon Gβγ [18]. Whether G proteins are directly activated through conformation changes in the PC1 C-terminus, initiated by changes in the extracellular domain or indirectly through another intracellular protein (nucleotide exchange factor), will require additional study. Alternatively, Gα subunits themselves may be activated in non-canonical ways by interacting with proteins that function as exchange factors (for example, Ric-8 [39]). Additional questions include whether the PC1 C-terminus interacts with more than one Gα subunit at a time, and how the PC1-Gα subunit interaction is regulated. Furthermore, Gα subunits themselves can also function as scaffolding proteins; Gαq was recently shown to form a ternary complex with PKCζ and MEK that is required for ERK activation [40]. Taken together, the PC1 C-terminus is likely to organize a macromolecular signaling complex. It is also feasible that constitutive binding of Gα12 to the PC1 C-terminus leads to changes in PC1 function through an inside-out signaling mechanism. However, in the absence of clearly defined and easily measured readouts of PC1 function, this possibility is difficult to examine at this time.
5. CONCLUSIONS
These studies demonstrate that direct binding of PC1 with a G protein α subunit is required for regulation of a specific signaling pathway.
NAAIRS mutants reveal key structural details modulating the interaction of Gα12 with the PC1 C-terminus.
Mutations of Gα12 in its PC1-binding regions do not affect ability to stimulate apoptosis yet are uncoupled from PC1 regulation.
The PC1 C-terminal region responsible for binding Gα12 is also essential for regulating apoptosis.
There is growing evidence that Gα12 binds PC1 with similar relative affinities in the active and inactive conformations. This has major implications for defining how PC1 regulates signaling and whether PC1 function(s) may be regulated by G proteins (inside out signaling similar to integrins). Ultimately, the discrete amino acid regions identified in these and related studies will provide novel tools to address these questions.
Acknowledgments
The authors would like to thank the members of the Harvard Polycystic Kidney Center and for helpful comments and suggestions. This work was supported by Polycystic Kidney Center P50 DK074030 (JZ, BMD) and GM55223 to BMD; North Carolina Biotechnology Center (BRG1229) and NIH (CA100869); Lineberger Comprehensive Cancer Center (Affiliate Award, University Cancer Research Fund) to TEM.
Footnotes
- ADPKD
- Autosomal Dominant Polycystic Kidney Disease
- PC1
- Polycystin-1
- MDCK
- Madin Darby canine kidney
- HEK
- human embryonic kidney
- NAAIRS
- asparagine-alanine-alanine-isoleucine-arginine-serine
- GST
- glutathione-S-transferase
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Wanfeng Yu, Renal Division Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA 02115.
Benjamin J. Ritchie, University of North Carolina at Asheville, Department of Biology, Asheville NC 28804
Xuefeng Su, Renal Division Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA 02115.
Jing Zhou, Renal Division Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA 02115.
Thomas E. Meigs, University of North Carolina at Asheville, Department of Biology, Asheville NC 28804
Bradley M. Denker, Renal Division Brigham and Women’s Hospital, and Harvard Medical School, Boston, MA 02115
REFERENCES
- [1].Parnell SC, Magenheimer BS, Maser RL, Rankin CA, Smine A, Okamoto T, Calvet JP. Biochem Biophys Res Commun. 1998;251(2):625–631. doi: 10.1006/bbrc.1998.9514. [DOI] [PubMed] [Google Scholar]
- [2].Parnell SC, Magenheimer BS, Maser RL, Zien CA, Frischauf AM, Calvet JP. J Biol Chem. 2002;277(22):19566–19572. doi: 10.1074/jbc.M201875200. [DOI] [PubMed] [Google Scholar]
- [3].Yuasa T, Takakura A, Denker BM, Venugopal B, Zhou J. Genomics. 2004;84(1):126–138. doi: 10.1016/j.ygeno.2004.02.008. [DOI] [PubMed] [Google Scholar]
- [4].Geng L, Segal Y, Pavlova A, Barros EJ, Lohning C, Lu W, Nigam SK, Frischauf AM, Reeders ST, Zhou J. Am J Physiol. 1997;272(4 Pt 2):F451–459. doi: 10.1152/ajprenal.1997.272.4.F451. [DOI] [PubMed] [Google Scholar]
- [5].Geng L, Segal Y, Peissel B, Deng N, Pei Y, Carone F, Rennke HG, Glucksmann-Kuis AM, Schneider MC, Ericsson M, Reeders ST, Zhou J. J Clin Invest. 1996;98(12):2674–2682. doi: 10.1172/JCI119090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, Ingber DE, Zhou J. Nat Genet. 2003;33(2):129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- [7].Takakura A, Contrino L, Beck AW, Zhou J. J Am Soc Nephrol. 2008;19(12):2351–2363. doi: 10.1681/ASN.2007101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Takakura A, Contrino L, Zhou X, Bonventre JV, Sun Y, Humphreys BD, Zhou J. Hum Mol Genet. 2009;18(14):2523–2531. doi: 10.1093/hmg/ddp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lanoix J, D’Agati V, Szabolcs M, Trudel M. Oncogene. 1996;13(6):1153–1160. [PubMed] [Google Scholar]
- [10].Ward CJ, Turley H, Ong AC, Comley M, Biddolph S, Chetty R, Ratcliffe PJ, Gattner K, Harris PC. Proc Natl Acad Sci U S A. 1996;93(4):1524–1528. doi: 10.1073/pnas.93.4.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lu W, Peissel B, Babakhanlou H, Pavlova A, Geng L, Fan X, Larson C, Brent G, Zhou J. Nat Genet. 1997;17(2):179–181. doi: 10.1038/ng1097-179. [DOI] [PubMed] [Google Scholar]
- [12].Zhou J. Annu Rev Physiol. 2009;71:83–113. doi: 10.1146/annurev.physiol.70.113006.100621. [DOI] [PubMed] [Google Scholar]
- [13].Kurbegovic A, Cote O, Couillard M, Ward CJ, Harris PC, Trudel M. Hum Mol Genet. 19(7):1174–1189. doi: 10.1093/hmg/ddp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Thivierge C, Kurbegovic A, Couillard M, Guillaume R, Cote O, Trudel M. Mol Cell Biol. 2006;26(4):1538–1548. doi: 10.1128/MCB.26.4.1538-1548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ong AC, Ward CJ, Butler RJ, Biddolph S, Bowker C, Torra R, Pei Y, Harris PC. Am J Pathol. 1999;154(6):1721–1729. doi: 10.1016/S0002-9440(10)65428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Woo D. N Engl J Med. 1995;333(1):18–25. doi: 10.1056/NEJM199507063330104. [DOI] [PubMed] [Google Scholar]
- [17].Nishio S, Hatano M, Nagata M, Horie S, Koike T, Tokuhisa T, Mochizuki T. J Clin Invest. 2005;115(4):910–918. doi: 10.1172/JCI22850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yu W, Kong T, Beaudry S, Tran M, Negoro H, Yanamadala V, Denker BM. J Biol Chem. 2010;285(14):10243–10251. doi: 10.1074/jbc.M109.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Delmas P, Nomura H, Li X, Lakkis M, Luo Y, Segal Y, Fernandez-Fernandez JM, Harris P, Frischauf AM, Brown DA, Zhou J. J Biol Chem. 2002;277(13):11276–11283. doi: 10.1074/jbc.M110483200. [DOI] [PubMed] [Google Scholar]
- [20].Luo Y, Vassilev PM, Li X, Kawanabe Y, Zhou J. Mol Cell Biol. 2003;23(7):2600–2607. doi: 10.1128/MCB.23.7.2600-2607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim E, Arnould T, Sellin L, Benzing T, Comella N, Kocher O, Tsiokas L, Sukhatme VP, Walz G. Proc Natl Acad Sci U S A. 1999;96(11):6371–6376. doi: 10.1073/pnas.96.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Meigs TE, Juneja J, DeMarco CT, Stemmle LN, Kaplan DD, Casey PJ. J Biol Chem. 2005;280(18):18049–18055. doi: 10.1074/jbc.M500445200. [DOI] [PubMed] [Google Scholar]
- [23].Zhu D, Tate RI, Ruediger R, Meigs TE, Denker BM. Mol Pharmacol. 2007;71(5):1268–1276. doi: 10.1124/mol.106.033555. [DOI] [PubMed] [Google Scholar]
- [24].Kreutz B, Yau DM, Nance MR, Tanabe S, Tesmer JJ, Kozasa T. Biochemistry. 2006;45(1):167–174. doi: 10.1021/bi051729t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wilson IA, Haft DH, Getzoff ED, Tainer JA, Lerner RA, Brenner S. Proc Natl Acad Sci U S A. 1985;82(16):5255–5259. doi: 10.1073/pnas.82.16.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Waheed AA, Jones TL. J Biol Chem. 2002;277(36):32409–32412. doi: 10.1074/jbc.C200383200. [DOI] [PubMed] [Google Scholar]
- [27].Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. Science. 1998;280(5372):2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- [28].Stemmle LN, Fields TA, Casey PJ. Mol Pharmacol. 2006;70(4):1461–1468. doi: 10.1124/mol.106.023705. [DOI] [PubMed] [Google Scholar]
- [29].Yamaguchi Y, Katoh H, Mori K, Negishi M. Curr Biol. 2002;12(15):1353–1358. doi: 10.1016/s0960-9822(02)01034-5. [DOI] [PubMed] [Google Scholar]
- [30].Boca M, Distefano G, Qian F, Bhunia AK, Germino GG, Boletta A. J Am Soc Nephrol. 2006;17(3):637–647. doi: 10.1681/ASN.2005050534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yanamadala V, Negoro H, Gunaratnam L, Kong T, Denker BM. J Biol Chem. 2007;282(33):24352–24363. doi: 10.1074/jbc.M702804200. [DOI] [PubMed] [Google Scholar]
- [32].Low SH, Vasanth S, Larson CH, Mukherjee S, Sharma N, Kinter MT, Kane ME, Obara T, Weimbs T. Dev Cell. 2006;10(1):57–69. doi: 10.1016/j.devcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- [33].Iiri T, Bell SM, Baranski TJ, Fujita T, Bourne HR. Proc Natl Acad Sci U S A. 1999;96(2):499–504. doi: 10.1073/pnas.96.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhu D, Kosik KS, Meigs TE, Yanamadala V, Denker BM. J Biol Chem. 2004;279(53):54983–54986. doi: 10.1074/jbc.C400508200. [DOI] [PubMed] [Google Scholar]
- [35].Fukuhara S, Chikumi H, Gutkind JS. FEBS Lett. 2000;485(2-3):183–188. doi: 10.1016/s0014-5793(00)02224-9. [DOI] [PubMed] [Google Scholar]
- [36].Oldham WM, Hamm HE. Nat Rev Mol Cell Biol. 2008;9(1):60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- [37].Markby DW, Onrust R, Bourne HR. Science. 1993;262(5141):1895–1901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- [38].Gundersen RE, You J, Rauch S, Farnham K, McCarty C, Willis N, Prince A. Biochim Biophys Acta. 2005;1722(3):262–270. doi: 10.1016/j.bbagen.2004.12.018. [DOI] [PubMed] [Google Scholar]
- [39].Tall GG, Krumins AM, Gilman AG. J Biol Chem. 2003;278(10):8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- [40].Garcia-Hoz C, Sanchez-Fernandez G, Diaz-Meco MT, Moscat J, Mayor F, Ribas C. J. Biol. Chem. 2010;285(18):13480–13489. doi: 10.1074/jbc.M109.098699. [DOI] [PMC free article] [PubMed] [Google Scholar]