Abstract
Objective
Systemic sclerosis (SSc) is an autoimmune disease characterized by fibrosis of the skin and internal organs. Dysregulation of the immune system, including the Th1/Th2 cytokine balance, is central to the pathogenesis of SSc. This study was undertaken to investigate the hypothesis that single-nucleotide polymorphisms (SNPs) in TBX21 and STAT4, both of which are critical transcription factors that regulate the Th1/Th2 balance, are associated with SSc susceptibility.
Methods
We tested SNPs in TBX21 and STAT4 for association with SSc in 2 independent cohorts, the SSc Registry cohort (880 SSc cases and 507 controls) and the University of Texas SSc cohort (522 cases and 531 controls). Additional white control genotypes were obtained from public repositories. We also investigated for gene–gene interactions. Plasma cytokines and whole blood gene expression profiles were examined to determine functional effects of these SNPs.
Results
Multiple SNPs in TBX21 and STAT4 were found to be associated with SSc. In a combined analysis of 902 SSc patients and 4,745 controls, TT genotyping of the TBX21 rs11650354 variant revealed a recessive pattern for disease susceptibility (Pcorr = 1.4 × 10−15, odds ratio 3.37, 95% confidence interval 2.4–4.6). In an analysis of 1,039 SSc patients and 3,322 controls, the A allele of the STAT4 variant rs11889341 was associated with increased SSc susceptibility in a dominant pattern (Pcorr = 2.4 × 10−5, odds ratio 1.29, 95% confidence interval 1.2–1.5). Furthermore, we identified gene–gene interaction among the TBX21 and STAT4 variants, such that the STAT4 genotype increased the risk of SSc only in the TBX21 CC genotype group. SSc patients carrying the TBX21 CC genotype had higher interleukin-6 (IL-6) and tumor necrosis factor α levels, and those with the TT genotype had elevated IL-2, IL-5, IL-4, and IL-13 (Th2) levels, compared with controls. Whole blood expression profiles revealed dysregulation of type I interferon pathways in the CC group and T cell pathways in the TT group of the TBX21 SNP.
Conclusion
The present results, from studies of 2 independent cohorts, indicate that SNPs in TBX21 and STAT4 contribute uniquely and interactively to SSc susceptibility, leading to altered cytokine balance and immune dysregulation.
Systemic sclerosis (SSc; scleroderma) is a chronic multisystem disease of unknown etiology, which is clinically characterized by progressive fibrosis of the skin and internal organs, widespread small vessel obliteration, and autoimmunity. Although SSc is relatively uncommon, affecting ~400,000 North Americans and Europeans, the lack of disease-modifying treatment results in significant morbidity and mortality to the individual as well as substantial economic cost (1).
Central to understanding the pathogenesis of SSc is defining the genes and pathways leading to autoimmunity and inflammation, vascular damage, and extra-cellular matrix production. Several genetic polymorphisms have been associated with scleroderma in multiple case–control studies and a few family studies (2–8). Some of these genetic variants are associated with susceptibility for development of scleroderma, while others act as disease modifiers.
There is substantial evidence indicating that dysregulation is a vital process in the pathogenesis of SSc, particularly early in the disease process. An indicator of immune dysregulation in SSc patients is the presence of disease-specific, mutually exclusive autoantibodies. These antibodies, most commonly anticentromere (ACA), anti–topoisomerase I (anti–topo I), and anti–RNA polymerase III (anti–RNAP III), identify relatively distinct clinical subgroups, (9–13). There have been conflicting reports regarding the role of T cells and Th1/Th2 cytokine balance in SSc (14). Some studies have provided evidence in support of the notion of Th1 activation in the peripheral blood with increased levels of interferon-γ (IFNγ) (15–17), while others indicate a preferential involvement of Th2 cells in SSc with increased levels of interleukin-4 (IL-4) and IL-13 (16,18,19). This variation could reflect the clinical diversity in SSc (e.g., SSc-associated autoantibody subsets). Another possibility could be the difference in the genetic backgrounds of patients, resulting in distinct alterations in immune balance.
The transcription factor T-bet (T-box expressed in T cells) (TBX21) is a key transcriptional activator of Th1 cell differentiation. T-bet plays an essential role in Th1/Th2 balance, where it is the master regulator of Th1 cell fate through promotion of Th1 cytokines and inhibition of Th2 cytokines (20,21). In a recent study, TBX21 polymorphisms were shown to be associated with rheumatoid arthritis (RA) (22), and in previous studies they were associated with asthma (a Th2-mediated disease characterized by overproduction of Th2 cytokines [IL-4, IL-5, and IL-13]) (25,26) and type 1 diabetes mellitus (DM) (27). Finally, the cytokine balance in mice deficient in T-bet is skewed toward Th2 cytokines, and tbx2-null mice have displayed increased sensitivity to bleomycin-induced dermal sclerosis (28,29).
STAT-4 is another critical transcription factor involved in regulation of the Th1/Th2 cytokine balance. It is expressed in activated peripheral blood monocytes, dendritic cells, and macrophages and is up-regulated by IL-12. Upon binding of IL-12 to the IL-12 receptor, STAT-4 is phosphorylated and forms a homodimer that translocates to the nucleus, where it enhances production of Th1 cytokines such as IFNγ and suppresses production of Th2 cytokines such as IL-4, IL-5, and IL-13. Interestingly, STAT4 also has been shown to be activated in response to type I IFNs, a cytokine network that is dysregulated in SSc (30). STAT4 polymorphisms have been found to be associated with SSc (31) and other autoimmune diseases, including RA (32), systemic lupus erythematosus (SLE) (32), asthma (33), type 1 DM (34), and Sjögren’s syndrome (SS) (35).
Given the potential importance of Th1/Th2 cytokine balance in SSc, we investigated the association of polymorphisms in the TBX21 and STAT4 genes with SSc. We demonstrated a significant association of both TBX21 and STAT4 polymorphisms with susceptibility to SSc in 2 large and independent cohorts. Further, we demonstrated gene–gene interaction between TBX21 and STAT4 variants. Moreover, the functional data suggested a Th2 cytokine profile in the TBX21 mutation group, a proinflammatory profile in the TBX21 wild-type group, and a Th1 profile in the TBX21 wild-type and STAT4 mutation groups.
PATIENTS AND METHODS
SSc patients and controls
Two independent cohorts of SSc patients and control subjects were used in the current study. The first cohort (SSc Registry cohort) consisted of 880 SSc patients and 507 healthy controls from the Scleroderma Family Registry and DNA Repository, a nationwide registry established in 1994 (36). The second cohort (UT Division cohort) consisted of 522 SSc patients and 531 healthy controls, mainly from Texas, evaluated at the University of Texas Health Science Center at Houston Rheumatology Division between 1986 and the present (10,37). All SSc patients fulfilled the American College of Rheumatology (formerly, the American Rheumatism Association) preliminary criteria for disease classification (38) or had at least 3 of the 5 features of CREST syndrome (calcinosis, Raynaud’s phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasias). SSc patients were further classified based on diffuse or limited skin involvement (39) and by autoantibody status. All subjects provided written informed consent, and the study was approved by the Committee for the Protection of Human Subjects of the University of Texas Health Science Center at Houston.
In order to increase the number of control subjects to attain a >1:1 case:control ratio, we used genotyping data from previously published genome-wide association studies in RA and SLE (32,40). Published data reported by Remmers et al (32) on the STAT4 rs11889341 polymorphism, collected for the New York Cancer Project from control subjects of self-reported European ancestry, (41) (n = 2,635), were used. Genotype data on the TBX21 rs11650354 single-nucleotide polymorphism (SNP) were obtained from 3,172 control samples (all self-described North Americans of European descent) from studies 64–67 in the publicly available iControlDB database (www.illumina.com/pages.ilmn?ID=231). Additionally, data on TBX21 rs11650354 SNP genotypes in 1,094 control subjects were obtained from the prostate cancer study (42) in the Cancer Genetic Markers of Susceptibility (CGEMS) project (http://cgems.cancer.gov).
To address the issue of population stratification between the 2 study cohorts, we compared the control frequencies in the SSc Registry cohort with control frequencies in the UT Division group, for all 13 TBX21 and 5 STAT4 polymorphisms. There were no statistically significant differences between the 2 control cohorts in any of the polymorphisms (see Supplementary Table A, available in the online version of this article at http://www3.interscience.wiley.com/journal/76509746/home).
Autoantibody analysis
All SSc patients were tested for antinuclear antibodies, using indirect immunofluorescence (Antibodies Inc., Davis, CA). ACAs were determined by their distinctive pattern on indirect immunofluorescence (43). Anti–topo I antibodies were determined by immunodiffusion (Inova Diagnostics, San Diego, CA). Anti–RNAP III antibodies were determined by enzyme-linked immunosorbent assay (ELISA; MBL, Nagoya, Japan).
SNP selection
SNPs were selected from the TBX21 gene–transcribed sequence and 8,000 bp upstream and downstream regions. SNPs with minor allele frequencies of >5% in the Centre d’Etude du Polymorphisme Humain from Utah population sample (Utah residents with ancestry from northern and western Europe) and a coefficient of determination of r2 = 0.8 were identified based on data from HapMap (44) (rs9910408, rs11079786, rs4794067, rs10514934, rs16946264, rs11650354, rs11657479, rs7502875, rs16947058, rs16947078, and rs17699436). Two additional coding region SNPs (rs2240017 and rs2074190) were also selected. SNPs on the STAT4 gene were selected based on the most significant STAT4 variants identified in the candidate gene study in RA and SLE (32) (rs11889341, rs7574865, rs8179673, rs10181656, and rs6752770).
SNP genotyping
Genomic DNA was extracted from peripheral blood with the PureGene genomic DNA isolation kit (Gentra Systems, Minneapolis, MN). The SNPs were genotyped with an ABI TaqMan SNP genotyping assay using a ABI 7900HT real-time thermocycler (Applied Biosystems, Foster City, CA). Automated allele calling was performed with allelic discrimination plots using SDS 2.3 software from Applied Biosystems. Multiple positive Centre d’Etude du Polymorphisme Humain DNA samples from Coriell Institute for Medical Research (Camden, NJ) and negative controls were used in each genotyping assay, and allele calls were verified with HapMap data for validation. Confirmation of the TaqMan genotyping was performed by bidirectional direct sequencing using 32 samples (24 SSc patient and 8 control samples). Visual inspection of sequences confirmed 100% concordance with results obtained by TaqMan assay (for detailed description of methods, see supplementary material, available in the online version of this article at http://www3.interscience.wiley.com/journal/76509746/home).
ELISA for cytokines
Plasma was collected in EDTA and stored at −80°C for bulk analysis using electrochemiluminescence multiplex assays (Meso Scale Discovery, Gaithersburg, MD) to determine levels of IFNγ, tumor necrosis factor α (TNFα), IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-13, IL-17, and IL-23 (45). Sample cytokine concentrations were determined with MSD Workbench 3.0 software.
Gene expression array of peripheral white blood cells and data analysis
Blood samples were drawn directly into PAXgene tubes (PreAnalytiX, Franklin Lakes, NJ), and total RNA was isolated. The amplified chromosomal RNA was hybridized on Human Ref-8 v2 arrays (Illumina, San Diego, CA), and the data were extracted with the Beadstudio software suite (Illumina). Raw data were also analyzed in BRB Array-Tools, version 3.7 (National Cancer Institute), developed by Dr. Richard Simon and Amy Peng Lam and the BRB Array-Tools Development Team (see detailed methods in supplementary material, http://www3.interscience.wiley.com/journal/76509746/home).
Statistical analysis
Statistical analyses were performed using SAS 9.1.3 (SAS Institute, Cary, NC). Allelic and genotypic associations were calculated using the standard Pearson’s chi-square test or, when appropriate, Fisher’s exact test. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated. The Mantel-Haenszel chi-square test was used to combine data from the 2 independent cohorts. P values less than 0.05 were considered significant. To account for multiple comparisons, Bonferroni correction was applied during the model-building phase. Logistic regression analysis was performed to identify significant independent risk factors for SSc. Measures of pairwise linkage disequilibrium (LD) were determined using Haploview (Whitehead Institute, Cambridge, MA). Classification and Regression Tree (CART) analysis (46) was performed to explore gene–gene interactions (CART 6.0; Salford Systems, San Diego, CA). Race, sex, and variants of TBX21 and STAT4 genes were used as nominal categorical variables to predict the outcome SSc. The cytokine values were compared using the Wilcoxon Mann-Whitney U test (detailed methods in supplementary material, http://www3.interscience.wiley.com/journal/76509746/home).
RESULTS
We conducted this candidate gene study using the SSc Registry cohort (36) and replicated it in an independent cohort (UT Division) (10,37). Clinical and serologic characteristics of the SSc patients from both of these cohorts are presented in Table 1. All TBX21 gene SNPs studied were in Hardy-Weinberg equilibrium in white controls. SNPs rs10514934 and rs16947078 in black controls and SNP rs17699436 in Hispanic controls were not in Hardy-Weinberg equilibrium and were removed from the analysis in these ethnic groups. All studied SNPs in the STAT4 gene were in HardyWeinberg equilibrium in all the 3 ethnic groups (see Supplementary Table 1, http://www3.interscience.wiley.com/journal/76509746/home).
Table 1.
Characteristics of the control subjects and the patients with systemic sclerosis (SSc)
| SSc Registry cohort |
UT Division cohort |
|||||
|---|---|---|---|---|---|---|
| White | Black | Hispanic | White | Black | Hispanic | |
| Controls | ||||||
| Sex | ||||||
| n* | 421 | 63 | 23 | 277 | 138 | 116 |
| Female, no. (%) | 198 (47.0) | 36 (57.1) | 15 (65.2) | 155 (56.6) | 106 (77.4) | 65 (56.0) |
| Male, no. (%) | 223 (53.0) | 27 (42.9) | 8 (34.8) | 119 (43.4) | 31 (22.6) | 51 (44.0) |
| SSc patients | ||||||
| Sex | ||||||
| n | 745 | 69 | 66 | 314 | 108 | 100 |
| Female, no. (%) | 676 (90.7) | 61 (88.4) | 58 (87.9) | 259 (82.5) | 90 (83.3) | 90 (90.0) |
| Male, no. (%) | 69 (9.3) | 8 (11.6) | 8 (12.1) | 55 (17.5) | 18 (16.7) | 10 (10.0) |
| Skin involvement | ||||||
| n | 712 | 67 | 60 | 292 | 97 | 97 |
| Limited cutaneous SSc, no. (%) | 465 (65.3) | 25 (37.3) | 28 (46.7) | 146 (50.0) | 28 (28.9) | 38 (39.2) |
| Diffuse cutaneous SSc, no. (%) | 247 (34.7) | 42 (62.7) | 32 (53.3) | 146 (50.0) | 69 (71.1) | 59 (60.8) |
| Antibodies | ||||||
| n | 509 | 43 | 45 | 158 | 43 | 60 |
| Anticentromere, no. (%) | 221 (43.4) | 4 (9.3) | 12 (26.7) | 79 (50.0) | 8 (18.6) | 17 (28.3) |
| Anti–topoisomerase I, no. (%) | 138 (27.1) | 22 (51.2) | 14 (31.1) | 36 (22.8) | 21 (48.8) | 25 (41.7) |
| Anti–RNA polymerase III, no. (%) | 150 (29.5) | 17 (39.5) | 19 (42.2) | 43 (27.2) | 14 (32.6) | 18 (30.0) |
Data missing in 3 white controls and 1 black control in the University of Texas SSc cohort (UT Division cohort).
LD and haplotype block structure in TBX21 and STAT4 genes
Pairwise LD was calculated by both D’ and r2 for the 13 TBX21 SNPs and 5 STAT4 SNPs typed in white, black, and Hispanic controls (see Supplementary Figure 1, http://www3.interscience.wiley.com/journal/76509746/home). LD in whites (r2 >0.8) was identified for the following pairs of TBX21 SNPs: rs11079786 and rs4794067, rs11079786 and rs2074190, rs4794067 and rs2074190, rs11657479 and rs7502875, rs11657479 and rs16947078, and rs7502875 and rs16947078. The 4 STAT4 SNPs rs11889341, rs7574865, rs8179673, and rs10181656 demonstrated high LD with one another in all 3 ethnic groups (r2 >0.95).
TBX21 and STAT4 SNP association analysis
Two of the 13 variants in the TBX21 gene, rs11650354 and rs17699436, demonstrated significant association with SSc in whites in both the SSc Registry and UT Division cohorts. These 2 variants exhibited association at the genotypic and allelic levels, respectively. When the 2 cohorts were combined using Mantel-Haenszel analysis, these 2 variations in TBX21 still exhibited strong association with SSc in whites, with rs11650354 being a susceptibility factor and rs17699436 playing a protective role (Table 2). Thus, we were able to demonstrate and replicate the association of the TBX21 gene variants rs11650354 and rs17699436 in the 2 independent cohorts.
Table 2.
Distribution of TBX21 and STAT4 SNP genotypes in North American white control subjects and patients with SSc*
| SSc Registry cohort |
UT Division cohort |
Mantel-Haenszel–combined cohorts |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP (minor allele) |
Position, bp |
MAF |
OR | P† | P‡ | MAF |
OR | P† | P‡ | OR (95% CI)† |
P† | OR (95% CI)‡ |
P‡ | ||
| Controls | SSc | Controls | SSc | ||||||||||||
| TBX21 § | |||||||||||||||
| rs9910408 (G) | 43157873 | 0.44 | 0.40 | 0.83 | 0.14 | 0.06 | 0.40 | 0.45 | 1.22 | 0.01 | 0.10 | 0.96 (0.8–1.2) | 0.65 | 0.97 (0.8–1.1) | 0.66 |
| rs11079786 (T) | 43160915 | 0.26 | 0.28 | 1.09 | 0.72 | 0.42 | 0.31 | 0.31 | 1.01 | 0.51 | 0.97 | 1.07 (0.9–1.3) | 0.52 | 1.05 (0.9–1.2) | 0.52 |
| rs4794067 (G) | 43163827 | 0.26 | 0.29 | 1.13 | 0.54 | 0.29 | 0.29 | 0.30 | 1.04 | 0.61 | 0.77 | 1.11 (0.9–1.4) | 0.33 | 1.09 (0.9–1.3) | 0.32 |
| rs2240017 (G) | 43165918 | 0.03 | 0.03 | 0.84 | 0.52 | 0.53 | 0.04 | 0.03 | 0.71 | 0.64 | 0.32 | 0.78 (0.5–1.2) | 0.27 | 0.78 (0.5–1.2) | 0.26 |
| rs2074190 (C) | 43166209 | 0.25 | 0.28 | 1.16 | 0.47 | 0.26 | 0.30 | 0.30 | 1.01 | 0.73 | 0.97 | 1.10 (0.9–1.4) | 0.41 | 1.08 (0.9–1.3) | 0.41 |
| rs10514934 (G) | 43167123 | 0.11 | 0.12 | 1.14 | 0.68 | 0.39 | 0.12 | 0.13 | 1.04 | 0.01 | 0.82 | 1.11 (0.9–1.4) | 0.43 | 1.10 (0.9–1.4) | 0.42 |
| rs16946264 (T) | 43168433 | 0.10 | 0.10 | 0.91 | 0.75 | 0.54 | 0.13 | 0.08 | 0.64 | 0.01 | 0.02 | 0.78 (0.6–1.01) | 0.06 | 0.79 (0.6–1.01) | 0.06 |
| rs11650354 (T) | 43177091 | 0.15 | 0.18 | 1.29 | 0.002 | 0.08 | 0.16 | 0.22 | 1.49 | 0.02 | 0.01 | 1.44 (1.1–1.8) | 0.006 | 1.38 (1.1–1.7) | 0.003 |
| rs11657479 (C) | 43177900 | 0.22 | 0.25 | 1.24 | 0.12 | 0.06 | 0.25 | 0.29 | 1.22 | 0.35 | 0.13 | 1.29 (1.1–1.6) | 0.016 | 1.23 (1.04–1.5) | 0.015 |
| rs7502875 (C) | 43178226 | 0.21 | 0.24 | 1.17 | 0.29 | 0.17 | 0.24 | 0.27 | 1.14 | 0.63 | 0.32 | 1.19 (0.97–1.5) | 0.10 | 1.16 (0.98–1.4) | 0.09 |
| rs16947058 (T) | 43180185 | 0.42 | 0.42 | 0.99 | 0.99 | 0.94 | 0.46 | 0.45 | 0.94 | 0.93 | 0.71 | 0.97 (0.8–1.2) | 0.78 | 0.97 (0.8–1.1) | 0.71 |
| rs16947078 (G) | 43180499 | 0.20 | 0.22 | 1.10 | 0.70 | 0.40 | 0.23 | 0.23 | 1.00 | 0.77 | 0.97 | 1.07 (0.9–1.3) | 0.51 | 1.06 (0.9–1.3) | 0.50 |
| rs17699436 (G) | 43183574 | 0.11 | 0.08 | 0.72 | 0.10 | 0.04 | 0.10 | 0.06 | 0.55 | 0.02 | 0.01 | 0.64 (0.5–0.8) | 0.002 | 0.66 (0.5–0.9) | 0.001 |
| STAT4 ¶ | |||||||||||||||
| rs11889341 (A) | 191651987 | 0.22 | 0.26 | 1.30 | 0.03 | 0.01 | 0.24 | 0.28 | 1.22 | 0.36 | 0.14 | 1.33 (1.1–1.6) | 0.005 | 1.27 (1.1–1.5) | 0.004 |
| rs7574865 (A) | 191672878 | 0.21 | 0.26 | 1.31 | 0.03 | 0.01 | 0.25 | 0.28 | 1.18 | 0.48 | 0.21 | 1.34 (1.1–1.6) | 0.005 | 1.26 (1.1–1.5) | 0.004 |
| rs8179673 (C) | 191677586 | 0.22 | 0.27 | 1.36 | 0.01 | 0.003 | 0.25 | 0.29 | 1.20 | 0.39 | 0.16 | 1.37 (1.1–1.7) | 0.002 | 1.30 (1.1–1.5) | 0.001 |
| rs10181656 (C) | 191678124 | 0.22 | 0.27 | 1.33 | 0.02 | 0.01 | 0.25 | 0.29 | 1.21 | 0.37 | 0.15 | 1.37 (1.1–1.6) | 0.002 | 1.29 (1.1–1.5) | 0.002 |
| rs6752770 (C) | 191681808 | 0.29 | 0.30 | 1.05 | 0.49 | 0.59 | 0.28 | 0.37 | 1.50 | 0.004 | 0.001 | 1.25 (1.1–1.5) | 0.017 | 1.20 (1.03–1.4) | 0.02 |
Odds ratios (ORs) and 95% confidence intervals (95% CIs) are for carriage of the minor allele genotype. Control subjects were used as reference for all comparisons. SNP = single-nucleotide polymorphism; SSc = systemic sclerosis; MAF = minor allele frequency; UT Division cohort = University of Texas SSc cohort.
Genotypic 3 × 2 chi-square comparison.
Allelic comparison.
Chromosome 17.
Chromosome 2.
All of the STAT4 variants exhibited increased minor allele frequencies in white SSc subjects in both cohorts. When the 2 cohorts were combined using Mantel-Haenszel analysis, all 5 variants remained significantly associated with SSc (Table 2).
The TBX21 and STAT4 SNP data in black and Hispanic subjects did not reach statistical significance. Data on these subjects are presented in Supplementary Tables 2 and 3.
TBX21 rs11650354-rs17699436 diplotype association analysis
Study of the haplotypes for the 2 replicating SNPs, rs11650354-rs17699436, revealed no significant difference in the CA (wild-type) haplotype frequency between SSc cases and controls. The CG haplotype was protective against SSc (7.0%; versus 10.7% in controls) whereas the TA haplotype was a susceptibility factor in SSc (19.0%; versus 15.4% in controls). Interestingly, the TG haplotype, carrying the mutation for both of the SNPs, was found to be a susceptibility factor in SSc, albeit in a very low frequency (0.6%; versus 0.01% in controls), suggesting that the 2 TBX21 variants were acting independently of one another.
CART analysis for gene–gene interaction
An established exploratory method (CART) (47) was used to explore for gene–gene interactions in TBX21 and STAT4 genes in the 2 cohorts combined. All of the TBX21 and STAT4 polymorphisms were entered asvariables in the CART model, along with race and sex. CART analysis interactively segregates subjects into 2 subgroups using the most powerful variable classifier for SSc. The variables partitioning out higher up in the decision tree suggest greater importance than the ones lower in the tree.
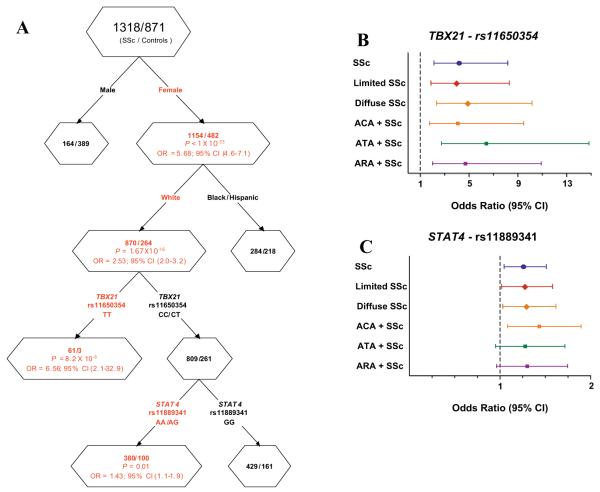
Figure 1A depicts female sex and white race as the first 2 splits. In white female subjects, the rs11650354 TT variant increased risk for SSc (OR 6.56 [95% CI 2.1–32.9]) consistent with a recessive effect. Individuals with the CC/CT genotype were further split into 2 subgroups based on the STAT4 variant (rs11889341) genotype. The AA/AG genotype of the STAT4 variant increased risk for SSc as compared with the GG wild-type genotype (OR 1.43 [95% CI 1.1–1.9]). Therefore, this analysis suggested a more prominent role of the TBX21 rs11650354 SNP and also revealed a potential gene–gene interaction between TBX21 and STAT4 genes among white female patients with SSc.
Figure 1.
A, Cartesian and Regression Tree analysis showing a gene–gene interaction between TBX21 and STAT4 polymorphisms in systemic sclerosis (SSc). Red text denotes SSc susceptibility factors. B and C, Estimated risk of TBX21 single-nucleotide polymorphism (SNP) rs11650354 (B) and STAT4 SNP rs11889341 (C)inSSc patients, by logistic regression analysis. The analysis was controlled for the confounding effects of sex and race. Control subjects were used as reference for all comparisons. The TBX21 SNP showed a recessive mode of inheritance, and the STAT4 SNP showed a dominant mode of inheritance. OR = odds ratio; 95% CI = 95% confidence interval; ACA = anticentromere antibody; ATA = anti–topoisomerase I antibody; ARA = anti–RNA polymerase III antibody.
Logistic regression analysis in white subjects after controlling for sex demonstrated significant P values for TBX21 CC/CT–STAT4 AA/AG (OR 1.36 [95% CI 1.1–1.7]) and TBX21 TT–STAT4 GG (OR 5.7 [95% CI 2.2–15.0]) as compared with the TBX21 CC/CT–STAT4 GG group. Results in the TBX21 TT–STAT4 AA/AG group were not statistically significant. Thus, the logistic regression analysis confirmed the gene–gene interaction observed in the CART analysis.
TBX21 and STAT4 polymorphisms in the SSc Registry and UT Division cohorts and additional controls
TBX21 SNP rs11650354 and STAT4 SNP rs11889341 were selected for further analysis based on their association with SSc as determined by CART, logistic regression, and chi-square analyses. As seen in Table 3, the TBX21 SNP rs11650354 depicted a recessive model and a significant association in both cohorts individually and combined.
Table 3.
Best fit model for distribution of the TBX21 SNP rs11650354 genotype in North American white control subjects and patients with SSc*
| SSc Registry cohort |
UT Division cohort |
Combined cohorts |
SSc Registry and UT Division cohorts + iControlDB and CGEMS controls† |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | TT, % |
CC/CT, % |
P | OR (95% CI) | n | TT, % |
CC/CT, % |
P | OR (95% CI) | n | P | OR (95% CI) | n | P | OR (95% CI) | |
| Control | 252 | 2.4 | 97.6 | 227 | 1.3 | 98.7 | 479 | 4,745 | ||||||||
| SSc patients | 596 | 8.6 | 91.4 | 0.003 | 3.84 (1.6–10.1) | 306 | 5.6 | 94.4 | 0.03 | 4.39 (1.2–19.1) | 902 | 3.9 × 10−5 | 4.26 (2.0–9.2) | 902 | 1.4 × 10−15 | 3.37 (2.4–4.6) |
| Limited cutaneous SSc | 364 | 9.1 | 90.9 | 0.002 | 4.09 (1.6–11.0) | 147 | 4.8 | 95.2 | 0.12 | 3.73 (0.8–18.5) | 511 | 4.8 × 10−5 | 4.44 (2.0–9.9) | 511 | 7.2 × 10−12 | 3.51 (2.4–5.1) |
| Diffuse cutaneous SSc | 189 | 9 | 91 | 0.006 | 4.05 (1.5–11.8) | 138 | 7.2 | 92.8 | 0.009 | 5.83 (1.4–27.2) | 327 | 5.1 × 10−5 | 4.70 (2.1–10.9) | 327 | 8.1 × 10−10 | 3.70 (2.3–5.8) |
| Antibodies | ||||||||||||||||
| Anticentromere | 178 | 10.7 | 89.3 | 0.0009 | 4.92 (1.8–14.0) | 88 | 4.5 | 95.5 | 0.24 | 3.56 (0.7–20.4) | 266 | 3.9 × 10−5 | 4.94 (2.1–11.7) | 266 | 2.2 × 10−9 | 3.92 (2.4–6.3) |
| Anti–topoisomerase I | 102 | 7.8 | 92.2 | 0.05 | 3.49 (1.1–11.7) | 34 | 5.9 | 94.1 | 0.21 | 4.67 (0.5–36.2) | 136 | 0.003 | 4.14 (1.5–11.4) | 136 | 0.0006 | 3.27 (1.6–6.6) |
| Anti-RNA polymerase III | 132 | 10.6 | 89.4 | 0.001 | 4.86 (1.7–14.6) | 34 | 5.9 | 94.1 | 0.21 | 4.67 (0.5–36.2) | 166 | 2.4 × 10−4 | 5.57 (2.2–14.0) | 166 | 2.2 × 10−8 | 4.41 (2.4–7.8) |
ORs and 95% CIs are for carriage of the TT genotype. Control subjects were used as reference for all comparisons. P values were corrected for multiple model testing using the Bonferroni adjustment. See Table 2 for other definitions.
The Illumina iControlDB database was queried to ascertain the SNP genotype frequencies in 3,172 white controls (CC 2,232 [70.4%], CT 863 [27.2%], TT 77 [2.4%]). The Cancer Genetic Markers of Susceptibility (CGEMS) database was queried to ascertain the SNP genotype frequencies in 1,094 white controls (CC 752 [68.7%], CT 316 [28.9%], TT 26 [2.4%]).
To further increase the ratio of cases to controls, additional genotypes from North American controls of European ancestry were obtained from public repositories (40,42). The TT genotype frequency in controls was similar between our combined cohort (1.9%) and the publicly available data on controls (iControlDB 2.4%, CGEMS 2.4%; P = 0.75 and P = 0.58, respectively), thus demonstrating that the controls in the publicly available databases were similar to our control cohorts and these could be combined for joint analyses. In a comparison of the genotype frequencies in 4,745 controls and 902 SSc cases, a highly significant association was observed (corrected P [Pcorr] = 1.4 × 10−15, OR 3.37, 95% CI 2.4–4.6) (Table 3). Logistic regression analysis revealed a recessive model as the best fit and confirmed that the TBX21 TT genotype was an independent risk factor for SSc and its subsets after controlling for the confounding effects of sex and race (Figure 1B).
Similarly, the frequency of the STAT4 variant rs11889341 in controls was obtained from published data on North Americans of European ancestry (32). The A allele frequency in controls was similar between our combined cohort (22.7%) and the published frequency (22%) (P = 0.57), thus suggesting that the controls in the publicly available databases were similar to our control cohorts and the two could be combined for joint analyses. On comparing the allelic frequencies in 3,322 controls and 1,039 SSc cases, a significant association was observed (Pcorr = 2.4 × 10−5, OR 1.29, 95% CI 1.2–1.5) (Table 4). Logistic regression analysis revealed a dominant model as the best fit and confirmed that the STAT4 AA/AG genotype was an independent risk factor for SSc and its subsets after controlling for the confounding effects of sex and race (Figure 1C).
Table 4.
Best fit model for distribution of the STAT4 SNP rs11889341 alleles in North American white control subjects and patients with SSc*
| SSc Registry cohort |
UT Division cohort |
SSc Registry + UT Division |
SSc Registry + UT Division + controls† |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2n | A, % |
P | OR (95% CI) | 2n | A, % |
P | OR (95% CI) | P | OR (95% CI) | 2n | A, % |
P | OR (95% CI) | |
| Control subjects | 826 | 21.7 | 548 | 24.3 | 6,644 | 22.2 | ||||||||
| SSc patients | 1,466 | 26.4 | 0.03‡ | 1.30 (1.1–1.6) | 612 | 28.1 | 0.14 | 1.22 (0.9–1.6) | 0.012‡ | 1.27 (1.1–1.5) | 2,078 | 26.9 | 2.4 × 10−5‡ | 1.29 (1.2–1.5) |
| Limited cutaneous SSc | 914 | 25.5 | 0.06 | 1.24 (0.99–1.5) | 284 | 30.6 | 0.15‡ | 1.38 (1.0–1.9) | 0.021‡ | 1.28 (1.1–1.5) | 1,198 | 26.7 | 1.7 × 10−3‡ | 1.28 (1.1–1.5) |
| Diffuse cutaneous SSc | 488 | 28.1 | 0.03‡ | 1.41 (1.1–1.8) | 284 | 26.8 | 0.43 | 1.14 (0.8–1.6) | 0.03‡ | 1.30 (1.1–1.6) | 772 | 27.6 | 2.0 × 10−3‡ | 1.34 (1.1–1.6) |
| Antibodies | ||||||||||||||
| Anticentromere | 430 | 26.3 | 0.07 | 1.29 (0.98–1.7) | 154 | 31.2 | 0.08 | 1.41 (0.95–2.1) | 0.03‡ | 1.33 (1.1–1.7) | 584 | 27.6 | 0.009‡ | 1.34 (1.1–1.6) |
| Anti–topoisomerase I | 272 | 26.5 | 0.10 | 1.30 (0.9–1.8) | 72 | 23.6 | 0.90 | 1.04 (0.6–1.8) | 0.17 | 1.21 (0.9–1.6) | 344 | 25.9 | 0.10‡ | 1.23 (0.95–1.6) |
| Anti–RNA polymerase III | 298 | 26.8 | 0.07 | 1.33 (0.98–1.8) | 82 | 26.8 | 0.62 | 1.14 (0.7–1.9) | 0.07 | 1.28 (0.98–1.7) | 380 | 26.8 | 0.09‡ | 1.29 (1.01–1.8) |
ORs and 95% CIs are for carriage of the A allele. Control subjects were used as reference for all comparisons. See Table 2 for definitions.
Published frequencies in controls were ascertained for 2,635 white control subjects from the New York Cancer Project.
Corrected for multiple model testing using the Bonferroni adjustment.
The associations of the TBX21 SNP rs11650354 and the STAT4 SNP rs11889341 were observed for SSc overall, and not for any specific clinical or autoantibody subset. We observed an association of the TBX21 rs11650354 variant with SSc both with and without pulmonary fibrosis, as compared with controls. There was no significant association between the TT genotype and the presence of pulmonary fibrosis when compared with patients without pulmonary fibrosis. These TBX21 and STAT4 SNPs were tested in black and Hispanic populations separately, and the results did not reach statistical significance in skin and autoantibody subset analyses (see Supplementary Tables 5 and 6, available in the online version of this article at http://www3.interscience.wiley.com/journal/76509746/home).
Association of the TBX21 variant rs11650354 with plasma cytokine levels
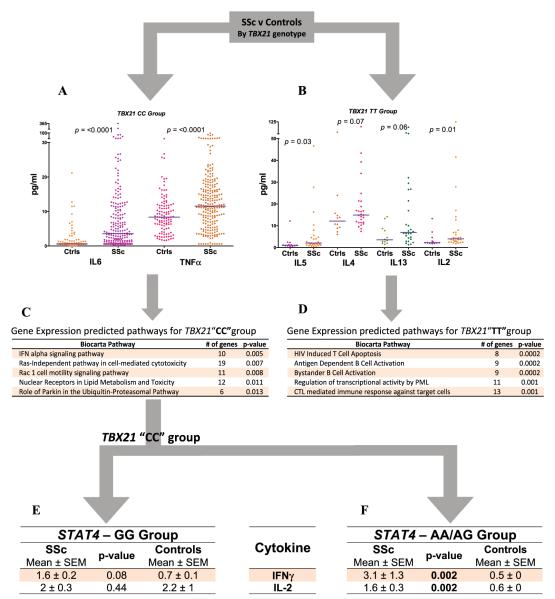
To better understand the immunologic significance of the TBX21 variant rs11650354, plasma levels of 13 cytokines were determined. SSc patient (n = 273) and control (n = 123) samples were stratified based on their genotypes (CC group and TT group). Compared with controls in the CC group, SSc patients in the CC group had higher levels of circulating IL-6 and TNFα (Figure 2A); however, no differences in levels of IL-4, IL-5, IL-13, or IL-2 were observed. In contrast, in the TT group, SSc patients had significantly elevated levels of circulating IL-2 and IL-5 and a trend toward an increase in IL-4 and IL-13 levels compared with controls (Figure 2B), with no increase in levels of IL-6 or TNFα. The remaining cytokines did not show a trend toward association in either of the groups. These findings suggest that patients in the TBX21 mutation group (TT) have a more prominent Th2 cytokine profile, while patients in the wild-type group (CC) have a more prominent proinflammatory cytokine profile.
Figure 2.
A–D, Plasma cytokine levels (A and B) and gene expression predicted pathways (C and D)by TBX21 single-nucleotide polymorphism (SNP) group in systemic sclerosis (SSc) patients compared with healthy controls (Ctrls). Bars in A and B show the means. P values for gene expression predicted pathways were determined by Fisher’s least significant statistic after 100,000 permutations (http://cgap.nci.nih.gov/Pathways/BioCarta/). E and F, Gene expression predicted pathways by STAT4 SNP group in SSc patients compared with healthy controls. IL-6 = interleukin-6; TNFα = tumor necrosis factor α; IFN = interferon; HIV = human immunodeficiency virus; PML = promyelocytic leukemia; CTL = cytotoxic T lymphocyte.
Whole blood RNA gene expression profiling
Next, we compared whole blood gene expression profiles of SSc patients and healthy controls stratified into CC (3 SSc patients and 3 controls) and TT (3 SSc and 2 controls) groups based on TBX21 genotype. We examined known biologic pathways to increase the study’s power to detect dysregulated gene networks. Of a total of 258 biologic pathways from the BioCarta database, 12 and 22 pathways in the CC and TT groups, respectively, were found to be differentially regulated at P < 0.05. The top 5 differentially regulated pathways in SSc are shown in Figures 2C and D. The most significantly differentially regulated pathway in the CC group was the IFNα signaling pathway. In the TT group, both T cell and B cell pathways were among those most significantly differentially regulated. These data support our observations in the studies of plasma cytokine networks and provide evidence suggesting a role of type I IFN and proinflammatory pathways in the CC group and T cell pathways in the TT group.
Association of the STAT4 variant rs11889341 with plasma cytokine levels
We observed a gene–gene interaction between the STAT4 variant rs11889341 and the CC/CT genotype of the TBX21 variant rs11650354, by CART analysis (Figure 1A). In the TBX21 wild-type(CC) subset, plasma IFNγ and IL-2 levels were increased in the AA/AG group of SSc patients as compared with healthy controls. Levels of these cytokines were not significantly different between SSc patients and controls in the GG group. TNFα and IL-6 were increased in SSc patients compared with controls in both of the STAT4 groups. The rest of the cytokines did not show a trend toward association in either of the groups. These data suggest that in SSc patients with the TBX21 wild-type genotype, the STAT4 variation is associated with alterations in circulating T cell cytokines, leading to a Th1 cytokine profile.
DISCUSSION
The present data, obtained using 2 independent cohorts, clearly demonstrate an association of TBX21 and STAT4 polymorphisms with SSc. We also showed a gene–gene interaction among the TBX21 and STAT4 variants, such that the STAT4 genotype increased the risk for SSc only in the group with the TBX21 wild-type genotype. Furthermore, we demonstrated distinct cytokine profiles based on the TBX21 and STAT4 genotypes suggestive of genetic influence on the complex immune balance and dysregulation of distinct gene expression pathways in each group.
Findings of recent genetic studies support the emerging concept that distinct clinical autoimmune diseases may share genetic susceptibility factors. Consistent with this, some of the genes implicated in SSc are also associated with susceptibility to other autoimmune diseases such as RA and SLE, including the HLA class II gene family (9,37,48), protein tyrosine phosphatase N22, and STAT4 (2). Associations of STAT4 polymorphisms with SSc (31), RA (32), SLE (32), type 1 DM (34), SS (35), and asthma (33), all of which are autoimmune disorders, have been reported. In the current study, we confirmed an association of STAT4 mutations with susceptibility to SSc. These data suggest that STAT4 may be an “autoimmune disease susceptibility gene” and support the concept of common dysregulated pathways across multiple autoimmune diseases. In contrast, other genes, such as those for allograft inflammatory factor 1 (3) and fibrillin 1 (8), have been reported in association with SSc only, with no reports to date of associations with RA and SLE.
The present report is the first to describe an association of TBX21 variants (rs11650354 and rs17699436) with SSc in North American whites. TBX21 polymorphisms have been recently reported to be associated with susceptibility to RA (22) and have also been found to be associated with asthma (25,26) and DM (27), although associations with SLE or SS have not been reported to date. These findings indicate that TBX21 might possibly be placed in the category of “autoimmune disease susceptibility gene,” and its potential role in other autoimmune diseases needs to be investigated.
An important feature of our study was the use of CART analysis to identify significant gene–gene interactions. CART analysis revealed a major gene–gene interaction between the TBX21 CC/CT genotype and the STAT4 rs11889341 variant, which was further confirmed with logistic regression analysis. The interaction between STAT4 and TBX21 is intriguing given the fact that both of these genes are involved in IL-12 signaling and regulation of the Th1/Th2 cytokine balance. Upon IL-12 stimulation of its receptor, STAT-4 forms an active homodimer that translocates to the nucleus to enhance TBX21 transcription, which subsequently serves as a major regulator of the Th1 pathway. We observed that a STAT4 mutation was important only in patients without the TBX21 mutation. This suggests that there may be independent perturbations at multiple points in the IL-12 pathway that contribute to SSc susceptibility. Thus, genetic alterations in TBX21 could lead to skewing of the Th1/Th2 cytokine balance toward Th2 cytokines. In the absence of TBX21 mutation, genetic alterations of STAT4, an upstream gene in the same pathway, could exert an effect leading to a Th1 cytokine profile.
Alterations in the Th1/Th2 cytokine balance have been demonstrated in some patients with SSc. These findings in humans are supported by the findings of 2 studies using the bleomycin-induced skin fibrosis model, which demonstrated the development of more severe disease in T-bet–deficient (Th2-prone) mice (28,49). Recent observations have suggested that in a subset of SSc patients, there is an increase in type I IFN pathways, similar to that observed in SLE (30). We hypothesize that in the presence of the TBX21 polymorphism, CD4+ T cells fail to differentiate into the Th1 lineage and default to a Th2 fate, leading to a Th2-predominant environment as suggested by the increase in levels of IL-4, IL-5, and IL-13 in plasma, while the STAT4 polymorphism group directs the CD4+ T cells toward a Th1 lineage, as highlighted by the increase in IL-2 and IFNγ levels. In contrast, the TBX21 CC variant is associated with alterations in type I IFNs, IL-6, and TNFα, suggesting a potential role of innate immune cells and proinflammatory pathways in this group of SSc patients. Our findings linking the TBX21 polymorphism with a Th2 cytokine profile in SSc are not only relevant to SSc, but could extend to other fibrotic and inflammatory diseases, e.g., asthma.
The potential influence of TBX21 polymorphisms in the Th1/Th2 cytokine balance observed in our study may also be of importance in asthma, a disease in which Th2 cytokines play an important role. Indeed, several studies have shown association of TBX21 polymorphisms with susceptibility to asthma and response to corticosteroids in children with asthma (50). It will be of interest to determine if similar alterations in Th1/Th2 cytokines are observed in asthma patients based on the polymorphisms in TBX21 as were observed in SSc patients in the current study. Interestingly, peripheralblood levels of T-bet messenger RNA (mRNA) have been reported to be lower in asthma patients than in controls (51). In the current study, we did not detect the TBX21 transcripts in the whole blood gene expression array in any of the samples, perhaps due to low expression levels. Further studies need to be undertaken to measure T-bet mRNA in SSc patients. If the mRNA levels are found to be dysregulated in SSc, this knowledge could be invaluable in developing therapies for a fibrotic disease driven by inflammation such as scleroderma (28,52).
The current study was limited by relatively small numbers of black and Hispanic subjects as compared with whites, who were from 2 large, independent, and well-established case–control cohorts. The SNP frequencies in blacks and Hispanics trended in the same direction as those in whites, and studies of larger cohorts of SSc patients and controls of these ethnic backgrounds will be necessary to carry these results further. The data-driven observation of a gene–gene interaction by CART analysis is an exploratory finding and is hypothesis generating, requiring confirmation in a much larger study. Another limitation is the lack of complete information regarding the causal polymorphisms and their exact functional roles. The SNPs in TBX21 and STAT4 are located in introns, which could be a site for potential splice variation and/or binding of regulatory elements. Overall, our results affirm the use of replicative cohorts along with multiple statistical approaches rather than a single methodology, as the optimal strategy to elucidate complex gene interactions in polygenic diseases.
In summary, the current study clearly demonstrates that TBX21 and STAT4 contributed uniquely and interactively to susceptibility to the development of SSc in 2 independent cohorts. Our data provide evidence that there is a unique subset of SSc patients defined by specific gene–polymorphisms, with a dysregulated Th1/Th2 pathway involved in inflammation, fibrosis, and autoimmunity. Identification of these subsets of patients could provide a focused population for rational selection of therapeutic options targeting these distinct pathways involved in the pathogenesis of SSc and other fibrotic disorders.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Julio Charles, Yasamin Salehi, and William Babu for excellent technical assistance. We would also like to thank Dr. Terry Mcnearney and Dr. Michael Fischbach for providing samples for study.
Supported in part by the NIH (National Institute of Arthritis and Musculoskeletal and Skin Diseases [NIAMS] Center of Research Translation in Scleroderma grant P50-AR-054144, NIAMS Scleroderma Family Registry and DNA Repository grant N01-AR-0-2251, General Clinical Research Center grants M01-RR-00073 and M01-RR-01346, National Center for Research Resources grant 3-UL-1-RR-024148 [University of Texas Health Science Center at Houston Center for Clinical and Translational Sciences], and NIAMS grant K08-AR-054404). Dr. Agarwal’s work was supported by a Scleroderma Foundation New Investigator Grant.
Footnotes
Dr. Mayes has received consulting fees, speaking fees, and/or honoraria from Actelion, United Therapeutics, and Gilead (more than $10,000 each).
REFERENCES
- 1.Bernatsky S, Joseph L, Pineau CA, Belisle P, Hudson M, Clarke AE. Scleroderma prevalence: demographic variations in a population-based sample. Arthritis Rheum. 2009;61:4–004. doi: 10.1002/art.24339. [DOI] [PubMed] [Google Scholar]
- 2.Gourh P, Tan FK, Assassi S, Ahn CW, McNearney TA, Fischbach M, et al. Association of the PTPN22 R620W polymorphism with anti–topoisomerase I– and anticentromere antibody–positive systemic sclerosis. Arthritis Rheum. 2006;54:3945–53. doi: 10.1002/art.22196. [DOI] [PubMed] [Google Scholar]
- 3.Alkassab F, Gourh P, Tan FK, McNearney T, Fischbach M, Ahn C, et al. An allograft inflammatory factor 1 (AIF1) single nucleotide polymorphism (SNP) is associated with anticentromere antibody positive systemic sclerosis. Rheumatology (Oxford) 2007;46:1248–51. doi: 10.1093/rheumatology/kem057. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal SK, Tan FK, Arnett FC. Genetics and genomic studies in scleroderma (systemic sclerosis) Rheum Dis Clin North Am. 2008;34:17–40. doi: 10.1016/j.rdc.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Wu SP, Leng L, Feng Z, Liu N, Zhao H, McDonald C, et al. Macrophage migration inhibitory factor promoter polymorphisms and the clinical expression of scleroderma. Arthritis Rheum. 2006;54:3661–9. doi: 10.1002/art.22179. [DOI] [PubMed] [Google Scholar]
- 6.Tan FK, Stivers DN, Arnett FC, Chakraborty R, Howard R, Reveille JD. HLA haplotypes and microsatellite polymorphisms in and around the major histocompatibility complex region in a Native American population with a high prevalence of scleroderma (systemic sclerosis) Tissue Antigens. 1999;53:74–80. doi: 10.1034/j.1399-0039.1999.530108.x. [DOI] [PubMed] [Google Scholar]
- 7.Tan FK, Stivers DN, Foster MW, Chakraborty R, Howard RF, Milewicz DM, et al. Association of microsatellite markers near the fibrillin 1 gene on human chromosome 15q with scleroderma in a Native American population. Arthritis Rheum. 1998;41:1729–37. doi: 10.1002/1529-0131(199810)41:10<1729::AID-ART5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Tan FK, Wang N, Kuwana M, Chakraborty R, Bona CA, Milewicz DM, et al. Association of fibrillin 1 single-nucleotide polymorphism haplotypes with systemic sclerosis in Choctaw and Japanese populations. Arthritis Rheum. 2001;44:893–901. doi: 10.1002/1529-0131(200104)44:4<893::AID-ANR146>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.McHugh NJ, Whyte J, Artlett C, Briggs DC, Stephens CO, Olsen NJ, et al. Anti-centromere antibodies (ACA) in systemic sclerosis patients and their relatives: a serological and HLA study. Clin Exp Immunol. 1994;96:267–74. doi: 10.1111/j.1365-2249.1994.tb06552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnett FC, Reveille JD, Goldstein R, Pollard KM, Leaird K, Smith EA, et al. Autoantibodies to fibrillarin in systemic sclerosis (scleroderma): an immunogenetic, serologic, and clinical analysis. Arthritis Rheum. 1996;39:1151–60. doi: 10.1002/art.1780390712. [DOI] [PubMed] [Google Scholar]
- 11.Douvas AS, Achten M, Tan EM. Identification of a nuclear protein (Scl-70) as a unique target of human antinuclear antibodies in scleroderma. J Biol Chem. 1979;254:10514–22. [PubMed] [Google Scholar]
- 12.Tan EM, Rodnan GP, Garcia I, Moroi Y, Fritzler MJ, Peebles C. Diversity of antinuclear antibodies in progressive systemic sclerosis: anti-centromere antibody and its relationship to CREST syndrome. Arthritis Rheum. 1980;23:617–25. doi: 10.1002/art.1780230602. [DOI] [PubMed] [Google Scholar]
- 13.Arnett FC. HLA and autoimmunity in scleroderma (systemic sclerosis) Int Rev Immunol. 1995;12:107–28. doi: 10.3109/08830189509056707. [DOI] [PubMed] [Google Scholar]
- 14.Kalogerou A, Gelou E, Mountantonakis S, Settas L, Zafiriou E, Sakkas L. Early T cell activation in the skin from patients with systemic sclerosis. Ann Rheum Dis. 2005;64:1233–5. doi: 10.1136/ard.2004.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molteni M, Della BS, Mascagni B, Bazzi S, Zulian C, Compasso S, et al. Increased interferon-γ (IFN-γ) levels produced in vitro by alloactivated T lymphocytes in systemic sclerosis and Raynaud’s phenomenon. Clin Exp Immunol. 1999;116:164–8. doi: 10.1046/j.1365-2249.1999.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Needleman BW, Wigley FM, Stair RW. Interleukin-1, interleukin-2, interleukin-4, interleukin-6, tumor necrosis factor α, and interferon-γ levels in sera from patients with scleroderma. Arthritis Rheum. 1992;35:67–72. doi: 10.1002/art.1780350111. [DOI] [PubMed] [Google Scholar]
- 17.Kahaleh MB, Leroy EC. Interleukin-2 in scleroderma: correlation of serum level with extent of skin involvement and disease duration. Ann Intern Med. 1989;110:446–50. doi: 10.7326/0003-4819-110-6-446. [DOI] [PubMed] [Google Scholar]
- 18.Mavalia C, Scaletti C, Romagnani P, Carossino AM, Pignone A, Emmi L, et al. Type 2 helper T-cell predominance and high CD30 expression in systemic sclerosis. Am J Pathol. 1997;151:1751–8. [PMC free article] [PubMed] [Google Scholar]
- 19.Giacomelli R, Cipriani P, Lattanzio R, Di Franco M, Locanto M, Parzanese I, et al. Circulating levels of soluble CD30 are increased in patients with systemic sclerosis (SSc) and correlate with serological and clinical features of the disease. Clin Exp Immunol. 1997;108:42–6. doi: 10.1046/j.1365-2249.1997.d01-991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O’Shea JJ, et al. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med. 2006;203:755–66. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 22.Chae SC, Shim SC, Chung HT. Association of TBX21 polymorphisms in a Korean population with rheumatoid arthritis. Exp Mol Med. 2009;41:33–41. doi: 10.3858/emm.2009.41.1.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finotto S, Hausding M, Doganci A, Maxeiner JH, Lehr HA, Luft C, et al. Asthmatic changes in mice lacking T-bet are mediated by IL-13. Int Immunol. 2005;17:993–1007. doi: 10.1093/intimm/dxh281. [DOI] [PubMed] [Google Scholar]
- 24.Finotto S, Neurath MF, Glickman JN, Qin S, Lehr HA, Green FH, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–8. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 25.Chung HT, Kim LH, Park BL, Lee JH, Park HS, Choi BW, et al. Association analysis of novel TBX21 variants with asthma phenotypes. Hum Mutat. 2003;22:257. doi: 10.1002/humu.9169. [DOI] [PubMed] [Google Scholar]
- 26.Ylikoski E, Kinos R, Sirkkanen N, Pykalainen M, Savolainen J, Laitinen LA, et al. Association study of 15 novel single-nucleotide polymorphisms of the T-bet locus among Finnish asthma families. Clin Exp Allergy. 2004;34:1049–55. doi: 10.1111/j.1365-2222.2004.01995.x. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki Y, Ihara K, Matsuura N, Kohno H, Nagafuchi S, Kuromaru R, et al. Identification of a novel type 1 diabetes susceptibility gene, T-bet. Hum Genet. 2004;115:177–84. doi: 10.1007/s00439-004-1146-2. [DOI] [PubMed] [Google Scholar]
- 28.Aliprantis AO, Wang J, Fathman JW, Lemaire R, Dorfman DM, Lafyatis R, et al. Transcription factor T-bet regulates skin sclerosis through its function in innate immunity and via IL-13. Proc Natl Acad Sci U S A. 2007;104:2827–30. doi: 10.1073/pnas.0700021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakos G, Melichian D, Wu M, Varga J. Increased bleomycininduced skin fibrosis in mice lacking the Th1-specific transcription factor T-bet. Pathobiology. 2006;73:224–37. doi: 10.1159/000098208. [DOI] [PubMed] [Google Scholar]
- 30.Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, et al. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology (Oxford) 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- 31.Rueda B, Broen J, Simeon C, Hesselstrand R, Diaz B, Sanchez H, et al. The STAT4 gene influences the genetic predisposition to systemic sclerosis phenotype. Hum Mol Genet. 2009;18:2071–7. doi: 10.1093/hmg/ddp119. [DOI] [PubMed] [Google Scholar]
- 32.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pykalainen M, Kinos R, Valkonen S, Rydman P, Kilpelainen M, Laitinen LA, et al. Association analysis of common variants of STAT6, GATA3, and STAT4 to asthma and high serum IgE phenotypes. J Allergy Clin Immunol. 2005;115:80–7. doi: 10.1016/j.jaci.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Zervou MI, Mamoulakis D, Panierakis C, Boumpas DT, Goulielmos GN. STAT4: a risk factor for type 1 diabetes? Hum Immunol. 2008;69:647–50. doi: 10.1016/j.humimm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Korman BD, Alba MI, Le JM, Alevizos I, Smith JA, Nikolov NP, et al. Variant form of STAT4 is associated with primary Sjögren’s syndrome. Genes Immun. 2008;9:267–70. doi: 10.1038/gene.2008.1. [DOI] [PubMed] [Google Scholar]
- 36.Mayes MD. The establishment and utility of a population-based registry to understand the epidemiology of systemic sclerosis. Curr Rheumatol Rep. 2000;2:512–6. doi: 10.1007/s11926-000-0029-3. [DOI] [PubMed] [Google Scholar]
- 37.Reveille JD, Fischbach M, McNearney T, Friedman AW, Aguilar MB, Lisse J, et al. Systemic sclerosis in 3 US ethnic groups: a comparison of clinical, sociodemographic, serologic, and immunogenetic determinants. Semin Arthritis Rheum. 2001;30:332–46. doi: 10.1053/sarh.2001.20268. [DOI] [PubMed] [Google Scholar]
- 38.Subcommittee for Scleroderma Criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee Preliminary criteria for the classification of systemic sclerosis (scleroderma) Arthritis Rheum. 1980;23:581–90. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 39.LeRoy EC, Medsger TA., Jr. Criteria for the classification of early systemic sclerosis. J Rheumatol. 2001;28:1573–6. [PubMed] [Google Scholar]
- 40.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D. The New York Cancer Project: rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81:301–10. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 43.Fritzler MJ, Kinsella TD. The CREST syndrome: a distinct serologic entity with anticentromere antibodies. Am J Med. 1980;69:520–6. doi: 10.1016/0002-9343(80)90462-3. [DOI] [PubMed] [Google Scholar]
- 44.The International HapMap Project Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 45.Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods. 2009;340:55–64. doi: 10.1016/j.jim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Bonney G. Use of classification trees for association studies. Genet Epidemiol. 2000;19:323–32. doi: 10.1002/1098-2272(200012)19:4<323::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 47.Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78:464–79. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnett FC, Howard RF, Tan F, Moulds JM, Bias WB, Durban E, et al. Increased prevalence of systemic sclerosis in a Native American tribe in Oklahoma: association with an Amerindian HLA haplotype. Arthritis Rheum. 1996;39:1362–70. doi: 10.1002/art.1780390814. [DOI] [PubMed] [Google Scholar]
- 49.Lakos G, Melichian D, Wu M, Varga J. Increased bleomycininduced skin fibrosis in mice lacking the Th1-specific transcription factor T-bet. Pathobiology. 2006;73:224–37. doi: 10.1159/000098208. [DOI] [PubMed] [Google Scholar]
- 50.Tantisira KG, Hwang ES, Raby BA, Silverman ES, Lake SL, Richter BG, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A. 2004;101:18099–104. doi: 10.1073/pnas.0408532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ko FW, Lun SW, Wong CK, Szeto CC, Lam CW, Leung TF, et al. Decreased T-bet expression and changes in chemokine levels in adults with asthma. Clin Exp Immunol. 2007;147:526–32. doi: 10.1111/j.1365-2249.2006.03315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park JW, Min HJ, Sohn JH, Kim JY, Hong JH, Sigrist KS, et al. Restoration of T-box-containing protein expressed in T cells protects against allergen-induced asthma. J Allergy Clin Immunol. 2009;123:479–85. doi: 10.1016/j.jaci.2008.10.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.