Abstract
Fundamental knowledge about how G protein-coupled receptors and their ligands interact is important for understanding receptor-ligand binding and the development of new drug discovery strategies. We have used cross-linking and tandem mass spectrometry analyses to investigate the interaction of the N terminus of the Saccharomyces cerevisiae tridecapeptide pheromone, α-factor (WHWLQLKPGQPMY), and Ste2p, its cognate G protein-coupled receptor. The Trp1 residue of α-factor was replaced by 3,4-dihydroxyphenylalanine (DOPA) for periodate-mediated chemical cross-linking, and biotin was conjugated to Lys7 for detection purposes to create the peptide [DOPA1,Lys7(BioACA),Nle12]α-factor, called Bio-DOPA1-α-factor. This ligand analog was a potent agonist and bound to Ste2p with ∼65 nanomolar affinity. Immunoblot analysis of purified Ste2p samples that were treated with Bio-DOPA1-α-factor showed that the peptide analog cross-linked efficiently to Ste2p. The cross-linking was inhibited by the presence of either native α-factor or an α-factor antagonist. MALDI-TOF and immunoblot analyses revealed that Bio-DOPA1-α-factor cross-linked to a fragment of Ste2p encompassing residues Ser251–Met294. Fragmentation of the cross-linked fragment and Ste2p using tandem mass spectrometry pinpointed the cross-link point of the DOPA1 of the α-factor analog to the Ste2p Lys269 side chain near the extracellular surface of the TM6-TM7 bundle. This conclusion was confirmed by a greatly diminished cross-linking of Bio-DOPA1-α-factor into a Ste2p(K269A) mutant. Based on these and previously obtained binding contact data, a mechanism of α-factor binding to Ste2p is proposed. The model for bound α-factor shows how ligand binding leads to conformational changes resulting in receptor activation of the signal transduction pathway.
Keywords: G Protein-coupled Receptors (GPCR), Mass Spectrometry (MS), Membrane Proteins, Peptide Hormones, Yeast, Cross-linking
Introduction
G protein-coupled receptors (GPCRs)4 comprise the largest family of cell surface proteins present in the human genome responsible for communication between the internal and external environments in response to signal molecules, including hormones, pheromones, and neurotransmitters. GPCRs are characterized by the presence of seven membrane-spanning helical segments separated by alternating intracellular and extracellular loop regions (1–3). Despite their diverse ligands, all GPCRs perform similar functions, coupling the binding of ligands to the activation of specific heterotrimeric guanine nucleotide-binding proteins (G proteins), leading to the modulation of downstream effector proteins and molecules. Due to their involvement in a multitude of physiological functions, GPCRs are targeted by ∼50% of the human drugs currently marketed (1, 4, 5). Detailed information about GPCR-ligand interactions is critical for our understanding of GPCR-ligand binding and function.
Despite the differences in the binding mechanisms of various ligand groups in the GPCR family, there are some similarities that span this superfamily of proteins (1, 3, 6–8). Ligand binding triggers conformational changes at the extracellular and transmembrane regions of the receptor, which are then propagated to the cytoplasmic surfaces, leading to G protein coupling and activation. Thus, ligand binding leads to the alteration of existing interhelical interactions, thereby promoting a set of new interactions that leads to a energetically favorable activated conformational state of the receptor (8, 9). Large ligands, such as proteins, bind to the extracellular loops of GPCRs, whereas small molecules like adrenergic agents bind within the transmembrane region of the receptor. Within the peptide-binding GPCRs, a combination of ligand binding to the extracellular loops followed by ligand penetration of the transmembrane region and interaction with receptor residues buried in the membrane has been suggested (8, 10–12). However, in comparison with details on the molecular interactions in the binding of GPCRs activated by small ligands, less information exists concerning the binding sites of peptide-responsive GPCRs.
The interaction between Ste2p (α-factor receptor encoded by the STE2 gene), a Saccharomyces cerevisiae pheromone receptor, and its ligand tridecapeptide α-factor (WHWLQLKPGQPMY) is one of the most extensively studied models for the peptide ligand-GPCR relationship (13, 14). Recently, we showed that the use of 3,4-dihydroxyphenylalanine (DOPA) incorporated into the ligand was very effective for specific cross-linking to Ste2p (15). An α-factor analog containing DOPA at position 13 cross-linked to a region of Ste2p composed of residues Phe55–Met69. Mutational analysis of this region suggested that position 13 (Tyr13) of α-factor is in close proximity to residues Arg58 and Cys59. A recent study using fluorinated phenylalanine α-factor analogs also showed that the α-factor Tyr13 side chain is involved in a cation-π interaction with the Ste2p-Arg58 guanidinium moiety (16).
In order to continue the investigation of the binding site of α-factor in Ste2p as a model for peptide ligand GPCR interactions, we investigated the cross-linking of an α-factor analog containing DOPA at position 1. We demonstrated that a Bio-DOPA1-α-factor analog was capable of binding and activating Ste2p similarly to native α-factor. Matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) analysis suggested that position 1 cross-linked into portions of Ste2p comprising residues Ser251–Met294. Tandem mass spectrometry fragmentation and sequencing of the cross-linked fragment pinpointed the cross-link to Lys269 of Ste2p. The site was confirmed using mutational studies. In this study, we propose a model of the bound α-factor peptide in the receptor based on the cross-linking studies and other information on pheromone-Ste2p interaction. The model provides insight into the mechanism of ligand-GPCR binding and how a peptide ligand initiates signal transduction.
EXPERIMENTAL PROCEDURES
Media, Reagents, Strains, and Plasmids
Saccharomyces cerevisiae strain LM102 (MATa, bar1, leu2, ura3, FUS1-lacZ::URA3, ste2Δ) (17) was used for growth arrest and binding assays and the protease-deficient strain BJS21 (MATa, prc1-407 prb1-1122 pep4-3 leu2 trp1 ura3-52 ste2::KanR) (18) was used for protein isolation and immunoblot analysis. The plasmid pBEC1 containing C-terminal FLAG and His-tagged STE2 (17) was transformed by the method of Turcatti et al. (19) into LM102 and BJS21 cells. Transformants were selected by growth on yeast medium (20) lacking tryptophan (MLT) to maintain selection for the plasmid. The cells were cultured in MLT and grown to mid-log phase at 30 °C with shaking (200 rpm) for all assays.
Site-directed Mutagenesis
The plasmid pBEC1 was engineered by site-directed PCR mutagenesis to replace lysine at position 269 in Ste2p with alanine, cysteine, or histidine residues, as described by Huang et al. (18). The product of the mutagenesis reaction mixture was transformed into Escherichia coli strain DH5α (Invitrogen), and transformants were selected on medium containing ampicillin. Plasmids were then isolated from transformants using the Wizard Miniprep kit (Promega, Madison, WI). After sequence confirmation, constructs were transformed into yeast strains LM102 and BJS21, and transformants were selected by their growth in MLT. Mutagenic and sequencing primers were purchased from Invitrogen. DNA sequencing was carried out in the Molecular Biology Resource Facility located on the campus of the University of Tennessee.
Synthesis and Characterization of DOPA-α-factor Analogs
The tridecapeptide pheromone α-factor analogs [DOPA1,Lys7(BioACA),Nle12]α-factor (Bio-DOPA1-α-factor) and [DOPA1,Nle12]α-factor (designated DOPA1-α-factor) were synthesized using automated solid-phase peptide synthesis with Fmoc/O-t-butyl protection schemes. The hydroxyl groups of DOPA were protected as the acetonide derivative. All protected amino reagents were purchased from Advanced Chem Tech (Louisville, KY), and all solvents and reagents were of the highest purity available. α-Factor and α-factor analogs used in this study all contain norleucine in place of the native Met12 residue. Norleucine is isosteric with methionine, and [Nle12]α-factor is isoactive and has the same receptor affinity as α-factor (21). The syntheses of [Nle12]α-factor, desTrp1,desHis2[Nle12]α-factor, and [Lys7(BioACA)]α-factor were done as described previously (14, 22).
FmocTyr(tert-butyl)-wang resin (0.1 mmol, 0.65 mmol/g) in DMF (10 ml) was loaded onto a model 433A solid phase peptide synthesizer (Applied Biosystems, Foster City, CA), and the chain was assembled using 0.1-mmol single coupling chemistry and 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate/N hydroxybenzotriazole as coupling reagents, as described previously (23). After completing the chain assembly, the resin was dried under vacuum. The weight gain was >90% of the expected value.
DOPA-HWLQLKPGQPNleY
For preparation of the free [DOPA1]α-factor, the dried resin was divided, and a portion was treated with piperidine to remove the Fmoc protecting group. The peptide-resin was washed and dried in vacuo, and ∼200 mg was resuspended in a solution of 0.75 g of phenol, 0.5 ml of EDT, 0.5 ml of thioanisole, and 0.5 ml of water in 10 ml of trifluroacetic acid. The resulting mixture was stirred at room temperature for 1.5 h in dark. After filtration, the filtrate was concentrated on a rotary evaporator, the residue was treated with cold diethyl ether (100 ml), and the precipitate was collected by centrifuge followed by washing with diethyl ether twice (30 ml × 2), dried on vacuum to give 70 mg of crude peptide. The crude peptide, DOPA-HWLQLKPGQPNleY (24.4 mg) was dissolved in 20% acetonitrile/water (0.1% TFA), and the solution was loaded on preparative HPLC (C4, Delta Pak, 19 × 300 mm) using a gradient from 20 to 50% acetonitrile (0.1% TFA) over 90 min. Two fractions were collected; one was homogeneous and the other was 96% pure. The recovery was about 30% of theoretical. MS: found, 1658.7; Calc., 1657.9.
Fmoc-DOPA-HWLQLKPGQPNleY
To 470 mg of the α-Fmoc protected peptide-resin was added a solution of 0.75 g of phenol, 0.5 ml of EDT, 0.5 ml of thioanisole, and 0.5 ml of water in 10 ml of trifluroacetic acid, and the resulting mixture was stirred at room temperature for 1.5 h in the dark. After filtration, the filtrate was concentrated on rotary evaporator, the residue was treated with 100 ml of cold diethyl ether, and the precipitate was collected by centrifuge followed by washing with diethyl ether twice (30 ml × 2), dried on vacuum to give 198 mg of crude Fmoc-DOPA-HWLQLKPGQPNleY. This protected peptide (24 mg) was dissolved in 33% acetonitrile/water (0.1% TFA), and the solution was loaded on preparative HPLC (C4, Delta Pak, 19 × 300 mm) using a gradient from 30 to 70% acetonitrile (0.1% TFA) over 90 min. The recovered Fmoc-DOPA-HWLQLKPGQPNleY peptide was homogeneous by analytical reverse-phase HPLC and had the expected MS (found, 1880.6; Calc., 1879.9).
DOPA-HWLQLK(AcaBiotin)PGQPNleY-OH
To a solution of Fmoc-DOPA-HWLQLKPGQPNleY-OH (26 mg, 12.4 μmol) in DMF (2 ml) and sodium borate (1 ml, 50 mm), was added biotinamidohexanoic acid N-hydroxysuccinimide ester (8.4 mg, 18.6 μmol) at 4 °C, and the solution was stirred for 2 h. After neutralized by 2% HCl, the solution was filtered, and the filtrate was loaded on preparative HPLC (C4 column, 19 × 300 mm) using a gradient from 20 to 70% acetonitrile (0.1% TFA)/water (0.1% TFA). Crude Fmoc-DOPA-HWLQLK(BiotinACA)PGQPNleY-OH (20 mg) was obtained. Yield: 69%. MS: Calc., 2219; found, 2220. This peptide was dissolved in DMF (1.0 ml), and piperidine (70 μl, 18% in DMF) was added at 4 °C. The solution was stirred at this temperature for 1 h and then was neutralized and filtered, and the filtrate was loaded on preparative HPLC using a gradient of 20–50% acetonitrile (0.1% TFA)/water (0.1% TFA) over 90 min. About 80% of a highly homogeneous DOPA-HWLQLK(AcaBiotin)PGQPNleY-OH (Bio-DOPA1-α-factor) was recovered. MW: Calc., 1998.2; found, 1998.2. The HPLC profiles and electron spray mass spectra of DOPA1-α-factor, Fmoc-DOPA1-α-factor, and Bio-DOPA1-α-factor are shown in the supplemental material.
Mass Spectrometric Analysis of Peptides
For MALDI analysis, the peptides were resuspended in 50:50 water/acetonitrile with 0.1% TFA at a final concentration of 0.1 μg/μl. A 20 mg/ml α-cyano-4-hydroxy-trans-cinnamic acid (Sigma-Aldrich) matrix was prepared by dissolving recrystallized α-cyano-4-hydroxy-trans-cinnamic acid in 50:50 water-acetonitrile with 0.1% TFA. An equal volume (0.5 μl) of peptide solution was mixed with matrix before spotting on the MALDI plate. The MADLI-TOF spectra were acquired on a Bruker Daltonics (Boston, MA) Microflex using the reflector methods. The tandem mass spectrometry (MS/MS) data were acquired by MALDI post-source decay on a Bruker Daltonics Microflex. The interpretation of the MS/MS data were processed using Bruker Daltonics BioTools software.
Growth Arrest Assays
LM102 cells expressing C-terminal FLAG and His-tagged Ste2p were grown at 30 °C in MLT, harvested, washed three times with water, and resuspended at a final concentration of 5 × 106 cells/ml (18). Cells (1 ml) were combined with 3.5 ml of agar noble (1.1%) and poured as a top agar lawn onto a medium lacking tryptophan agar plate. Filter disks (BD Biosciences) impregnated with α-factor or various α-factor analogs were placed on the top agar. The plates were incubated at 30 °C for 18 h and then observed for clear halos around the discs. The experiment was repeated at least three times, and reported values represent the mean of these tests. The reproducibility of the inhibition zones was always within 2 mm for a given concentration.
Fus1-LacZ Assays
LM102 cells expressing Ste2p wild type and K269A, K269C, and K269H mutants were grown at 30 °C, harvested, washed three times with fresh media, and resuspended at a final concentration of 5 × 107 cells/ml. Cells (0.5 ml) were combined with α-factor pheromone (final concentration of 1 μm) and incubated at 30 °C for 90 min. The cells were transferred to a 96-well flat bottom plate in triplicates and permeabilized with 0.5% Triton X-100 in 25 mm PIPES buffer, and then β-galactosidase assays were carried out using fluorescein di-β-galactopyranoside (Molecular Probes, Inc., Eugene, OR) as a substrate as described previously (17). The reaction mixtures were incubated at 37 °C for 60 min, and 1.0 m Na2CO3 was added to stop the reaction. The fluorescence signal (excitation of 485 nm and emission of 530 nm) was determined using a 96-well plate reader Synergy2 (BioTek Instruments, Inc., Winooski, VT). The data were analyzed using Prism software (GraphPad Software, San Diego, CA). The experiments were repeated at least three times, and reported values represent the means of these tests.
Binding Competition Assays
This assay was performed using LM102 cells expressing C-terminal FLAG and His-tagged Ste2p. Tritiated [3H]α-factor (10.2 Ci/mmol, 12 μm) prepared as described previously (18, 21) was used in competition binding assays on whole cells. The cells were grown at 30 °C in MLT, harvested, washed three times with YM1 (0.5 m potassium phosphate (pH 6.24) containing 10 mm tert-amyl-methyl ether, 10 mm sodium azide, 10 mm potassium fluoride, and 1% BSA), and adjusted to a final concentration of 2 × 107 cells/ml in YM1 plus protease inhibitors (YM1i) (24). For the competition binding studies, cells (600 μl) were combined with 150 μl of ice-cold 5× YM1i supplemented with 6 nm [3H]α-factor in the presence or absence of α-factor or α-factor analogs and incubated at room temperature for 30 min. The final concentrations of α-factor and α-factor analogs ranged from 0.5 × 10−10 to 1 × 10−6 m. After incubation, triplicate samples of 200-μl aliquots were filtered and washed over glass fiber filter mats using the Standard Cell Harvester (Skatron Instruments, Sterling, VA) and placed in scintillation vials. The 3H radioactivity on the filter was counted by liquid scintillation spectroscopy. The binding data were analyzed by non-linear regression analysis for one-site competition binding using Prism software (GraphPad Software) to determine the binding affinity (Kd) and potency (EC50) for each peptide. The Ki values were calculated by using the equation of Cheng and Prusoff, where Ki = EC50/(1 + [ligand]/Kd) (25). The final values represent the binding constants from at least three independent experiments.
DOPA Chemical Cross-linking
BJS21 cells expressing C-terminal FLAG and His-tagged STE2 were grown, and total cell membranes were isolated as described previously (18). Protein concentration was determined by a Bio-Rad protein assay (17). The membranes were re-suspended in NE-Buffer (20 mm HEPES, 20% glycerol, 100 mm KCl, 12.5 mm EDTA, 0.5 mm DTT) (26) incubated with Bio-DOPA-α-factor (1 μm) in the presence or absence of 100 μm α-factor (WHWLQLKPGQPNle12Y), 100 μm α-factor antagonist (desW1desH2WLQLKPGQPNle12Y) (22), or 2 μg of BSA for 120 min at 4 °C. For periodate-mediated cross-linking, a final concentration of 1.0 mm NaIO4 was added to the mixture and incubated for 2 min. A final concentration of 100 mm DTT was used to quench the reaction (26). The cross-linked membranes were washed three times with CAPS buffer (Sigma) (10 mm, pH 10) by centrifugation to remove non-bound Bio-DOPA. The washed, cross-linked samples were fractionated by SDS-PAGE and then immunoblotted. The blots were probed with FLAGTM antibody (Sigma-Aldrich) to detect Ste2p or with NeutrAvidin-HRP conjugate (Pierce) to detect the biotin tag on Bio-DOPA pheromone covalently linked to the Ste2p. The signals generated were analyzed using Quantity One software (version 4.5.1) on a Chemi-Doc XRS photodocumentation system (Bio-Rad).
Purification of Intact Cross-linked Ste2p
The cross-linked Ste2p was enriched using His-SelectTM high capacity-nickel affinity gel (Sigma-Aldrich) following the manufacturer's directions. Approximately 10 mg of cell membrane containing cross-linked Ste2p were resuspended in ice-cold solubilization buffer (50 mm Tris HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100) with protease inhibitors (phenylmethylsulfonyl fluoride, pepstatin A, and leupeptin) and incubated overnight at 4 °C with end-over-end mixing and then centrifuged at 15,000 × g for 30 min to remove non-soluble material. The solubilized proteins were then mixed with His-HC-nickel gel and incubated at 4 °C with end-over-end mixing for 1 h. The gel was collected by centrifugation at low speed (500 × g, 1 min) and resuspended and collected four times in wash buffer (50 mm sodium phosphate, pH 8.0, 0.3 m sodium chloride, and 5 mm imidazole). Ste2p was eluted by resuspending the resin in 1 ml of ice-cold elution buffer (50 mm sodium phosphate, pH 8.0, 0.3 m sodium chloride, and 250 mm imidazole) and incubated at 4 °C with end-over-end mixing for 10 min. The resin was pelleted by centrifugation (2000 × g, 1 min), and the supernatant, containing the eluted Ste2p, was transferred to a fresh tube. Purity and concentration of samples were estimated by Coomassie Blue and silver staining of SDS-polyacrylamide gels (data not shown). The samples were also analyzed by immunoblotting using an antibody (anti-FLAGTM) directed against the FLAG epitope tag in Ste2p and with NeutrAvidin-HRP conjugate to detect the biotin tag on Bio-DOPA1-α-factor.
Digestion of Cross-linked Ste2p
The enriched cross-linked Ste2p samples eluted from the His-nickel column were digested with cyanogen bromide (CNBr). The eluted samples containing Ste2p (∼20 μg) were dried by vacuum centrifugation (Thermo Scientific, Waltham, MA) and then dissolved in 100% TFA containing 10 mg/ml CNBr. Deionized distilled water was then added to adjust the final TFA concentration to 80%, and the samples were incubated at 37 °C in the dark for 18 h (18, 27). The samples were dried by vacuum centrifugation and washed three times with deionized distilled water, and then 1 m Tris (pH 8.0) was added to neutralize the acidic mixture.
Purification of Cross-linked Ste2p Fragments
Fragments from the CNBr digestion of cross-linked Ste2p were resuspended in PBS buffer (0.1 m sodium phosphate, 0.15 m sodium chloride, pH 7), mixed with monomeric avidin resin (Pierce), and incubated for 6 h at 4 °C with end-over-end mixing (28). The resin was collected by centrifugation at low speed (1000 × g, 1 min) and resuspended and collected four times in PBS buffer. The cross-linked Ste2p fragments were eluted by resuspending the resin in 200 μl of ice-cold elution buffer (0.1 m glycine, pH 2.5) and incubating at 4 °C with end-over-end mixing for 5 min. The resin was pelleted by centrifugation (2000 × g, 1 min), and the supernatant, containing the eluted cross-linked Ste2p fragments, was transferred to a fresh tube containing 20 μl of TBS (0.5 m Tris-HCl, pH 7.4, 1.5 m NaCl).
MALDI-TOF Analysis of Cross-linked Samples
The eluted samples from the avidin resin were further washed and concentrated using a pipette with C18 chromatographic media (ZipTipC18 pipette tips, Millipore Corp., Billerica, MA) following the manufacturer's directions and resuspended in 60% acetonitrile, 40% water (0.1% TFA). For MALDI-TOF analysis, α-CHCA (20 mg/ml (in 50:50 acetone/isopropyl alcohol)) was used as the matrix. The samples (0.5 μl) were mixed with 0.5 μl of matrix before spotting on the target, or 1.0 μl of matrix was spotted and allowed to dry before applying 1.0 μl of samples (18). MALDI-TOF spectra were acquired on a Bruker Daltonics Microflex using the reflector method. Masses were calculated using PROWL peptide mass prediction tools (29) and also based on the chemistry of the DOPA cross-linking (30).
Desalting CNBr-digested Cross-linking Samples and MS/MS Analysis
The CNBr-digested samples eluted from the avidin columns or free Bio-DOPA1-α-factor peptide were desalted using a reverse phase C18 trap system (Michrom Bioresources, Inc.). The C-18 cartridge was equilibrated with 2% acetonitrile, 0.1% TFA as described previously (31). Prior to desalting, the samples were centrifuged at 14,000 × g for 1 min to remove any solid particulates and loaded onto the equilibrated desalting cartridge using a 250-μl Hamilton syringe. The cartridge was then washed with 500 μl of 2% acetonitrile, 0.1% TFA. The peptides were eluted using 500 μl of 95% acetonitrile, 0.1% TFA. The eluted samples were then frozen on dry ice and lyophilized to dryness in a SpeedVac. The dried peptides were then resuspended in 10 μl of 0.5% formic acid solution.
The desalted cross-linked samples were analyzed by reverse-phase microcapillary LC-ESI-MS/MS. A fritless, microcapillary column (100-μm inner diameter) was packed with 10 cm of 5-μm C18 reverse-phase material (Synergi 4u Hydro RP80a, Phenomenex). The CNBr-digested peptides were loaded onto the reverse-phase column equilibrated in buffer A (0.1% formic acid and 5% acetonitrile). The column was placed in line with a nanoESI-LTQ-Orbitrap mass spectrometer (Thermo Scientific, Inc.). Peptides were eluted using a 100-min linear gradient from 0 to 55% buffer B (0.1% formic acid, 80% acetonitrile) at a flow rate of 0.3 μl/min. Eluting peptides were electrosprayed into the mass spectrometer with an applied spray voltage of 2.2 kV. During the gradient, the eluting ions were analyzed by one full precursor MS scan (400–2000 m/z), followed by five MS/MS scans on the five most abundant ions detected in the precursor MS scan while operating under dynamic exclusion. During the LC/MS/MS analysis of the sample, the precursor ions were analyzed in the Orbitrap, and MS/MS data were analyzed in the ion trap analyzer. The ion trap is more sensitive, enabling the detection of the MS/MS fragment ions, but it has somewhat lower precision than the Orbitrap. All the acquired MS/MS data were searched against the S. cerevisiae protein data base using the SEQUEST algorithm (32). To identify the MS/MS spectra of the cross-linked Bio-DOPA1-α-factor fragment, we compared the free Bio-DOPA1-α-factor peptide spectra with the spectra generated from the cross-linked samples eluted from the avidin column. The Ste2p fragment ions were identified on the cross-linked spectra by comparing the masses of fragment ions that did not correspond to any of the Bio-DOPA1-α-factor fragment ions with predicted masses of Ste2p fragment ions using PROWL MS/MS peptide and protein fragmentation tools (29).
RESULTS
Synthesis and Characterization of DOPA-α-factor Analogs
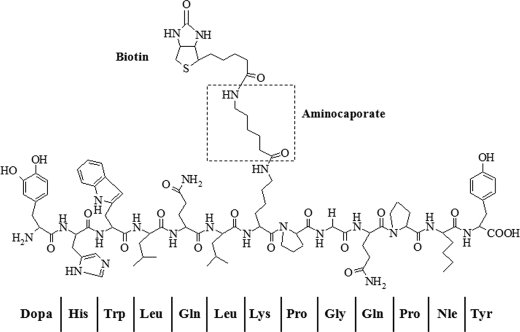
The syntheses of analogs of α-factor containing DOPA were carried out using automated solid phase synthesis on a WANG resin. The intermediate Fmoc-protected peptide was obtained in high yield and purity and was used in the hydroxysuccinimide-mediated addition of biotinylamido caproate (14). The final peptides used in bioassays and cross-linking studies were virtually homogeneous as judged by gradient HPLC and had the expected molecular weights (supplemental Figs. S1 and S2). The structure of Bio-DOPA1-α-factor is shown in Fig. 1, and MALDI-TOF/MS analyses of the synthesized peptide are provided in supplemental Fig. S3). The observed masses of the α-factor and Bio-DOPA1-α-factor as determined by MALDI-TOF were similar to masses predicted by the PROWL peptide mass prediction tools (29); α-factor was observed as 1666.09 Da (predicted, 1665.96 Da), and Bio-DOPA1 was 1998.76 Da (predicted, 1998.22 Da). Nine of the 13 b ion and five of the 13 y ion fragments were observed on the Bio-DOPA1 spectrum (supplemental Fig. S1). A mass shift of about 333 Da was observed due to the tagging of biotinylaminocaproate onto Lys7 and the replacement of Trp1 with DOPA (supplemental Fig. S3).
FIGURE 1.
Structures of Bio-DOPA1-α-factor analogs. Trp1 of α-factor is replaced by DOPA, and biotin is conjugated through its carboxyl group to the ϵ-amine of Lys7 using aminocaproate as a linker.
Bioactivity and Binding of Bio-DOPA1-α-factor
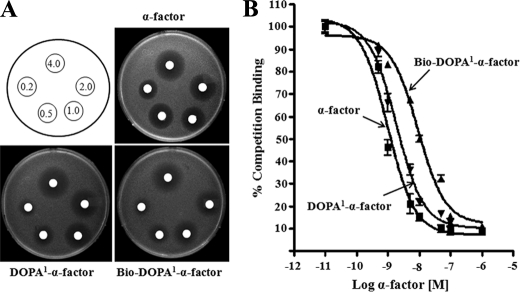
The replacement of DOPA at position 1 without biotin at position 7 (DOPA1-α-factor) (DOPA-HWLQLKPGQPNleY) or with biotin at position 7 (Bio-DOPA1-α-factor) ([DOPA-HWLQLK(BiotinACA)PGQPNleY]α-factor) led to a 2- and 4-fold lower ability, respectively, to induce growth arrest as compared with native α-factor (Fig. 2A). DOPA1-α-factor and Bio-DOPA1-α-factor displayed binding affinities (Kd values) of 19 ± 1.2 and 66.4 ± 2.8 nm, respectively, compared with that of the native α-factor (WHWLQLKPGQPNleY), which was 12 ± 1.6 nm (Fig. 2B). Thus, the replacement of Trp at position 1 with DOPA in α-factor (DOPA1-α-factor) did not greatly affect the affinity for Ste2p, suggesting that this chemically reactive pheromone can be used for receptor cross-linking (see below). The relative affinities of the DOPA1-α-factor and Bio-DOPA1-α-factor are consistent with previous studies that showed that the addition of biotin to position 7 of native α-factor resulted in a 3–5-fold reduction in α-factor binding (15).
FIGURE 2.
Biological and binding assays of DOPA-containing α-factor analogs. A, growth arrest assay to determine the ability of the peptides to activate α-factor receptor, Ste2p. Different amounts (0.25–4.0 μg) of the peptides were spotted on filter disks on a lawn of cells. The images of the plates shown were taken after 16 h. B, the binding of the peptides was determined by competition binding with [3H]α-factor for Ste2p as described in detail under “Experimental Procedures.” Error bars, S.E.
Bio-DOPA-α-factor Analogs Cross-link into Ste2p
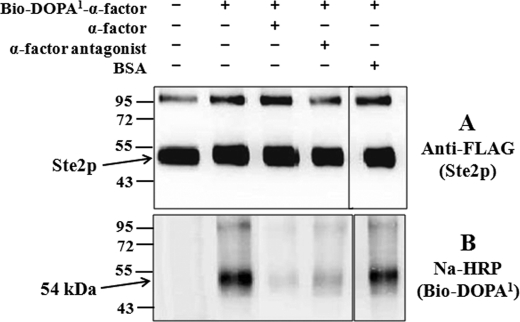
Membranes from cells expressing Ste2p (His- and FLAG-tagged) were incubated with Bio-DOPA1 α-factor and cross-linked by periodate oxidation, as described previously (15). Cross-linking was carried out in the absence or presence of α-factor, a high affinity α-factor antagonist, or BSA as controls to evaluate the specificity of the cross-linking. The Bio-DOPA1-α-factor-treated membranes were mixed with a His-HC nickel resin after the cross-linking reaction to enrich (purify) the Ste2p. The enriched cross-linked samples were resolved on 10% SDS-PAGE and silver-stained to check the purity. The His-purified samples showed high purity of the Ste2p-Bio-DOPA1-α-factor cross-linked receptors, as observed on the silver-stained SDS-PAGE, which revealed the receptor at ∼52–54 kDa and several minor bands that were not stained by either anti-FLAG or avidin probes (supplemental Fig. S4). The samples were probed with anti-FLAG antibody to detect Ste2p (Ste2p tagged with FLAG epitope tag) or NeutrAvidin-HRP (Na-HRP) to detect biotin attached to Bio-DOPA1-α-factor analogs (Fig. 3). Bands of similar intensity were observed at ∼52–54 kDa, corresponding to the size of Ste2p, in all lanes on blots probed with the anti-FLAG (Fig. 3A). Weaker bands were observed on these blots at ∼100–105 kDa corresponding to Ste2p dimers. When the immunoblots were probed with Na-HRP (Fig. 3B), a distinct band at 54 kDa was detected in the lane where Bio-DOPA1-α-factor analog was oxidized in the presence of Ste2p. This band was absent in samples that were not treated with Bio-DOPA1-α-factor, indicating that the ∼54 kDa band is a Ste2p-Bio-DOPA1-α-factor (∼52 kDa + ∼2 kDa) cross-linked product, as previously observed for Bio-DOPA13-α-factor (15). The cross-linking was nearly eliminated in the presence of excess (100-fold) α-factor and significantly reduced in the presence of a 100-fold excess of the α-factor antagonist. However, the presence of BSA had no effect on the level of cross-linked product. The cross-linking pattern of the putative Ste2p dimer paralleled that of the monomer. The results suggest that Bio-DOPA1-α-factor remained linked to Ste2p and Ste2p dimer even after the harsh treatment for purifying Ste2p, indicating that a stable cross-link existed between Ste2p and Bio-DOPA1-α-factor and that the cross-linking could be inhibited by the native α-factor and the antagonist.
FIGURE 3.
Bio-DOPA-α-factor analog chemical cross-linking into Ste2p. Cell membranes containing wild-type Ste2p were incubated with Bio-DOPA analog alone or with Bio-DOPA analog in the presence of 100-fold wild-type α-factor, antagonist, or BSA. The samples were resolved on 10% SDS-PAGE and probed with anti-FLAG (A) to detect Ste2p and with Na-HRP (B) to detect the biotin tag on Bio-DOPA1-α-factor.
Ste2p-Lys269 Cross-linked to Bio-DOPA1-α-Factor
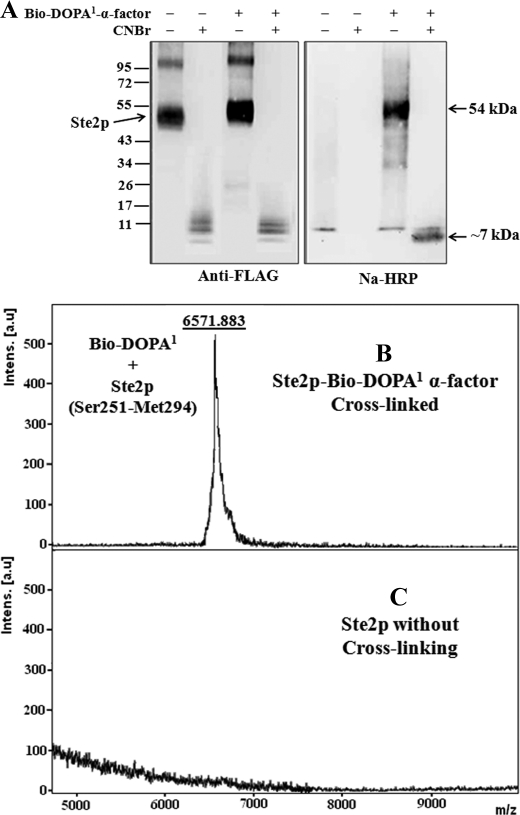
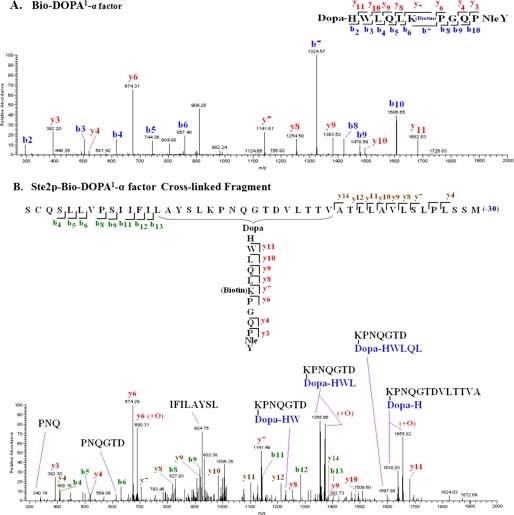
The purified cross-linked Ste2p-Bio-DOPA1-α-factor and Ste2p (without cross-linking) samples were treated with CNBr to fragment Ste2p. The samples were resolved on 4–20% gradient SDS-PAGE, blotted, and then probed with anti-FLAG antibody or Na-HRP (Fig. 4A). The anti-FLAG blot shows that Ste2p was fragmented, as indicated by the small fragment signals (∼8–10 kDa) in both the cross-linked and non-cross-linked samples. An incremental change in size of the cross-linked Ste2p (∼54 kDa) relative to the non-cross-linked receptor (∼52 kDa) was observed on the 4–20% gradient SDS-PAGE (Fig. 4A). This difference was not observed on 10% SDS-PAGE (Fig. 3) due to the higher resolution of proteins in the gradient gel. A signal of about 7 kDa was observed on the Na-HRP blot in the cross-linked sample but not the sample without cross-linking. Thus, the results suggest that the Ste2p-Bio-DOPA1 cross-linked fragment is about 7 kDa. The size of the Ste2p-Bio-DOPA1 cross-linked fragment was determined by MALDI-TOF mass spectrometry after mixing the CNBr-fragmented samples with avidin beads to capture the cross-linked Ste2p-Bio-DOPA1-α-factor fragment. The samples eluted from the avidin beads were then concentrated using a Millipore ZipTip to remove detergent and compounds that might interfere with mass spectrometry analysis as described previously (15). MALDI-TOF analysis of the samples shows that the Ste2p-Bio-DOPA1-α-factor cross-linked sample eluted from the avidin resins contained a 6571.883-Da fragment (Fig. 4B), but the Ste2p without cross-linking had no detectable fragment (Fig. 4C). The 6571.883 Da peak corresponds to the CNBr Ste2p fragment Ser251–Met294 (4572.455 Da) cross-linked to Bio-DOPA1-α-factor (1998.221 Da) because the observed mass (6571.883) on the MALDI-TOF spectrum is similar to the calculated mass (6568.676 Da).
FIGURE 4.
MALDI-TOF analysis of Ste2p-Bio-DOPA1 cross-linked CNBr fragment. A, immunoblot analysis of Ste2p-Bio-DOPA1 cross-linked sample after CNBr treatment. The samples were resolved on 4–20% gradient SDS-PAGE and probed with anti-FLAG to detect Ste2p and with Na-HRP to detect the biotin tag on Bio-DOPA1-α-factor. A band at about 7 kDa is observed in the Bio-DOPA1-treated lane. B, MALDI-TOF spectrum of fragmented Ste2p- Bio-DOPA1 cross-linked to peptide eluted from avidin resin. A 6571.883-Da peak corresponding to Ste2p fragment Ser251–Met294 (4572.455 Da) cross-linked to Bio-DOPA1 α-factor (1998.221 Da) was observed. C, spectrum of sample eluted from avidin mixed with CNBr-treated Ste2p (without cross-linking).
To further analyze the Ste2p-Bio-DOPA1-α-factor cross-linked fragment, an LC/MS/MS analysis was carried out on the eluted samples from the avidin beads. The MS/MS results indicated that whereas both the b and y ions of the Bio-DOPA1-α-factor were observed in the spectrum from the free peptide Bio-DOPA1-α-factor (Fig. 5A), in the spectrum of cross-linked Ste2p-Bio-DOPA1-α-factor samples, only the y ions and not the b ions of Bio-DOPA1-α-factor were observed (Fig. 5B). The b (b4–b13) and y (y4–y14) ions corresponding to residues Ser254–Ile263 and Ala281–Leu291, respectively, in the Ste2p CNBr fragment Ser251–Met294 were observed in the cross-linked Ste2p-Bio-DOPA1-α-factor spectrum. Significantly, fragments corresponding to linkage between the DOPA peptide and the Ste2p CNBr fragment, Ser251–Met294, such as K(DOPA-HWL)PNQGTD and K(DOPA-H)PNQGTDVLTTVA, were also observed in the MS/MS spectrum. Many of these fragments are consistent with the Ste2p portion containing Lys269 cross-linked to Bio-DOPA1-α-factor. Summaries of all of the observed masses in both spectra compared with theoretical masses are shown in Tables 1–3.
FIGURE 5.
MS/MS analysis of Bio-DOPA1-Ste2p cross-linked fragments. Inserts are one-letter amino acid residue abbreviations of the fragment labeled with the identified ion types. A, MS/MS spectrum of fragment Bio-DOPA1 α-factor. The spectrum shows nine and eight of 13 b and y ion fragments, respectively. B, MS/MS spectrum of the cross-linked fragment. Only the y ions of Bio-DOPA1 α-factor were identified. In the Ste2p fragment (Ser251–Met294), eight b ions representing the N terminus and eight y ions representing the C terminus were identified. Fragment ions corresponding to internal fragments of the cross-linked peptide are labeled with their amino acid sequences. Lys269 in Ste2p is observed to cross-link to Bio-DOPA1 α-factor in some of the fragments. The oxidized form (+O) of some of the fragments were also observed. A list of observed masses (m/z) of the fragment ions in all the spectra compared with the theoretical masses are shown in Table 1.
TABLE 1.
A list of masses (m/z) of Bio-DOPA1-α-factor fragment ions observed in the Bio-DOPA1-α-factor spectrum (Fig. 5A) compared with theoretical values
| Ion number |
b ions (m/z) |
y ions (m/z) |
||
|---|---|---|---|---|
| Expected | Observed | Expected | Observed | |
| 2 | 317.33 | 317.15 | ||
| 3 | 503.54 | 503.31 | 392.47 | 392.20 |
| 4 | 616.70 | 616.21 | 520.61 | 520.39 |
| 5 | 744.833 | 744.36 | ||
| 6 | 857.99 | 857.40 | 674.77 | 674.31 |
| 7 | 1325.46 | 1324.57 | 1142.24 | 1141.61 |
| 8 | 1422.58 | 1421.70 | 1255.41 | 1254.50 |
| 9 | 1479.64 | 1478.59 | 1383.54 | 1383.52 |
| 10 | 1607.77 | 1606.65 | 1496.70 | 1496.92 |
| 11 | 1682.91 | 1682.83 | ||
TABLE 2.
A list of masses (m/z) of Ste2p-Bio-DOPA1-α-factor cross-linked fragment ions observed in the Bio-DOPA1-α-factor Bio-DOPA1 spectrum (Fig. 5B) compared with theoretical values
| Fragment ion number | Ste2p fragments (Ser251–Met294) |
Bio-DOPA1-α-factor fragments; y ions (m/z) |
||||
|---|---|---|---|---|---|---|
| Expected |
b ions (m/z) |
y ions (m/z) |
||||
| Observed | Expected | Observed | Expected | Observed | ||
| 3 | 392.47 | 392.30 | ||||
| 4 | 406.44 | 406.48 | 407.54 | 408.18 | 520.61 | 520.20 |
| 5 | 519.60 | 519.20 | ||||
| 6 | 632.76 | 631.36 | 674.77 | 674.29 | ||
| 7 | 704.89 | 704.47 | 1142.24 | 1141.49 | ||
| 8 | 829.01 | 827.93 | 818.05 | 819.21 | 1255.41 | 1254.66 |
| 9 | 916.09 | 916.68 | 917.18 | 918.01 | 1383.54 | 1382.73 |
| 10 | 988.26 | 988.82 | 1496.70 | 1497.92 | ||
| 11 | 1142.41 | 1142.65 | 1101.42 | 1102.53 | 1682.91 | 1681.60 |
| 12 | 1289.58 | 1289.02 | 1214.58 | 1213.59 | ||
| 13 | 1402.74 | 1402.19 | ||||
| 14 | 1386.76 | 1386.48 | ||||
TABLE 3.
A list of masses (m/z) of Ste2p-Bio-DOPA1-α-factor cross-linked fragments observed in the Bio-DOPA1-α-factor Bio-DOPA1 spectrum (Fig. 5B) compared with theoretical values
| Residues | Fragment size (m/z) |
|
|---|---|---|
| Expected | Observed | |
| PNQ | 339.37 | 340.18 |
| PNQGTD | 612.61 | 613.45 |
| IFILAYSL | 922.81 | 924.75 |
| (DOPA-HW) + KPNQGTD | 1243.32 | 1244.41 |
| (DOPA-HWL) + KPNQGTD | 1356.48 | 1355.66 |
| (DOPA-HWL) + KPNQGTD (+O) | 1374.56 | 1373.76 |
| (DOPA-HWLQL) + KPNQGTD | 1597.77 | 1597.88 |
| (DOPA-H) + KPNQGTDVLTTVA | 1639.82 | 1638.03 |
| (DOPA-H) + KPNQGTDVLTTVA (+O) | 1657.01 | 1655.82 |
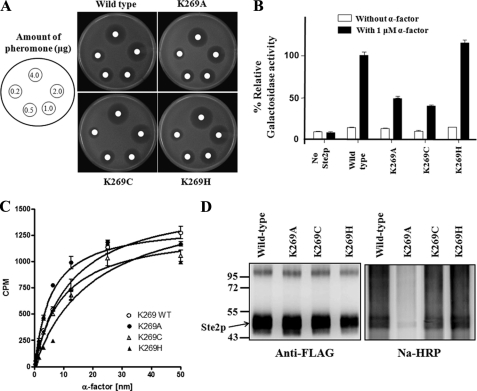
Reduced Cross-linking in Ste2p K269A Mutant
To further examine the reactivity of Lys269 with DOPA at position 1 of α-factor, Lys269 was replaced with alanine, cysteine, or histidine. Ste2p(K269A), Ste2p(K269C), and Ste2p(K269H) were expressed and displayed binding affinity to α-factor similar to wild-type Ste2p (Fig. 6, A–C). Ste2p K269A and K269C both displayed reduced biological activity, whereas K269H showed activity similar to that of the wild type, as indicated by the Fus1-LacZ assay (Fig. 6B). Membranes were prepared from cells expressing Ste2p mutants K269A, K269C, and K269H, and cross-linking with Bio-DOPA1-α-factor was carried out as described above. The immunoblot analysis (Fig. 6D) showed that whereas Ste2p(K269H) receptor cross-linked similarly to wild-type receptor, the cross-linking of the Ste2p(K269A) mutant was significantly reduced (∼20%), and the K269C mutant showed about ∼75% cross-linking of the wild type, although similar levels of Ste2p signal were detected in all samples, as shown in blot probed with anti-FLAG (to detect Ste2p).
FIGURE 6.
Biological activities and cross-linking of Ste2p K269A, K269C, and K269H mutants. A, growth inhibition induced by α-factor. Different amounts (0.25–4.0 μg) of α-factor were spotted on filter disks on a lawn of cells. The images of the plates shown were taken after 16 h. B, induction of transcription of the FUS1 gene was carried in the absence or presence of 1 μm α-factor. The percentages of β-galactosidase activity of the various mutants were determined by normalizing with the wild type (100%). C, binding affinity of Ste2p mutants analyzed by saturation binding with tritium-labeled α-factor. Both mutants displayed binding affinity similar to wild type. D, immunoblot images of Bio-DOPA1-α-factor cross-linking into Ste2p K269A, K269C, and K269H mutants. Purified Ste2p-Bio-DOPA1-α-factor cross-linked samples were resolved on 10% SDS-PAGE and probed with anti-FLAG and Na-HRP. Error bars, S.E.
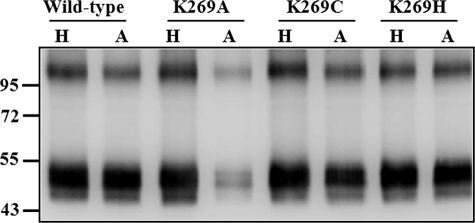
To quantify the amount of cross-linked receptors, the cross-linked membrane samples were divided into two equal parts. The first part was mixed with His-SelectTM HC-nickel affinity gel beads to enrich Ste2p receptors (both cross-linked and non-cross-linked receptors), and the second part was mixed with avidin affinity beads to enrich only cross-linked receptors. The samples were eluted from the His affinity and avidin affinity beads and then probed with anti-FLAG antibody to detect Ste2p (Fig. 7). The band intensities on the blot were analyzed using Quantity One software (version 4.5.1) on a Chemi-Doc XRS photodocumentation system (Bio-Rad). The percentage of cross-linked receptors was determined by dividing the band intensity of NeutrAvidin-purified samples by the band intensity of the His-purified samples and then multiplied by 100. The results suggested that most receptors (∼90%) of the wild type and K269H mutant were cross-linked, whereas about 25 and 75% of the K269A and K269C receptors, respectively, were cross-linked. It has previously been reported that the periodate-activated DOPA system often results in high yields (30). The residual amount of cross-linking found in the wild-type receptor in the presence of α-factor (Fig. 3) was also seen for the K269C, K269A, and K269H mutants (data not shown), indicating that a small amount of nonspecific cross-linking occurred in the mutants. The results are consistent with the conclusion that Lys269 in Ste2p is responsible for the cross-linking signal observed in the Ste2p-Bio-DOPA1-α-factor reaction.
FIGURE 7.
Quantitation of Bio-DOPA1 cross-linking of Ste2p Lys269 mutant receptors by immunoblot analysis. Cross-linked membrane samples were divided into two parts; one part was mixed with His-SelectTM HC-nickel affinity gel (H) to enrich Ste2p receptors, and the other part was mixed with avidin affinity beads (A) to enrich only cross-linked receptors. The samples eluted from the His affinity and avidin affinity beads were resolved on 10% SDS-PAGE and probed with anti-FLAG antibody to detect Ste2p. The band intensities were quantitated using a Chemi-Doc XRS photodocumentation system, and the percentage of cross-linking was calculated as described under “Experimental Procedures.” H, nickel affinity-purified receptors; A, avidin affinity-purified receptors.
DISCUSSION
Although Ste2p and α-factor interactions have been studied extensively and used as a model system to understand the binding mechanism of other peptide-responsive GPCR systems, the contact sites between α-factor and Ste2p are still not fully defined. We had reported that α-factor analogs containing benzoyl-phenylalanine at position 1 cross-linked into portions of Ste2p comprising residues Ser251–Met294, but those studies with benzoyl-phenylalanine analogs did not identify the specific residue of Ste2p that interacted with position 1 of α-factor. Previously, we showed that DOPA13-α-factor cross-linking into Ste2p was more effective and specific than benzoyl-phenylalanine-mediated cross-linking. Using mass spectroscopy and the known reactivities of DOPA, we were able to identify specific residue-to-residue interactions between ligand and Ste2p (15). In the present study, the periodate-mediated chemical cross-linking of Bio-DOPA1-α-factor into Ste2p was investigated. This analog was a potent agonist. Comparison with the α-factor control indicated that the replacement of Trp at position 1 with DOPA did not significantly affect the ability of the peptide to bind Ste2p. Moreover, cross-linking of Bio-DOPA1-α-factor can be competed by both α-factor and an α-factor antagonist, indicating that this pheromone cross-links to the α-factor binding pocket in its cognate GPCR. Thus, it is not unreasonable to conclude that the DOPA side chain in Bio-DOPA1-α-factor is in a similar spatial orientation as the side chain of the native Trp residue.
Affinity purification of biotinylated peptides (33) together with MS analysis is widely used for detection of cross-linked products with high sensitivity and accuracy (34). Methods using LC-MS/MS allow direct identification of peptides (31, 35). The MS/MS analysis of Ste2p-Bio-DOPA1-α-factor cross-linked product confirmed that the region Ser251–Met294 cross-linked with Bio-DOPA1-α-factor. Whereas both the b and y ion fragments of Bio-DOPA1-α-factor were observed in the Bio-DOPA1-α-factor spectrum, only the y ions of the ligand were identified in the cross-linked spectrum, suggesting that the N terminus of the ligand containing DOPA is cross-linked to a fragment of Ste2p. The b ions of residues Ser251–Ile263 and y ions of residues Leu283–Met294 were also identified in the Ste2p fragment, Ser251–Met294. The identification of the N and C termini of the Ste2p fragment, Ser251–Met294, and the C terminus of Bio-DOPA1-α-factor in one spectrum indicated that all of these fragments are covalently linked (cross-linked fragment). However, the instrument used for the MS/MS has a mass/charge ratio (m/z) limit of 2000; therefore, it was not possible to obtain a fragment ion that was larger than 2000 m/z on the spectrum examined. Hence, we do not expect to see all of the fragment ions of the cross-linked product in the spectrum. Nevertheless, the results implied that Bio-DOPA1-α-factor was cross-linked with one of the residues in Ser251–Met294 of Ste2p. Detailed examination of fragment ions observed in the cross-linked spectrum, in particular the observation of fragments with molecular weights corresponding to K(DOPA-HWL)PNQGTD and K(DOPA-H)PNQGTDVLTTVA, led us to the conclusion that Bio-DOPA1-α-factor was cross-linked to Lys269 located at the boundary between the sixth transmembrane domain and extracellular loop 3 (TM6-EL3). This conclusion was further examined using Ste2p mutated at position 269.
It was previously shown that the ϵ-amino of lysine, the imidazole of histidine, and the thiol of cysteine were capable of attacking the ortho-quinone obtained on periodate oxidation of DOPA. However, to efficiently couple to DOPA, the side groups of these amino acids and DOPA must be in very close proximity (30). Based on these findings, we investigated the cross-linking of Bio-DOPA1-α-factor in Ste2p mutants K269A, K269C, and K269H that have lysine replaced with alanine, cysteine, and histidine, respectively. These mutant receptors were expressed well and had binding affinity similar to that of wild-type receptor. Immunoblotting analyses showed that Bio-DOPA1-α-factor cross-linking was significantly reduced in K269A compared with wild type and K269C and K269H mutants (Fig. 6D). In addition, quantification of the amount of receptors cross-linked suggested that whereas wild type and K269H receptors displayed about 90% labeling by the Bio-DOPA1-α-factor, the K269A and K269C receptors showed 25 and 75% labeling, respectively (Fig. 7). Kodadek and co-workers (30) have previously reported that the periodate-activated DOPA system often results in high yields. Alanine is not expected to react with DOPA because it lacks a nucleophilic side chain. Therefore, the minor amount of cross-linking (∼25%) in the Ste2p K269A observed may be due to other nucleophiles in proximity to position 269 of Ste2p or some small amount of nonspecific cross-linking. This conclusion is supported by the observation that the residual amount of cross-linking was not reduced in the presence of α-factor. From this perspective, we note that a large excess of α-factor eliminates most but not all of the DOPA-mediated cross-linking in the WT (Fig. 3). Nevertheless, it seems clear that the large majority of the cross-linking is with Lys269, supporting the conclusion that the ϵ-amino functional group of Lys269 was cross-linked to Bio-DOPA1-α-factor and that Lys269 is in close proximity with position 1 of α-factor. Cysteine has been shown to react more strongly with periodate-treated DOPA than Lys or His (30). In this study, the results obtained showed that wild-type Ste2p (Ste2p-Lys269) and K269H mutant reacted better than Ste2p K269C, suggesting that the DOPA1 residue may be in closer proximity to the ϵ-amino of lysine (wild-type Lys269) and the imidazole of histidine (K269H) than it is to the sulfhydryl of cysteine (K269C). This probably reflects the three additional methylene groups in the Lys side chain that can change the position of the nucleophile by about 4.5 Å.
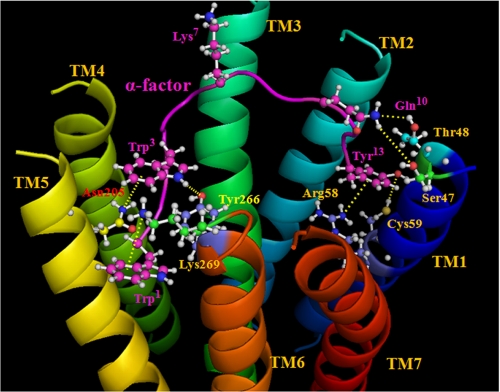
The results presented in this study, combined with the work of others including receptor mutagenesis analyses and structure-activity relationships of α-factor analogs, lead us to propose a refined binding model for the ligand-receptor complex. The model (Fig. 8) includes a well documented bend in the α-factor around the Pro8-Gly9 sequence (36), with the Lys7 side chain facing away from the TM domains and interacting with a binding pocket formed by the extracellular loops (37, 38). Studies of the N terminus of α-factor indicated a strong preference for a large hydrophobic residue at the N-terminal position of the pheromone (39–41); analyses of Ala-scanned α-factor analogs and fluorescent α-factor analogs indicated that the side chain of Trp3 is in a hydrophobic pocket when bound to the receptor (14, 42) interacting with Asn205 at the TM5-EL2 boundary and Tyr266 at the TM6-EL3 boundary of Ste2p (41, 43). Therefore, the bound α-factor model incorporates a hydrophobic region where Trp1 and Trp3 side chains cluster with the Tyr266 residue of Ste2p. The indole side chain of Trp1 of α-factor also interacts with Lys269 of Ste2p, possibly through a cation-π interaction, although we cannot rule out other interactions involving hydrogen bonds. The C terminus of α-factor interacts with TM1, where Tyr13 of the pheromone is in close proximity to Arg58 and Cys59 of Ste2p. Recently, the α-factor Tyr13 side chain has been shown to be involved in a cation-π interaction with the Ste2p-Arg58 guanidinium moiety (16) and a hydrogen bond with the Ste2p-Cys59 sulfhydryl group (15). Gln10 of α-factor is suggested to be in close proximity to residues Ser47 and Thr48 of Ste2p when α-factor is bound to Ste2p (25). When the positions of the loop residues are determined, these constraints may prove useful in attempts to calculate a bound structure of the pheromone.
FIGURE 8.
Proposed model of the interactions of α-factor with residues of Ste2p. The TMs are labeled in different colors, and the α-factor backbone is shown as a purple ribbon. Selected residues of Ste2p and α-factor are shown in a ball-and-stick representation. All atoms in the selected α-factor residues are colored purple, except for nitrogen (blue) and oxygen (red), and atoms of Ste2p residues are in different colors to differentiate them from the α-factor atoms. The model shows the interactions of the N terminus and C terminus of α-factor with Ste2p TM5-TM6 and TM1, respectively. A bend in α-factor around the Pro8-Gly9, with the Lys7 side chain facing away from the TM domains, is shown. The Trp1 indole side chain of α-factor is proposed to form a cation-π (or hydrogen bonding) interaction with the Lys269 ϵ-amino group. Trp3 is in a hydrophobic pocket interacting with Asn205 (TM5) and Tyr266 (TM6). Gln10 of α-factor is suggested to be in close proximity to residues Ser47 and Thr48 (TM1). The Tyr13 side chain phenyl ring of α-factor is proposed to be involved in a cation-π interaction with the Arg58 (TM1) guanidinium moiety, and the phenolic OH (Tyr13) forms a hydrogen bond with the Cys59 (TM1) sulfhydryl group. All suggested interactions between α-factor residues and Ste2p residues are shown by the dashed lines in yellow.
It remains unclear how the binding of a peptide ligand to a GPCR leads to changes in interactions of the ground state of a receptor that propagate a signal to the cytoplasmic ends of the receptor to initiate signal transduction. However, many elegant studies with rhodopsin have indicated that the mechanism of activation may involve multiple switches on the extracellular side of rhodopsin, which trigger structural changes that converge to disrupt the ionic lock between helices H3 and H6 on the intracellular side of the receptor (44). Comparisons of crystal structures of rhodopsin in its inactive and active conformations suggest that binding pocket residues change conformation during the light activation of this GPCR. For instance, the interaction between Lys296 (on the TM7 region-EL3 boundary) and the Schiff base counterion Glu113 (on the TM3-EL1 boundary) of rhodopsin is disrupted, and the binding pocket becomes slightly wider in opsin (45). Such changes in residue interactions on the extracellular face of a GPCR may be coupled to conformational changes that are observed at the cytoplasmic ends of TM5, TM6, and TM7, which result in the activation of the G protein (2). By analogy, the interactions of residues at the N terminus of α-factor with the TM5 and TM6 regions of Ste2p may induce conformational changes at these TMs that are transduced to the cytoplasmic ends of these domains. TM6 of Ste2p, like that of most GPCRs, plays an important role in receptor activation. For example, Ste2p TM6 mutations of P258L and P258L/S259L resulted in constitutive activation (46), and Y266A and Y266C were deficient in signaling activity (41, 47). Moreover, Ste2p Tyr266 (on the extracellular end of TM6) interacts with Asn205 (on the extracellular end of TM5) only in the active conformation of the receptor (48), suggesting that these extracellular regions of the Ste2p undergo conformational changes upon α-factor binding.
In the current study, we showed that although Ste2p K269A and K269C mutants maintained high ligand binding, they displayed reduced (40–45% of wild-type levels) signaling activity as judged by the Fus-LacZ assay (Fig. 6D), suggesting that Lys269 is important for Ste2p activation but not for binding of α-factor to Ste2p. On the other hand, the replacement of lysine with histidine at position 269 (K269H) of Ste2p did not have any observable effects on the biological activities of Ste2p. The results suggest that the presence of a positively charged amino acid at position 269 may be important for ligand interaction and Ste2p activation. Given that Lys269 is located at the TM6-EL3 boundary, near Try266, we speculate that Lys269 of Ste2p may be involved in different interactions with other Ste2p residues in inactive (ligand-free) and active (ligand-bound) states. Similar to the protonated rhodopsin Schiff base involving Lys296, we propose that in the inactive conformation of Ste2p, Lys269 may be involved in interactions with counterions, such as Asp201 (on the EL2-TM5 boundary) or Asp275 (on the EL3-TM7 boundary), and these interactions may be disrupted and replaced by a cation-π or hydrogen bond interaction involving the indole side chain of Trp1 of α-factor. Triple mutant cycle analysis concluded that the interaction of Trp3 of α-factor with Tyr266 and Asn205 of Ste2p was strongly influenced by residues Trp1, His2, and Leu4 of the pheromone, suggesting that the N terminus of α-factor interacts cooperatively with residues at the TM5 and TM6 extracellular surface (43). Thus, binding of the tridecapeptide might disrupt and transform existing networks of interactions between various Ste2p residues to form a new network of interactions between the α-factor residues and Ste2p, resulting in the propagation of a conformational change across the receptor and G protein signaling. That residues at the N terminus of the pheromone are critical for receptor and G protein activation as shown by the fact that the deletion of the first two residues (Trp1 and His2) of α-factor results in a potent antagonist that binds well but cannot activate the receptor (22).
In conclusion, we have identified a specific residue-to-residue interaction between position 1 of the α-factor pheromone and its GPCR, Ste2p, and also proposed a binding mechanism that incorporates all of the contact information available from chemical cross-linking, photochemical cross-linking, double mutant cycle analysis, ligand analogs, and receptor mutagenesis. The use of the tandem mass spectrometry employed in our study to pinpoint the actual cross-link is a powerful method that can be applied to other peptide-activated GPCRs, in particular those that are involved in various pathologies. To our knowledge, this is the first time that the actual cross-link point between a peptide ligand and its GPCR has been determined directly by MS/MS sequencing approaches. Therefore, the information provided in this report will be important for understanding other peptide-GPCR interactions and may influence development of drugs to modulate the activities of GPCRs.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health, NIGMS, Grant GM22087.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- GPCR
- G protein-coupled receptor
- BioACA
- biotinyl-aminocaproic acid
- DOPA
- 3,4-dihydroxylphenylalanine
- Bio-DOPA-α-factor
- an analog of α-factor with DOPA replacing tryptophan 1 and biotin attached to lysine 7 ([Lys7(BioACA),Nle12,DOPA1]α-factor)
- DMF
- dimethylformamide
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- MLT
- medium lacking tryptophan
- Nle
- norleucine
- TM
- transmembrane domain
- EL
- extracellular loop
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- Na-HRP
- NeutrAvidin-HRP
- ESI
- electrospray ionization
- MLT
- medium lacking tryptophan
- Ste2p
- α-factor receptor encoded by the STE2 gene.
REFERENCES
- 1.De Amici M., Dallanoce C., Holzgrabe U., Tränkle C., Mohr K. (2009) Med. Res. Rev. 30, 463–549 [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum D. M., Rasmussen S. G., Kobilka B. K. (2009) Nature 459, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams C., Hill S. J. (2009) Methods Mol. Biol. 552, 39–50 [DOI] [PubMed] [Google Scholar]
- 4.Lundstrom K. (2009) Methods Mol. Biol. 552, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapter M. C., White C. M., DeRidder A., Chadwick W., Martin B., Maudsley S. (2010) Pharmacol. Ther. 125, 39–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory K. J., Sexton P. M., Christopoulos A. (2007) Curr. Neuropharmacol. 5, 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leach K., Sexton P. M., Christopoulos A. (2007) Trends Pharmacol. Sci. 28, 382–389 [DOI] [PubMed] [Google Scholar]
- 8.Eglen R. M., Reisine T. (2009) Methods Mol. Biol. 552, 1–13 [DOI] [PubMed] [Google Scholar]
- 9.Dinger M. C., Bader J. E., Kobor A. D., Kretzschmar A. K., Beck-Sickinger A. G. (2003) J. Biol. Chem. 278, 10562–10571 [DOI] [PubMed] [Google Scholar]
- 10.Blake A. D., Bot G., Reisine T. (1996) Chem. Biol. 3, 967–972 [DOI] [PubMed] [Google Scholar]
- 11.Gurrath M. (2001) Curr. Med. Chem. 8, 1605–1648 [DOI] [PubMed] [Google Scholar]
- 12.Kolb P., Rosenbaum D. M., Irwin J. J., Fung J. J., Kobilka B. K., Shoichet B. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6843–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naider F., Becker J. M. (2004) Peptides 25, 1441–1463 [DOI] [PubMed] [Google Scholar]
- 14.Son C. D., Sargsyan H., Naider F., Becker J. M. (2004) Biochemistry 43, 13193–13203 [DOI] [PubMed] [Google Scholar]
- 15.Umanah G. K., Son C., Ding F., Naider F., Becker J. M. (2009) Biochemistry 48, 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tantry S. J., Ding F. X., Dumont M. E., Becker J. M., Naider F. R. (2010) Biochemistry 49, 5007–5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauser M., Kauffman S., Lee B. K., Naider F., Becker J. M. (2007) J. Biol. Chem. 282, 10387–10397 [DOI] [PubMed] [Google Scholar]
- 18.Huang L. Y., Umanah G., Hauser M., Son C., Arshava B., Naider F., Becker J. M. (2008) Biochemistry 47, 5638–5648 [DOI] [PubMed] [Google Scholar]
- 19.Turcatti G., Nemeth K., Edgerton M. D., Meseth U., Talabot F., Peitsch M., Knowles J., Vogel H., Chollet A. (1996) J. Biol. Chem. 271, 19991–19998 [DOI] [PubMed] [Google Scholar]
- 20.Sherman F. (1991) Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 21.Raths S. K., Naider F., Becker J. M. (1988) J. Biol. Chem. 263, 17333–17341 [PubMed] [Google Scholar]
- 22.Eriotou-Bargiota E., Xue C. B., Naider F., Becker J. M. (1992) Biochemistry 31, 551–557 [DOI] [PubMed] [Google Scholar]
- 23.Abel M. G., Zhang Y. L., Lu H. F., Naider F., Becker J. M. (1998) J. Pept. Res. 52, 95–106 [DOI] [PubMed] [Google Scholar]
- 24.David N. E., Gee M., Andersen B., Naider F., Thorner J., Stevens R. C. (1997) J. Biol. Chem. 272, 15553–15561 [DOI] [PubMed] [Google Scholar]
- 25.Lee B. K., Khare S., Naider F., Becker J. M. (2001) J. Biol. Chem. 276, 37950–37961 [DOI] [PubMed] [Google Scholar]
- 26.Burdine L., Gillette T. G., Lin H. J., Kodadek T. (2004) J. Am. Chem. Soc. 126, 11442–11443 [DOI] [PubMed] [Google Scholar]
- 27.Kraft P., Mills J., Dratz E. (2001) Anal. Biochem. 292, 76–86 [DOI] [PubMed] [Google Scholar]
- 28.Son C. D., Sargsyan H., Hurst G. B., Naider F., Becker J. M. (2005) J. Pept. Res. 65, 418–426 [DOI] [PubMed] [Google Scholar]
- 29.Beavis R., Fenyo D. (2004) Curr. Protoc. Bioinformatics, Chapter 13, Unit 13.12 [DOI] [PubMed] [Google Scholar]
- 30.Liu B., Burdine L., Kodadek T. (2006) J. Am. Chem Soc. 128, 15228–15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Link A. J., Fleischer T. C., Weaver C. M., Gerbasi V. R., Jennings J. L. (2005) Methods 35, 274–290 [DOI] [PubMed] [Google Scholar]
- 32.Lundgren D. H., Martinez H., Wright M. E., Han D. K. (2009) Curr. Protoc. Bioinformatics, Chapter 13, Unit 13.13 [DOI] [PubMed] [Google Scholar]
- 33.Bauer A., Kuster B. (2003) Eur. J. Biochem. 270, 570–578 [DOI] [PubMed] [Google Scholar]
- 34.Wilm M. (2000) Adv. Protein Chem. 54, 1–30 [DOI] [PubMed] [Google Scholar]
- 35.Graumann J., Dunipace L. A., Seol J. H., McDonald W. H., Yates J. R., 3rd, Wold B. J., Deshaies R. J. (2004) Mol. Cell. Proteomics 3, 226–237 [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y. L., Marepalli H. R., Lu H. F., Becker J. M., Naider F. (1998) Biochemistry 37, 12465–12476 [DOI] [PubMed] [Google Scholar]
- 37.Ding F. X., Lee B. K., Hauser M., Davenport L., Becker J. M., Naider F. (2001) Biochemistry 40, 1102–1108 [DOI] [PubMed] [Google Scholar]
- 38.Bajaj A., Connelly S. M., Gehret A. U., Naider F., Dumont M. E. (2007) Biochim. Biophys. Acta 1773, 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y. L., Lu H. F., Becker J. M., Naider F. (1997) J. Pept. Res. 50, 319–328 [DOI] [PubMed] [Google Scholar]
- 40.Henry L. K., Khare S., Son C., Babu V. V., Naider F., Becker J. M. (2002) Biochemistry 41, 6128–6139 [DOI] [PubMed] [Google Scholar]
- 41.Lee B. K., Lee Y. H., Hauser M., Son C. D., Khare S., Naider F., Becker J. M. (2002) Biochemistry 41, 13681–13689 [DOI] [PubMed] [Google Scholar]
- 42.Ding F. X., Lee B. K., Hauser M., Patri R., Arshava B., Becker J. M., Naider F. (2002) J. Pept. Res. 60, 65–74 [DOI] [PubMed] [Google Scholar]
- 43.Naider F., Becker J. M., Lee Y. H., Horovitz A. (2007) Biochemistry 46, 3476–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornak V., Ahuja S., Eilers M., Goncalves J. A., Sheves M., Reeves P. J., Smith S. O. (2010) J. Mol. Biol. 396, 510–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 46.Konopka J. B., Margarit S. M., Dube P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6764–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommers C. M., Dumont M. E. (1997) J. Mol. Biol. 266, 559–575 [DOI] [PubMed] [Google Scholar]
- 48.Lee Y. H., Naider F., Becker J. M. (2006) J. Biol. Chem. 281, 2263–2272 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.