Abstract
The PhoP-PhoQ two-component system is commonly used by bacteria to sense environmental factors. Here we show that the PhoP-PhoQ system of Edwardsiella tarda detects changes in environmental temperature and Mg2+ concentration as well as regulates the type III and VI secretion systems through direct activation of esrB. Protein secretion is activated from 23 to 35 °C or at low Mg2+ concentrations, but it is suppressed at or below 20 °C, at or above 37 °C, or at high Mg2+ concentrations. The effects of temperature and Mg2+ concentration are additive. The PhoQ sensor domain has a low Tm of 37.9 °C, and it detects temperatures through a conformational change of its secondary structure. Mutation of specific Pro or Thr residues increased the stability of the PhoQ sensor drastically, altering its temperature-sensing ability. The PhoQ sensor detects Mg2+ concentration through the direct binding of Mg2+ to a cluster of acidic residues (DDDSAD) and through changes that likely affect its tertiary structure. Here, we describe for the first time the use of PhoP-PhoQ as a temperature sensor for bacterial virulence control.
Keywords: Bacterial Signal Transduction, Bacterial Transcription, Circular Dichroism (CD), DNA-binding Protein, Protein Stability, PhoP-PhoQ, Type III and VI Secretion Systems, Temperature Sensing, Two-component System
Introduction
Edwardsiella tarda is a Gram-negative bacterial pathogen that is associated with septicemia and fatal infections in a wide variety of animals including fish and humans (1, 2). Using a functional genomics approach (3, 4), a type III and a type VI secretion system (T3SS and T6SS) were identified as the two most important virulence mechanisms in E. tarda PPD130/91 (5, 6). The expression and secretion of both T3SS proteins, such as EseB and EseD, and T6SS proteins, such as EvpA and EvpC, were suppressed at a growth temperature of 37 °C. Furthermore, the virulence of E. tarda PPD130/91 grown at this temperature was drastically reduced compared with that grown at 25 °C (4). A subsequent study suggested that temperature affected the expression of T3SS and T6SS through the regulation of the expression of the two-component system, ersA-esrB and esrC (7). The identity of the regulator involved in sensing temperature changes and its mechanism of action, however, were not determined.
The two-component system PhoP-PhoQ, in which PhoQ is the sensor histidine kinase and PhoP is the response regulator, senses environmental stimuli and regulates those responses that are essential for the survival and virulence of the bacteria and is one of the most studied bacterial signaling systems. In Salmonella, the intracellular level of Mg2+ is tightly regulated by the PhoP-PhoQ system for Mg2+ homeostasis and avoidance of metal toxicity (8). In addition, the PhoP-PhoQ system of Salmonella typhimurium is essential for virulence by allowing the pathogen to sense divalent cations, mildly acidic pH, and antimicrobial peptides, all of which provide cues that the bacterium is inside the phagosome of a macrophage. In response to these conditions, the PhoP-PhoQ system regulates the expression of hundreds of genes encoding virulence proteins with various properties, including intracellular survival, invasion, lipid A structure, resistance to antimicrobial peptides, and phagosome alteration (9). The highly acidic surface of the PhoQ sensor domain has been proposed to bind both divalent cations and antimicrobial peptides. Depletion of Mg2+ or binding to antimicrobial peptides displaces the divalent cations located between PhoQ metal binding sites and the membrane phospholipid to initiate signal transduction (10). As of yet, there has been no report on the detection of temperature, an important environmental factor for bacterial survival and virulence, by the PhoP-PhoQ system.
Environmental temperature detection is essential for the survival and virulence of many pathogenic bacteria, especially with respect to the recognition of a suitable host, and many pathogens have the ability to sense temperature changes. As an example, the secretion of EspA and EspB by human enteropathogenic Escherichia coli (EPEC) is maximal at 36 °C, decreased at 39 °C, and abolished at 42 °C. In contrast, the secretion of these proteins by rabbit EPEC (RDEC-1) is maximal at 39 °C and still occurs at 42 °C (11). Two-component systems for the detection of environmental temperatures have been reported previously, such as the DesK-DesR system of Bacillus subtilis that regulates membrane fluidity according to temperature (12). The membrane domain of DesK is the temperature-sensing element (13), but its mechanism of action is unknown.
Here, we report the characterization of PhoP-PhoQ, a two-component system in E. tarda that is involved in sensing environmental temperatures and Mg2+ concentrations for the regulation of virulence through EsrB. The periplasmic sensor domain of PhoQ is responsible for sensing temperatures by a conformational change that is highly sensitive to temperature. Using thermal and urea denaturations combined with site-directed mutagenesis, we identified residues in the PhoQ sensor domain that are essential for temperature sensing and Mg2+ binding. Our findings provide an understanding of the novel mechanism of temperature and Mg2+ detection by a protein sensor and also shed light on the regulation of virulence in pathogenic bacteria by sensing temperature and Mg2+ concentration in the surrounding environment.
EXPERIMENTAL PROCEDURES
Cloning of the PhoP-PhoQ Two-component System in E. tarda PPD130/91
Bacterial genomic DNA was extracted using the Wizard genomic DNA purification kit (Promega, Madison, WI). PCR amplification (2 min at 94 °C, 30 cycles each of 10 s at 94 °C, 30 s at 56 °C, and 1 min at 72 °C, a final extension of 5 min at 72 °C) was carried out using the Advantage 2 polymerase mix (Clontech, Mountain View, CA) with two pairs of degenerate primers, phoPdeg and phoQdeg (supplemental Table 1). The PCR products were cloned with the pGEM-T Easy vector system (Promega) and transformed into E. coli DH5α cells. The cloned fragments were sequenced using the PRISMTM 3100 automated DNA sequencer with the ABI Prism Big Dye termination cycle sequence kit (Applied Biosystems, Foster City, CA). This approach identified a 400-bp fragment of the phoP gene that was later used to design primers for genome walking. To obtain the full-length sequences of phoP and phoQ, genome walking libraries of E. tarda PPD130/91 were created and digested with SmaI, ScaI, EcoRV, StuI, and PvuII according to the procedure described in the Universal Genome Walker kit manual (Clontech). PCR amplification (7 cycles each of 25 s at 94 °C and 3 min at 72 °C, 32 cycles each of 25 s at 94 °C and 3 min at 65 °C, and a final extension of 5 min at 65 °C) was carried out using primers specific for sequences of phoP and phoQ, with the adaptor primer 1 (Clontech). The complete sequences of phoP and phoQ, including flanking sequences comprising the 1260 bp upstream of phoP and 1296 bp downstream of phoQ, were obtained by this method.
LacZ Reporter Gene System
For the construction of the ersB LacZ reporter gene system, the primer pair pRWesrb (supplemental Table 1), derived from E. tarda PPD130/91 genomic DNA, was used to amplify the putative promoter region of esrB containing a PhoP box. The PCR product was digested with restriction enzymes and then ligated with the pRW50 plasmid (14). The ligation mixture was introduced into either the wild type or mutant E. tarda by electroporation. The transformants were screened for resistance to both colistin (12.5 μg/ml) and tetracycline (10 μg/ml). Similar procedures were followed to generate the phoP LacZ reporter gene system using the primer pair pRWphoP (supplemental Table 1). For the β-galactosidase assays, E. tarda cells were grown in N-minimal medium (15) overnight at 20, 23, 25, 30, 35, or 37 °C with 1 or 10 mm Mg2+. The overnight culture (5%) was inoculated into fresh medium and grown at the specified temperatures until the cell density reached 0.5, as measured by optical density at 600 nm (A600). Cells were permeabilized (16) by the addition of 25 μl of 0.1% SDS and 25 μl of chloroform to a solution containing the cell pellet from a 1 ml culture resuspended with 600 μl of Z-buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, and 1 mm MgSO4, pH 7.0). To start the reaction, 100 μl of ortho-nitrophenyl-β-galactosidase (4 mg/ml) was added. When the suspension started to turn yellow, the reaction was terminated by adding 250 μl of 1 m Na2CO3. The mixture was centrifuged for 5 min at 13,000 × g, and the supernatant was transferred to a cuvette to measure the absorbance at 420 nm. The cell density was assessed by the A600, and the unit of activity was calculated as (absorbance at 420 nm)/(reaction time in min × A600).
Electrophoretic Mobility Shift Assay (EMSA)
The full-length PhoP protein at various concentrations (0, 0.5, 1, 1.5, or 2 μm) was mixed with purified esrB or phoP promoter DNA fragments (2 μg) labeled 5′ with 6-carboxyfluorescein tag (1st BASE, Singapore) in a reaction mixture (50 μl) containing 50 mm Tris-HCl, pH 7.5, 200 mm KCl, 0.1 mm EDTA, and 5% v/v glycerol. The mixture was incubated at 25 °C for 2 h before loading (15 μl) onto each lane of a 5% native polyacrylamide gel for electrophoresis (0.5× Tris borate-EDTA buffer, 1 mA/cm for 4 h).
Western Blot Analysis
For SDS-PAGE (12% separation gel) used for Western blot analyses, 5 ml of bacterial culture at A600 = 1.0 was used for isolation of extracellular proteins (ECPs)2 for each lane. Proteins were transferred to a PVDF membrane with a semidry system and examined by using the SuperSignal WestPico Chemiluminescent substrate (Pierce) under the conditions recommended by the manufacturer. EseB and EseC were detected by the addition of diluted anti-EseB (1:10,000) and anti-EseC (1:10,000) polyclonal antisera, respectively, followed by a 1:5,000 dilution of mouse anti-rabbit IgG HRP (Santa Cruz Biotechnology, Santa Cruz, CA).
Cloning, Expression, and Purification of the PhoQ Sensor Domain
The PhoQ periplasmic sensor domain (from residue Phe-47 to Glu-186) was amplified by using the primer pair phoQs (supplemental Table 1) derived from E. tarda PPD130/91 genomic DNA. The PCR product was subcloned into pET-M, a modified pET-32a vector (Novagen, Darmstadt, Germany) (pETM-phoQs), and then transformed into E. coli BL21(DE3) cells for expression. To generate pETM-phoQf, the full-length phoQ gene was amplified using the primer pair phoQf (supplemental Table 1) and subcloned into pET-M. An overnight culture of pETM-phoQs in BL21(DE3) cells was inoculated into LB with 100 μg/ml ampicillin, and the cells were grown until the cell density reached 0.8 at A600. Protein expression was induced with 0.5 mm of isopropyl 1-thio-β-d-galactopyranoside, and the cells were grown at 30 °C overnight. Cells were harvested by centrifugation and resuspended in 50 mm sodium phosphate, pH 8.0, and 300 mm NaCl. The cell suspension was sonicated and then centrifuged to obtain the inclusion bodies as a pellet. The inclusion bodies were resuspended in 20 mm sodium phosphate, pH 8.0, 100 mm NaCl, and 8 m urea and lysed by sonication. The lysate was centrifuged, and the supernatant was purified using a nickel-nitrilotriacetic acid affinity column in buffer containing 8 m urea. Refolding was performed by rapidly diluting the eluate into an ice-cold solution of 20 mm sodium phosphate, pH 8.0, containing 0.1 or 10 mm Mg2+ with stirring. The refolded sample was dialyzed overnight against a buffer containing 20 mm sodium phosphate, pH 6.5, and 100 mm NaCl and in the presence or absence of 10 mm Mg2+. The sample was further purified by a Superdex-75 gel filtration column (GE Healthcare) equilibrated with the dialysis buffer. Fractions containing PhoQ sensor were pooled and concentrated. A similar procedure was followed for the purification of the various PhoQ mutants.
CD Monitoring of the Thermal and Urea Denaturation of PhoQ Sensor
Circular dichroism (CD) measurements were performed using a J-810 spectropolarimeter (Jasco, Easton, MD) with a 1 mm path length cuvette (Hellma, Müllheim, Germany). For thermal denaturation, a 300 μl sample of 15 μm PhoQ sensor in 20 mm sodium phosphate, pH 6.5, and 100 mm NaCl and in the presence or absence of 10 mm Mg2+ was used. Thermal denaturation was monitored by changes in CD ellipticity at 218 or 206.5 nm as a function of temperature from 5 to 80 οC with a heating rate of 2 οC/min. Urea denaturation was performed by monitoring changes in the CD ellipticity at 210 nm. Samples containing 20 μm PhoQ sensor and various amounts of urea were prepared in a buffer containing 20 mm sodium phosphate, pH 6.5, and 100 mm NaCl in the presence or absence of 10 mm Mg2+. The protein sample was equilibrated for at least 1 h at 20, 30, or 37 °C before measurement.
Fluorescence Spectra and Urea Denaturation of the PhoQ Sensor
Fluorescence spectra were performed using 2 μm purified PhoQ in 20 mm sodium phosphate, pH 6.5, and 100 mm NaCl in the presence of different concentrations of Mg2+ at 20, 30, or 37 °C. The sample was excited at 280 nm, and the emission spectra were recorded at 0.2 nm intervals from 300 to 400 nm. Urea denaturation was monitored by changes in fluorescence at 350 nm. Samples containing 2 μm PhoQ sensor and different concentrations of urea were prepared in a buffer containing 20 mm sodium phosphate, pH 6.5, and 100 mm NaCl in the presence or absence of 10 mm Mg2+. The protein sample was equilibrated at 20, 30, or 37 °C for at least 1 h before measurement.
Generation of phoPi and phoQi Mutants and Complementation Experiments
Insertional mutants of phoP (phoPi) and phoQ (phoQi) in E. tarda were constructed with the suicide plasmid pRE112 (17). For the construction of phoPi, an internal fragment of phoP was amplified from E. tarda genomic DNA with the primer pair phoPmut (supplemental Table 1), which contains a KpnI restriction enzyme site. The PCR product was digested by restriction enzymes and ligated into the pRE112 plasmid, and the resulting plasmid was transformed into E. coli MC1061 λpir. After sequencing, the recombinant plasmid was then transformed into E. coli SM10 λpir. These transformants were used to conjugate with wild type E. tarda to obtain defined mutants by selecting colonies resistant to both chloramphenicol (30 μg/ml) and colistin (12.5 μg/ml). The insertion of the plasmid into chromosomal DNA was confirmed by sequence analysis. The primer pair phoQmut (supplemental Table 1) was used for the construction of phoQi. Complementation of the phoPi and phoQi mutants was performed with a pACYC184-based system. To obtain phoPi + phoP, the complete phoP gene was prepared by PCR using the primer pair phoPfull (supplemental Table 1) from E. tarda PPD130/91 genomic DNA. The PCR product was digested with restriction enzymes and ligated into the digested pACYC184 plasmid. The obtained ligation mixture was transformed into E. coli DH5α, and then the plasmid DNA was extracted and transformed into competent cells of E. tarda phoPi by electroporation. The same procedure was followed to obtain phoQi + phoQ mutant using the primer pair phoQfull and the phoQi + phoQEPEC mutant using the primer pair phoQEPEC_full (supplemental Table 1).
Preparation of ECPs
Overnight cultures of E. tarda grown at different temperatures in DMEM (A600 = 0.8) were diluted 1:200 into fresh DMEM and incubated for 24 h at the indicated temperatures. For SDS-PAGE (12% separation gel) silver staining, 20 ml of bacterial culture at A600 = 1.0 was used for isolation of ECPs for each lane. Cells were removed from the culture by centrifugation (5500 × g, 20 min, 4 °C), and the supernatant was filtered through a 0.22 μm pore size small-protein binding filter (Millipore, Billerica, MA). The ECP fraction was isolated by trichloroacetic acid precipitation, and the protein pellet was washed three times with −20 °C acetone and then air dried. ECP protein pellets were solubilized in Ready Prep reagent 3 (5 m urea, 2 m thiourea, 2% (w/v) CHAPS, 2% (w/v) SB 3–10, 40 mm Tris, and 0.2% (w/v) Bio-Lyte 3/10 ampholyte (Bio-Rad)) and stored at −80 °C until analysis. The protein concentration was determined with a Bio-Rad protein assay kit using bovine serum albumin as the standard.
RESULTS
Identification of the PhoP-PhoQ Two-component System
Using genome walking with degenerate primers derived from the conserved nucleotide sequences of related bacterial species, the phoP and phoQ genes of E. tarda PPD130/91 (GenBankTM accession code GU324976) were identified and sequenced. Using RT-PCR experiments on RNA isolated from E. tarda PPD130/91, the phoP and phoQ genes are found to be transcribed as an operon (supplemental Fig. S1). Sequence alignment of the periplasmic PhoQ sensor domain of E. tarda showed a 61.9% identity to Yersinia pestis KIM (18) followed by a 51.8% identity to both S. typhimurium LT2 (19) and E. coli CFT073 (20) (Fig. 1). In contrast, the sequence identity between the PhoQ sensor domains of E. coli and S. typhimurium is 82.0%. The relatively low sequence identity between the PhoQ sensor domain of E. tarda and those of E. coli and S. typhimurium suggests that this domain may sense different extracellular stimuli for each of these bacteria.
FIGURE 1.
Sequence alignment of the PhoQ sensor domains. The sequence of the PhoQ periplasmic sensor region from E. tarda (PhoQ_ET; GU324976) was compared with those from Y. pestis (PhoQ_YP; NP_669110), S. typhimurium (PhoQ_ST; NP_460200), and E. coli (PhoQ_EC; NP_753417). The secondary structural elements from the crystal structure of the E. coli PhoQ sensor are shown underneath the sequences, whereas the predicted secondary structural elements of the E. tarda PhoQ sensor are shown on top of the sequences. Asterisks below the sequences indicate identical residues. Negatively charged residues in the acidic patches are in bold. Pro and Thr residues in the E. tarda PhoQ sensor that are not conserved with those in E. coli and S. typhimurium and were selected for mutation studies are boxed.
The crystal structures of the PhoQ sensor domains of S. typhimurium (PDB ID 1YAX) (21) and E. coli (PDB ID 3BQ8) (22) adopt a fold characteristic of the PAS (Per-Arnt-Sim) domain superfamily (23). The predicted secondary structure of the PhoQ sensor domain of E. tarda using PsiPred (24, 25) is similar to those seen in the three-dimensional structures from E. coli and S. typhimurium, suggesting that it may also assume a PAS-fold (Fig. 1).
PhoP-PhoQ Regulates T3SS and T6SS through EsrB
To investigate a possible contribution of the PhoP-PhoQ system to the virulence of E. tarda, two E. tarda mutants having insertions at phoP (phoPi) and phoQ (phoQi), respectively, were constructed. The growth and protein secretion profiles of the wild type PPD130/91, phoPi, and phoQi, were measured at the optimal growth temperature of 35 °C. SDS-PAGE analysis showed that both the phoPi and phoQi mutants were deficient in the production of ECPs, including EseB, EseC, and EseD from T3SS as well as EvpC and EvpP from T6SS (Fig. 2A). At 35 °C, the ECPs secreted by the wild type strain reached a concentration of 2.50 ± 0.09 μg/ml (n = 3) after 24 h. Significantly lower levels of ECPs were observed in the phoPi (0.35 ± 0.04 μg/ml, n = 3) and phoQi (0.40 ± 0.06 μg/ml, n = 3) mutants. Cell densities of the mutants were comparable with that of the wild type bacteria, suggesting that the reduced protein secretion in the mutants was not due to growth deficiencies (data not shown). Complementation of phoPi and phoQi in trans with a plasmid-borne wild type copy of phoP and phoQ, respectively, restored the secretion of ECPs to levels comparable with that of the wild type bacteria (Fig. 2A). These results indicated that the PhoP-PhoQ system is involved in regulation of the T3SS and T6SS in E. tarda PPD130/91.
FIGURE 2.
PhoP-PhoQ regulates T3SS and T6SS through direct binding of PhoP to the promoter region of ersB. A, silver-stained SDS-PAGE shows the ECP (T3SS: EseB, EseC and EseD; T6SS: EvpC and EvpP) secretion profiles of E. tarda PPD130/91 (WT), E. tarda carrying an insertion mutation in phoP (phoPi) or phoQ (phoQi), and the mutant E. tarda with complementation of the corresponding wild type gene (phoPi + phoP or phoQi + phoQ). The incubation temperature was 35 °C. Lane BL contains a blank control with only DMEM and no bacterial cells. B, shown is the promoter region of the E. tarda 130/91 esrB gene, a putative PhoP box. The sequences corresponding to the (G/T)GTTTA direct repeats are shaded. The classic PhoP box from the mgtA promoter and the imperfect but orthodox PhoP box from the orgB promoter of S. typhimurium are shown for comparison. T and A residues that are conserved among PhoP boxes are in bold. C, shown is an electrophoretic mobility shift assay of PhoP protein on a 5′ 6-carboxyfluorescein-labeled DNA fragment (470 bp, from nt −467 to +3) from the promoter region of esrB (upper panel) and another DNA fragment with the same boundaries but with the PhoP box (nt −311 to −295) removed (lower panel). D, shown is an electrophoretic mobility shift assay of PhoP protein on a 5′ 6-carboxyfluorescein-labeled DNA fragment (454 bp, from nt −451 to +3) from the promoter region of phoP (upper panel) and another DNA fragment with the same boundaries but with the PhoP box (nt −192 to −176) removed (lower panel). E, shown are transcription levels of the esrB-LacZ reporter gene fusion measured by β-galactosidase activity from bacterial cells cultured at 30 °C in TSB medium. WT, E. tarda PPD130/91; phoPi, E. tarda with insertion mutation in the phoP gene; phoPi + phoP, E. tarda phoPi mutant complemented with wild type phoP.
Based on the previous observation that an esrB mutant of E. tarda showed an ECP profile similar to that of either the phoPi or phoQi mutant (7), we speculated that PhoP regulates the secretion of T3SS and T6SS proteins by modulating the expression of EsrB. To confirm this hypothesis, the interaction of PhoP with the promoter region of esrB was examined. A sequence motif comprising a repeat of two hexanucleotide sequences (T/G) GTTTA separated by five bases has been defined as the high affinity PhoP binding region in E. coli and was named the “PhoP box” (26, 27). Inspection of the DNA sequence upstream of esrB revealed a putative PhoP box of the sequence 5′-ACTCCA AAGGG CATTTA-3′ between nt −311 and −295 upstream of the start codon of esrB (Fig. 2B). This sequence is considered to be an imperfect but orthodox match to the consensus PhoP box sequence. Using DNase I footprinting, a similar imperfect but orthodox PhoP box of the sequence 5′-TATTGA GGAGG CATTGA-3′ has been identified between nt −42 and −26 relative to the PhoP-induced transcriptional start site of the orgBC promoter in S. typhimurium (28). Electrophoretic mobility shift assay showed that the His-tagged PhoP can bind to a 470 bp DNA fragment (from nt −467 to +3) derived from the promoter region of esrB containing a putative PhoP box (Fig. 2C). However, there was no interaction observed between PhoP and a DNA fragment derived from the same promoter region of esrB in the absence of the PhoP box region (Fig. 2C). Inspection of the promoter region of the phoP-phoQ genes also revealed a putative PhoP box, with the sequence 5′-AGTTTA CCTAC CGTTGA-3′, located upstream between nt −192 and −176. Electrophoretic mobility shift assay showed that PhoP can also bind to a 454 bp DNA fragment (from nt −451 to +3) derived from the promoter region of phoP and at an affinity even higher than that between PhoP and the promoter region of esrB (Fig. 2D).
To confirm that the expression of the esrB gene was controlled by PhoP, an esrB-LacZ reporter gene fusion (pRWesrB130/91) under the control of the putative esrB promoter (from nt −900 to +150) was created, and the activity of the β-galactosidase reporter genes (LacZ) for the esrB promoter region was measured in both the wild type strain and the phoPi mutant. The esrB-LacZ activity for the phoPi mutant was significantly reduced compared with that for the wild type (Fig. 2E). When the pACYC184 plasmid carrying the wild type phoP gene was introduced into the phoPi strain for complementation, the β-galactosidase activity recovered to a level similar to that of the wild type (Fig. 2E). These results indicated that PhoP, a global regulator that binds upstream of esrB, controls the transcription of esrB by modulating its promoter activity.
Secretion of T3SS and T6SS Proteins Is Dependent on Both the Temperature and Mg2+ Concentration
To gain insight into the effects of temperature and Mg2+ concentration on the secretion of proteins by E. tarda, wild type bacterial cultures were grown at different temperatures and Mg2+ concentrations in TSB, and the identities and amounts of secreted ECPs were investigated. SDS-PAGE analysis showed that both T3SS (EseB, EseC, and EseD) and T6SS (EvpC and EvpP) proteins were secreted by E. tarda. The secretion of these ECPs is prominent at growth temperatures ranging from 23 to 35 °C (2.75 ± 0.06 μg/ml at 23 °C, 3.00 ± 0.11 μg/ml at 25 °C, 2.75 ± 0.10 μg/ml at 30 °C, and 2.50 ± 0.09 μg/ml at 35 °C) but significantly reduced at 20 °C (0.40 ± 0.06 μg/ml) and 37 °C (0.45 ± 0.08 μg/ml) (Fig. 3A). These results clearly show that protein secretion by E. tarda T3SS and T6SS is highly temperature-dependent. Mg2+ concentration also affected the amounts of protein secreted by E. tarda T3SS (EseB) and T6SS (EvpC). Protein secretion is slightly reduced in the presence of 10 mm Mg2+ concentration, at least at 37 °C (Fig. 3B), suggesting that the effects of temperature and Mg2+ concentration may be additive.
FIGURE 3.
The secretion of T3SS and T6SS ECPs by E. tarda is temperature- and Mg2+ concentration-dependent through transcriptional activation of esrB, phoP, and phoQ. A, silver-stained SDS-PAGE shows T3SS and T6SS ECP secretion by E. tarda PPD130/91 at different incubation temperatures as indicted above the lanes. The lane BL contains a blank control with only DMEM and no bacterial cells. B, Western blot analysis shows the secretion of EseB from T3SS (upper panel) and EvpC from T6SS (lower panel) by E. tarda at the temperatures of 20, 30, and 37 °C in the presence of 1 mm (L) or 10 mm (H) Mg2+ using N-minimal medium. C and D, the β-galactosidase activities of the reporter genes esrB-LacZ and phoP-LacZ were determined in N-minimal medium under different incubation temperatures in the presence of 1 mm (striped bars) or 10 mm (empty bars) Mg2+. Both genes showed reduced activity at 20 and 37 °C compared with 23, 25, and 35 °C. The activities of both genes are also repressed at higher concentrations of Mg2+, and this effect is additive with that of the temperature.
To verify the effects of temperature and Mg2+ concentration on the PhoP activation of esrB, the β-galactosidase activities of the reporter gene (LacZ) for the esrB promoter region was measured in the wild type strain under different temperatures in the presence of high (10 mm) or low (1 mm) concentrations of Mg2+ using N-minimal medium. The expression levels of esrB-LacZ were ∼30–40% higher at the low Mg2+ concentration when compared with those at the high Mg2+ concentration (Fig. 3C). The difference is even more significant at temperatures that suppress the ECP secretion (e.g. 60% higher at 20 °C and 80% higher at 37 °C). In general, the expression levels of esrB-LacZ were significantly higher for the temperature range of 23 to 35 °C when compared with those at 20 and 37 °C. This result is in agreement with the E. tarda ECP secretion profiles observed at these respective temperatures and Mg2+ concentrations and further confirms that EsrB is a positive regulator of protein secretion by T3SS and T6SS in E. tarda. In addition to EsrB, the effects of temperature and Mg2+ concentration on the expression levels of PhoP was studied using a new strain harboring lacZ fusion genes under the control of the putative promoters for phoP (from nt −1460 to +150). LacZ transcriptional fusion with the PhoP (pRWphoP130/91) protein showed temperature and Mg2+ concentration responsive phenomena similar to those seen for pRWesrB130/91 (Fig. 3D).
PhoQ Sensor Domain Undergoes a Conformational Change at Low Temperatures
We hypothesized that PhoQ is responsible for sensing environmental temperature and Mg2+ concentration and that it regulates T3SS and T6SS through EsrB. To verify these hypotheses, the sensor domain of E. tarda PhoQ was cloned and expressed, and its thermal denaturation was studied in the presence or absence of Mg2+ using CD. The N-terminal sensor domain of PhoQ (PhoQs) from residues 45 to 187 is located between two predicted transmembrane helices and is speculated to be periplasmic. Although the PhoQs protein was expressed in inclusion bodies, it could be refolded properly in a buffer containing a minimal amount of 0.1 mm Mg2+, as shown by CD. Thermal denaturation followed by CD at 206.5 nm showed a sharp conformational transition between 30 and 40 °C for PhoQs in the absence of Mg2+, with a Tm of around 37.9 °C after curve-fitting according to the equation by Ruiz-Sanz et al. (29). The addition of 10 mm Mg2+ stabilized the PhoQs only slightly, with the Tm shifting to about 40.2 °C (Fig. 4B). A thermal denaturation experiment carried out on the homologous PhoQ sensor domain from E. coli obtained a Tm of 59.2 and 65.4 °C in the absence and presence of 10 mm Mg2+, respectively (30). This sharp conformational transition in the PhoQs of E. tarda at a relatively low temperature is consistent with the temperature transition (35 to 37 °C) that regulates the secretion of proteins by T3SS and T6SS, suggesting that this conformational change could be the mechanism employed by PhoQ to sense the environmental temperature. No conformational transition, however, was detected by CD between 16 and 30 °C, a range within which another temperature transition (20 to 23 °C) was supposed to regulate protein secretions by T3SS and T6SS in E. tarda.
FIGURE 4.
Thermal and urea denaturation of the E. tarda PhoQ sensor domain. A, shown is a far-UV CD spectra of the PhoQ sensor domain at 20 °C (open circle), 30 °C (closed squares), 35 °C (open squares), and 37 °C (closed circles). B, shown is thermal denaturation of the E. tarda PhoQ sensor domain monitored by CD at 206.5 nm in the presence (closed circles) or absence (open circles) of 10 mm Mg2+. C, shown is urea denaturation of the E. tarda PhoQ sensor domain monitored by CD at 210 nm at 20 °C (circles), 30 °C (squares), and 37 °C (triangles) in the presence (open symbols) or absence (closed symbols) of 10 mm Mg2+. D, shown is urea denaturation of the E. tarda PhoQ sensor domain monitored by fluorescence at 350 nm at 20 °C (circles), 30 °C (squares), and 37 °C (triangles) in the presence (open symbols) or absence (closed symbols) of 10 mm Mg2+.
To determine whether there was any conformational change of PhoQs at 20 °C, the equilibrium urea denaturation of E. tarda PhoQs was monitored by CD at 210 nm at 20, 30, and 37 °C in the absence or presence of 10 mm Mg2+. The data were curve-fitted according to the equation of Mok et al. (31) to obtain the ΔG values. In the absence of Mg2+, PhoQs had a higher stability toward urea at 30 °C (ΔG = 8.5 kcal/mol) than at 37 °C (ΔG = 7.6 kcal/mol) (Fig. 4C). In the presence of 10 mm Mg2+, the stability at both 30 °C (ΔG = 9.4 kcal/mol) and 37 °C (ΔG = 8.6 kcal/mol) increased slightly. The lower stability of PhoQs at 37 °C toward urea when compared with that at 30 °C is consistent with the thermal denaturation data. Interestingly, there was a similar drop in the stability of PhoQs toward urea at 20 °C (ΔG = 7.3 kcal/mol and 8.0 kcal/mol in the absence and presence of 10 mm Mg2+, respectively) compared with that at 30 °C, suggesting that PhoQ may also sense the temperature transition between 20 and 23 °C and regulate T3SS and T6SS of E. tarda within this temperature range.
Fluorescence spectra were used to determine whether there were any tertiary structure changes after the urea denaturation of PhoQs at different temperatures in the absence or presence of 10 mm Mg2+ (Fig. 4D). Surprisingly, there were no significant changes in the stability of the PhoQs tertiary structure at the temperatures of 20, 30, and 37 °C. The addition of 10 mm Mg2+ stabilized the PhoQs domain by ∼1–2 kcal/mol, comparable with what had been observed previously in the E. coli PhoQ sensor domain (30). During urea denaturation, the secondary structure of PhoQs starts to melt before the tertiary structure (Fig. 4, C and D), suggesting that in PhoQs conformational changes due to temperature transitions mainly involve secondary structures and that the secondary structures appear to change more readily than the tertiary structure.
Thr and Pro Residues Are Responsible for Temperature Sensing by PhoQ
To understand the mechanism underlying the temperature-dependent conformational changes of PhoQs secondary structures, the sequence of PhoQs from E. tarda was compared with those from other bacteria. The results showed that some residues at the turn regions of PhoQs of S. typhimurium and E. coli were replaced with either Pro or Thr residues in PhoQs of E. tarda (Fig. 1). Based on this observation, these Pro and Thr residues were selected for site-specific mutation studies. Six single point mutants of E. tarda PhoQs (T76E, P77L, P79E, P120N, P140H, and T167P) were generated by converting selected Pro or Thr residues to the corresponding residues in S. typhimurium and E. coli.
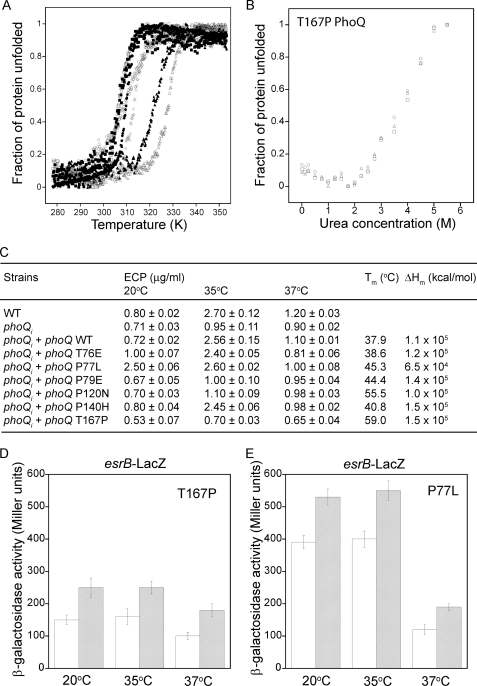
Thermal denaturation studies monitored by far-UV CD showed that PhoQs mutants P120N and T167P had Tm temperatures of 55.5 and 59.0 °C, respectively, which were both significantly higher than that of the wild type PhoQs (Tm = 37.9 °C) but comparable with that of the E. coli PhoQ sensor domain (Tm = 65.4 °C in 10 mm Mg2+) (30) (Fig. 5A). In contrast, the PhoQs mutants T76E (Tm = 38.6 °C) and P140H (Tm = 40.8 °C) had Tm values similar to that of the wild type protein, and the PhoQs mutants P77L (Tm = 45.3 °C) and P79E (Tm = 44.4 °C) had Tm values slightly higher than that of the wild type protein. The ΔHm value (related to the slope of transition) of P77L (6.5 × 104 kcal/mol), however, was much smaller than that of P79E (1.4 × 105 kcal/mol). Analysis of the urea stability (monitored by CD at 210 nm) of the most thermally stable mutant, T167P, revealed that unlike the wild type protein, temperature (at 20, 30, or 37 °C) had no effect on its stability toward urea (Fig. 5B), suggesting that mutation of a single Pro or Thr residue within the turn region of E. tarda PhoQs could abolish the temperature transition responses (20 to 23 °C and 35 to 37 °C) that occur in the wild type protein.
FIGURE 5.
Thermal denaturation of Thr or Pro residue mutants of the E. tarda PhoQ sensor domain. A, shown is thermal denaturation of the wild type and mutant E. tarda PhoQ sensor domains monitored by CD at 202 nm for P79E (open rhombuses) and P120N (filled triangles), at 206.5 nm for T167P (open triangles), and at 218 nm for wild type (open squares), T76E (closed squares), P77L (open circles), and P140H (closed circles). All samples had a concentration of 10 mm Mg2+. B, shown is urea denaturation of the E. tarda PhoQ T167P mutant sensor domain monitored by CD at 210 nm at 20 °C (open circles), 30 °C (open squares), and 37 °C (open triangles) in the presence of 10 mm Mg2+. C, shown is the amount of ECP obtained at the incubation temperatures of 20, 35, and 37 °C from wild type E. tarda, the E. tarda phoQi mutant, and the E. tarda phoQi mutant complemented with various phoQ gene mutations. The Tm and ΔHm values obtained from the curve-fitting of the thermal denaturation data from the wild type and mutant PhoQs are also listed. D and E, the β-galactosidase activity of the reporter gene esrB-LacZ was determined in N-minimal medium under different incubation temperatures in the presence of 1 mm (striped bars) or 10 mm (empty bars) Mg2+ for E. tarda phoQi + phoQ T167P or phoQi + phoQ P77L.
Based on our hypothesis that the temperature transition responses were responsible for temperature sensing by PhoQs, the mutants P120N and T167P were predicted to lose their ability to sense temperature and affect ECP secretion through T3SS and T6SS. On the other hand, mutants T76E and P140H were predicted to exhibit little effect, whereas P77L and P79E were predicted to exhibit an intermediate effect on the temperature-regulated secretion by E. tarda T3SS and T6SS. To verify these predictions, the E. tarda strain carrying the phoQi insertion mutation was complemented with a full-length phoQ gene carrying a single site mutation at various Pro and Thr residues. Complementation with the thermally stable mutants P79E, P120N, and T167P could not recover the effect of phoQi (Fig. 5C), indicating that they are “loss-of-function” mutations in which the PhoQ mutant cannot activate PhoP and promote the secretion of ECPs from E. tarda T3SS and T6SS at any of the temperatures tested. In contrast, complementation with PhoQ mutants that have thermal stabilities similar to that of the wild type protein (e.g. T76E and P140H) could completely recover the effect of phoQi. Interestingly, complementation with the mutant P77L rendered the E. tarda phoQi strain “temperature-blind” and resulted in the constitutive secretion of ECPs at 20 °C. The PhoQ P77L mutant was, however, still functional in its ability to suppress ECP secretion at 37 °C. The PhoQ P77L mutant had a Tm value (45.3 °C) close to that of the loss-of-function P79E mutant but with a much lower ΔHm value (6.5 × 104 kcal/mol), suggesting that these two mutants could respond differently to changes in temperature. For E. tarda phoQi mutants complemented with phoQ T167P or phoQ P77L, further esrB-LacZ assays were performed at different environmental temperatures and Mg2+ concentrations (Fig. 5, D and E). Similar trends were observed for the esrB-LacZ and the ECPs assay, and both PhoQ T167P and P77L mutants were still able to sense Mg2+ concentration, although their temperature sensing properties were changed.
To confirm that it was the difference in the PhoQ sensor domain that caused this temperature sensing activity, the amount of ECPs from E. tarda phoQi mutant complemented with the phoQ gene from EPEC 2348/69 was determined (supplemental Fig. S3A). As a human pathogen, EPEC 2348/69 secreted translocators such as EspA, EspB, and EspD from the T3SS at 37 °C but not at 30 °C (Ref. 32 and supplemental Fig. S3B). The E. tarda phoQi mutant, when complemented with phoQ from EPEC 2348/69, secreted similar levels of ECPs at 37 °C (2.7 ± 0.07 μg/ml) as compared with the wild type E. tarda at 30 °C. At 30 °C, a much reduced amount of ECPs was secreted (1.4 ± 0.06 μg/ml) by this mutant (supplemental Fig. S3C).
The PhoQ Sensor Domain Binds Mg2+ through a Patch of Acidic Residues
As our results showed that Mg2+ and temperature had additive effects on the regulation of E. tarda T3SS and T6SS, we attempted to characterize the binding of Mg2+ to PhoQs. The binding of Mg2+ to PhoQs had no effect on the far-UV CD spectra of the protein (supplemental Fig. S2), similar to the observation seen for the PhoQ sensor domain of E. coli (30). Therefore, the intrinsic fluorescence (excitation at 280 nm and emission at 350 nm) of the E. tarda PhoQs was used instead to study the direct binding of Mg2+ to the protein (Fig. 6A). This assay showed that Mg2+ can bind directly to PhoQs with a Kd value of ∼95 μm. Temperature affected the secondary structure of PhoQs but had little effect on the affinity of Mg2+ binding by PhoQs from 20 to 37 °C (Fig. 6A).
FIGURE 6.
The cluster of acidic residues in PhoQ is responsible for sensing Mg2+ concentration and antimicrobial peptide but not acidic pH. A, binding of Mg2+ to the PhoQ sensor domain was monitored by the absolute change in fluorescence at 350 nm at 20 °C (filled rhombuses), 30 °C (closed squares), and 37 °C (closed circles). Mg2+ binding by the acidic cluster mutant, PhoQ NNNSANA, was monitored at 30 °C (filled triangles). B, Western blot analysis shows the secretion of EseB from T3SS (upper panel) and EvpC from T6SS (lower panel) by the E. tarda phoQi mutant strain complemented with either phoQ NNNSANA or wild type phoQ in the absence (L) or presence (H) of 10 mm Mg2+ at 30 °C. C, the activity of the reporter gene esrB-LacZ at different temperatures in the presence of 1 mm (striped bars) or 10 mm (empty bars) Mg2+ using the E. tarda phoQi mutant complemented with phoQ NNNSANA is shown. D, Western blot analysis shows the secretion of proteins from T3SS (EseB) and T6SS (EvpC) by E. tarda phoQi mutant complemented with either phoQ NNNSANA or wild type phoQ grown in minimal medium at pH 5.5 (100 mm MES) or pH 7.5 (100 mm Tris-HCl) and in the absence or presence of 5 μg/ml antimicrobial peptide KR-20 (AMP). E, the activity of the reporter gene esrB-LacZ using E. tarda phoQi mutant complemented with either phoQ NNNSANA (striped bars) or wild type phoQ (empty bars) grown at pH 5.5 or 7.5 and in the absence or presence of 5 μg/ml antimicrobial peptide KR-20 (AMP).
A previous study showed that divalent cations may bind to a cluster of acidic residues (EDDDDAE) and stabilize PhoQ in an inactive conformation to prevent PhoP-mediated transcription in response to divalent cation starvation in vivo (30). A similar cluster of acidic residues with a slightly different sequence (DDDSADA) was also observed in the E. tarda PhoQs (Fig. 1). Replacement of these acidic residues with conservative uncharged residues (NNNSANA) completely abolished Mg2+ binding by the E. tarda PhoQs (Fig. 6A), indicating that this acidic cluster is the putative Mg2+ binding site. The acidic cluster mutant has a slightly higher stability than the wild type PhoQs, as monitored by CD at 218 nm (Tm = 47.4 °C), which is in agreement with a similar observation for the E. coli PhoQ sensor domain. Unlike the wild type E. tarda PhoQs, the addition of 10 mm Mg2+ did not provide additional stabilization to the acidic cluster mutant (supplemental Fig. S4).
To confirm that the acidic cluster residues of PhoQ are responsible for in vivo Mg2+ sensing in E. tarda, the phoQi mutant was complemented with the phoQ gene carrying NNNSANA mutations, and the amounts of EseB (T3SS) and EvpC (T6SS) that were secreted as well as expression level of esrB-LacZ in the absence or presence of 10 mm Mg2+ were determined. In the presence of 10 mm Mg2+, the phoQi mutant complemented with the wild type phoQ showed a reduction in the secretion levels of both EseB and EvpC. In contrast, the phoQi mutant complemented with phoQ carrying NNNSANA mutations showed similar secretion levels of both EseB and EvpC (Fig. 6B) and expression levels of esrB-LacZ (Fig. 6C) in the absence or presence of 10 mm Mg2+, suggesting that this cluster of acidic residues in PhoQ is essential for Mg2+ binding and concentration sensing for E. tarda.
In addition to sensing Mg2+ concentration, the PhoQ sensor of S. typhimurium is also activated by acidic pH (33) and antimicrobial peptides (10). To confirm if E. tarda can also sense acidic pH and antimicrobial peptides, the amounts of ECPs as well as expression level of ersB-LacZ in acidic culture medium, pH 5.5, and in the presence of an antimicrobial peptide (KR-20 or C-terminal 20 residues of cathelicidin LL-37) (34) were determined. In acidic pH or the presence of antimicrobial peptide, E. tarda showed an increase in the secretion of ECPs from both T3SS and T6SS (Fig. 6D) as well as an increase in the expression level of esrB-LacZ (Fig. 6E). The effect of antimicrobial peptide is more prominent than that of acidic pH, and both effects are additive, suggesting that E. tarda can sense both acidic pH and antimicrobial peptides in addition to temperature and Mg2+ concentration. As residues from the acidic cluster of Salmonella PhoQ were previously shown to be involved in recognizing antimicrobial peptide (10), the effects of antimicrobial peptide and acidic pH on the secretion of ECPs as well as expression levels of esrB-LacZ were determined using E. tarda phoQi mutant complemented with phoQ NNNSANA. Mutation of the acidic cluster residues activated secretion of ECPs and expression of esrB-LacZ, but the addition of antimicrobial peptide had no further effect (Fig. 6, D and E). In contrast, the secretion of ECPs and expression of esrB-LacZ in this E. tarda mutant are further activated by acidic pH (Fig. 6, D and E), suggesting that the cluster of acidic residues in PhoQ is responsible for sensing antimicrobial peptide but not acidic pH.
DISCUSSION
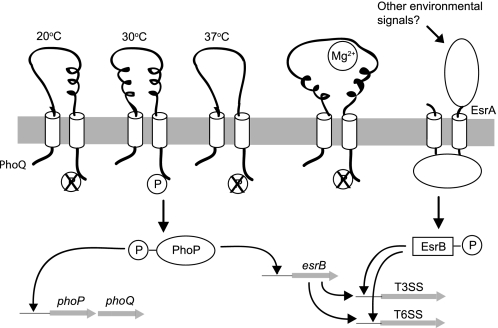
The EsrB regulator is encoded within the T3SS gene cluster and belongs to the response regulator of another two-component system called EsrA-EsrB. A functional EsrA-EsrB system is required to regulate the expression of EsrC, an AraC family transcriptional regulator that controls the expression of proteins encoded by T3SS and T6SS in E. tarda (7). Our EMSA results showed that PhoP binds to the promoter regions of both esrB and phoP, suggesting that it may be involved in regulating the expression levels of both itself and EsrB. Transcriptional autoregulation was also observed in the phoPQ operon of S. typhimurium (35). The data herein demonstrated that the PhoP-PhoQ system senses temperature and Mg2+ concentration. In turn, the response regulator PhoP communicates the status of these environmental conditions to the EsrA-EsrB two-component system (Fig. 7). This kind of “dual regulation” has also been observed in S. typhimurium, in which the SsrA-SsrB two-component system is regulated together with the two-component system OmpR-EnvZ, which responds to osmolarity through the direct binding of OmpR to the promoter region of ssrA (36, 37). Transcriptional regulation by a cascade of two-component systems allows pathogenic bacteria to express their virulence determinants in response to a broad spectrum of environmental cues. The PhoQ sensor in E. tarda can sense both temperature and Mg2+ concentration in an additive manner. We propose that it may integrate these signals with the signal detected by the EsrA-EsrB system to regulate the expression of proteins from T3SS and T6SS.
FIGURE 7.
Model illustrating the temperature and Mg2+ regulation of T3SS and T6SS by the PhoP-PhoQ system. The PhoQ sensor senses changes in temperature through conformational changes in its thermally unstable secondary structures (coils in the diagram). There should be less secondary structure at 37 °C as compared with 30 °C, and the conformational change at 20 °C should be different from that at 37 °C to distinguish the different temperatures. The overall tertiary structure, or shape, of the PhoQ sensor remains unchanged at different temperatures. In addition to temperature, the PhoQ sensor can also detect changes in Mg2+ concentration through direct binding of Mg2+ to a cluster of acidic residues that likely change the tertiary structure of the protein. The signals from environmental temperature and Mg2+ concentration are additive with each other. Activation of the PhoQ histidine kinase over the temperature range from 23 to 35 °C at low Mg2+ concentrations leads to autophosphorylation and the transfer of a phosphate group from PhoQ to PhoP. The phosphorylated PhoP binds directly to the PhoP box within the promoter region of esrB to activate its transcription. PhoP also self-regulates by binding to another PhoP box within its own promoter region to up-regulate the expression of both PhoP and PhoQ. The EsrB protein then integrates the signal from another two-component system, EsrA-EsrB, to activate the transcription of genes from both T3SS and T6SS.
There was a significant decrease in the secondary structure (raw ellipticity from the CD spectra) of PhoQs as the temperature increased (Fig. 4A), suggesting that temperature shift-induced conformational change occurs mainly on the level of the secondary structure rather than of the tertiary structure. PhoQs likely has less secondary structure at 37 °C than at 30 °C, but it still maintains its tertiary structure throughout this range of temperatures. The data also indicated that the conformations of PhoQs at 20 and 37 °C could be different even though both would have the ability to maintain PhoQs in its inactive state. A sequence comparison with homologues from S. typhimurium and E. coli revealed that residues at positions 76, 77, 79, 120, 140, and 167 of PhoQs are replaced with either Pro or Thr in E. tarda. The branched side chain of a Thr residue has been shown to generate instability at turn regions. Mutation of a Thr residue (Thr-22) in the diverging β-turn of the drkN SH3 domain to a Gly residue results in a dramatic stabilization of the protein, with its Tm increased by 20 °C (38). In contrast, a Pro residue is more rigid than other naturally occurring amino acids and could stabilize or destabilize a protein depending on its location (39). The use of a phoQi mutant complemented with the wild type or mutant phoQ genes allowed those residues that are essential for thermal stability and temperature sensing to be delineated. Mutation of Pro-79, Pro-120, or Thr-167 to their corresponding residues in the E. coli or S. typhimurium PhoQ sequence significantly stabilized the PhoQs in E. tarda. The T167P mutant (Tm = 59.0 °C) of E. tarda PhoQs was thermally stable, and its stability toward urea did not change at temperatures between 20 and 37 °C. These loss-of-function mutations improved the thermal stability of PhoQs but rendered it unable to make the conformational changes necessary for the activation of PhoP. Consequently, no ECPs were detected at any of the tested temperatures, which was similar to results seen in the phoQi mutant without any complementation. In contrast, the P77L mutant became temperature-blind at 20 °C. This mutation prevented the secondary structure conformational change necessary to inhibit PhoQ activity at 20 °C but not at 37 °C. So far, no mutation that rendered the PhoQ sensor temperature-blind at 37 °C or at both 20 and 37 °C has been identified. The observation that the P77L mutation only had an effect on temperature-sensing at 20 °C, but not at 37 °C, also supported the notion that the conformations of the PhoQ sensor are different at these two temperatures.
In this study, high Mg2+ concentration (10 mm) reduced the transcription of phoP, phoQ, and esrB and, thus, reduced protein secretion from T3SS and T6SS in E. tarda. Mg2+ binding likely changed the tertiary structure, but not the secondary structure, of the PhoQ sensor. This explains the observation that Mg2+ binding by the PhoQ sensor was not affected by temperature at 20, 30, or 37 °C, which only resulted in changing the secondary structure of the protein as well as the additive nature of the effects of Mg2+ and temperature on the PhoQ sensor, as the conformational changes caused by these factors could occur simultaneously and independently of each other. In S. typhimurium, Mg2+ is proposed to bind to the acidic patch EDDDDAE in the PhoQ sensor and form a metal bridge with negatively charged groups of the inner membrane (21). A similar but distinct acidic patch with the sequence DDDSADA is found within the PhoQ sensor domain of E. tarda. Our data confirmed that this cluster of acidic residues likely represents the only Mg2+ binding site on PhoQ because mutation of these residues to NNNSANA completely abolished Mg2+ binding by the PhoQ sensor and because the phoQi mutant complemented with this NNNSANA mutant is rendered Mg2+-blind. Mutations of this cluster of acidic residues did not affect the ability of PhoQ to activate PhoP and protein secretion from T3SS and T6SS or the ability of PhoQ to sense temperatures. Our study showed that Mg2+ bound to the PhoQ sensor domain of E. tarda at a relatively high affinity, with a Kd value of ∼95 μm, comparable with the value obtained for the PhoQ sensor domain of Pseudomonas aeruginosa (Kd = 37 μm for Ca2+; Kd = 207 μm for Mg2+) (40) but distinct from that of the PhoQ of S. typhimurium (Kd = 250 μm for Ca2+; Kd = 7 μm for Mg2+) (41). Interestingly, the P. aeruginosa PhoQ lacks this acidic patch of residues, and unlike the E. coli protein, the P. aeruginosa PhoQ sensor domain undergoes changes in its CD and fluorescence spectra in response to divalent cations (42). These data suggested that the PhoQ sensors of E. tarda, S. typhimurium, and P. aeruginosa may have very different mechanisms of signal detection. The high affinity binding of Mg2+ also increased the stability of the PhoQ sensor slightly, as monitored by both CD and fluorescence spectra. This increased stability is likely due to the neutralization of electrostatic repulsion among residues within the acidic patch. Unlike the situation with the T167P mutation, this slight increase in stability due to Mg2+ binding is not sufficient to disrupt temperature detection by PhoQ.
E. tarda infects many different fish species, such as blue gourami fish and channel catfish. The body temperature of catfish fluctuates with and approximates the surrounding water temperature. Their active metabolic rate increases as a hyperbolic function of temperature, reaching a peak at around 28–30 °C. Reduced feeding by the fish should occur at the higher temperature of 35 °C, with none at 36–38 °C. Alternatively, there should also be reduced feeding by the fish at the lower temperature of 15 °C and no feeding at 8–10 °C (43). This growth temperature profile of fish is in agreement with our findings on the temperature dependence of protein secretion from the E. tarda T3SS and T6SS, suggesting that the PhoP-PhoQ system of E. tarda is only activated at the optimum growth temperature of the host to ensure the highest level of virulence and the survival of the bacteria. These findings also agree with the observation that outbreaks of acute E. tarda infection are mostly found in channel catfish culture systems when the temperature rises due to overcrowding (44). On the other hand, Mg2+ concentration detection by the PhoP-PhoQ system seems to provide E. tarda with cues that it located inside the host body. At a physiological level of around 1–2 mm, the Mg2+ concentration inside the host body is generally lower than that of the external environment (10). Direct measurement of Mg2+ within the Salmonella-containing vacuole using nanosensor particles showed that during the initial period of phoP activation, the concentration of the divalent cation was rapidly regulated and stabilized at ∼1 mm (45). In addition to temperature and Mg2+ concentration, E. tarda can also sense both acidic pH and the presence of antimicrobial peptides, which are characteristics of environment inside the phagosome of macrophage.
In conclusion, we cloned the PhoP-PhoQ two-component system of E. tarda and confirmed that the PhoQ sensor domain senses changes in temperature through conformational changes in its thermally unstable secondary structures. The PhoQ sensor can also simultaneously detect changes in Mg2+ concentrations through the direct binding of Mg2+ to a cluster of acidic residues, which likely results in a change in the tertiary structure of the protein. Both the temperature and Mg2+ signals are integrated with the signal detected by another two-component system, EsrA-EsrB, through the direct activation of the transcription of the EsrB response regulator by PhoP. The data herein provides a basic understanding of the mechanism underlying the temperature and Mg2+-sensing abilities in bacteria and could aid future structural studies of the system as well as control of the pathogen.
Supplementary Material
This work was supported by Biomedical Research Council of the Agency for Science Technology and Research (A*STAR) of Singapore Grant 07/1/21/19/495 (to Y.-K. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S4.
- ECP
- extracellular protein
- bp
- base pairs.
REFERENCES
- 1.Janda J. M., Abbott S. L. (1993) Clin. Infect. Dis. 17, 742–748 [DOI] [PubMed] [Google Scholar]
- 2.Thune R. L., Stanley L. A., Cooper R. K. (1993) Annu. Rev. Fish Dis. 3, 37–68 [Google Scholar]
- 3.Srinivasa Rao P. S., Lim T. M., Leung K. Y. (2003) Infect. Immun. 71, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivasa Rao P. S., Yamada Y., Tan Y. P., Leung K. Y. (2004) Mol. Microbiol. 53, 573–586 [DOI] [PubMed] [Google Scholar]
- 5.Tan Y. P., Zheng J., Tung S. L., Rosenshine I., Leung K. Y. (2005) Microbiology 151, 2301–2313 [DOI] [PubMed] [Google Scholar]
- 6.Zheng J., Leung K. Y. (2007) Mol. Microbiol. 66, 1192–1206 [DOI] [PubMed] [Google Scholar]
- 7.Zheng J., Tung S. L., Leung K. Y. (2005) Infect. Immun. 73, 4127–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamnongpol S., Groisman E. A. (2002) Mol. Microbiol. 44, 561–571 [DOI] [PubMed] [Google Scholar]
- 9.Prost L. R., Miller S. I. (2008) Cell. Microbiol. 10, 576–582 [DOI] [PubMed] [Google Scholar]
- 10.Bader M. W., Sanowar S., Daley M. E., Schneider A. R., Cho U., Xu W., Klevit R. E., Le Moual H., Miller S. I. (2005) Cell 122, 461–472 [DOI] [PubMed] [Google Scholar]
- 11.Abe A., Kenny B., Stein M., Finlay B. B. (1997) Infect. Immun. 65, 3547–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cybulski L. E., Albanesi D., Mansilla M. C., Altabe S., Aguilar P. S., de Mendoza D. (2002) Mol. Microbiol. 45, 1379–1388 [DOI] [PubMed] [Google Scholar]
- 13.Hunger K., Beckering C. L., Marahiel M. A. (2004) FEMS Microbiol. Lett. 230, 41–46 [DOI] [PubMed] [Google Scholar]
- 14.Lodge J., Fear J., Busby S., Gunasekaran P., Kamini N. R. (1992) FEMS Microbiol. Lett. 95, 271–276 [DOI] [PubMed] [Google Scholar]
- 15.Nelson D. L., Kennedy E. P. (1971) J. Biol. Chem. 246, 3042–3049 [PubMed] [Google Scholar]
- 16.Miller S. (1972) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 17.Edwards R. A., Keller L. H., Schifferli D. M. (1998) Gene 207, 149–157 [DOI] [PubMed] [Google Scholar]
- 18.Deng W., Burland V., Plunkett G., 3rd, Boutin A., Mayhew G. F., Liss P., Perna N. T., Rose D. J., Mau B., Zhou S., Schwartz D. C., Fetherston J. D., Lindler L. E., Brubaker R. R., Plano G. V., Straley S. C., McDonough K. A., Nilles M. L., Matson J. S., Blattner F. R., Perry R. D. (2002) J. Bacteriol. 184, 4601–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Hou S., Layman D., Leonard S., Nguyen C., Scott K., Holmes A., Grewal N., Mulvaney E., Ryan E., Sun H., Florea L., Miller W., Stoneking T., Nhan M., Waterston R., Wilson R. K. (2001) Nature 413, 852–856 [DOI] [PubMed] [Google Scholar]
- 20.Welch R. A., Burland V., Plunkett G., 3rd, Redford P., Roesch P., Rasko D., Buckles E. L., Liou S. R., Boutin A., Hackett J., Stroud D., Mayhew G. F., Rose D. J., Zhou S., Schwartz D. C., Perna N. T., Mobley H. L., Donnenberg M. S., Blattner F. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho U. S., Bader M. W., Amaya M. F., Daley M. E., Klevit R. E., Miller S. I., Xu W. (2006) J. Mol. Biol. 356, 1193–1206 [DOI] [PubMed] [Google Scholar]
- 22.Cheung J., Bingman C. A., Reyngold M., Hendrickson W. A., Waldburger C. D. (2008) J. Biol. Chem. 283, 13762–13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor B. L., Zhulin I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryson K., McGuffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. (2005) Nucleic Acids Res. 33, W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones D. T. (1999) J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 26.Kato A., Tanabe H., Utsumi R. (1999) J. Bacteriol. 181, 5516–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minagawa S., Ogasawara H., Kato A., Yamamoto K., Eguchi Y., Oshima T., Mori H., Ishihama A., Utsumi R. (2003) J. Bacteriol. 185, 3696–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguirre A., Cabeza M. L., Spinelli S. V., McClelland M., García Véscovi E., Soncini F. C. (2006) J. Bacteriol. 188, 6889–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz-Sanz J., de Prat Gay G., Otzen D. E., Fersht A. R. (1995) Biochemistry 34, 1695–1701 [DOI] [PubMed] [Google Scholar]
- 30.Waldburger C. D., Sauer R. T. (1996) J. Biol. Chem. 271, 26630–26636 [DOI] [PubMed] [Google Scholar]
- 31.Mok Y. K., de Prat Gay G., Butler P. J., Bycroft M. (1996) Protein Sci. 5, 310–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M., Rosenshine I., Tung S. L., Wang X. H., Friedberg D., Hew C. L., Leung K. Y. (2004) Appl. Environ. Microbiol. 70, 5274–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prost L. R., Daley M. E., Le Sage V., Bader M. W., Le Moual H., Klevit R. E., Miller S. I. (2007) Mol. Cell 26, 165–174 [DOI] [PubMed] [Google Scholar]
- 34.Murakami M., Lopez-Garcia B., Braff M., Dorschner R. A., Gallo R. L. (2004) J. Immunol. 172, 3070–3077 [DOI] [PubMed] [Google Scholar]
- 35.Soncini F. C., Véscovi E. G., Groisman E. A. (1995) J. Bacteriol. 177, 4364–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng X., Oropeza R., Kenney L. J. (2003) Mol. Microbiol. 48, 1131–1143 [DOI] [PubMed] [Google Scholar]
- 37.Lee A. K., Detweiler C. S., Falkow S. (2000) J. Bacteriol. 182, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mok Y. K., Elisseeva E. L., Davidson A. R., Forman-Kay J. D. (2001) J. Mol. Biol. 307, 913–928 [DOI] [PubMed] [Google Scholar]
- 39.Prajapati R. S., Das M., Sreeramulu S., Sirajuddin M., Srinivasan S., Krishnamurthy V., Ranjani R., Ramakrishnan C., Varadarajan R. (2007) Proteins 66, 480–491 [DOI] [PubMed] [Google Scholar]
- 40.Prost L. R., Daley M. E., Bader M. W., Klevit R. E., Miller S. I. (2008) Mol. Microbiol. 69, 503–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Véscovi E. G., Ayala Y. M., Di Cera E., Groisman E. A. (1997) J. Biol. Chem. 272, 1440–1443 [DOI] [PubMed] [Google Scholar]
- 42.Lesley J. A., Waldburger C. D. (2001) J. Biol. Chem. 276, 30827–30833 [DOI] [PubMed] [Google Scholar]
- 43.Hargreaves J. A., Tomasso J. R., Jr. (2004) in Biology and Culture of Channel Catfish (Tucker C. S., Hargreaves J. A. eds) pp. 42–43, Elsevier Science Publishers B. V., Amsterdam [Google Scholar]
- 44.Mohanty B. R., Sahoo P. K. (2007) J. Biosci. 32, 1331–1344 [DOI] [PubMed] [Google Scholar]
- 45.Martin-Orozco N., Touret N., Zaharik M. L., Park E., Kopelman R., Miller S., Finlay B. B., Gros P., Grinstein S. (2006) Mol. Biol. Cell 17, 498–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.