Abstract
Histone deacetylase 9 (HDAC9), like most Class II HDACs, catalyzes the removal of acetyl moieties from the ϵ-amino groups of conserved lysine residues in the N-terminal tail of histones. Biologically, HDAC9 regulates a wide variety of normal and abnormal physiological functions, including cardiac growth, T-regulatory cell function, neuronal disorders, muscle differentiation, development, and cancer. In a biochemical approach to identify non-histone substrates of HDAC9, we found that HDAC9 co-purifies specifically with the ataxia telangiectasia group D-complementing (ATDC; also called TRIM29) protein. HDAC9 deacetylates ATDC, alters the ability of ATDC to associate with p53, and consequently inhibits the cell proliferation-promoting activity of ATDC. These results implicate the importance of non-histone deacetylation by HDAC9 and confirm and further extend the multifunctions of this Class II deacetylase.
Keywords: Gene Expression, Gene Regulation, Histone Deacetylase, Protein-Protein Interactions, Transcription
Introduction
In humans, histone deacetylases (HDACs)2 are divided into three categories (reviewed in Refs. 1 and 2): the Class I RPD3-like proteins (HDAC1, HDAC2, HDAC3, and HDAC8); the Class II HDA1-like proteins (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10); and the Class III SIR2-like proteins (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7). The Class II enzymes can be further divided into two subclasses: IIa (HDAC4, HDAC5, HDAC7, and HDAC9) and IIb (HDAC6 and HDAC10). HDAC11 uniquely shares sequence homology to the catalytic domains of both Class I and II HDAC enzymes.
The myocyte enhancer-binding factor 2-interacting transcriptional repressor (MITR), which possesses amino acid sequence similarity to Class II HDACs but lacks an HDAC catalytic domain, was first discovered as a binding partner and a negative regulator of MEF-2 (myocyte enhancer-binding factor 2) in a yeast two-hybrid screen (3). MITR binds to the N-terminal MADS region of MEF-2 and recruits HDAC1 to repress transcription (3). In addition to HDAC1, MITR represses MEF-2 activity through recruitment of multicomponent co-repressor complexes that include CtBP and HP1 (4, 5). MITR can inhibit skeletal muscle myogenesis, and the ability of MITR to block muscle differentiation depends on its phosphorylation status (6). MITR was independently discovered as histone deacetylase-related protein (HDRP) through database search and sequence analyses (7). It was found to share 50% identity in amino acid sequence to the noncatalytic N terminus of HDAC4 and HDAC5. HDRP forms a complex with HDAC1 and HDAC3 and represses transcription independent of deacetylase activity.
HDAC9 was first described from a result of a database search for proteins with similarity to the deacetylase domain of other Class II HDACs (8). The human HDAC9 gene is located on chromosome 7p21 (9), a region that has been implicated in neurological disorders and a variety of cancers, including colorectal cancer, fibrosarcoma, childhood acute lymphoblastic leukemia, Wilms tumor, peripheral nerve sheath tumors, and gynecological tumors. The HDAC9 gene encodes multiple protein isoforms due to alternative splicing, and one of these isoforms, an N-terminal splice variant, is MITR/HDRP (also called HDAC9ΔCD) (8, 10). Some HDAC9 isoforms display distinct cellular localization patterns, suggesting potential different biological functions. For simplicity, we will refer to the first reported full-length HDAC9 isoform as HDAC9 (8). This isoform contains 1011 amino acids and has a predicted molecular mass of 111.3 kDa and an isoelectric point of 6.41. Like all Class I and II HDACs, HDAC9 possesses a conserved deacetylase domain. Also, similar to most HDACs, HDAC9 represses gene transcription activity when recruited to a promoter and deacetylates histones H3 and H4 in vitro and in vivo (10).
HDAC9 is highly expressed in cardiac muscle but does not affect normal heart growth. In a series of studies, Olson and colleagues (6, 11) showed that activation of the cardiac myocyte fetal gene program by a range of potent hypertrophic inducers could be blocked by expressing mutated HDAC9. Furthermore, mutant mice lacking HDAC9 are sensitized to hypertrophic signals and exhibit stress-dependent cardiomegaly, suggesting that HDAC9 is a negative regulator of cardiomyocyte hypertrophy.
Another important function of HDAC9 is to control the fate of regulatory T cells (Treg cells) (12, 13). HDAC9 is expressed in higher levels in Treg than non- Treg cells. Foxp3 associates with HDAC9 (14). Treatment of Treg cells with an HDAC inhibitor increases Foxp3 gene expression and causes hyperacetylation of the forkhead domain of Foxp3. Acetylation of Foxp3 enhances Foxp3 binding to the IL2 promoter and consequently represses IL-2 production. CD4+ Foxp3+ T cells in lymphoid tissues of Hdac9−/− mice increase by about 50% compared with wild type mice. Also, Treg cells derived from Hdac9−/− mice show enhanced expression of Foxp3, CTLA4, and GITR.
In addition to its roles in heart development and in T cell functions, HDAC9 has a role in limb patterning by suppression of the sonic hedgehog signaling pathway (15). HDAC9/HDRP knock-out mice exhibit post-axial polydactyly. Other functions of HDAC9 include mediating the effect of neuron-elicited electrical activity (16), regulating activity-dependent gene expression and dendritic growth in developing cortical neurons (17), regulating the expression of fatty acid synthase enzyme through deacetylation of the transcription factor USF-1 (18), and possibly regulating Ras-mediated epigenetic silencing of Fas (19).
As a step toward understanding how HDAC9 regulates many seemingly unrelated cellular functions, we have undertaken the identification of proteins that interact with HDAC9. Using a biochemical approach, we found that one of the proteins that associates with HDAC9 is ATDC (ataxia telangiectasia group D-complementing, also known as TRIM29). ATDC is a member of the tripartite motif (TRIM) protein family (also known as the RBCC family), which is characterized by three zinc-binding domains, a RING, a B-box type 1, and a B-box type 2, followed by a coiled-coil region (20, 21). Some TRIM proteins homomultimerize through their coiled-coil region, and the integrity of the TRIM is required for proper subcellular localization of TRIM proteins (22). ATDC (TRIM29) possesses a B-box type 1, a B-box type 2, and a coiled-coil domain but not a RING finger. Previously, Wang et al. (23) showed that ATDC possesses oncogenic functions in pancreatic cancer through Wnt pathway activation and β-catenin stabilization. More recently, we have shown that ATDC binds p53, and this interaction is potentially fine tuned by posttranslational acetylation of lysine 116 on ATDC (24). ATDC inhibits p53 nuclear activities; represses expression of p53-regulated genes, including p21 and NOXA; and increases cell proliferation. Here, we report that ATDC is a novel HDAC9 substrate. Deacetylation of ATDC by HDAC9 changes the ability of ATDC to bind p53 and consequently affects expression of p53-regulated genes and reverses the cell proliferation-promoting activity of ATDC.
EXPERIMENTAL PROCEDURES
Plasmids, siRNA, shRNA, Antibodies, and Viruses
Plasmids that express the following proteins have been described previously: FLAG-HDAC9 (8), HA-p300 (25), HA-ATDC (24), HA-ATDC (K116R) (24), FLAG-HDAC7 (26), FLAG-p53 (27), and FLAG-HDAC1 (28). The luciferase reporter plasmid BP-100-GL2 has also been described (27). pGL3-control was purchased from Promega. pEGFP-C3, a GFP expression plasmid, was purchased from Clontech. pAdTrack-CMV-FLAG-HDAC9-HA was subcloned from pFLAG-HDAC9 using standard recombinant DNA and PCR techniques. Expression plasmids encoding FLAG-tagged deletions of HDAC9 (1–350, 351–600, and 601–1011) were derived from pFLAG-HDAC9 using PCR and standard subcloning. HDAC9 shRNA and control shRNA were purchased from Open Biosystems. ATDC siRNA and control siRNA have been described previously (24).
Mouse affinity-purified monoclonal anti-FLAG M2, rabbit affinity-purified polyclonal anti-HA, and mouse monoclonal anti-β-actin antibodies were purchased from Sigma. Goat polyclonal anti-ATDC, goat polyclonal anti-p53, and mouse monoclonal anti-p53 (clone DO-1) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Rabbit polyclonal anti-acetyl-lysine antibody was purchased from Upstate (Millipore). Rabbit polyclonal anti-acetylated Lys382-p53 was purchased from Cell Signaling Technology. Mouse monoclonal anti-p21 antibody was purchased from Neomarkers (Thermo Scientific). A rabbit polyclonal antibody that uniquely recognizes acetylated Lys116 of ATDC (anti-Ac-Lys116-ATDC) was generated using a mixture of lysine-acetylated peptides corresponding to the acetylated Lys116 region of ATDC. Serum samples were first tested by ELISA against non-acetylated and acetylated peptides to determine the extent of antibody cross-reactivity. Two animals with specific anti-acetyl-Lys116-ATDC antibody response were identified, and they were purified over Protein A-agarose followed by non-acetylated and acetylated peptide-Sepharose and analyzed by protein dot blots and by Western blots.
Cell Culture and Transfection
Mouse embryonic fibroblasts were generated from Hdac9 nullizygote (−/−) and wild type (+/+) mouse embryos using standard methods. 293T, HeLa, SiHa, U2OS, and mouse embryonic fibroblast cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and penicillin/streptomycin. The AT5BIVA cell line (GM05849) was obtained from the Coriell Cell Repository and grown in minimum essential medium with 10% FCS and penicillin/streptomycin. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, and all transfections were normalized with equal amounts of parental vector DNA.
Purification and Analysis of HDAC9-containing Complexes
Recombinant adenoviruses that express double-tagged FLAG- and HA-HDAC9 were generated using the AdEasy system (29). HeLa cells were then infected with adenovirus that expresses either FLAG-HDAC9-HA or the GFP protein (control). Affinity purification of HDAC9-containing complex was performed according to our previously published method (30). Purified complexes were concentrated, resolved by SDS-PAGE, and analyzed by silver staining. A colloidal blue-stained sample was prepared in parallel, and bands corresponding to HDAC9-associated proteins were excised and subjected to proteolytic digestion. The protein sequence analysis was performed at the Harvard Microchemistry Facility by microcapillary reverse-phase HPLC nanoelectrospray tandem spectrometry (μLC/MS/MS) on a Finnigan LCQ DECA XP Plus quadrupole ion trap mass spectrometer.
Immunoprecipitation and Western Blot Analysis
For immunoprecipitations, cells were lysed in buffer (50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 0.5% Nonidet P-40, and a protease inhibitor mixture) containing either 500 mm NaCl (high stringency) or 150 mm NaCl (low stringency). The lysates were incubated with the primary antibody overnight at 4 °C. The resultant immunocomplexes were collected, washed four times in lysis buffer, and resolved by SDS-PAGE. For immunoblotting, samples were transferred onto nitrocellulose membranes. Membranes were probed with the appropriate antibodies, and proteins of interest were visualized using the Chemiluminescent Detection Kit (Pierce).
Colony Survival Assay
Colony survival assays were performed as described previously (31) with minor modifications. Briefly, 24 h after transfection, cells were plated in quadruplicate (1,000 cells/60-mm tissue culture dish). The cells were treated with γ-irradiation, and after 2 weeks, dishes were washed with PBS, fixed in ice-cold methanol for 15 min, and then stained with Giemsa stain for 30 min. Colonies on each plate were quantified and expressed as the percentage of the unirradiated control.
Luciferase Assay
The luciferase assay was performed as described previously (32). Briefly, cells were plated in 6-well dishes at a density of 4 × 105 cells/well. Twenty-four h after transfection, cells were treated as indicated before harvesting with 250 μl of passive lysis buffer (Promega). Protein concentrations of all samples were determined using Bradford reagent (Bio-Rad), and the relative light units were measured with firefly assay reagent (Promega) and a luminometer.
Cell Apoptotic Assay
The apoptotic assay was performed as described previously (33) with minor modifications. Briefly, 4 × 105 cells were co-transfected with 0.5 μg of pFLAG-HDAC9 and 0.05 μg of pEGFP-C3. One day after transfection, apoptosis was induced with 5 Gy of γ-irradiation. Two days after treatment, cells were stained with DAPI, and apoptotic nuclei were counted in GFP-expressing cells under a fluorescence microscope (300 cells were counted for each experiment).
RESULTS
ATDC Interacts with HDAC9
To further understand the functions and mechanisms of action of HDAC9, we sought to identify proteins that associate with HDAC9. To facilitate the purification of the HDAC9 complex, we first infected HeLa cells with adenovirus expressing a FLAG- and HA-tagged HDAC9 protein. FLAG-HDAC9-HA was purified from a whole cell extract by affinity chromatography using anti-FLAG antibody-conjugated agarose. The bound polypeptides were then eluted with the FLAG peptide under native conditions. The FLAG affinity-purified fraction was further purified by immunoaffinity chromatography using an anti-HA antibody. As a control, we performed parallel purifications using extracts from HeLa cells expressing the GFP protein.
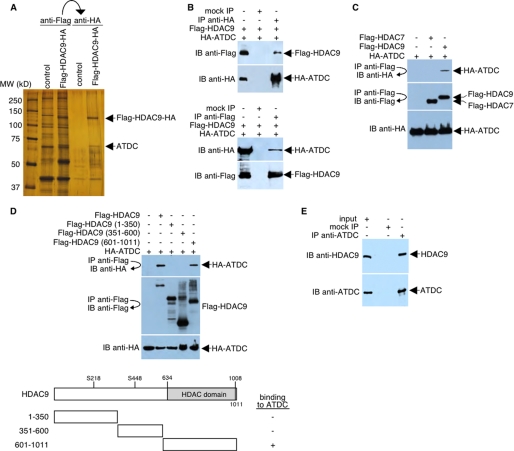
After two rounds of the affinity purification, the gel pattern, based on silver staining coupled with Western blot analyses (using antibodies directed against proteins known to interact with HDAC9 and MITR) indicated a distinct novel HDAC9 complex. Four polypeptides that specifically associate with HDAC9 were resolved by SDS-PAGE (Fig. 1A). Mass spectrometry analysis indicated that the 66-kDa protein that co-purified with HDAC9 is the ATDC protein (also known as TRIM29; NM_012101). We are currently working to confirm the identification and characterization of the remaining three proteins that co-purified with HDAC9.
FIGURE 1.
HDAC9 interacts with ATDC. A, silver-stained SDS-PAGE of the immunopurified FLAG- and HA-tagged HDAC9-containing complexes. Control, immunopurified samples prepared in parallel from HeLa cells expressing the GFP protein. B, HeLa cells were transfected with plasmids encoding FLAG-tagged HDAC9 and HA-tagged ATDC. Whole cell lysates were immunoprecipitated and probed with FLAG-specific or HA-specific antibodies as indicated. C, HeLa cells were transfected with plasmids encoding FLAG-tagged HDAC7 or HDAC9 and HA-tagged ATDC. Whole cell lysates were immunoprecipitated and probed with FLAG-specific or HA-specific antibodies as indicated. D, top, HeLa cells were transfected with plasmids encoding full-length or deletion mutants of FLAG-tagged HDAC9 and HA-tagged ATDC. Western blot analysis was performed on the anti-FLAG immunoprecipitates with antibodies specific for HA. Levels of FLAG-tagged HDAC9 and HA-tagged ATDC were determined by Western blot analysis of cell extracts using antibodies specific for FLAG or HA, respectively. Bottom, a schematic diagram (not drawn to scale) of HDAC9 and three HDAC9 deletion mutants. Ser218 and Ser448 (S218 and S448) are serine phosphorylation sites. For simplicity, the FLAG portions of the fusion proteins are not shown. The ability of each FLAG-HDAC9 fusion protein to bind HA-ATDC is indicated (plus and minus signs). E, HeLa cell lysates were incubated with preimmune serum (mock IP) or the anti-ATDC antibody. Precipitated materials were analyzed by Western blotting using either anti-HDAC9 or anti-ATDC antibodies. IP, immunoprecipitation; IB, immunoblot.
To confirm that HDAC9 associates with ATDC, we prepared a lysate from HeLa cells that expressed FLAG-HDAC9 and HA-ATDC. HA-ATDC was immunoprecipitated with an anti-HA antibody, and the resulting precipitate was analyzed by Western blot with an anti-FLAG antibody. As shown in Fig. 1B (top), FLAG-HDAC9 was co-precipitated with HA-ATDC. In the negative control, HA-ATDC was not precipitated when no primary antibody was used. In a reciprocal experiment, HA-ATDC was co-precipitated with FLAG-HDAC9 using an anti-FLAG antibody followed by Western blot analysis with an anti-HA antibody (Fig. 1B, bottom). To demonstrate specificity for the HDAC9-ATDC interaction, we overexpressed FLAG-tagged HDAC7 (another Class IIa HDAC) in HeLa cells. HDAC7-containing protein complexes were then immunoprecipitated from HeLa extracts using a FLAG-specific antibody. The presence of ATDC in these immunoprecipitates was analyzed by Western blotting. As shown in Fig. 1C, the anti-FLAG antibody specifically co-precipitated HA-ATDC from cells expressing FLAG-tagged HDAC9 but not from cells expressing FLAG-tagged HDAC7. This finding suggests that the ATDC-HDAC9 interaction is highly specific. Analyses of three different HDAC9 deletions indicated that ATDC interacts with the C-terminal (residues 601–1011) of HDAC9 (Fig. 1D).
To determine whether HDAC9 interacts with ATDC under normal physiologic conditions, we examined whether the two proteins could be co-immunoprecipitated from an extract in which neither protein was overexpressed. Our results indicated that a significant fraction of HDAC9 could be co-precipitated with an anti-ATDC antibody but not with preimmune serum (Fig. 1E).
ATDC is an HDAC9 Substrate
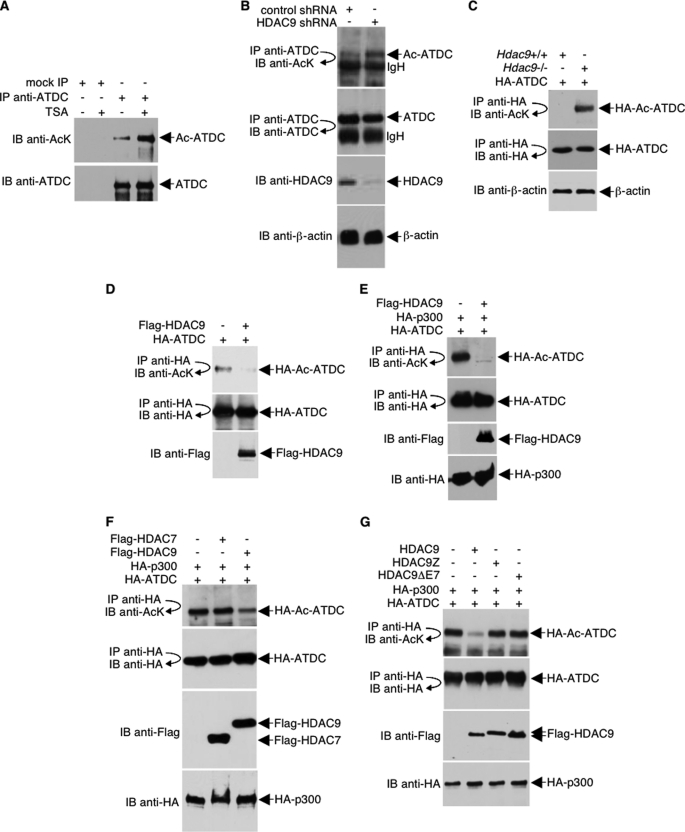
Previously, we found that ATDC is an acetylated protein (24). Our data that ATDC associates with HDAC9 strongly suggests that ATDC may be a deacetylation target of HDAC9. Treatment of cells with the HDAC inhibitor, trichostatin A (TSA), revealed that the acetylation of ATDC is indeed regulated by HDAC(s) (Fig. 2A). To examine the possibility that ATDC can be deacetylated by HDAC9, we examined the effect of shRNA-mediated HDAC9 knockdown on ATDC acetylation. HDAC9 shRNA efficiently silenced expression of HDAC9 but not ATDC or the control protein β-actin, as monitored by Western blot analysis (Fig. 2B). In agreement with our hypothesis, acetylation of ATDC noticeably increased as a result of HDAC9 knockdown. In a similar experiment, we examined the acetylation of overexpressed HA-ATDC in Hdac9−/− mouse embryonic fibroblasts and found that ATDC acetylation dramatically increased in the absence of Hdac9 (Fig. 2C).
FIGURE 2.
ATDC is deacetylated by HDAC9. A, HeLa cells were treated with TSA (400 ng/ml) or left untreated overnight. Cell lysates were then immunoprecipitated under high stringency conditions with an anti-ATDC antibody. Endogenous acetylated ATDC was analyzed by Western blotting with an anti-acetyl-lysine antibody. A Western blot was also performed with anti-ATDC to assess the ATDC immunoprecipitation efficiency. B, 293T cells were transfected with either HDAC9 shRNA or control shRNA. Following immunoprecipitation with anti-ATDC antibodies, Western blots were performed with the indicated antibodies to assess the acetylation of endogenous ATDC and ATDC immunoprecipitation efficiency. Western blots were also performed with the indicated antibodies to assess the levels of HDAC9 and β-actin expression. C, murine Hdac9+/+ and Hdac9−/− fibroblasts were transfected with HA-ATDC expression plasmids, and cell lysates were collected and analyzed by immunoprecipitation with an anti-HA antibody and Western blotting with an anti-acetyl-lysine antibody. The blot was stripped and reprobed with anti-HA to confirm equal HA-ATDC protein levels. A Western blot was also performed with an anti-β-actin antibody to confirm equal loading. D–G, HeLa cells were co-transfected with plasmids that express HA-ATDC and HA-p300 (or HA vector) in the presence or absence of co-transfection with FLAG-HDAC7 or different FLAG-HDAC9 plasmids. The acetylation of HA-ATDC and the expression levels of different proteins were determined with direct Western blot or immunoprecipitations and then followed by Western blotting using the indicated antibodies. IP, immunoprecipitation; IB, immunoblot.
In complementary experiments, HA-ATDC was deacetylated by overexpression of HDAC9 (Fig. 2D). Because p300/KAT3B promotes ATDC acetylation in mammalian cells (24), we showed that HA-ATDC that was hyperacetylated by p300 is similarly deacetylated by HDAC9 (Fig. 2E). Deacetylation of ATDC by HDAC9 is highly specific because overexpression of neither HDAC7 (Fig. 2F) nor HDAC9 mutants that do not bind ATDC (HDAC9Z and HDAC9ΔE7) (Fig. 2G) (data not shown) had an effect on ATDC acetylation. Collectively, our data convincingly argue that HDAC9 regulates the acetylation status of ATDC.
HDAC9 Regulates ATDC Functions
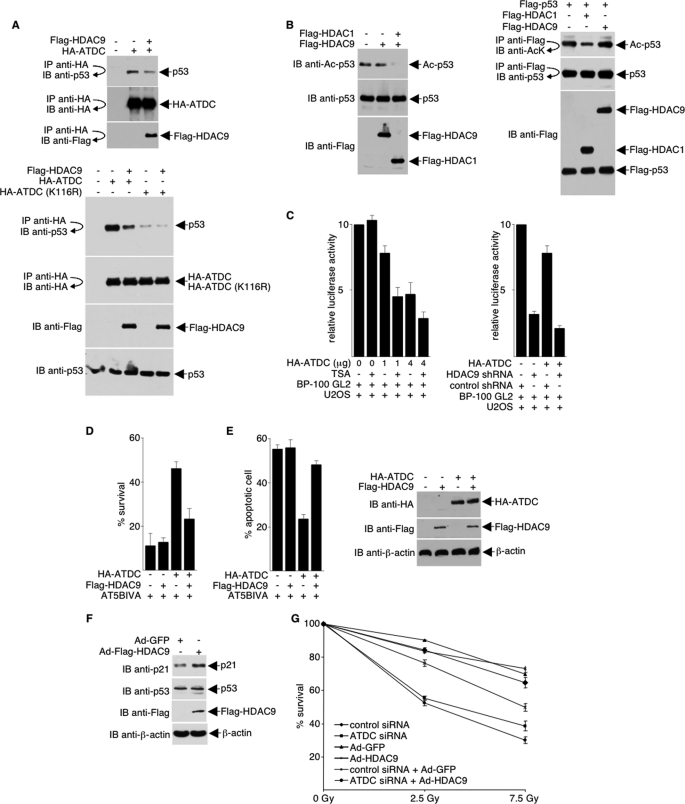
ATDC is overexpressed in a wide variety of different cancers, and its overexpression induces cell survival or confers cell growth advantage. Previously, we have shown that ATDC increases cell proliferation via inhibition of p53 nuclear activities (24). ATDC binds p53 and represses expression of p53-regulated genes, such as p21. To determine if HDAC9 deacetylation of ATDC affects ATDC-p53 interaction and subsequently alters ATDC and p53 functions, we assayed the association of ATDC and p53 in the presence and absence of HDAC9. As shown in Fig. 3A, in the presence of HDAC9, the ATDC-p53 interaction is markedly decreased in wild type but not K116R ATDC mutant (top, compare lanes 2 and 3; bottom, compare lanes 2 and 3 and lanes 4 and 5). Acetylation of p53 is not affected by HDAC9 (Fig. 3B), suggesting that the reduced ATDC-p53 interaction in the presence of HDAC9 is a direct result of ATDC deacetylation by HDAC9.
FIGURE 3.
HDAC9 regulates ATDC-p53 interactions and ATDC functions. A, 293T cells were co-transfected with plasmids encoding the FLAG-HDAC9 and HA-ATDC (or HA-ATDC-K116R) fusion proteins. Anti-HA immunoprecipitates obtained under low stringency conditions were analyzed by Western blotting with an anti-p53 antibody. The blot was stripped and sequentially reprobed with anti-HA and anti-FLAG antibodies, or separately with anti-FLAG and anti-p53, to confirm protein expressions. B, left, 293T cells were transfected with expression plasmids either for FLAG-HDAC1 (as a positive control for p53 deacetylation) or FLAG-HDAC9. Cell lysates were subjected to Western blot analysis using an anti-acetyl-p53 antibody. The blot was stripped and reprobed with the indicated antibodies to confirm equal protein expressions and loading. Right, 293T cells were transfected with expression plasmids for FLAG-p53 plus either FLAG-HDAC1 or FLAG-HDAC9. Cell lysates were immunoprecipitated (IP) under high stringency conditions using an anti-FLAG antibody. Immunoprecipitates were subjected to Western blot (IB) analysis using an anti-acetyl-lysine (AcK) antibody. The blot was stripped and reprobed with anti-p53. A separate blot was probed with the anti-FLAG antibody to confirm equal transfection efficiency and loading. C, pBP100-GL2 reporter plasmid was transfected into U2OS cells with either the pcDNA3.1HA vector or the plasmid expressing HA-ATDC. Experiments displayed on the left were either treated or untreated with 400 ng/ml TSA, and experiments shown on the right were transfected with either HDAC9 shRNA or control shRNA. Luciferase activity was determined 24 h after transfection, and values were normalized by protein concentration. Results from the average of three independent experiments ± S.D. are shown. D, AT5BIVA cells that were transfected with plasmids expressing the indicated proteins were irradiated (2 Gy of irradiation) or left unirradiated. The surviving colonies were counted after 2 weeks. Fractional cell survival is the fraction of colonies surviving irradiation divided by the total number of colonies in the unirradiated parallel culture. E, left, to examine the effects of ATDC and HDAC9 on cell death, cells were co-transfected with pEGFP to identify transfected cells. One day later, cells were stained with Hoechst dye, and apoptotic nuclei were counted in GFP-expressing cells under a fluorescence microscope. For each experiment, 300 cells were counted. Protein expression was assessed by direct Western blot analyses. Representative blots are shown in the right panel. The data shown are the average values ± S.D. from three separate experiments. F, extracts prepared from cells expressing HDAC9 or the control GFP protein were immunoblotted with the indicated antibodies to assess endogenous p21 protein expression. G, SiHa cells were transfected with either the control or ATDC siRNA and infected with adenoviruses expressing either GFP (control) or HDAC9. Forty-eight h later, cells were unirradiated or were irradiated with 2.5 or 7.5 Gy. Surviving colonies were counted 10 days later, and the fraction of colonies surviving irradiation divided by the total number of colonies in the unirradiated parallel culture is shown. Results from the average of two independent experiments ± S.D. (error bars) are shown.
Previously, we have shown that overexpression of wild type, but not K116R, ATDC in U2OS cells represses luciferase expression from a promoter containing the p53-binding site (24). Because HDAC9 deacetylation of ATDC leads to a decrease in ATDC-p53 interaction, we reason that HDAC9 concentration can alter transcription of p53-targeted promoters. As shown in Fig. 3C, trichostatin A or knockdown of HDAC9, which increased ATDC-p53 interaction, significantly increased the ability of ATDC to down-regulate transcription from pBP100-GL2, a plasmid with a p53-responsive promoter upstream of a luciferase reporter (Fig. 3C).
Clonogenic cell survival assays were employed to examine the importance of HDAC9 deacetylation of ATDC. AT5BIVA cells, which lack functional ATDC, were exposed to a sublethal dose of γ-IR, and colonies were counted 2 weeks later. Consistent with our findings that ATDC is a substrate for HDAC9, HDAC9-deacetylated ATDC was much less effective in complementing the IR sensitivity of AT5BIVA cells (Fig. 3D). Because ATDC affects cellular apoptotic pathways (24), we further examined whether HDAC9 regulates the function of ATDC on cell death. As shown in Fig. 3E, overexpression of ATDC, but not overexpression of HDAC9 alone, protected AT5BIVA cells from apoptosis following irradiation. Interestingly, HDAC9-deacetylated ATDC was significantly less effective in protecting AT5BIVA cells from apoptosis.
One of the functions of ATDC is to repress p21 gene expression through p53 (24). Consistent with our data indicating that HDAC9 deacetylates ATDC, decreases ATDC-p53 interaction, and increases transcription of p53-targeted promoters, overexpression of HDAC9 led to an increase of p21 (Fig. 3F). Also consistent with the finding that HDAC9 deacetylates ATDC and regulates functions of ATDC, overexpression of HDAC9 and knockdown of ATDC sensitize cells to γ-irradiation (Fig. 3G).
HDAC9 Deacetylates Lys116 of ATDC
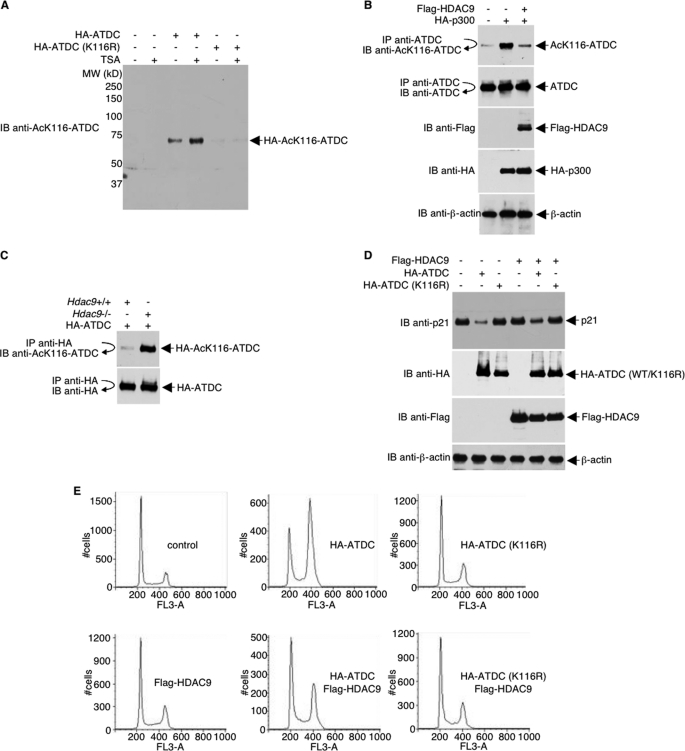
Because the interaction between p53 and ATDC is fine tuned by posttranslational acetylation of lysine 116 on ATDC (24), it is reasonable to speculate that HDAC9 regulates ATDC by deacetylation of Lys116 on ATDC. To further study the role of acetylation/deacetylation on Lys116 of ATDC, we produced a rabbit polyclonal antibody that uniquely recognizes acetylated Lys116 of ATDC (Fig. 4A). This antibody allowed us to unambiguously test the hypothesis that, in addition to its role in histone deacetylation, HDAC9 has the distinct function of converting Lys116 of ATDC into a hypoacetylated state. As shown in Fig. 4B, p300 acetylated ATDC can be deacetylated by HDAC9, as detected with the anti-Ac-Lys116-ATDC antibody, suggesting that of the nine identified acetylation sites on ATDC, at least Lys116 is deacetylated by HDAC9. The ability of HDAC9 to deacetylate ATDC was further confirmed by comparing the acetylation of Lys116 in HA-ATDC overexpressed in Hdac9−/− versus Hdac9+/+ mouse embryonic fibroblasts (Fig. 4C).
FIGURE 4.
HDAC9 deacetylates Lys116 of ATDC and alters ATDC functions. A, lysates were prepared from 293T cells transfected with plasmids that express wild type or mutated ATDC or vector alone. Proteins were separated by electrophoresis, transferred onto a membrane, and analyzed by Western blot. B, HeLa cells were co-transfected with plasmids that express FLAG-HDAC9, HA-p300, or vector. Endogenous ATDC Lys116 acetylation and the expression levels of different proteins were determined with direct Western blot or immunoprecipitations and then followed by Western blotting using the indicated antibodies. C, murine Hdac9+/+ and Hdac9−/− fibroblasts were transfected with HA-ATDC expression plasmids, and cell lysates were collected and analyzed by immunoprecipitation with an anti-HA antibody and Western blotting with the anti-acetyl-Lys116-ATDC antibody. The blot was stripped and reprobed with anti-HA to confirm equal HA-ATDC protein levels. D, U2OS cells were transfected with plasmids expressing the indicated proteins. Thirty-six h after transfection, cells were subjected to 10 Gy of irradiation. One h post-irradiation, cell lysates were prepared and analyzed by Western blots with the indicated antibodies. E, U2OS cells were co-transfected with a GFP expression plasmid together with either vector or plasmids expressing HA-ATDC or FLAG-HDAC9. Twenty-four h after transfection, cells were subjected to 10 Gy of irradiation. Sixteen h post-irradiation, cells were fixed briefly with ethanol and subjected to FACS analysis for cell cycle distribution. IP, immunoprecipitation; IB, immunoblot.
Previously, we found that ATDC reduces p53 recruitment on the p21 promoter and represses p21 expression. Overexpression of ATDC decreased p21 expression in U2OS cells, and depletion of ATDC in SiHa cells with siRNA resulted in an increase of p21 expression (24). Consistent with these previous results, overexpression of wild type ATDC, but not Lys116 mutant, inhibited p21 expression. Interestingly, the repression of p21 expression by wild type (but not Lys116 mutant) ATDC was diminished in the presence of HDAC9 (Fig. 4D). The effects of HDAC9 on ATDC and consequently on p21 expression are reflected in the increase in the percentage of cells in G1 and the reduction of S-, G2-, and M-phase cells in ATDC-HDAC9-overexpressing U2OS cells (Fig. 4E). This effect of HDAC9 on ATDC is mediated by deacetylation of Lys116 because K116R mutant is refractory to the change. Thus, the data unequivocally demonstrate that deacetylation of Lys116 of ATDC by HDAC9 alters ATDC function.
DISCUSSION
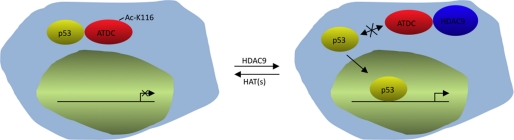
Identifications of HDAC-associated proteins have been tremendously useful in understanding the functions and mechanisms of action of HDACs. This is particularly true for three Class I HDACs, HDAC1, HDAC2, and HDAC3 (e.g. Refs. 34–38). Using similar strategies, we have successfully identified several novel HDAC-binding proteins and characterized non-histone substrates for HDAC6 and HDAC11 (39, 40). Here, in a biochemical approach to identify proteins that interact with HDAC9, we discovered that the ATDC protein binds HDAC9. We demonstrated that HDAC9 deacetylates Lys116 on ATDC, prevents ATDC from binding p53, and consequently leads to activation of p53-inducible genes (illustrated in Fig. 5). This finding expands the already complex mechanisms of action and functions of HDAC9.
FIGURE 5.
A model illustrating that deacetylation of Lys116 of ATDC leads to inhibition of p53-ATDC interaction and activation of p53-responsive genes.
Our finding that an HDAC inhibitor, TSA, induces ATDC hyperacetylation, coupled with the observation that acetylation of ATDC antagonizes p53 nuclear functions, suggests that the anti-tumor activity of HDAC inhibitors may vary depending on the status of p53 and ATDC within a cell. In addition to inactivation of p53, ATDC has oncogenic functions through Wnt pathway activation and β-catenin stabilization (23). At this time, we do not know if ATDC hyperacetylation activates ATDC upstream of the Wnt pathway.
One of the best characterized mechanisms of action of HDAC9 is its ability to partner with MEF-2 and repress MEF-2 activity. Here we describe an alternative, although non-mutually exclusive, mechanism by which HDAC9 alters gene transcription. By partnering with and deacetylating ATDC, HDAC9 inhibits ATDC-p53 interaction and thus increases the transcription activation function of p53. In addition to HDAC9, MEF-2 binds to HDAC3, HDAC4, HDAC5, and HDAC7 (41–47). In our hands, HDAC3, HDAC4, HDAC5, and HDAC7 did not co-precipitate with ATDC. This is somewhat surprising, given that the ATDC-interacting region on HDAC9 (residues 601–1011) is fairly conserved in HDAC4, HDAC5, and HDAC7. A finer deletion mapping of the ATDC-interacting domain on HDAC9, coupled with point mutation analysis, will perhaps help to clarify this issue. Nevertheless, we did find that ATDC interacted with HDAC11 in overexpression and co-immunoprecipitation studies.3 At this time, we do not yet know if HDAC11 similarly regulates ATDC by deacetylation of acetylated Lys116 of ATDC.
HDAC9 appears to have many seemingly unrelated and even contradictory functions. The results presented in our current study suggest that the multiple functions of HDAC9 may be attributed to the expression pattern of HDAC9 target proteins. For instance, HDAC9 may impart different functional consequences on different cells, depending on ATDC expression patterns. We predict that many p53-inducible genes are non-responsive to HDAC9 in cells that lack ATDC (such as U2OS), whereas these same p53-inducible genes can be activated by HDAC9 in cells that express abundant ATDC (such as SiHa).
In addition to MEF-2 and Foxp3, a number of other cellular proteins have been reported to associate with HDAC9 in vitro and in vivo. For example, the ankyrin repeat proteins ANKRA and RFXANK that were initially found to interact with HDAC4 also complex with HDAC9 (48). In future studies, it would be useful to determine if ANKRA and other HDAC9-interacting proteins undergo acetylation and serve as HDAC9 substrates and, if so, whether deacetylation of these HDAC9-interacting proteins alters their functions.
To date, there is no structural information available for HDAC9. A recent report on the structure of HDAC4 catalytic domain reveals a conformationally flexible structural zinc-binding domain conserved in all Class IIa enzymes (49). Similarly, crystal structure of the catalytic domain of human HDAC7 uncovered a novel zinc-binding motif adjacent to the active site, which is conserved in Class IIa HDACs (50). Predictably, the HDAC-specific zinc binding CCHC motif in HDAC9 may participate in ATDC recognition.
Our results clearly showed that deacetylation of Lys116 of ATDC by HDAC9 is a critical step in the regulation of ATDC functions by HDAC9. However, in a previous analysis of purified ATDC by LC tandem mass spectrometry (LC-MS/MS), eight additional lysine residues were found to be acetylated (24), and at this time, we have not ruled out that HDAC9 might also deacetylate these other acetylated lysines and regulate the function of ATDC through modification of these other residues. Further experiments to address this issue are in progress.
Acknowledgments
We thank Eric Olson (Southwestern Medical Center) for Hdac9−/− mice, Paul Marks (Sloan-Kettering Institute) for the HDAC9 cDNA, Arthur Zelent (Institute of Cancer Research, University of London) for the HDAC9 isoform expression plasmids, Eric Verdin (University of California, San Francisco) for the FLAG-HDAC7 expression plasmid, Jiandong Chen (Moffitt Cancer Center) for the FLAG-p53 expression plasmid, Bill Lane (Harvard University) for LC-MS/MS, Rockland Immunochemicals for generating the anti-Ac-Lys116-ATDC antibody, and the Moffitt Cancer Center Core Facility for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM081650 (to E. S.). This work was also supported by American Heart Association Grant 0755298 and a grant from the Kaul Foundation (to E. S.).
N. Rezai-Zadeh and E. Seto, unpublished data.
- HDAC
- histone deacetylase
- TRIM
- tripartite motif
- MITR
- myocyte enhancer-binding factor 2-interacting transcriptional repressor
- HDRP
- histone deacetylase-related protein
- Gy
- grays
- TSA
- trichostatin A.
REFERENCES
- 1.Yang X. J., Seto E. (2003) Curr. Opin. Genet. Dev. 13, 143–153 [DOI] [PubMed] [Google Scholar]
- 2.Yang X. J., Seto E. (2008) Nat. Rev. Mol. Cell. Biol. 9, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sparrow D. B., Miska E. A., Langley E., Reynaud-Deonauth S., Kotecha S., Towers N., Spohr G., Kouzarides T., Mohun T. J. (1999) EMBO J. 18, 5085–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C. L., McKinsey T. A., Lu J. R., Olson E. N. (2001) J. Biol. Chem. 276, 35–39 [DOI] [PubMed] [Google Scholar]
- 5.Zhang C. L., McKinsey T. A., Olson E. N. (2002) Mol. Cell. Biol. 22, 7302–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C. L., McKinsey T. A., Olson E. N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7354–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X., Richon V. M., Rifkind R. A., Marks P. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1056–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X., Marks P. A., Rifkind R. A., Richon V. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10572–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahlknecht U., Schnittger S., Will J., Cicek N., Hoelzer D. (2002) Biochem. Biophys. Res. Commun. 293, 182–191 [DOI] [PubMed] [Google Scholar]
- 10.Petrie K., Guidez F., Howell L., Healy L., Waxman S., Greaves M., Zelent A. (2003) J. Biol. Chem. 278, 16059–16072 [DOI] [PubMed] [Google Scholar]
- 11.Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. (2002) Cell 110, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao R., de Zoeten E. F., Ozkaynak E., Chen C., Wang L., Porrett P. M., Li B., Turka L. A., Olson E. N., Greene M. I., Wells A. D., Hancock W. W. (2007) Nat. Med. 13, 1299–1307 [DOI] [PubMed] [Google Scholar]
- 13.de Zoeten E. F., Wang L., Sai H., Dillmann W. H., Hancock W. W. (2010) Gastroenterology 138, 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B., Samanta A., Song X., Iacono K. T., Bembas K., Tao R., Basu S., Riley J. L., Hancock W. W., Shen Y., Saouaf S. J., Greene M. I. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4571–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison B. E., D'Mello S. R. (2008) Exp. Biol. Med. 233, 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Méjat A., Ramond F., Bassel-Duby R., Khochbin S., Olson E. N., Schaeffer L. (2005) Nat. Neurosci. 8, 313–321 [DOI] [PubMed] [Google Scholar]
- 17.Sugo N., Oshiro H., Takemura M., Kobayashi T., Kohno Y., Uesaka N., Song W. J., Yamamoto N. (2010) Eur. J. Neurosci. 31, 1521–1532 [DOI] [PubMed] [Google Scholar]
- 18.Wong R. H., Chang I., Hudak C. S., Hyun S., Kwan H. Y., Sul H. S. (2009) Cell 136, 1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gazin C., Wajapeyee N., Gobeil S., Virbasius C. M., Green M. R. (2007) Nature 449, 1073–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonhardt E. A., Kapp L. N., Young B. R., Murnane J. P. (1994) Genomics 19, 130–136 [DOI] [PubMed] [Google Scholar]
- 21.Reddy B. A., Etkin L. D., Freemont P. S. (1992) Trends Biochem. Sci. 17, 344–345 [DOI] [PubMed] [Google Scholar]
- 22.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P. G., Ballabio A. (2001) EMBO J. 20, 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Heidt D. G., Lee C. J., Yang H., Logsdon C. D., Zhang L., Fearon E. R., Ljungman M., Simeone D. M. (2009) Cancer Cell 15, 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Z., Villagra A., Peng L., Coppola D., Glozak M., Sotomayor E. M., Chen J., Lane W. S., Seto E. (2010) Mol. Cell. Biol. 30, 3004–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aizawa H., Hu S. C., Bobb K., Balakrishnan K., Ince G., Gurevich I., Cowan M., Ghosh A. (2004) Science 303, 197–202 [DOI] [PubMed] [Google Scholar]
- 26.Dequiedt F., Kasler H., Fischle W., Kiermer V., Weinstein M., Herndier B. G., Verdin E. (2003) Immunity 18, 687–698 [DOI] [PubMed] [Google Scholar]
- 27.Peng Y., Li C., Chen L., Sebti S., Chen J. (2003) Oncogene 22, 4478–4487 [DOI] [PubMed] [Google Scholar]
- 28.Yang W. M., Yao Y. L., Sun J. M., Davie J. R., Seto E. (1997) J. Biol. Chem. 272, 28001–28007 [DOI] [PubMed] [Google Scholar]
- 29.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rezai-Zadeh N., Tsai S. C., Wen Y. D., Yao Y. L., Yang W. M., Seto E. (2004) Methods Enzymol. 377, 167–179 [DOI] [PubMed] [Google Scholar]
- 31.Zhao S., Renthal W., Lee E. Y. (2002) Nucleic Acids Res. 30, 4815–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Wharton W., Yuan Z., Tsai S. C., Olashaw N., Seto E. (2004) Mol. Cell. Biol. 24, 5106–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawada M., Sun W., Hayes P., Leskov K., Boothman D. A., Matsuyama S. (2003) Nat. Cell. Biol. 5, 320–329 [DOI] [PubMed] [Google Scholar]
- 34.Hassig C. A., Fleischer T. C., Billin A. N., Schreiber S. L., Ayer D. E. (1997) Cell 89, 341–347 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. (1997) Cell 89, 357–364 [DOI] [PubMed] [Google Scholar]
- 36.Wade P. A., Gegonne A., Jones P. L., Ballestar E., Aubry F., Wolffe A. P. (1999) Nat. Genet. 23, 62–66 [DOI] [PubMed] [Google Scholar]
- 37.Tsai S. C., Valkov N., Yang W. M., Gump J., Sullivan D., Seto E. (2000) Nat. Genet. 26, 349–353 [DOI] [PubMed] [Google Scholar]
- 38.Wen Y. D., Perissi V., Staszewski L. M., Yang W. M., Krones A., Glass C. K., Rosenfeld M. G., Seto E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7202–7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Yuan Z., Zhang Y., Yong S., Salas-Burgos A., Koomen J., Olashaw N., Parsons J. T., Yang X. J., Dent S. R., Yao T. P., Lane W. S., Seto E. (2007) Mol. Cell. 27, 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glozak M. A., Seto E. (2009) J. Biol. Chem. 284, 11446–11453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miska E. A., Karlsson C., Langley E., Nielsen S. J., Pines J., Kouzarides T. (1999) EMBO J. 18, 5099–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinsey T. A., Zhang C. L., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miska E. A., Langley E., Wolf D., Karlsson C., Pines J., Kouzarides T. (2001) Nucleic Acids Res. 29, 3439–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang A. H., Yang X. J. (2001) Mol. Cell. Biol. 21, 5992–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu J., McKinsey T. A., Nicol R. L., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4070–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kao H. Y., Verdel A., Tsai C. C., Simon C., Juguilon H., Khochbin S. (2001) J. Biol. Chem. 276, 47496–47507 [DOI] [PubMed] [Google Scholar]
- 47.Grégoire S., Xiao L., Nie J., Zhang X., Xu M., Li J., Wong J., Seto E., Yang X. J. (2007) Mol. Cell. Biol. 27, 1280–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang A. H., Grégoire S., Zika E., Xiao L., Li C. S., Li H., Wright K. L., Ting J. P., Yang X. J. (2005) J. Biol. Chem. 280, 29117–29127 [DOI] [PubMed] [Google Scholar]
- 49.Bottomley M. J., Lo Surdo P., Di Giovine P., Cirillo A., Scarpelli R., Ferrigno F., Jones P., Neddermann P., De Francesco R., Steinkühler C., Gallinari P., Carfí A. (2008) J. Biol. Chem. 283, 26694–26704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuetz A., Min J., Allali-Hassani A., Schapira M., Shuen M., Loppnau P., Mazitschek R., Kwiatkowski N. P., Lewis T. A., Maglathin R. L., McLean T. H., Bochkarev A., Plotnikov A. N., Vedadi M., Arrowsmith C. H. (2008) J. Biol. Chem. 283, 11355–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]