Abstract
One mechanism by which mammalian cells regulate the uptake of glucose is the number of glucose transporter proteins (GLUT) present at the plasma membrane. In insulin-responsive cells types, GLUT4 is released from intracellular stores through inactivation of the Rab GTPase activating protein Tre-2/USP6-BUB2-Cdc16 domain family member 4 (TBC1D4) (also known as AS160). Here we describe that TBC1D4 forms a protein complex with protein kinase WNK1 in human embryonic kidney (HEK293) cells. We show that WNK1 phosphorylates TBC1D4 in vitro and that the expression levels of WNK1 in these cells regulate surface expression of the constitutive glucose transporter GLUT1. WNK1 was found to increase the binding of TBC1D4 to regulatory 14-3-3 proteins while reducing its interaction with the exocytic small GTPase Rab8A. These effects were dependent on the catalytic activity because expression of a kinase-dead WNK1 mutant had no effect on binding of 14-3-3 and Rab8A, or on surface GLUT1 levels. Together, the data describe a pathway regulating constitutive glucose uptake via GLUT1, the expression level of which is related to several human diseases.
Keywords: Cell Surface Protein, Glucose Transport, Intracellular Trafficking, Protein Phosphorylation, Signal Transduction, AS160, GLUT1, TBC1D4, WNK1
Introduction
WNK1 is one of four human members of the WNK2 subfamily of serine/threonine protein kinases (1) and is the only family member with a ubiquitous expression pattern (1–4). The WNK1 protein contains 2382 amino acids with a predicted molecular mass of 251 kDa and has its catalytic domain near the N terminus, including the unique sequence variation around catalytically important lysine residues that characterize the WNK subfamily (5, 6). WNK1 contains coiled-coil domains but the bulk sequence reveals no homology to other known protein domains.
Currently, the knowledge on WNK1-regulated cellular pathways is still limited (7). On the one hand, WNK1 can affect multiple signaling pathways related to cell proliferation, including the epidermal growth factor-dependent stimulation of ERK5 (8) and the inhibition of Smad2/TGFβ signaling (9) in HeLa cells. Additionally, insulin-like growth factor 1 treatment of human embryonic kidney (HEK293) cells or adipocytes stimulates protein kinase AKT to phosphorylate WNK1 at threonine residue 60, however, the physiological relevance of this event is unclear because phosphorylation did not affect WNK1 kinase activity, or its subcellular localization (10–12). On the other hand, WNK1 is involved in restoring osmotic homeostasis. It directly phosphorylates and activates SPAK and OSR1 protein kinases, which interact with and stimulate the activity of the cation chloride co-transporters NKCC1 (13–16) or NCC (14, 17) in HEK293 or HeLa cells. The expression of WNK1 appears also to be part of a pathway involved in inhibitory phosphorylation events of co-transporter KCC (18).
Mutations in the WNK1 and WNK4 genes have been linked to pseudo-hypoaldosteronism type II (or Gordon's syndrome), a rare familial form of hypertension (2), which is characterized by increased renal salt reabsorption and decreased potassium secretion. The identified WNK1 mutations increase WNK1 expression levels (2, 19) and in vitro data indicate that WNK1 inhibits WNK4, which itself regulates the renal sodium co-transporter NCC and the potassium transporter ROMK1, in particular the amount of these channel proteins present at the surface of collecting duct cells (20–22). In addition, WNK1 was shown to have WNK4-independent effects on ion channels: it stimulates clathrin-dependent endocytosis of ROMK1 by interaction with the scaffold protein intersectin 1 and this does not require WNK1 kinase activity (23, 24). Moreover, WNK1 activates the serum- and glucocorticoid-inducible protein kinase (SGK1), which phosphorylates the ubiquitin-protein isopeptide ligase Nedd4-2 so that less epithelial sodium channel is endocytosed from the plasma membrane of HEK293 cells (25).
Beside its role in ion channel regulation, WNK1 has also been implicated in protein secretion. In pancreatic β cells WNK1 binds to synaptotagmin 2 (26) and Munc18c (27), two proteins involved in the fusion of secretory membrane vesicles.
Altogether, these data suggest that WNK1 regulates the retention or insertion of various transmembrane channel proteins and that different downstream effector mechanisms are involved. Here we describe a role for WNK1 in regulating glucose transporter GLUT1 (SLC2A1 gene) in HEK293 cells. The constitutive expression of this transmembrane protein at the cell surface ensures glucose uptake in most tissues. We found that WNK1 through interaction with TBC1D4, a Rab GTPase-activating protein (GAP) involved in regulated exocytosis of glucose transporters, promotes GLUT1 surface expression.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
HEK293 (human embryonic kidney) cells were maintained in Dulbecco's minimal essential medium (DMEM) supplemented with 10 units/ml of penicillin, 10 μg/ml of streptomycin, and 10% fetal calf serum (Invitrogen), and regularly checked for absence of mycoplasm infection.
For ectopic expression of plasmid cDNAs, HEK293 cells were transfected at 80–90% confluence using Metafectene (Biontex, Martinsried/Planegg, Germany) according to the manufacturer's instructions. Transfection efficiencies were found to be 90% for HEK293 cells, as determined microscopically using a GFP expression vector. Total amounts of transfected plasmid DNA were kept constant at 6 μg/60-mm dish or 2 μg/35-mm dish and adjusted with empty vector if required. Cells were analyzed after 20 h for biochemical assays or after 16 h for immunofluorescence experiments.
For gene silencing, HEK293 at 30% confluence were transfected with 200 pmol of small interfering RNA oligonucleotides (siRNAs) per 35-mm dish or 400 pmol/60-mm dish using Lipofectamine 2000 (Invitrogen). Cells transfected with siRNAs were analyzed after 24 or 48 h and the achieved reduction in target gene expression was determined in each experiment by removing a 40-μl aliquot from the cell lysate for Western blot analysis. All results were confirmed in at least three independent experiments.
Expression Constructs and Small Interfering RNA Oligonucleotides
The previously described WNK1 full-length cDNA encoding amino acids 1–2382 (1) was subcloned as an EcoRI fragment into expression plasmids pcDNA3-Myc or pEGFP-c2. The kinase-dead WNK1 mutant K233M was made by site-directed mutagenesis of codon AAG to ATG using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). WNK1 fragments were generated as follows: WNK1(1–538) was amplified with primer pair PK12-5-FL1 (5′-TCT GGC GGC GCC GCA GAG AA) and PK12-R2 (5′-CTT GTG CAA CAT CTT CTG GGA CA-3′), cloned into pCR2.1-Topo (Invitrogen), and then transferred with EcoRI into pEGFP; WNK1(1–1340) was obtained by digestion of pEGFP-C2-WNK1 with AflII to delete an internal fragment and religation of the remaining vector; for WNK1(1307–2382), pEGFP-C2-WNK1 was digested with StuI and SalI, the fragment cloned into pBluescript and transferred as an EcoRI fragment into pEGFP; for WNK1(2030–2382), pEGFP-C2-WNK1 was digested with SacI and EcoRI and the fragment recloned into pEGFP. Previously published constructs used in this study were Myc-tagged hnRNPA1 (28) and pCR3.1/AS160–2myc (29). All constructs were verified by automated DNA sequencing.
Pools of three siRNA oligonucleotides were obtained from Santa Cruz Biotechnologies (Santa Cruz, CA) to deplete expression of endogenous TBC1D4 (sc-61654), Rab8A (sc-41828), or Rab8B (sc-106474). The siRNA targeting WNK1 was the previously validated siWNK1-a (5′-GCA GGA GUG UCU AGU UAU ATT) (30). As controls, pre-designed luciferase GL2 siRNA (5′-CGU ACG CGG AAU ACU UCG ATT) or a GFP-specific siRNA (5′-GGC UAC GUC CAG GAG CGC ACC TT) were used (all obtained from MWG-Biotech AG, Ebersberg, Germany).
Immunoprecipitation and Western Blot Procedures
For co-immunoprecipitation experiments, cells were grown in either 60- (transfected cells) or 100-mm dishes (endogenous proteins), lysed on ice in 250 μl of nondenaturing lysis buffer (50 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 100 mm NaCl, 10% glycerol, 10 mm MgCl2) supplemented with a protease inhibitor mixture composed of 1 mm PMSF, 1 mm 1,10-phenanthroline, 1 mm EGTA, 10 μm E64, and 10 μg/ml of each aprotinin, leupeptin, and pepstatin A (all from Sigma). The cell lysates were incubated for 2 h at 4 °C with the specified antibodies (2.5 μg/ml of anti-GFP ab1218 or anti-TBC1D4 ab24469 (Abcam, Cambridge, UK), anti-Myc clone 9E10 (M5546, Sigma), rabbit anti-Myc A14 (sc-789 from Santa Cruz Biotechnologies), or affinity-purified rabbit anti-WNK1 serum 308), then further incubated for 1 h with protein G-agarose beads (Roche Applied Science), and finally washed three times in cold lysis buffer containing 300 mm NaCl. Proteins were solubilized from the beads in 2× SDS sample buffer, separated in a 10% SDS-PAGE Protean III mini-gels (Bio-Rad).
For proteomic identification of proteins, the gel was stained in standard Coomassie Brilliant Blue R-250, destained and a digital image taken over a light box. Candidate bands were excised from the gel using a stereomicroscope, placed into sterile water, and shipped for mass spectrometry as indicated by the service provider Alphalyse (Odense, Denmark).
For detection of specific proteins, the polyacrylamide gel was transferred onto a PVDF membrane (Bio-Rad) using a Mini Trans-Blot cell (Bio-Rad; 100 V for 1 h) followed by Coomassie staining to check for equal transfer. Membranes were blocked in TBS, 0.1% Triton X-100, 5% milk powder, probed using the indicated antibodies, and then incubated with a secondary peroxidase-conjugated antibody (Bio-Rad) followed by chemiluminescence detection. Primary antibodies used for Western blots were rabbit anti-WNK1 serum 308 (affinity-purified polyclonal antibody raised and purified by Eurogentec (Ougree, Belgium) against peptide TSKDRPVSQPSLVGSKEC) (1), rabbit anti-Myc A14 (sc-789), rabbit anti-pan 14-3-3 K19 (sc-629), and anti-GLUT1 H43 (sc-7903) from Santa Cruz Biotechnologies, rabbit anti-GFP ab290 and rabbit anti-TBC1D4 ab24469 from Abcam (Cambridge, UK), mouse anti-Rab8 (clone 4/Rab4) and mouse anti-Rab4 (clone 7/Rab4) from BD Biosciences Europe (Erembodegem, Belgium), mouse anti-T7 tag and mouse anti-PCNA ab-1 (clone PC10) from Merck Biosciences (Nottingham, UK), mouse anti-α-tubulin (clone B5-1-2) from Sigma, and mouse anti-human transferrin receptor (clone H68.4, Invitrogen).
Biotinylation of Cell Surface Proteins
HEK293 cells were transfected as described above, washed twice with warm culture medium, and placed on ice in a cold room. Cells were incubated for 5 min with cold PBS-CM (PBS, pH 8.0, containing 0.1 mm CaCl2 and 1 mm MgCl2) and then for 30 min with 0.5 mg/ml of EZlink sulfo-NHS-SS-biotin (Pierce Biotechnology; catalogue number 21331) in PBS-CM before being rinsed twice and left for 10 min on ice with ice-cold Tris glycine (100 mm Tris/HCl, pH 8.0, 150 mm NaCl, 0.1 mm CaCl2, 1 mm MgCl2, 10 mm glycine, 1% BSA) to terminate the biotinylation reaction. Cells were again washed 3 times with cold PBS-CM and lysed in 250 μl of pulldown buffer (50 mm Tris/HCl, pH 7.5, 100 mm NaCl, 10% glycerol, 1% Nonidet P-40) in the presence of the aforementioned protease inhibitor mixture. The cell lysates were harvested by scraping and cleared by centrifugation at 16,000 × g at 4 °C for 5 min. An aliquot of 40 μl representing the total GLUT1 level was removed and added to 5× SDS sample buffer, whereas 200 ml of lysate was added to 50 μl of streptavidin-agarose beads (Sigma) previously incubated for 1 h in 1 ml of cold pulldown buffer containing 2% nonfat milk powder and washed 3 times in pulldown buffer. For purification of biotinylated proteins, lysate and beads were incubated under rotation for 1 h at 4 °C, the beads were collected by centrifugation (1 min at 3000 × g) and washed 3 times in cold wash buffer (100 mm Tris/HCl, pH 7.5, 300 mm NaCl, 1% Triton X-100). Captured proteins were recovered by boiling the beads in 25 μl of 2× SDS sample buffer plus 2 μl of 1 m DTT. Total and biotinylated proteins were analyzed alongside by SDS-PAGE followed by Western blot.
Immunoprecipitation from Cell Lysates Stabilized by Cross-linking
For experiments using a cleavable chemical cross-linker, HEK293 cells were seeded in 60-mm dishes, transfected as described, rinsed 3 times in PBS-CM at room temperature, and incubated with 0.5 mm dithiobis(succinimidyl propionate (D3669 from Sigma) on ice for 30 min. Subsequently, cells were rinsed twice and left for 15 min on ice with ice-cold Tris glycine buffer (see above) to terminate the cross-linking reaction. Cells were again washed 3 times with cold PBS-CM and lysed in 500 μl of RIPA buffer (50 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with protease inhibitors (see above). To reduce extract viscosity 500 units/ml of benzonase (Sigma) and 5 mm MgCl2 were added. An aliquot of 80 μl representing the total protein levels was removed and added to 5× SDS sample buffer, whereas 400 μl of lysate were used for immunoprecipitation with anti-Myc antibodies, as described above. Proteins were solubilized from the beads in 2× SDS sample buffer supplemented with 5% β-mercaptoethanol to reverse the introduced cross-links and analyzed by SDS-PAGE followed by Western blot.
Production of Recombinant OSR1 and in Vitro Protein Kinase Assays
For the production of recombinant OSR1 (oxidative stress responsive-1 kinase), a reported WNK1 substrate (31), its coding sequence was amplified from cDNA clone IRAUp969A0428D (imaGenes, Berlin Germany) by PCR with primers Bam-Osr1-F (5′-GGA TCC ATG TCC GAG GAC TCG AG) and Hind-Osr1-R (5′-TTC GAA TTA GCT GAT GCT GAG CTG), cloned into pCR2.1 Topo, and then subcloned into the BamHI and HindIII sites of T7/His-tagged pET28 vector (Novagen). Subsequently, codon 164 of the cDNA was mutated from GAC to GCC using the QuikChange mutagenesis kit (Stratagene) to generate the kinase-dead OSR1-D164A mutant. The protein was expressed in the Escherichia coli BL21 strain under isopropyl 1-thio-β-d-galactopyranoside induction and the bacterial pellets were harvested at 1400 × g for 20 min and frozen. For protein extraction, pellets were resuspended in 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 1 mm DTT in the presence of the protease inhibitor mixture described above and then sonicated on ice in 10 cycles of 30 s with 10-s intervals (Sonics Vibra Cell sonicator, set at 40% amplitude). Following centrifugation of the extract at 16,000 × g, the supernatant was incubated with nickel-nitrilotriacetic acid-agarose beads (Qiagen, Hilden Germany) for 1 h at 4 °C. Beads were washed twice with cold lysis buffer containing 20 mm imidazole and protease inhibitors. Recombinant OSR1 was eluted in cold lysis buffer containing 250 mm imidazole, quantified, and stored in aliquots at −80 °C.
For in vitro protein kinase assays, cells were lysed in RIPA buffer supplemented with protease inhibitors and treated with benzonase (see above). Following immunoprecipitation of WNK1 as described above, the resulting beads were washed three times in cold lysis buffer containing 300 mm NaCl and then resuspended in 20 μl of kinase reaction buffer (30 mm Tris/HCl, pH 7.5, 10% glycerol, 1 mm DTT, 1 mm Na3VO4, 10 mm MgCl2, and 100 μm ATP), mixed with their substrate (either beads containing immunoprecipitated Myc-TBC1D4 or 500 ng of recombinant kinase-dead OSR1), and incubated in the presence of 5 μCi of [γ-32P]ATP at 30 °C for 30 min. Finally, 2× SDS sample buffer was added, samples were boiled and separated by SDS-PAGE followed by Western blot. Membranes were exposed to x-ray films and subsequently incubated with the indicated antibodies to document protein quantities.
Immunofluorescence Microscopy
HEK293 cells were grown on coverslips, transfected as indicated, fixed after 16 h with 4% formaldehyde (Merck Chemicals, 104003) in PBS, and permeabilized with 0.2% Triton X-100 in PBS. Cells were then immunolabeled for 1 h with rabbit anti-GLUT1 (United States Biologicals, G3900-03D), washed 3× 5 min in PBS, 0.01% Tween, and incubated with a secondary Alexa 532-conjugated antibody (Invitrogen). Coverslips were mounted on microscope slides with Vectashield (Vector Laboratories, Burlingame, CA), images were recorded on a Leica TCS-SPE confocal microscope and processed with Adobe Photoshop software.
RESULTS
WNK1 Exists in a Complex with TBC1D4
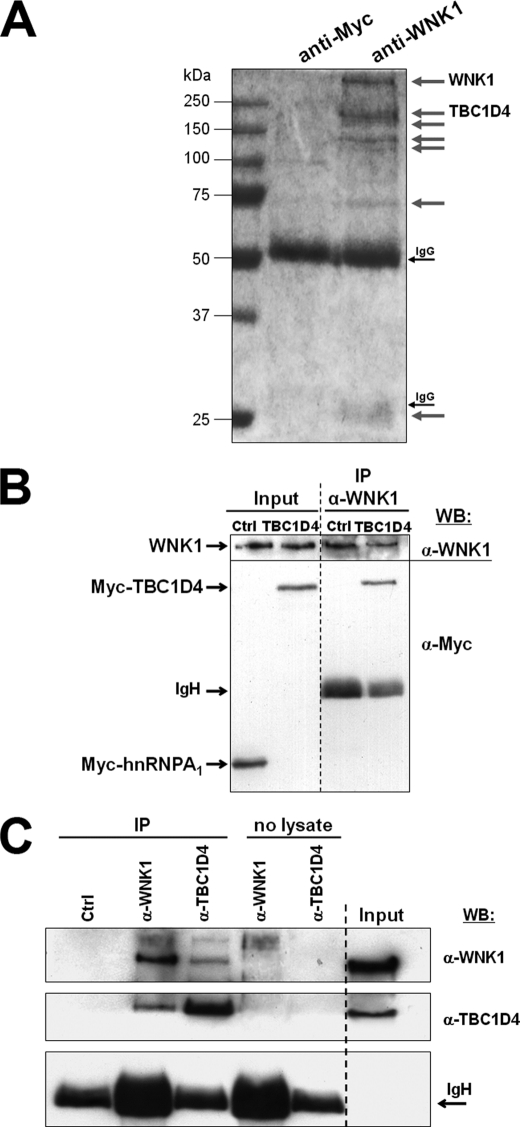
To identify proteins interacting under physiological conditions, endogenous WNK1 was immunoprecipitated under nondenaturing conditions from HEK293 cells using a previously described anti-WNK1 serum (1). The obtained protein fraction was then separated by gel electrophoresis and stained for the presence of co-immunoprecipitated proteins (Fig. 1A), which were excised from the gel and identified by mass spectrometry. Besides WNK1 itself, we identified a major co-purifying protein migrating above 150 kDa as the TBC1 (Tre-2/USP6, BUB2, Cdc16) domain family, member 4 (TBC1D4), also known as AS160 (AKT substrate of 160 kDa), a protein involved in insulin-stimulated glucose transport in adipocytes (32, 33).
FIGURE 1.
WNK1 forms a protein complex with TBC1D4 in HEK293 cells. A, cells were lysed under nondenaturing conditions and incubated with affinity-purified antipeptide antibodies against either human WNK1 or the Myc-epitope (negative control). Shown is the digital image of a Coomassie-stained gel and gray arrows indicate the bands that specifically co-precipitated with the anti-WNK1 antibodies. These bands were excised from the gel for mass spectrometric identification and those identified as WNK1 and TBC1D4 are indicated. Note that endogenous WNK1 protein has been described to migrate at an apparent molecular mass above 251 kDa (1, 10). B, HEK293 cells were transfected with Myc-TBC1D4 or Myc-hnRNP A1 (negative control), then lysed under nondenaturing conditions and incubated with anti-WNK1 to immunoprecipitate endogenous WNK1. The total cell lysate (Input) and the precipitate (IP) were analyzed by Western blot (WB) with the indicated antibodies. Note that Myc-TBC1D4 but not Myc-hnRNP A1 co-immunoprecipitates with endogenous WNK1. C, co-immunoprecipitation of endogenous TBC1D4 and WNK1. HEK293 cells were lysed under nondenaturing conditions, incubated for immunoprecipitation with a control (Ctrl) rabbit antibody, or anti-WNK1 or anti-TBC1D4 (IP) and the resulting fraction analyzed by Western blot. In addition, anti-WNK1 and anti-TBC1D4 antibodies were incubated in the absence of cell lysate (no lysate) to exclude that nonspecific antiserum-derived bands would confound the results. Note the presence of TBC1D4 in the sample precipitated with anti-WNK1 and the detection of WNK1 in the sample precipitated with anti-TBC1D4. The lane marked as Input shows the expression levels observed in whole cell lysates, and immunoglobulin heavy chains (IgH) are indicated to document success and comparability of the immunoprecipitation reactions.
For confirmation of the observed interaction, Myc-TBC1D4 was transfected into HEK293 cells and endogenous WNK1 was immunoprecipitated as above. Anti-Myc antibodies clearly detected the presence of TBC1D4 in the precipitate. As a control, the unrelated nuclear protein Myc-hnRNP A1 was expressed but did not co-immunoprecipitate with WNK1 (Fig. 1B). Moreover, both endogenous proteins co-immunoprecipitated, i.e. using specific anti-TBC1D4 or anti-WNK1 antibodies we could demonstrate by Western blot that endogenous TBC1D4 co-immunoprecipitated with endogenous WNK1, or vice versa (Fig. 1C).
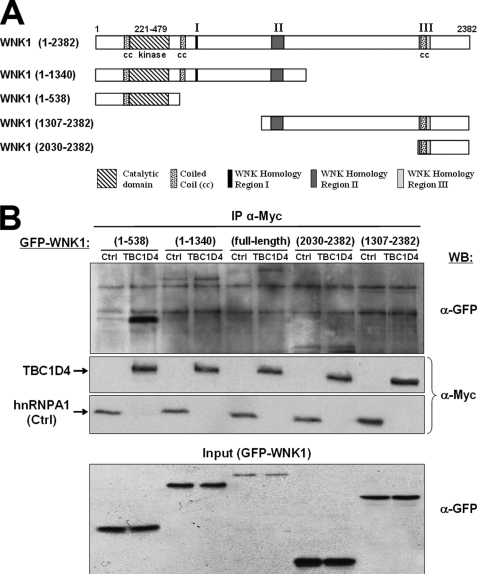
To determine which WNK1 domain mediates the interaction with TBC1D4, several GFP-tagged WNK1 deletion mutants were expressed and tested for their ability to co-immunoprecipitate with Myc-TBC1D4. The results showed that besides full-length WNK1(1–2382) the N-terminal WNK1(1–538) fragment had the strongest affinity to TBC1D4 (Fig. 2). The weak affinity for intermediate fragment WNK1(1–1340) suggests that intramolecular interactions exist that can modulate access to the N terminus. Together, these data validate the existence of a protein complex containing WNK1 and TBC1D4 in HEK293 cells.
FIGURE 2.
The N-terminal WNK1 region is required for protein complex formation with TBC1D4. A, schematic representation of full-length human WNK1 and the four fragments tested with the respective amino acid numbers in parentheses. B, HEK293 cells were co-transfected with one of the indicated GFP-WNK1 constructs and either Myc-TBC1D4 or Myc-hnRNP A1 (negative control). Following cell lysis under nondenaturing conditions, anti-Myc antibodies were added and immunoprecipitated proteins were analyzed by Western blot (WB). The lower panel shows the expression levels of GFP-WNK1 constructs in the whole cell lysate (Input), the middle panels show the quantities of precipitated Myc-tagged TBC1D4 and hnRNP A1, and the upper panel shows the amounts of co-precipitated GFP-WNK1. Note the specific co-immunoprecipitation of full-length WNK1 and its N-terminal fragment (1–538).
WNK1 Phosphorylates TBC1D4 in Vitro
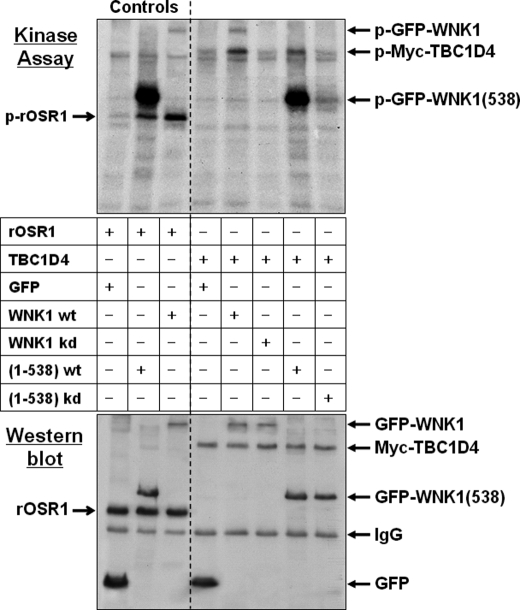
Given the preferential interaction with the N-terminal WNK1 fragment, which contains the catalytic domain, we next tested whether TBC1D4 served as a substrate for WNK1 kinase activity. For this, WNK1 was immunoprecipitated from HEK293 cells transfected with either GFP-WNK1 or the N-terminal GFP-WNK1(1–538) fragment. Cells were lysed in RIPA buffer and the immunoprecipitated WNK1 beads washed in high salt buffer so that contamination with other kinases from the cell lysate was minimized. The immunoprecipitated kinases were analyzed in an in vitro kinase assay followed by electrophoretic protein separation and transfer to PVDF membranes. As a positive control we added recombinant OSR1 to the assay, a recently described WNK1 substrate (31). As shown in Fig. 3, both WNK1 and its N-terminal fragment autophosphorylated and phosphorylated OSR1. To test TBC1D4 as a substrate, Myc-TBC1D4 was immunoprecipitated from transfected cells in RIPA buffer, the beads were washed in high-salt buffer and added as substrate to the kinase assay. We observed that both WNK1 and its N-terminal fragment phosphorylated TBC1D4. In contrast, the respective kinase-dead WNK1 mutants did not phosphorylate it (Fig. 3), confirming the absence of contaminant kinase activities.
FIGURE 3.
WNK1 phosphorylates TBC1D4 in vitro. HEK293 cells were transfected with either GFP empty vector, GFP-WNK1, fragment GFP-WNK1(1–538), or the respective kinase-dead (kd) WNK1-K233M mutants. WNK1 was immunoprecipitated using anti-GFP antibodies and tested in an in vitro protein kinase assay. The substrates were either recombinant OSR1 a previously described physiological substrate (31) (Controls), or beads containing immunoprecipitated Myc-TBC1D4. After 30 min of incubation the samples were denatured by adding SDS sample buffer, then separated by gel electrophoresis and transferred to PVDF blotting membranes. Incorporated radioactive [32P]phosphate was detected by exposing the membranes for 24 h to x-ray films (Kinase Assay). Subsequently, the amounts of recombinant T7-OSR1 or immunoprecipitated proteins were documented by Western blot using anti-GFP and then reprobed with either anti-T7 or anti-Myc antibodies. Note that both WT full-length WNK1 and its fragment (1–538) autophosphorylate and also phosphorylate the physiological substrates OSR1 and TBC1D4. Whereas fragment 1–538 shows stronger autophosphorylation activity, possibly due to partial deletion of the inhibitory domain (63), WT full-length WNK1 has higher activity toward TBC1D4. Beads containing the respective kinase-dead mutants revealed no phosphorylation activity.
WNK1 Catalytic Activity Increases the Association of Endogenous TBC1D4 with 14-3-3 Proteins
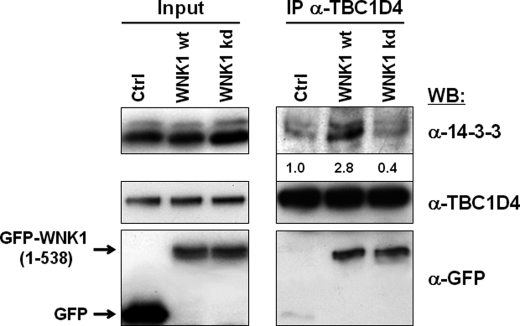
Previously, it has been described that TBC1D4 is a phosphoprotein that interacts with 14-3-3 (34–36), a family of proteins that bind phosphoproteins and modulate the corresponding signaling activities (37). We thus tested the effect of WNK1 expression on the interaction of TBC1D4 with 14-3-3 proteins. Endogenous TBC1D4 was then immunoprecipitated and the precipitate analyzed by Western blot. It was found that the protein complex formed between the endogenous TBC1D4 and WNK1 contained considerably increased amounts of 14-3-3 proteins. In contrast, cells expressing the kinase-dead WNK1 mutant showed only co-immunoprecipitation between WNK1 and TBC1D4 and no change in the association with 14-3-3, which remained at background levels comparable with that observed in GFP-expressing control cells (Fig. 4).
FIGURE 4.
Expression of WNK1 increases the association of endogenous TBC1D4 with 14-3-3 proteins. HEK293 cells were transfected with either GFP empty vector, or fragment GFP-WNK1(1–538) or its respective kinase-dead mutant. Endogenous TBC1D4 was immunoprecipitated from cell lysates prepared under nondenaturing conditions and probed by Western blot (WB). Shown are the levels of GFP-WNK1 and endogenous TBC1D4 and 14-3-3 proteins in total cell lysates (Input; left panel) and in the immunoprecipitate (IP; right panel). The quantification of the 14-3-3 band intensities is given below the blot as fold over control values. Note that the amount of 14-3-3 co-precipitating with TBC1D4 increased considerably in the presence of WNK1, not, however, of kinase-dead WNK1.
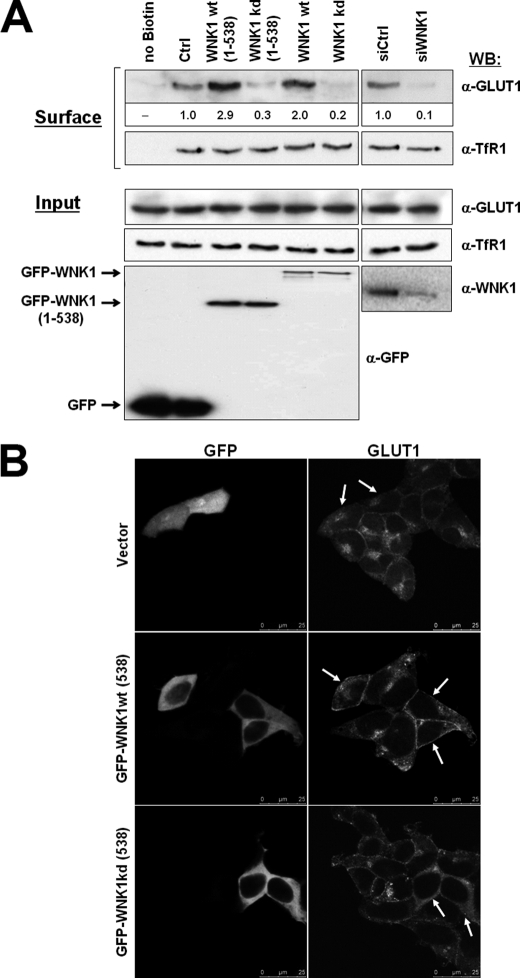
WNK1 Leads to Increased Expression of GLUT1 at the Cell Surface
In insulin-sensitive adipocytes and skeletal muscle cells TBC1D4 was shown to control the translocation of the insulin-responsive glucose transporter GLUT4 from intracellular storage vesicles to the plasma membrane (33, 38). Most other cell types, however, express the transporter GLUT1 for their constitutive uptake of glucose (39). We thus tested whether the observed phosphorylation of TBC1D4 by WNK1 affected the expression of endogenous GLUT1 at the cell surface. HEK293 cells were transfected with either full-length WNK1, or WNK1(1–538) or their kinase-dead mutants K233M and after 20 h of expression transferred to 4 °C for biotinylation of cell surface proteins. Following cell lysis, biotinylated proteins were captured using streptavidin-agarose beads and analyzed by Western blot using specific antibodies. As positive and negative controls, we confirmed that the biotinylated protein fraction contained the surface protein E-cadherin but not cytosolic tubulin or the nuclear protein PCNA (data not shown). Under these experimental conditions we found increased GLUT1 cell surface expression in the presence of catalytically active WNK1 (either full-length or fragment 1–538) (Fig. 5A). In contrast, expression of the kinase-dead mutants inhibited the level of surface GLUT1 compared with control cells. As further support for the role of WNK1, we depleted endogenous WNK1 from HEK293 cells by RNA interference and analyzed endogenous GLUT1 surface expression. In cells depleted of WNK1 a significant reduction in biotinylated GLUT1 was detected (Fig. 5A). Under these experimental conditions the cell surface levels of the endogenous transferrin receptor (TfR1) did not change (Fig. 5A).
FIGURE 5.
Expression of WNK1 affects the amount of GLUT1 expressed at the cell surface. A, HEK293 cells were transfected as indicated with either GFP empty vector (Ctrl), full-length GFP-WNK1, fragment GFP-WNK1(1–538), or the respective kinase-dead mutants, as indicated. In separate experiments, HEK293 cells were transfected with either control (siCtrl) or WNK1-specific (siWNK1) small interfering oligonucleotides. Following 20 h cell surface proteins were biotinylated and captured from the cell lysate using streptavidin beads. Shown are Western blots (WB) detecting GLUT1 or transferrin receptor TfR1 in the biotinylated protein fraction (top panels, Surface). The corresponding quantification of GLUT1 band intensities is given as fold over control values below the blot. Shown also are the expression levels in whole cell lysates (Input) of endogenous GLUT1 and TfR1 (middle panels) and of GFP-tagged proteins or endogenous WNK1 (bottom panels). Note that GLUT1 surface levels increase upon expression of catalytically active WNK1 but decrease in WNK1-depleted cells, whereas surface levels of TfR1 remained unaffected. B, visualization of endogenous GLUT1 protein by immunofluorescence microscopy. HEK293 cells were transfected as indicated with GFP empty vector, GFP-WNK1(1–538), or the respective kinase-dead mutant. Cells were fixed after 16 h of expression, stained with anti-GLUT1 followed by a secondary Alexa 532-conjugated antibody and images were recorded on a Leica SPE confocal microscope. Transfected cells are marked by white arrows. Note the increased surface staining of GLUT1 in cells expressing WNK1 in contrast to increased intracellular GLUT1 staining upon expression of kinase-dead WNK1.
The cell surface expression of endogenous GLUT1 was also determined by immunofluorescence microscopy. In HEK293 cells expressing catalytically active WNK1 as above, we observed an increased surface staining with anti-GLUT1 antibodies compared with nontransfected cells (Fig. 5B). On the contrary, expression of kinase-dead WNK1 reduced GLUT1 staining at the cell surface, whereas increasing the intracellular GLUT1 signal (Fig. 5B).
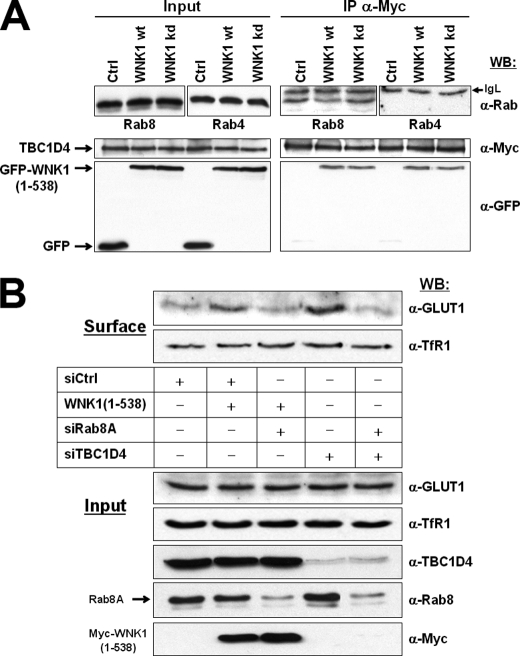
Rab8A Is Required for GLUT1 Expression Downstream of WNK1
The above data are compatible with the idea that phosphorylation of TBC1D4 by WNK1 increases its association with 14-3-3 proteins and inhibits TBC1D4 as a negative regulator of GLUT1 surface expression. TBC1D4 was described to act as a GAP for Rab2A, Rab8A, Rab10, and Rab14 (40, 41). Because Rab8 is involved in constitutive biosynthetic trafficking from the trans-Golgi network to the plasma membrane (42, 43), we tested its involvement in GLUT1 surface expression. We co-expressed GFP-WNK1(1–538) or its kinase-dead mutant and Myc-TBC1D4 in HEK293 cells. Due to the transient nature of GAP-GTPase interactions, cells were briefly treated with the mild cross-linking reagent dithiobis(succinimidyl propionate) before cell lysis, then Myc-TBC1D4 was immunoprecipitated and the precipitated fraction analyzed for the presence of either endogenous Rab8 or Rab4 (as negative control). Whereas no Rab4 was detected, the endogenous Rab8 clearly co-immunoprecipitated. In addition, the amount of Rab8 in the complex with TBC1D4 was reduced in the presence of WNK1, but increased when the kinase-dead WNK1 mutant was expressed (Fig. 6A). These data indicate that the Rab-GAP TBC1D4 interacts transiently with Rab8 in HEK293 cells and that phosphorylation of TBC1D4 by WNK1 affects the amount of Rab8 engaged.
FIGURE 6.
Rab8A is required for GLUT1 expression downstream of WNK1. A, HEK293 cells were transfected as indicated and treated briefly with the mild reversible cross-linking reagent dithiobis(succinimidyl propionate) before lysis. Myc-TBC1D4 was immunoprecipitated and analyzed by Western blot (WB). Shown are the expression levels of GFP-WNK1(1–538), Myc-TBC1D4, and the endogenous Rab8 and Rab4 proteins in total cell lysates (left side panels, Input) as well as the immunoprecipitated fractions (right side panels, IP). Note that Rab8 but not Rab4 co-precipitated with TBC1D4 and that the amount decreased in the presence of WNK1, but increased in the presence of kinase-dead WNK1. B, HEK293 cells were transfected with control (siGFP) or Rab8A-specific (siRab8A) small interfering RNAs, either alone or in the presence of Myc-WNK1(1–538). In parallel, cells were transfected with TBC1D4-specific siRNAs (siTBC1D4), either alone or in combination with siRab8A. As described in Fig. 5A, the biotinylated cell surface proteins (Surface) and whole cell lysates (Input) were analyzed by Western blot to document the amounts of endogenous GLUT1, TfR1, TBC1D4, or Rab8, as well as of transfected Myc-WNK1(1–538). Note the successful depletion of TBC1D4 (lanes 4 and 5) and Rab8A (lanes 3 and 5). GLUT1 surface expression was promoted by either expression of WNK1 or depletion of TBC1D4 and the simultaneous knockdown of Rab8A impaired both effects. Under these conditions the surface levels of endogenous TfR1 remained unchanged.
To substantiate that GLUT1 traffic is regulated by WNK1/TBC1D4 through Rab8, endogenous Rab8A and Rab8B were first depleted by specific small interfering oligonucleotides. These experiments (data not shown) identified the slower migrating band as Rab8A and revealed it to be the predominantly expressed form in HEK293 cells. To provide evidence that Rab8A operates downstream of WNK1, HEK293 cells were first transfected with siRab8A and 24 h later transfected with WNK1(1–538). Under these conditions of Rab8A depletion, the transfection of WNK1 no longer promoted GLUT1 surface expression (Fig. 6B). Also, the depletion of endogenous TBC1D4 by siRNAs clearly increased GLUT1 levels at the surface but the simultaneous transfection with siRab8A completely impaired the increase in GLUT1 observed upon depletion of TBC1D4 alone. Again, under these conditions the cell surface levels of endogenous TfR1 remained unaffected (Fig. 6B). Together, these experiments clearly revealed that Rab8A is involved downstream of WNK1 and TBC1D4 in the control of GLUT1 expression at the surface of HEK293 cells.
DISCUSSION
The main conclusion from the present work is that protein kinase WNK1 forms a complex with the Rab-GAP TBC1D4 that is involved in regulating the amount of glucose transporter GLUT1 expressed at the cell surface.
The complex formation between TBC1D4 and WNK1 was initially identified using an unbiased proteomic approach but then confirmed by immunoprecipitation using various antibodies. In particular, anti-Myc antibodies detected co-immunoprecipitation of epitope-tagged Myc-TBC1D4 together with GFP-WNK1 (Fig. 2) as well as with endogenous WNK1 (Fig. 1B). Furthermore, GFP-tagged WNK1 was pulled down when antibodies against endogenous TBC1D4 were used (Fig. 4) and the co-immunoprecipitation of both the endogenous proteins could also be demonstrated in HEK293 cells (Fig. 1C). Together, these experiments provide solid support for the specificity of the observed interaction between WNK1 and TBC1D4.
TBC1D4 was previously identified to regulate GLUT4 trafficking to the cell surface following insulin stimulation of 3T3L1 adipocytes or skeletal muscle cells. This pathway involves phosphorylation of TBC1D4 by protein kinase AKT and have led to its alternative designation as AKT substrate of 160 kDa (AS160) (32, 33, 38, 40, 44). Experimental suppression of TBC1D4 leads to leaky and insulin-independent release of GLUT4 into the plasma membrane (45). There is considerable evidence that in the absence of insulin the GAP domain of TBC1D4 keeps a critical Rab protein in its inactive GDP-bound state to prevent GLUT4 vesicle release (46). Upon insulin treatment of adipocytes, AKT-phosphorylated TBC1D4 associates with 14-3-3 proteins (35, 41, 44) and becomes inhibited.
In agreement with a previous report, TBC1D4 can act also as a negative regulator of ubiquitously expressed GLUT1 and not only of GLUT4 in adiopocytes (47). Our results show that protein kinase WNK1 is involved in the regulation of GLUT1, which secures glucose transport in noninsulin target cells. WNK1 forms a complex with TBC1D4 and phosphorylation by WNK1 leads to increased interaction of TBC1D4 with 14-3-3 proteins (Fig. 4). Interestingly, a previous proteomic analysis in HEK293 cells used mass spectrometry to identify proteins that associate with FLAG-tagged 14-3-3 proteins and identified both TBC1D4 and WNK1 among a list of 170 unique proteins (34).
We found that TBC1D4 interacted preferentially with an N-terminal WNK1 fragment containing the catalytic domain (Fig. 2). Consistent with this observation, we detected the phosphorylation of TBC1D4 by WNK1 in vitro (Fig. 3). For these experiments WNK1 was precipitated under stringent conditions including SDS-containing RIPA lysis buffer and 300 mm NaCl containing washing steps. Because no phosphorylation was obtained when a kinase-dead WNK1 mutant was prepared under these conditions, our data provide strong evidence that TBC1D4 is a specific WNK1 substrate. The observed phosphorylation activity was comparable with that of the described physiological model substrate OSR1 (31).
TBC1D4 is known to be a phosphoprotein and eight sites (Ser318, Ser341, Thr568, Ser570, Ser588, Thr642, Ser666, and Ser751) have been identified that can be phosphorylated in vivo (36, 44). Further in vitro studies have revealed that at least four kinases (AKT, SGK1, RSK1, and AMPK) can phosphorylate TBC1D4 in distinct stimulus-dependent patterns (36), however, the functional difference between the individual phosphorylation sites is unclear and requires more detailed studies.
The WNK1-mediated phosphorylation of TBC1D4 resulted in increased amounts of GLUT1 at the surface of HEK293 cells. Under these experimental conditions we also observed a slight increase in cell number (data not shown), indicative of stimulated metabolic activity and cell growth. In contrast, expression of kinase-dead WNK1 mutants inhibited the level of surface GLUT1 compared with control cells. Moreover, the suppression of endogenous WNK1 by small interfering RNA oligonucleotides also decreased surface amounts of GLUT1. These effects were demonstrated by biotinylation of cell surface proteins (Fig. 5A) and by immunofluorescence microscopy (Fig. 5B). Under these experimental conditions no changes in the surface levels of the endogenous transferrin receptor 1 were observed (Fig. 5A), demonstrating the specificity of WNK1/TBC1D4 activity on GLUT1 surface expression.
The WNK1 substrate TBC1D4 has been found to exhibit selective GAP activity toward Rab2A, -8A, -8B, -10, and -14 in vitro. In adipocytes, specific depletion of Rab10 was identified to reduce GLUT4 translocation (40, 48, 49), whereas in L6 muscle cell lines Rab8A and Rab14 appeared to be more important for regulating GLUT4 (41, 50). Thus, depending on the cell type, TBC1D4 may control distinct Rab GTPases.
In HEK293 cells we provide evidence that Rab8A is involved in the WNK1-regulated GLUT1 traffic. Rab8 is known to participate in exocytic events (42, 43) and exists in two isoforms: Rab8A and Rab8B. Our studies show that Rab8A is the predominantly expressed form in the HEK293 cell and that endogenous Rab8A associates with TBC1D4 in a complex, the amount of which depends on the catalytic activity of WNK1 (Fig. 6A). When HEK293 cells were transfected with Rab8A-specific siRNAs, the WNK1-induced surface expression of GLUT1 was significantly reduced (Fig. 6B). Similarly, depletion of Rab8A specifically prevented the increase in GLUT1 surface expression observed following depletion of TBC1D4 by siRNA treatment. Under the same experimental conditions the surface levels of TfR1 did not change (Fig. 6B). These results demonstrate the requirement of Rab8A for GLUT1 regulation in HEK293 cells and that it operates downstream of WNK1 and TBC1D4.
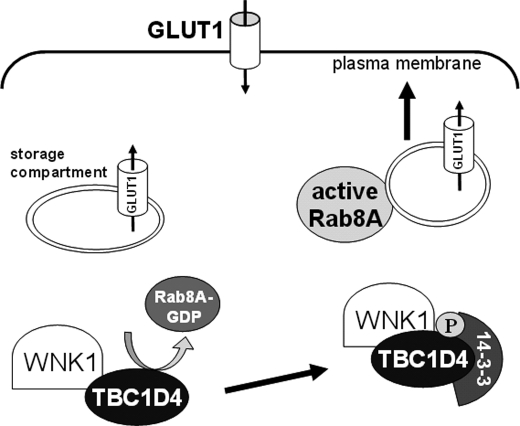
These data are in agreement with the current view in the literature that TBC1D4 maintains an intracellular storage compartment for glucose transporter proteins under basal conditions (51–53). This compartment can be mobilized upon phosphorylation of TBC1D4, by promoting its binding to 14-3-3 proteins and subsequent inactivation of its Rab-GAP activity. This permits the forward trafficking of the transporters to the apical membrane. Based on this view, the pathway involving WNK1 in the control of GLUT1 levels at the surface of HEK293 cells is schematically depicted in Fig. 7 (see figure legend for details).
FIGURE 7.
Proposed model for the role of the WNK1-TBC1D4 complex in regulating GLUT1 traffic in HEK293 cells. A fraction of GLUT1 is kept in an endosomal storage compartment in the cytosol. In the storage compartment TBC1D4 keeps Rab8A in the inactive GDP-bound conformation and prevents vesicle trafficking to the plasma membrane. Upon activation of WNK1, TBC1D4 becomes phosphorylated and associates with the phosphoprotein-binding 14-3-3 adaptor molecules so that the GAP activity of TBC1D4 is inhibited. Following Rab8A activation by GDP/GTP exchange, delivery of GLUT1-containing vesicles to the plasma membrane can occur promoting increased cellular glucose uptake. The upstream stimuli regulating WNK1 activation remain to be determined.
Taken together, our data describe a pathway regulating surface expression of GLUT1 that is of considerable biomedical interest. First, malignant cells have accelerated metabolism and satisfy their increased requirements for ATP production by aerobic glycolysis (54). Especially during the initiation of tumor formation, cells experience suboptimal supply of oxygen and nutrients due to diffusion limits. One mechanism that tumor cells use to adapt to these conditions is GLUT1 overexpression (55) and our data suggest that changes in expression or regulation of WNK1 may be an alternative route for adaptation. The WNK catalytic domain represents a unique target site for the development of small molecule kinase inhibitors, because it is characterized by a typical sequence variation in which a lysine from subdomain I substitutes for a usually highly conserved catalytic lysine in subdomain II (hence their name with no [K] = lysine) (5, 6). Second, the described pathway may be an interesting drug target in certain diseases linked to epilepsy and mental retardation that are characterized by insufficient GLUT1 activity (56, 57). Finally, hypertensive glomerular injury has been linked to pathological overexpression of GLUT1 in renal cells (58). This is intriguing because some patients with familial hypertension (Gordon syndrome) carry germline WNK1 mutations that lead to WNK1 overexpression. Our data clearly suggest that increased expression of WNK1 promotes GLUT1 surface expression in a kidney-derived cell line. The clinical features of the WNK1 mutant families have been suggested to reflect a form of hypertension that is not exclusively salt dependent, as opposed to families with mutant WNK4 (59, 60). Thus, WNK1 may in part affect renal salt balance by glucose overload causing glomerular insufficiency.
In the future it will be important to elucidate upstream mechanisms involved in the WNK1-mediated activation of GLUT1 trafficking. Previous studies reported that hypertonic stress, including glucose can increase WNK1 activity. Treatment of distal collecting duct cells with 0.5 m glucose, for example, promoted a significant increase in WNK1 activity (61). Very recent evidence also revealed that TBC1D4 regulates other transporter proteins. Following aldosterone stimulation of cortical collecting duct cells, TBC1D4 regulated trafficking of the epithelial sodium channel epithelial sodium channel to the apical membrane leading to increased sodium absorption (62). Similar to our results, this involved phosphorylation of TBC1D4 and increased association with 14-3-3 proteins but was mediated by another kinase, SGK1. Intriguingly, WNK1 has been proposed to serve as a scaffold required for efficient SGK1 activation and there is evidence that SGK1 may in turn phosphorylate WNK1 on threonine 60 (12). Taken together, these data indicate TBC1D4 as a central player in the surface expression levels of GLUT4, GLUT1, and epithelial sodium channel and it will be interesting to determine whether phosphorylation by WNK1, SGK1, or AKT represent cell type-specific pathways or a common regulatory kinase cascade.
Acknowledgments
We thank A. Albiston (Melbourne, Australia) and H. König, (Karlsruhe, Germany) for kindly providing vectors pCR3.1/AS160–2myc and Myc-hnRNP A1, respectively.
This work was supported by Portuguese Fundação para a Ciência e Tecnologia through Grant POCI_PPCDT/SAU-MMO/56439/04, Programa de Financiamento Plurianual do CIGMH, and Fellowship BD 23001/05 (to A. I. M.).
- WNK
- With-No-K (=lysine) protein kinase
- GLUT1
- solute carrier family 2 (facilitated glucose transporter) member 1
- TBC1D4
- TBC1 (Tre-2/USP6, BUB2, Cdc16) domain family member 4
- OSR1
- oxidative stress responsive-1 kinase
- TfR
- transferrin receptor
- GAP
- GTPase-activating protein.
REFERENCES
- 1.Veríssimo F., Jordan P. (2001) Oncogene 20, 5562–5569 [DOI] [PubMed] [Google Scholar]
- 2.Wilson F. H., Disse-Nicodème S., Choate K. A., Ishikawa K., Nelson-Williams C., Desitter I., Gunel M., Milford D. V., Lipkin G. W., Achard J. M., Feely M. P., Dussol B., Berland Y., Unwin R. J., Mayan H., Simon D. B., Farfel Z., Jeunemaitre X., Lifton R. P. (2001) Science 293, 1107–1112 [DOI] [PubMed] [Google Scholar]
- 3.Choate K. A., Kahle K. T., Wilson F. H., Nelson-Williams C., Lifton R. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delaloy C., Hadchouel J., Imbert-Teboul M., Clemessy M., Houot A. M., Jeunemaitre X. (2006) Am. J. Pathol. 169, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu B., English J. M., Wilsbacher J. L., Stippec S., Goldsmith E. J., Cobb M. H. (2000) J. Biol. Chem. 275, 16795–16801 [DOI] [PubMed] [Google Scholar]
- 6.Min X., Lee B. H., Cobb M. H., Goldsmith E. J. (2004) Structure 12, 1303–1311 [DOI] [PubMed] [Google Scholar]
- 7.Moniz S., Jordan P. (2010) Cell. Mol. Life Sci. 67, 1265–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu B. E., Stippec S., Lenertz L., Lee B. H., Zhang W., Lee Y. K., Cobb M. H. (2004) J. Biol. Chem. 279, 7826–7831 [DOI] [PubMed] [Google Scholar]
- 9.Lee B. H., Chen W., Stippec S., Cobb M. H. (2007) J. Biol. Chem. 282, 17985–17996 [DOI] [PubMed] [Google Scholar]
- 10.Vitari A. C., Deak M., Collins B. J., Morrice N., Prescott A. R., Phelan A., Humphreys S., Alessi D. R. (2004) Biochem. J. 378, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Z. Y., Zhou Q. L., Holik J., Patel S., Leszyk J., Coleman K., Chouinard M., Czech M. P. (2005) J. Biol. Chem. 280, 21622–21628 [DOI] [PubMed] [Google Scholar]
- 12.Xu B. E., Stippec S., Lazrak A., Huang C. L., Cobb M. H. (2005) J. Biol. Chem. 280, 34218–34223 [DOI] [PubMed] [Google Scholar]
- 13.Vitari A. C., Deak M., Morrice N. A., Alessi D. R. (2005) Biochem. J. 391, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moriguchi T., Urushiyama S., Hisamoto N., Iemura S., Uchida S., Natsume T., Matsumoto K., Shibuya H. (2005) J. Biol. Chem. 280, 42685–42693 [DOI] [PubMed] [Google Scholar]
- 15.Anselmo A. N., Earnest S., Chen W., Juang Y. C., Kim S. C., Zhao Y., Cobb M. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10883–10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitari A. C., Thastrup J., Rafiqi F. H., Deak M., Morrice N. A., Karlsson H. K., Alessi D. R. (2006) Biochem. J. 397, 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson C., Rafiqi F. H., Karlsson H. K., Moleleki N., Vandewalle A., Campbell D. G., Morrice N. A., Alessi D. R. (2008) J. Cell. Sci. 121, 675–684 [DOI] [PubMed] [Google Scholar]
- 18.Rinehart J., Maksimova Y. D., Tanis J. E., Stone K. L., Hodson C. A., Zhang J., Risinger M., Pan W., Wu D., Colangelo C. M., Forbush B., Joiner C. H., Gulcicek E. E., Gallagher P. G., Lifton R. P. (2009) Cell 138, 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delaloy C., Elvira-Matelot E., Clemessy M., Zhou X. O., Imbert-Teboul M., Houot A. M., Jeunemaitre X., Hadchouel J. (2008) Hypertension 52, 1149–1154 [DOI] [PubMed] [Google Scholar]
- 20.Gamba G. (2005) Am. J. Physiol. Renal Physiol. 288, F245–252 [DOI] [PubMed] [Google Scholar]
- 21.Kahle K. T., Ring A. M., Lifton R. P. (2008) Annu. Rev. Physiol. 70, 329–355 [DOI] [PubMed] [Google Scholar]
- 22.Richardson C., Alessi D. R. (2008) J. Cell. Sci. 121, 3293–3304 [DOI] [PubMed] [Google Scholar]
- 23.Cope G., Murthy M., Golbang A. P., Hamad A., Liu C. H., Cuthbert A. W., O'Shaughnessy K. M. (2006) J. Am. Soc. Nephrol. 17, 1867–1874 [DOI] [PubMed] [Google Scholar]
- 24.He G., Wang H. R., Huang S. K., Huang C. L. (2007) J. Clin. Invest. 117, 1078–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu B. E., Stippec S., Chu P. Y., Lazrak A., Li X. J., Lee B. H., English J. M., Ortega B., Huang C. L., Cobb M. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10315–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee B. H., Min X., Heise C. J., Xu B. E., Chen S., Shu H., Luby-Phelps K., Goldsmith E. J., Cobb M. H. (2004) Mol. Cell. 15, 741–751 [DOI] [PubMed] [Google Scholar]
- 27.Oh E., Heise C. J., English J. M., Cobb M. H., Thurmond D. C. (2007) J. Biol. Chem. 282, 32613–32622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matter N., Marx M., Weg-Remers S., Ponta H., Herrlich P., König H. (2000) J. Biol. Chem. 275, 35353–35360 [DOI] [PubMed] [Google Scholar]
- 29.Peck G. R., Ye S., Pham V., Fernando R. N., Macaulay S. L., Chai S. Y., Albiston A. L. (2006) Mol. Endocrinol. 20, 2576–2583 [DOI] [PubMed] [Google Scholar]
- 30.Moniz S., Veríssimo F., Matos P., Brazão R., Silva E., Kotelevets L., Chastre E., Gespach C., Jordan P. (2007) Oncogene 26, 6071–6081 [DOI] [PubMed] [Google Scholar]
- 31.Zagórska A., Pozo-Guisado E., Boudeau J., Vitari A. C., Rafiqi F. H., Thastrup J., Deak M., Campbell D. G., Morrice N. A., Prescott A. R., Alessi D. R. (2007) J. Cell. Biol. 176, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson R. T., Pessin J. E. (2006) Trends Biochem. Sci. 31, 215–222 [DOI] [PubMed] [Google Scholar]
- 33.Sakamoto K., Holman G. D. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin J., Smith F. D., Stark C., Wells C. D., Fawcett J. P., Kulkarni S., Metalnikov P., O'Donnell P., Taylor P., Taylor L., Zougman A., Woodgett J. R., Langeberg L. K., Scott J. D., Pawson T. (2004) Curr. Biol. 14, 1436–1450 [DOI] [PubMed] [Google Scholar]
- 35.Ramm G., Larance M., Guilhaus M., James D. E. (2006) J. Biol. Chem. 281, 29174–29180 [DOI] [PubMed] [Google Scholar]
- 36.Geraghty K. M., Chen S., Harthill J. E., Ibrahim A. F., Toth R., Morrice N. A., Vandermoere F., Moorhead G. B., Hardie D. G., MacKintosh C. (2007) Biochem. J. 407, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackintosh C. (2004) Biochem. J. 381, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koumanov F., Holman G. D. (2007) Biochem. J. 403, e9–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao F. Q., Glimm D. R., Kennelly J. J. (1993) Int. J. Biochem. 25, 1897–1903 [DOI] [PubMed] [Google Scholar]
- 40.Mîinea C. P., Sano H., Kane S., Sano E., Fukuda M., Peränen J., Lane W. S., Lienhard G. E. (2005) Biochem. J. 391, 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikura S., Bilan P. J., Klip A. (2007) Biochem. Biophys. Res. Commun. 353, 1074–1079 [DOI] [PubMed] [Google Scholar]
- 42.Zerial M., McBride H. (2001) Nat. Rev. Mol. Cell. Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 43.Stenmark H. (2009) Nat. Rev. Mol. Cell. Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 44.Sano H., Kane S., Sano E., Mîinea C. P., Asara J. M., Lane W. S., Garner C. W., Lienhard G. E. (2003) J. Biol. Chem. 278, 14599–14602 [DOI] [PubMed] [Google Scholar]
- 45.Eguez L., Lee A., Chavez J. A., Miinea C. P., Kane S., Lienhard G. E., McGraw T. E. (2005) Cell. Metab. 2, 263–272 [DOI] [PubMed] [Google Scholar]
- 46.Stöckli J., Davey J. R., Hohnen-Behrens C., Xu A., James D. E., Ramm G. (2008) Mol. Endocrinol. 22, 2703–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ngo S., Barry J. B., Nisbet J. C., Prins J. B., Whitehead J. P. (2009) Mol. Cell. Endocrinol. 302, 33–40 [DOI] [PubMed] [Google Scholar]
- 48.Sano H., Eguez L., Teruel M. N., Fukuda M., Chuang T. D., Chavez J. A., Lienhard G. E., McGraw T. E. (2007) Cell. Metab. 5, 293–303 [DOI] [PubMed] [Google Scholar]
- 49.Sano H., Roach W. G., Peck G. R., Fukuda M., Lienhard G. E. (2008) Biochem. J. 411, 89–95 [DOI] [PubMed] [Google Scholar]
- 50.Ishikura S., Klip A. (2008) Am. J. Physiol. Cell. Physiol. 295, C1016-C1025 [DOI] [PubMed] [Google Scholar]
- 51.Pessin J. E., Thurmond D. C., Elmendorf J. S., Coker K. J., Okada S. (1999) J. Biol. Chem. 274, 2593–2596 [DOI] [PubMed] [Google Scholar]
- 52.Dugani C. B., Klip A. (2005) EMBO Rep. 6, 1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leney S. E., Tavaré J. M. (2009) J. Endocrinol. 203, 1–18 [DOI] [PubMed] [Google Scholar]
- 54.Gatenby R. A., Gillies R. J. (2004) Nat. Rev. Cancer. 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 55.Macheda M. L., Rogers S., Best J. D. (2005) J. Cell. Physiol. 202, 654–662 [DOI] [PubMed] [Google Scholar]
- 56.Klepper J., Leiendecker B. (2007) Dev. Med. Child Neurol. 49, 707–716 [DOI] [PubMed] [Google Scholar]
- 57.Brockmann K. (2009) Brain Dev. 31, 545–552 [DOI] [PubMed] [Google Scholar]
- 58.Gnudi L., Viberti G., Raij L., Rodriguez V., Burt D., Cortes P., Hartley B., Thomas S., Maestrini S., Gruden G. (2003) Hypertension 42, 19–24 [DOI] [PubMed] [Google Scholar]
- 59.Xie J., Craig L., Cobb M. H., Huang C. L. (2006) Pediatr. Nephrol. 21, 1231–1236 [DOI] [PubMed] [Google Scholar]
- 60.Hadchouel J., Delaloy C., Fauré S., Achard J. M., Jeunemaitre X. (2006) J. Am. Soc. Nephrol. 17, 208–217 [DOI] [PubMed] [Google Scholar]
- 61.Lenertz L. Y., Lee B. H., Min X., Xu B. E., Wedin K., Earnest S., Goldsmith E. J., Cobb M. H. (2005) J. Biol. Chem. 280, 26653–26658 [DOI] [PubMed] [Google Scholar]
- 62.Liang X., Butterworth M. B., Peters K. W., Frizzell R. A. (2010) Mol. Biol. Cell. 21, 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu B. E., Min X., Stippec S., Lee B. H., Goldsmith E. J., Cobb M. H. (2002) J. Biol. Chem. 277, 48456–48462 [DOI] [PubMed] [Google Scholar]