Abstract
Vesicle fusion in eukaryotes is thought to involve the assembly of a highly conserved family of proteins termed soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) into a highly stable parallel four-helix bundle. We have used intermolecular single-molecule fluorescence resonance energy transfer to characterize preassembled neuronal SNARE complexes consisting of syntaxin, synaptobrevin, and synaptosome-associated protein of 25 kDa on deposited lipid bilayers. Surprisingly, we found a mixture of parallel as well as antiparallel configurations involving the SNARE motifs of syntaxin and synaptobrevin as well as those of syntaxin and synaptosome-associated protein of 25 kDa. The subpopulation with the parallel four-helix bundle configuration could be greatly enriched by an additional purification step in the presence of denaturant, indicating that the parallel configuration is the energetically most favorable state. Interconversion between the configurations was not observed. From this observation, we infer the conversion rate to be <1.5 h–1. The existence of antiparallel configurations suggests a regulatory role of chaperones, such as N-ethylmaleimide-sensitive factor, or the membrane environment during SNARE complex assembly in vivo, and it could be a partial explanation for the relatively slow rates of vesicle fusion observed by reconstituted fusion experiments in vitro.
All membrane fusion events in eukaryotes are thought to involve a highly conserved family of proteins termed soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) (1, 2). The neuronal SNAREs syntaxin, synaptosome-associated protein of 25 kDa (SNAP-25), and synaptobrevin are involved in the Ca2+-dependent fusion of synaptic vesicles with the presynaptic plasma membrane (3). Before synaptic vesicle docking, the individual SNAREs, syntaxin and SNAP-25, are primarily found on the plasma membrane, where they may exist as unfolded monomers, preformed binary complexes, or in complex with chaperones. At some point during docking and fusion of synaptic vesicles, heterotrimeric SNARE complexes form between syntaxin, SNAP-25, and the synaptic vesicle SNARE, synaptobrevin. SNARE complex formation is thought to occur first in trans with proteins on opposite membranes, and end with formation of a cis complex with proteins residing in the same membrane. Thus, SNARE complex crystal structures (4–7) probably represent the state of the SNARE complex at the endpoint of the fusion reaction (8).
The inhibition of neurotransmitter release after the specific cleavage of any one of the SNAREs by clostridial neurotoxin proteases supports the fundamental role of the SNARE proteins in synaptic vesicle fusion (9, 10). However, whereas high concentrations of SNAREs have been shown to be sufficient to fuse synthetic liposomes in vitro (11), the molecular relationship between SNARE complex formation and Ca2+-triggered vesicle–membrane fusion in vivo remains unclear (12).
Syntaxin, SNAP-25, and synaptobrevin have been the subject of extensive biochemical characterization that has generally focused on the soluble cytoplasmic fragments of these proteins that form the protease-resistant core of the complex (13). These fragments contain heptad repeats, the so-called SNARE motifs, that have a high propensity for coiled-coil formation (14, 15). Studies of the assembly reaction by circular dichroism and NMR revealed that the SNAREs are mostly unstructured in isolation and acquire α-helicity on complex formation (16–20). In binary combinations, the SNAREs associate with each other relatively weakly, with dissociation constants among the various combinations estimated to range from one-half to a few μM (21). However, in a heterotrimer combination (1:1:1 ratio), the resulting complex gains noteworthy stability (17): SDS resistance, a >90°C melting temperature, and enhanced resistance to proteolysis, including to the aforementioned clostridial neurotoxin proteases (22).
Structural investigations of purified neuronal SNARE complexes using deep-etch electron microscopy (23), electron spin resonance (24), and x-ray crystallography (4, 6, 7) revealed parallel configurations for the SNARE motifs, which implies that the transmembrane domains at the C termini of syntaxin and synaptobrevin have to be in close proximity. The parallel configuration thus strongly restrains the maximum distance possible between the target and vesicle membranes. Taken together, these results supported a model of vesicle–membrane fusion that is now commonly referred to as the zipper model (1, 11, 23, 25), wherein directional folding of parallel SNAREs, from the N to the C terminus, drives membrane fusion.
In addition to the well studied heterotrimeric neuronal and yeast plasma membrane SNARE complexes, other homo- and heteromeric interactions between individual SNAREs have been found and characterized (26–28). These alternate complexes are less stable than the heterotrimeric SNARE complex, but the relevance of these interactions is unknown. Additionally, SNARE interactions have been shown to be promiscuous in that SNAREs from different transport pathways, which would not be expected to form complexes in vivo, readily form noncognate complexes in vitro with stability similar to that of cognate SNARE complexes (29, 30).
Here, we sought to identify and characterize the different states and configurations produced by SNARE complex assembly by using single-molecule techniques. We have used fluorescence resonance energy transfer (FRET) (31, 32), which is capable of determining the overall molecular configuration of the SNARE complexes. To identify the mixture of complexes present, it was crucial to resolve subpopulations within a sample by single-molecule methods (33–37). We find that the unregulated assembly of SNAREs results in significant fractions of both parallel and antiparallel configurations of the individual SNARE motifs.
Methods
cDNA for full-length syntaxin-1A was cloned from a rat brain cDNA library (Clontech) by using standard methods and was subcloned into pet28a (Novagen). Rat SNAP-25A cDNA and rat synaptobrevin-II, residues 1–96, subcloned into pet28a, were described (13). For the purpose of site-specific labeling, a series of point mutations was introduced. First, the cysteines occurring in wild-type SNAREs were mutated to serine (syntaxin, Cys271Ser and Cys272Ser; synaptobrevin, Cys103Ser; and SNAP-25A, Cys84Ser, Cys85Ser, Cys90Ser, and Cys92Ser) by using the Quick Change mutagenesis kit (Stratagene). Single cysteines were then introduced into these constructs to produce a series of proteins for site-specific labeling. The mutations in syntaxin were Ser193Cys and Ser249Cys. For synaptobrevin, the mutations were Ser28Cys and Ser61Cys. For SNAP-25A, the mutations were Gln20Cys and Lys76Cys.
Full-length syntaxin-1A, full-length SNAP-25A, and the cytosolic domain of synaptobrevin-II were expressed and purified separately. All proteins were expressed in Escherichia coli BL21 (DE3) in Terrific Broth media as described (13, 16, 17). SNAP-25A and synaptobrevin-II were purified by using Ni-nitrilotriacetic acid (NTA) agarose. Syntaxin was purified on Ni-NTA agarose in PBS containing 0.5% Triton X-100 and 0.5% sarcosyl (38) and eluted in PBS containing 1 mM dodecyl maltoside and 250 mM imidazole. Syntaxin was further purified by anion exchange chromatography on monoQ resin (Amersham Biosciences) in Tris·HCl, pH 8.2, with 1 mM dodecyl maltoside. Purified proteins were labeled with maleimide derivatives of Cy dye (Amersham Biosciences) or Alexa dye (Molecular Probes, Eugene, OR) after gel filtration on PD-10 columns (Amersham Biosciences) into labeling buffer (PBS, pH 7.4, containing 0.1 mM Tris(2-carboxyethyl)phosphine). Labeling buffer for syntaxin included 1 mM dodecyl maltoside. SNAP-25 and synaptobrevin 1–96 were purified away from free dye by gel filtration, followed by dialysis. Syntaxin was purified away from free dye by using Ni-NTA agarose chromatography. Histidine tags were removed by incubation with thrombin overnight at 4°C.
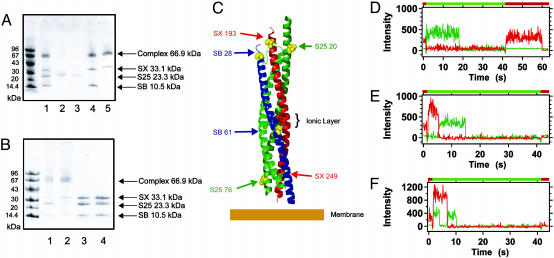
SNARE complexes were formed by adding SNAP-25 to syntaxin followed by the addition of synaptobrevin (13, 16, 17). It has been shown that fusion-competent SNARE complexes are obtained by mixing SNARE components (39). The reaction was allowed to proceed at 4°C. Formation of SNARE complex was complete by 48 h (data not shown). SNARE complexes were assembled in 20 mM Tris·HCl, pH 8.2/200 mM NaCl/1 mM dodecyl maltoside (Tris-DM buffer) unless indicated otherwise. Proteins were mixed at a ratio of 1:2:5 (syntaxin:SNAP-25:synaptobrevin) unless indicated otherwise. Mixing ratios do effect the observed populations: for example, 1:1:1 yields a population with ≈54% FRET = 1 and 46% FRET = 0 for the syntaxin Ser193Cys-acceptor(Cy5), unlabeled SNAP-25, synaptobrevin 1–96 Ser28Cys–donor(Cy3) complex. For the purposes of additional purification, the histidine tag was left on synaptobrevin 1–96. The assembly reaction was rebound to Ni-NTA agarose and washed extensively at room temperature with Tris-DM buffer. Additional washing with Tris-DM buffer containing 7.5 M urea was used if indicated. When urea was used, it was washed out of the column with Tris-DM buffer and then the sample was eluted. All samples were further purified on a monoQ resin (Amersham Biosciences) in 20 mM Tri·HCl, pH 8.2, containing 100 mM β-d-octyl-glucoside. An SDS/PAGE gel of this assembly reaction without boiling is shown in Fig. 1A, lane 1. SDS-stable SNARE complex is clearly present, as are residual monomeric SNAREs. The reaction was incubated with Ni-NTA agarose and washed extensively, but only SNAP-25 was visible in the flow-through (Fig. 1 A, lane 2). Samples were further purified by anion exchange chromatography (peak fraction is shown in Fig. 1 A, lane 5). The peak fractions containing ternary SNARE complexes were used for further analysis. Representative unboiled and boiled SDS/PAGE of purified ternary SNARE complexes in the presence and absence of urea treatment are shown in Fig. 1B. Positions of labels are shown in Fig. 1C.
Fig. 1.
Purification and characterization of SNARE complexes. (A) SNARE complex purification by using denaturant. Unboiled samples from intermediate stages of SNARE complex purification were analyzed by SDS/PAGE on 8–25% gradient Phast gels (Amersham Biosciences) using Coomassie staining. Markers are Amersham Biosciences low molecular mass standards. Lane 1, unpurified SNARE complex assembly reaction. Lane 2, unbound material after incubation with Ni-NTA agarose. Only the synaptobrevin cytoplasmic domain retains a histidine tag to allow direct interaction with the beads. Lane 3, 7.5 M urea wash. Lane 4, imidazole elution from Ni-NTA agarose. Lane 5, peak fraction from anion exchange using a monoQ column. Samples bound to Ni-NTA agarose were further washed with buffer containing 7.5 M urea, which displaced additional SNAP-25 and some syntaxin (lane 3). The column was subsequently washed with urea-free buffer and eluted with imidazole (lane 4). (B) Unboiled and boiled SDS/PAGE of purified ternary SNARE complexes in the presence and absence of urea treatment. For SDS/PAGE, 8–25% gradient Phast gels (Amersham Biosciences) were used with Coomassie staining. Markers are Amersham Biosciences low molecular mass markers; lanes 1 and 2 show unboiled samples, and lanes 3 and 4 show boiled samples. Lanes 1 and 3 represent the purification with denaturant step, and lanes 2 and 4 represent samples with intervening denaturant step. The gels were normalized to an absorbance of 0.3 absorbance units at 280 nm in a 1-cm pathlength cuvette. Both samples contain all three SNARE components in roughly the same stoichiometric ratio (compare lanes 3 and 4 corresponding to the boiled samples). However, the amount of SDS-resistant complex is somewhat diminished for the samples purified in the presence of denaturant (compare lanes 1 and 2). (C) Ribbon diagram of the core SNARE complex with labeling sites and the ionic layer shown as spheres. SB, syntapobrevin; SX, syntaxin; S25, SNAP-25. The complex used in this study included additional residues (not shown): the flexible linker between the two helices of SNAP25, the transmembrane and regulatory three-helix bundle domains of syntaxin, and the N-terminal disordered region of synaptobrevin. Note that experiments were carried out on selected pairs of the indicated sites. The membrane is not drawn to scale, but it serves to illustrate the relationship between the cis complex and the phospholipid bilayer. Graphics were generated by using pymol (43). (D–F) Single-molecule time traces of donor (green) and acceptor (red) dye fluorescence intensities from SNARE complexes in a supported lipid bilayer. For the first second, 635-nm illumination was used to directly excite the acceptor dye. Between 1.5 and 42 sec, the illumination was switched to 532 nm, which directly excites the donor dye. During this time, the samples were analyzed for the presence of FRET. Red laser light (635 nm) was used for times >42.5 sec to determine whether the donor dye had been photobleached. (D) This time trace shows a FRET = 0 example: the acceptor is present with red light, the donor emits only under green light with eventual donor photobleaching, and, finally, the acceptor is present during the second red-light phase with eventual photobleaching. (E) This time trace shows typical FRET = 1 type behavior: the acceptor presence is verified with red light, followed by high acceptor and low donor intensity under green light, accompanied with anticorrelated photobleaching of the acceptor and recovery of the donor, and, finally, photobleaching of the donor. (F) An example of aggregation: a single FRET = 0 donor coincides with a complete FRET donor/acceptor pair. The presence of multiple donor and acceptor dyes was deduced from such multiple plateaus and single-step photobleaching events. Shown are representative examples from the system syntaxin–Ser193Cys-acceptor(Cy5)–SNAP-25 Gln20Cys-donor(Cy3)–synaptobrevin 1–96 without urea treatment, but all combinations showed similar behavior.
Egg phosphatidylcholine (Avanti Polar Lipids) was dried from chloroform, resuspended at 30 mg/ml in 20 mM Tris·HCl, pH 8.2/200 mM NaCl, and passed 29 times through a 50-nm pore membrane (Whatman) by using an Avanti miniextruder. Protein samples in 100 mM β-octyl glucoside were mixed at a 1:4 ratio with liposomes for 30 min on ice. After a 1:1 ratio dilution in detergent-free buffer, which drops β-octyl glucoside below the reported critical micelle concentration, the samples were loaded onto a Sepharose CL4B (Amersham Biosciences) column and the void volume fraction was collected. Given the long off rates for the SNARE complex, we do not expect the reconstitution protocol to affect the assembled SNARE complexes.
Quartz microscope slides were cleaned by 15-min incubations in a bath sonicator in the following sequence of solvents: Alconox (White Plains, NY) detergent/water, acetone, ethanol, 1 M KOH, ethanol, 1 M KOH, and deionized water. Cleaned slides were stored in deionized water. Before use, the slides were flamed in a propane torch, and a channel was constructed on their surface by using a no. 1.5 coverslip, UV curing optical adhesive (Norland, Cranbury, NJ), and Scotch tape (3M Co.) as a spacer. Buffers could be exchanged in the channel using holes drilled through the slide into the channel.
Supported lipid bilayers containing the SNAREs were formed on the quartz by incubating appropriate proteoliposomes at room temperature in the channel at 3 mg/ml for 10 min, followed by protein-free liposomes at 15 mg/ml for 1 h. No difference was observed when observations were made on SNARE complexes inserted into intact, biotinylated liposomes immobilized through a biotin–streptavidin interaction on a biotin-polyethylene glycol-coated surface, rather than condensation into a supported bilayer (ref. 37 and data not shown).
All microscopy was performed at room temperature in 20 mM Tris·HCl, pH 7.8/200 mM NaCl/1 mM DTT/2% glucose (wt/vol), supplemented with an enzymatic oxygen scavenger mixture consisting of glucose oxidase (Sigma) at 100 units/ml and catalase (Sigma) at 1,000 units/ml. The microscopy buffer additionally contained the triplet state quenchers diazabicyclo[2.2.2]octane (DABCO) (10 mM) and cyclooctatetraene (100 μM) to further decrease the rate of dye photobleaching.
Dyes were excited with focused laser light at 532 and 635 nm introduced to the bilayer by prism-type total internal reflection at the quartz–water interface. Fluorescence emission was collected with a 1.2-n.a. water immersion objective. The ratio of protein to lipid was adjusted to provide a protein density of 20–30 per 1,000 μm2 in the supported bilayer, thus allowing individual dye molecules to be distinctly imaged by the microscope. The mobility of the SNARE complexes in deposited bilayers was sufficiently low to allow observation for 1–2 min without losing track of the identity of individual complexes. A 650-nm dichroic mirror spectrally split the image, and each of the two spectral bands was further filtered to pass dye fluorescence and block the laser colors. The two spectrally resolved copies of the microscope image were relayed onto halves of an intensified Pentamax charged-coupled device camera (Roper Scientific, Ottobrunn, Germany) and recorded at 10 frames per second. Measured emission intensity values were corrected for background fluorescence as Ĩ = Imeasured – Ibackground. FRET efficiency was calculated from the background corrected measurements of the acceptor emission intensity (Ĩacceptor) and the donor emission intensity (Ĩdonor) as FRET = Ĩacceptor/(Ĩacceptor + Ĩdonor). Colocalization and FRET measurements were made on the same sets of molecules in the bilayer by using a shuttering system to switch the two laser colors and alternately excite the donor and acceptor dyes.
Despite attempts to optimize the labeling reactions, variable fractions ranging from 20% to 85% of the proteins failed to incorporate a dye molecule as assessed by absorption spectroscopy. Therefore, complexes containing an acceptor dye were first identified by briefly flashing a red laser at 635 nm that directly excites only the acceptor. Illumination was then switched to a green laser at 532 nm to excite the donor (Fig. 1D). Donor-only emission indicated colocalization with FRET = 0 (Fig. 1D), whereas emerging acceptor emission indicated FRET between the dyes (Fig. 1E). Single-molecule observation allowed only the subset of complexes that had been labeled with both a donor and acceptor to be considered. Illumination by the green laser was carried out continuously for 40 sec to ensure eventual photobleaching of the donors. Finally, illumination was switched back to the red laser to photobleach the acceptor dyes. By counting plateaus in time traces, we could quantify the presence of multiple dyes due to aggregation of SNARE complexes (Fig. 1F). Our specific labeling strategy made it very unlikely that multiple copies of donors or acceptors are attached to a single SNARE complex. Furthermore, the single-molecule approach allowed individual instances of aggregation where there are multiple dyes seen to be rejected.
Anticorrelated intensity jumps within a single time bin (100 msec) provided definite proof of the existence of FRET between donor and acceptor dyes (Fig. 1E). If an anticorrelated event permanently diminished the acceptor intensity and simultaneously increased the donor intensity, we interpreted the event as photobleaching of the acceptor dye followed by increase of the donor intensity due to the removal of FRET. However, there are other possible behaviors that were still counted as FRET. For instance, if the donor undergoes photobleaching, it causes the high acceptor emission to vanish, but without anticorrelated time trace behavior. The fact that intensity levels for these and other possible behaviors are consistent with the levels seen for the anticorrelated traces was used as evidence for the interpretation of such cases as acceptable FRET events.
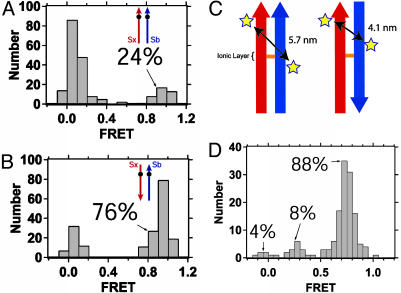
For all accepted spots with a single donor and a single acceptor, the FRET value was calculated from the average intensities during the first 0.5 sec of green illumination and compiled into a histogram. The numbers tagging each histogram were derived by finding the fraction with FRET >0.5 from the histograms for the extreme N and C label sites where the data naturally partitions into peaks at low and high FRET (e.g., Fig. 2 A and B). The numbers tagging each peak in Fig. 2D reflect the fraction of events in each of the Gaussian-shaped peaks in that histogram.
Fig. 2.
Distributions of single-molecule FRET values for ternary SNARE complexes containing a donor-labeled synaptobrevin and acceptor-labeled syntaxin. Bars in the histogram represent the number of donor and acceptor pairs at a given FRET efficiency. The orientation of the complexes and the position of the dye are illustrated schematically by arrows. The arrowheads are placed at the N-termini. Syntaxin is red and synaptobrevin is blue. Positions of the labels are denoted as filled circles. The numbers tagging each histogram indicate the fraction of the population in the FRET = 1 state. Complexes are: Syntaxin Ser193Cys–acceptor(Cy5), unlabeled SNAP-25, and synaptobrevin 1–96 Ser28Cys–donor(Cy3) (A); and syntaxin Ser249Cys–acceptor(Alexa647), unlabeled SNAP-25, and synaptobrevin 1–96 Ser28Cys–donor(Cy3) (B). (C) A schematic illustration of the change in the position of the synaptobrevin Ser-61 middle labeling site relative to syntaxin Ser-193 in the parallel (Left) and antiparallel (Right) configurations. Positions of the labeling sites are denoted by stars. (D) Ser193Cys-donor (Alexa 555), SNAP-25, and synaptobrevin 1–96 Ser61Cys-acceptor(Cy5). FRET values clustered around 0.3 and 0.75 apparent in the population distribution are consistent with a parallel and antiparallel incorporation of the α-helix for synaptobrevin while maintaining the alignment of the ionic layer. The antiparallel population dominates, strongly suggesting that a major mode of misfolding is antiparallel incorporation of the synaptobrevin helix relative to syntaxin. The value of 88% antiparallel incorporation of synaptobrevin labeled at residue 61 is somewhat larger than the 76% antiparallel incorporation of synaptobrevin labeled at residue 28. This difference could be due to the different ratio of SNAREs during this assembly reaction [a 1:1.5:1.5 ratio (syntaxin:SNAP-25:synaptobrevin) as opposed to a 1:2:5 ratio], because the assembly conditions can affect the resulting distribution. Furthermore, EPR studies of SNARE complexes containing a spin label at residue 61 of synaptobrevin observed pronounced immobile peaks, suggesting that labels at this residue may have interactions with surrounding residues (26). These interactions could change the relative stability of the parallel and antiparallel complexes, thus altering the resultant distribution.
The combination of incomplete dye labeling of the protein samples and the presence of protein aggregation leads to an overcounting of colocalized complexes that have FRET = 0. A dimer containing one complex with one donor and another complex with one acceptor is indistinguishable from a monomeric complex with one donor and one acceptor where the dyes are separated sufficiently to provide FRET = 0. There are various ways to estimate corrections for this effect. Measurements using two N-terminal labels are expected to yield complementary results to corresponding measurements using one N- and one C-terminal label. For pairs of experiments, such as reported in Figs. 2 A and B and 3, we would expect our observations to be consistent in the absence of measurement errors. For data that are not internally consistent, we can thus estimate the fraction of incompletely labeled aggregates: Fig. 2 A and B, aggregation = 0%; Fig. 3, aggregation = 24%. Furthermore, in Fig. 2D, one can directly estimate that aggregation = 4%. Errors designating individual time trace into categories during the sorting of thousands of time traces will alter the absolute values of fractional populations. In cases with low labeling efficiencies (donor · acceptor = 0.36 · 0.68 = 25% and 0.15 · 0.65 = 9% for our best and worst cases, respectively), such errors become more significant. Thus, incomplete labeling adds additional uncertainties to our estimates of aggregation and FRET assignments.
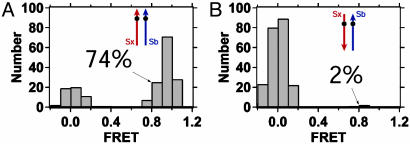
Fig. 3.
Distributions of single-molecule FRET values for SNARE complexes resistant to denaturation. An additional purification step was used to select for SNARE complexes stable in 7.5 M urea for samples used in Fig. 2 A and B. For both samples, the parallel configuration predominates after urea treatment, indicating a higher stability relative to the antiparallel configuration.
A simple model using the measured labeling efficiencies to estimate aggregation factors from observed ratios of spots with single donor-labeled synaptobrevins to spots with two donor-labeled synaptobrevins (data not shown) yields estimates of the falsely colocalized population of complexes ranging from 5% to 13% for the samples considered above. Thus, the absolute values of the fractional populations reported in Figs. 2, 3, 4 are inaccurate by typically 3–13%.
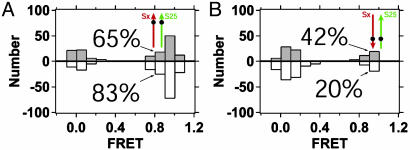
Fig. 4.
Distributions of single-molecule FRET values for ternary SNARE complexes containing a donor-labeled N-terminal helix of SNAP-25 and acceptor-labeled syntaxin. Syntaxin is red and SNAP-25 is green. The upward, shaded bars represent complexes assembled in solution and reconstituted directly for observation. The downward, white bars represent the same samples after selection for stability in 7.5 M urea (described in the text and in Methods). Complexes are as follows: Syntaxin Ser193Cys–acceptor(Cy5), SNAP-25 Gln20Cys–donor(Cy3), and synaptobrevin 1–96 (A); and syntaxin Ser193Cys–acceptor(Cy5), SNAP-25-Lys76Cys–donor(Cy3), and synaptobrevin 1–96 (B).
Results
Full-length syntaxin-1A, including the transmembrane domain, full-length SNAP-25A, and the cytosolic domain of synaptobrevin-II were expressed and purified separately, the neuronal SNARE complex was assembled (Fig. 1 A and B), and was then reconstituted into proteoliposomes. Supported lipid bilayers containing the SNARE complex were formed on a quartz slide by condensation of the proteoliposomes. One SNARE motif in each complex was labeled with a donor dye (Cy3 or Alexa 555) and another SNARE motif was labeled with an acceptor dye (Cy5 or Alexa 647) before assembly and purification of the complexes. The Förster radius, R0, for these donor-acceptor pairs is ≈5 nm. Therefore, with guidance from the x-ray crystal structure of the neuronal SNARE complex (4, 7), specific surface-exposed labeling sites were selected to produce either FRET = 1 or FRET = 0 by being at the same or opposite ends of the 12-nm-long helical bundle, respectively (Fig. 1C). In addition, a synaptobrevin-labeling site located near the center of the complex was used.
Single-molecule fluorescence techniques were used to monitor both donor and acceptor emission intensity as a function of time for complexes reconstituted in a single deposited bilayer. Thousands of intensity time traces were analyzed and categorized into instances of FRET = 0, FRET = 1, and aggregation (Fig. 1 D–F). Based on the crystal structure of the neuronal complex (4, 7), if the donor and acceptor dyes are at the same end of the helical bundle, the labeled residues are 1- to 2-nm apart and should result in FRET = 1. If the labeled residues are at the opposite ends of the bundle, the dyes are 7- to 9-nm apart and FRET = 0. Fig. 2 A shows the distribution of FRET values for individual SNARE complexes with a donor on residue 28 near the N terminus of the synaptobrevin core helix and an acceptor on residue 193 near the N terminus of the syntaxin core helix (see Fig. 1C.). The histogram shows two populations clustering around 0 and 1, indicating FRET = 0 and FRET = 1, respectively. Only 24% of colocalized donor and acceptor dyes are giving raise to FRET = 1, corresponding to a parallel configuration of syntaxin and synaptobrevin. Consequently, the remaining 76% of the complexes with FRET = 0 consist of antiparallel configurations along with a smaller portion of aggregates of singly labeled complexes.
To obtain a more accurate estimate of the size of the antiparallel population, the labeling site for syntaxin was moved to residue 249 at the C terminus of the syntaxin core helix. An antiparallel configuration should then produce FRET = 1 for the synaptobrevin 28 · syntaxin 249 labeling pair and can be distinguished from aggregates of singly labeled SNARE complexes. Seventy-six percent of the resulting complexes show FRET = 1 (Fig. 2B) corresponding to an antiparallel configuration of syntaxin and synaptobrevin. That both labeling experiments result in the same percentage of antiparallel configurations is an indication that dye labeling does not affect the assembly reaction. By inspection of the intensity time traces, we do not observe any events that would indicate an interconversion between the parallel and antiparallel configurations (no interconversions in 427 FRET = 1 antiparallel complexes within 1- to 10-min observation implies the rate of interconversion is <1.5 h–1). Thus, on the hour time scale, both the parallel and antiparallel configurations are stable.
To further confirm the observed antiparallel configuration for synaptobrevin relative to syntaxin, we formed SNARE complexes containing the acceptor at residue 61 of synaptobrevin and the donor at residue 193 of syntaxin (see Fig. 1C.). For the parallel configuration observed in the crystal structure (4, 7), the two labeling sites separated by 5.7 nm would produce a calculated FRET value of 0.31. If synaptobrevin and syntaxin were in an antiparallel configuration, while maintaining the register of the buried ionic layer, the distance would decrease to 4.1 nm, producing a calculated FRET value of 0.77 (Fig. 2C). The measured FRET values cluster into groups centered at 0.3 and 0.75, which is consistent with parallel and antiparallel configurations, respectively, along with a smaller subpopulation of complexes with FRET = 0 (Fig. 2D). Thus, our results are consistent with the maintenance of the ionic layer for the antiparallel configuration.
A hallmark of the neuronal SNARE complex is its high thermal and chemical stability (16, 17). The existence of both parallel and antiparallel configurations of SNARE complexes on unregulated assembly implies an energy landscape with two stable local minima. To investigate the relative stability of the configurations, we determined the population distributions after adding a denaturant step during purification (Fig. 1 A, lanes 3 and 4, and Fig. 1B). These samples were then reconstituted and analyzed for single-molecule FRET. For the synaptobrevin 28 · syntaxin 193 labeling pair, 74% of complexes resistant to urea denaturation show FRET = 1, which is in agreement with a parallel configuration (Fig. 3A). On reversal of the syntaxin-labeling site, only 2% of the urea-resistant complexes show FRET = 1 (Fig. 3B), corresponding to the antiparallel configuration. The observation that urea treatment results in an increase in complexes showing FRET values in agreement with the crystal structure suggests that the antiparallel configuration does not have the hallmark stability of the parallel configuration.
Next, we examined the orientation of the N-terminal helix of SNAP-25 by placing the donor at residue 20 of SNAP-25, while the acceptor remained at residue 193 of syntaxin (Fig. 4A). Without urea treatment, 65% of the complexes showed FRET = 1, as expected for a parallel configuration (Fig. 4A, shaded bars). Of the complexes stable in urea, 83% showed FRET = 1 (Fig. 4A, white bars), which is consistent with a parallel configuration. Unlike synaptobrevin, the majority of the complexes adopt a parallel configuration of the N-terminal helix of SNAP-25 relative to syntaxin. This trend is also observed for the binary syntaxin–SNAP-25 complex for both SNAP-25 helices (data not shown). Similarly, when the donor was located at residue 76 near the C terminus of the first SNAP-25 core helix and the acceptor was at residue 193 of syntaxin, 42% of complexes obtained without urea treatment showed FRET = 1, whereas only 20% of the urea-resistant complexes retained FRET = 1 (Fig. 4B). That the antiparallel configuration of SNAP-25 relative to syntaxin could not be reduced <20%, suggests that the stability of the complex is not as directly linked to the SNAP-25 configuration as was observed for synaptobrevin.
Discussion
We found that SNARE proteins spontaneously assemble into mixtures of parallel and antiparallel configurations. This result was most pronounced for synaptobrevin relative to syntaxin (Fig. 2 A and B), and to a somewhat lesser degree for the N-terminal SNARE motif of SNAP-25 relative to syntaxin (Fig. 4 A and B). However, the parallel configuration of the complex is significantly more stable than antiparallel configurations because the parallel syntaxin–synaptobrevin subpopulation was significantly enhanced on urea treatment during purification (Fig. 3). We did not observe interconversion between the antiparallel and parallel subpopulations on the hour time scale.
The observed distribution of parallel and antiparallel configurations of SNARE complexes on unregulated assembly suggests an energy landscape with several local minima. The parallel helix bundle is one of many complexes that can form. It presumably represents the lowest free-energy state. This landscape complicates studies of the reversibility of folding transitions involving highly purified SNARE complexes, because purification takes the system far from mixing equilibrium (40). In vitro, the folding energy landscape can be manipulated through the use of denaturants to remove local minima. This finding should have significant effects on the observed kinetics and products of the assembly reaction. Indeed, bulk FRET studies of the SNARE complex, performed without extensive purification, were inconsistent with the crystal structure of the core complex, because FRET was observed both between the N termini as well as between the N terminus of synaptobrevin and the C terminus of syntaxin (25), as one would expect for a mixture of parallel and antiparallel configurations. Likewise, the nonzero FRET between syntaxin C-terminally fused to blue fluorescent protein and synaptobrevin N-terminally fused to GFP in 20 S particles N-ethylmaleimide-sensitive factor ((NSF), α-SNAP, and SNARE complexes; ref. 41), can now be explained by the partial presence of antiparallel SNARE complexes. Interestingly, when SNARE complex was obtained from brain extracts, all SNARE complexes appeared to be parallel, as observed in electron micrographs (41), suggesting that the purification scheme selected for the parallel configuration or, more likely, that the presence of cofactors or chaperones promoted the parallel configuration in vivo.
The zipper model for SNARE-catalyzed membrane fusion proposes that the energy from directional folding of the SNARE complex, from the N-terminal toward the C-terminal transmembrane domains of syntaxin and synaptobrevin, is used to overcome the repulsive forces between opposing membranes (1, 11, 23, 25). When the proteins are in opposing membranes, the parallel configuration can begin to form when the membranes are within 20 nm (N-terminal · N-terminal contact for fully extended SNAREs) and the membranes must then proceed to contacting each other on complex formation. In contrast, the antiparallel configuration can form completely when the membranes are separated by 10 nm. Presumably, the antiparallel configuration would not lead to vesicle–membrane fusion. The existence of both parallel and antiparallel configurations could be a partial explanation for the slow rates of vesicle fusion observed by reconstituted fusion experiments in vitro (11, 42). The parallel complex could be selected for in vivo by factors that regulate vesicle distance, such as the membrane environment (38), or by chaperones that affect SNARE complex assembly. Thus, the antiparallel configuration could represent an off state that could be regulated as a means of controlling release probability.
Acknowledgments
We thank James Ernst, Barry Wilk, and Anton Rozenbaum for technical assistance, and Richard Scheller, R. Bryan Sutton, and Richard Tsien for discussions. This work was supported in part by National Institutes of Health Grant 1-RO1-MH63105-01 (to A.T.B.), and grants from the National Science Foundation, the Air Force Office of Scientific Research, and the National Aeronautics and Space Administration (S.C.). K.W. acknowledges a Career Award at the Scientific Interface from the Burroughs Wellcome Foundation.
Abbreviations: SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptors; SNAP-25, synaptosome-associated protein of 25 kDa; FRET, fluorescence resonance energy transfer.
References
- 1.Jahn, R. & Südhof, T. C. (1999) Annu. Rev. Biochem. 68, 863–911. [DOI] [PubMed] [Google Scholar]
- 2.Chen, Y. A. & Scheller, R. H. (2001) Nat. Rev. Mol. Cell Biol. 2, 98–106. [DOI] [PubMed] [Google Scholar]
- 3.Südhof, T. C. (2000) Neuron 28, 317–320. [DOI] [PubMed] [Google Scholar]
- 4.Sutton, R. B., Fasshauer, D., Jahn, R. & Brunger, A. T. (1998) Nature 395, 347–353. [DOI] [PubMed] [Google Scholar]
- 5.Brunger, A. T. (2001) Curr. Opin. Struct. Biol. 11, 163–173. [DOI] [PubMed] [Google Scholar]
- 6.Antonin, W., Fasshauer, D., Becker, S., Jahn, R. & Schneider, T. R. (2002) Nat. Struct. Biol. 9, 107–111. [DOI] [PubMed] [Google Scholar]
- 7.Ernst, J. A. & Brunger, A. T. (2003) J. Biol. Chem. 278, 8630–8636. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y. A., Scales, S. J., Patel, S. M., Doung, Y. C. & Scheller, R. H. (1999) Cell 97, 165–174. [DOI] [PubMed] [Google Scholar]
- 9.Jahn, R., Hanson, P. I., Otto, H. & Ahnert-Hilger, G. (1995) Cold Spring Harbor Symp. Quant. Biol. 60, 329–335. [DOI] [PubMed] [Google Scholar]
- 10.Schiavo, G., Rossetto, O. & Montecucco, C. (1994) Semin. Cell. Biol. 5, 221–229. [DOI] [PubMed] [Google Scholar]
- 11.Weber, T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., Söllner, T. H. & Rothman, J. E. (1998) Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- 12.Ungermann, C., Sato, K. & Wickner, W. (1998) Nature 396, 543–548. [DOI] [PubMed] [Google Scholar]
- 13.Fasshauer, D., Eliason, W. K., Brunger, A. T. & Jahn, R. (1998) Biochemistry 37, 10354–10362. [DOI] [PubMed] [Google Scholar]
- 14.Chapman, E. R., An, S., Barton, N. & Jahn, R. (1994) J. Biol. Chem. 269, 27427–27432. [PubMed] [Google Scholar]
- 15.Fasshauer, D., Sutton, R. B., Brunger, A. T. & Jahn, R. (1998) Proc. Natl. Acad. Sci. USA 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fasshauer, D., Otto, H., Eliason, W. K., Jahn, R. & Brunger, A. T. (1997) J. Biol. Chem. 272, 28036–28041. [DOI] [PubMed] [Google Scholar]
- 17.Fasshauer, D., Bruns, D., Shen, B., Jahn, R. & Brunger, A. T. (1997) J. Biol. Chem. 272, 4582–4590. [DOI] [PubMed] [Google Scholar]
- 18.Rice, L. M., Brennwald, P. & Brunger, A. T. (1997) FEBS Lett. 415, 49–55. [DOI] [PubMed] [Google Scholar]
- 19.Fiebig, K. M., Rice, L. M., Pollock, E. & Brunger, A. T. (1999) Nat. Struct. Biol. 6, 117–123. [DOI] [PubMed] [Google Scholar]
- 20.Hazzard, J., Südhof, T. C. & Rizo, J. (1999) J. Biomol. NMR 14, 203–207. [DOI] [PubMed] [Google Scholar]
- 21.Pevsner, J., Hsu, S. C., Braun, J. E., Calakos, N., Ting, A. E., Bennett, M. K. & Scheller, R. H. (1994) Neuron 13, 353–361. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi, T., McMahon, H., Yamasaki, S., Binz, T., Hata, Y., Südhof, T. C. & Niemann, H. (1994) EMBO J. 13, 5051–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson, P. I., Roth, R., Morisaki, H., Jahn, R. & Heuser, J. E. (1997) Cell 90, 523–535. [DOI] [PubMed] [Google Scholar]
- 24.Poirier, M. A., Xiao, W., Macosko, J. C., Chan, C., Shin, Y. K. & Bennett, M. K. (1998) Nat. Struct. Biol. 5, 765–769. [DOI] [PubMed] [Google Scholar]
- 25.Lin, R. C. & Scheller, R. H. (1997) Neuron 19, 1087–1094. [DOI] [PubMed] [Google Scholar]
- 26.Margittai, M., Fasshauer, D., Pabst, S., Jahn, R. & Langen, R. (2001) J. Biol. Chem. 276, 13169–13177. [DOI] [PubMed] [Google Scholar]
- 27.Misura, K. M., Scheller, R. H. & Weis, W. I. (2001) J. Biol. Chem. 276, 13273–13282. [DOI] [PubMed] [Google Scholar]
- 28.Misura, K. M., Gonzalez, L. C., Jr., May, A. P., Scheller, R. H. & Weis, W. I. (2001) J. Biol. Chem. 276, 41301–41309. [DOI] [PubMed] [Google Scholar]
- 29.Fasshauer, D., Antonin, W., Margittai, M., Pabst, S. & Jahn, R. (1999) J. Biol. Chem. 274, 15440–15446. [DOI] [PubMed] [Google Scholar]
- 30.Yang, B., Gonzalez, L. C., Jr., Prekeris, R., Steegmaier, M., Advani, R. J. & Scheller, R. H. (1999) J. Biol. Chem. 274, 5649–5653. [DOI] [PubMed] [Google Scholar]
- 31.Stryer, L. & Haugland, R. P. (1967) Proc. Natl. Acad. Sci. USA 58, 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalet, X., Kapanidis, A. N., Laurence, T., Pinaud, F., Doose, S., Pflughoefft, M. & Weiss, S. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 161–182. [DOI] [PubMed] [Google Scholar]
- 33.Weiss, S. (1999) Science 283, 1676–1683. [DOI] [PubMed] [Google Scholar]
- 34.Kelley, A. M., Michalet, X. & Weiss, S. (2001) Science 292, 1671–1672. [DOI] [PubMed] [Google Scholar]
- 35.Zhuang, X., Bartley, L. E., Babcock, H. P., Russell, R., Ha, T., Herschlag, D. & Chu, S. (2000) Science 288, 2048–2051. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang, X., Kim, H., Pereira, M. J., Babcock, H. P., Walter, N. G. & Chu, S. (2002) Science 296, 1473–1476. [DOI] [PubMed] [Google Scholar]
- 37.Ha, T., Rasnik, I., Cheng, W., Babcock, H. P., Gauss, G. H., Lohman, T. M. & Chu, S. (2002) Nature 419, 638–641. [DOI] [PubMed] [Google Scholar]
- 38.Kweon, D.-H., Kim, C. S. & Shin, Y.-K. (2003) Nat. Struct. Biol. 6, 440–447. [DOI] [PubMed] [Google Scholar]
- 39.Hu, K., Carroll, J., Fedorovich, S., Rickman, C., Sukhodub, A. & Davletov, B. (2002) Nature 415, 646–650. [DOI] [PubMed] [Google Scholar]
- 40.Fasshauer, D., Antonin, W., Subramaniam, V. & Jahn, R. (2002) Nat. Struct. Biol. 9, 144–151. [DOI] [PubMed] [Google Scholar]
- 41.Hohl, T. M., Parlati, F., Wimmer, C., Rothman, J. E., Söllner, T. H. & Engelhardt, H. (1998) Mol. Cell 2, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelham, H. R. (2001) Trends Cell Biol. 11, 99–101. [DOI] [PubMed] [Google Scholar]
- 43.Delano, W. L. (2002) The PyMol User's Manual (DeLano Scientific, San Carlos, CA).