Abstract
Suitable alterations in gene expression are believed to allow animals to survive drastic changes in environmental conditions. Drosophila melanogaster larvae cease eating and exit moist food to search for dry pupation sites after the foraging stage in what is known as the wandering stage. Although the behavioral change from foraging to wandering causes desiccation stress, the mechanism by which Drosophila larvae protect themselves from desiccation remains obscure. Here, we identified a gene, CG14686 (designated as Desiccate (Desi)), whose expression was elevated during the wandering stage. The Desi expression level was reversibly decreased by transferring wandering larvae to wet conditions and increased again by transferring them to dry conditions. Elevation of Desi expression was also observed in foraging larvae when they were placed in dry conditions. Desi encoded a 261-amino acid single-pass transmembrane protein with notable motifs, such as SH2 and PDZ domain-binding motifs and a cAMP-dependent protein kinase phosphorylation motif, in the cytoplasmic region, and its expression was observed mainly in the epidermal cells of the larval integuments. Overexpression of Desi slightly increased the larval resistance to desiccation stress during the second instar. Furthermore, Desi RNAi larvae lost more weight under dry conditions, and subsequently, their mortalities significantly increased compared with control larvae. Under dry conditions, consumption of carbohydrate was much higher in Desi RNAi larvae than control larvae. Based on these results, it is reasonable to conclude that Desi contributes to the resistance of Drosophila larvae to desiccation stress.
Keywords: Development, Drosophila Genetics, Gene Expression, Insect, Membrane Proteins, Desiccation, Larvae, Stress, Survival, Wandering
Introduction
A wide variety of stressful stimuli change patterns of gene expression, which enables animals to adapt to stress, and such changes in gene expression are believed to allow them to survive drastic environmental changes. Activation of heat shock protein genes (hsp) is a typical example; all organisms express a particular set of hsp genes in response to stressors such as temperature extremes, aversive chemical application, anoxia, and many other environmental injuries (1, 2). Hsp proteins are generally divided into three families: the 90-kDa, 70-kDa, and small heat shock proteins (3). It has been reported that a nonlethal desiccation at 0% relative humidity (RH)2 enhanced transcriptional levels of the two hsp genes, hsp70 and hsp23, in pupae of the flesh fly Sarcophaga crassipalpis (4). Although the two hsp transcripts were up-regulated in response to desiccation, the up-regulation was less dramatic than that elicited by heat shock, and desiccation failed to generate tolerance to high or low temperatures. Recently, it has been also reported that dehydration elicited expression of hsp70 in three mosquito species, Aedes aegypti, Anopheles gambiae, and Culex pipiens, but hsp90 expression levels remained fairly constant. Furthermore, injection of dsRNA to knock down expression of hsp70 and hsp90 significantly decreased survival rates of A. aegypti under dehydration (5). Exposure of adult male Drosophila melanogaster to desiccation enhanced transcriptional levels of Frost and senescence marker protein-30 (smp-30) but did not change those of hsp70Aa and hsp23 (6). Although these previous studies demonstrated desiccation-induced gene expression, we do not know whether the up-regulation of the gene expression levels confers desiccation resistance on animals.
Drosophila melanogaster, like all holometabolous insects, undergoes complete metamorphosis to reach adulthood. Each phase of the life cycle is characterized by a coordinated program of developmental events and behavioral transitions that have evolved to promote fitness and survival (7, 8). In Drosophila larvae, the essential midthird instar transition from foraging (feeding) to wandering (nonfeeding) behavior occurs prior to pupariation and metamorphosis. Although this behavioral transition imposes desiccation stress on the larvae, it is unknown not only whether there is a specific mechanism responsible for providing the larvae with desiccation tolerance but also how the tolerance of the larvae is enhanced during the wandering stage. In this study, we sought a gene whose expression was elevated by desiccation stress and identified Desiccate (Desi). Desi expression in the larvae is dependent on relative humidity; desiccation enhanced its expression, and conversely, humidification repressed its expression. Desi encoded a single-pass transmembrane protein and expressed its transcripts actively in epidermal cells of the integument. Furthermore, Desi expression specifically increased during the wandering stage and the survival rates of Desi RNAi larvae significantly declined under the dry condition.
EXPERIMENTAL PROCEDURES
Drosophila Culture and Stocks
Fly cultures and crosses were grown on cornmeal-glucose-yeast medium at 25 °C. The RNAi stocks (UAS-dsDesi (dsCG14686)) strains were obtained from the Vienna Drosophila RNAi Center (Vienna, Austria).
UAS-Desi Constructions and Transgenic Fly Generations
A 786-bp Desi cDNA fragment containing the signal peptide and Desi-coding sequence was obtained by PCR and was cloned into NotI/KpnI sites of pUAST vector to yield the pUAST-Desi construct. After verifying the construct by DNA sequencing, it was injected into embryos, and UAS-Desi transgenic lines were generated by P-element-mediated germ line transformation of a w− strain (9).
Differential Display RT-PCR and Sequence Analysis of Cloned Gene
Total RNAs were prepared from whole bodies of control and stressed Drosophila larvae, which were put under a desiccated condition (15% RH) for 5 h. Differential display RT-PCR was performed on total RNAs by using the GeneFishingTM DEG Premix kit according to the manufacturer's instructions (Seegene, Inc., Rockville, MD). Five micrograms of total RNA were converted to cDNA by reverse transcription using 2 μl of 10 μm anchor primer (dT-annealing control primer 1). Synthesized cDNA was amplified with a combination of arbitrary primers (annealing control primers 1–120) in the above kit (10). The cDNA bands preferentially expressed in either control or stressed larval samples were directly cloned into pGEM-T vector (pGEM-T Vector System I, Promega, Madison, WI) and sequenced by a 310 DNA sequencer (Applied Biosystems, Foster City, CA). To obtain full-length cDNA, 5′ rapid amplification of cDNA ends (5′ rapid amplification of cDNA ends) was performed using 5′ rapid amplification of cDNA ends kit (Invitrogen) as described previously (11). Computer-assisted sequence analyses were performed by GENETYX-MAC (version 13.1.7; Software Development Co., Tokyo, Japan).
Quantitative RT-PCR
Two micrograms of total RNA isolated from whole bodies or indicated tissues of control, and test larvae was reverse transcribed with oligo(dT) primer using ReverTra Ace (Toyobo, Osaka, Japan). Real-time quantitative PCR was carried out with the reverse transcription products in a 20-μl reaction volume of LightCycler Fast DNA Master SYBR Green I (Roche Applied Science), using the Light-Cycler 1.2 instrument and software (Roche Applied Science,). RT-PCR data were normalized to rp49 expression, and the message abundance of each gene was compared with that in control samples (12). Specific primers used for the PCR analysis were as follows: Desi-Fw, GATAGCCATAAGTTCTATGCG; Desi-Rv, GCCTCCTTAATAGCCGTTCC; rp49-Fw, GATCGTGAAGAAGCGCACCAAG; and rp49-Rv, CCGGATTCAAGAAGTTCCTGGTG.
In Situ Hybridization
Digoxigenin-labeled RNA fragment synthesized from a Desi cDNA template (coding region, 1–782) was used as probes for in situ hybridization. Hybridization and washes were carried out as described previously (13).
Preparation of Anti-Desi IgG
The cDNA containing the ORF of Desi (residues 1–781) was cloned into pET32a(+) (Novagen) and expressed in Escherichia coli strain BL21(DE3)pLys. The recombinant GST-Desi fusion protein was purified by a glutathione-Sepharose column (GE Healthcare). The purified protein was emulsified by Titer Max Gold (CytRx Corp.) and injected to immunize a rabbit. Anti-Desi IgG was precipitated by adding ammonium sulfate to 40% saturation and further purified by an affinity column of protein G-Sepharose (GE Healthcare).
Western Blotting and Immunocytochemistry
Western blotting and immunocytochemical analyses using anti-Desi IgG were performed as described previously (14).
Extraction and Analysis of Epicuticular Hydrocarbon
Epicuticular hydrocarbons were extracted from 100 third instar larvae with hexane for 5 min. The extract was concentrated to dryness with N2, dissolved in hexane, and applied to gas chromatograph (GC-2014, Shimadzu) with capillary column (inner diameter, 25 m × 0.25 mm, Ulbon HR-SS-10, Shinwa Chemical Industries, Ltd.). Helium was used as the column carrier gas. The column oven temperature was held at 50 °C for 2 min, increased to 150 °C at 36 °C/min, increased at 8 °C/min to 220 °C, and held at 220 °C for 10 min (15).
Quanatitative Analyses of Carbohydrate and ATP Concentrations
After washing Drosophila larvae with phosphate-buffered saline (8 mm Na2HPO4, 1.5 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl, pH 7.2), larvae were put in ice-cold 1% (w/v) trichloroacetic acid (10 μl/larva) and homogenized in Artek Sonic Dismembrator (20 pulses at 50 watts). The homogenate was centrifuged at 20,000 × g for 5 min at 4 °C, and the supernatant was collected. Carbohydrate concentration in the supernatant was determined by the anthrone/sulfuric acid method as described previously (16). ATP concentration was measured with luciferin-luciferase reagent by the method of Stanley and William (17).
RESULTS
Survival Rates of Drosophila Larvae under Dry Condition
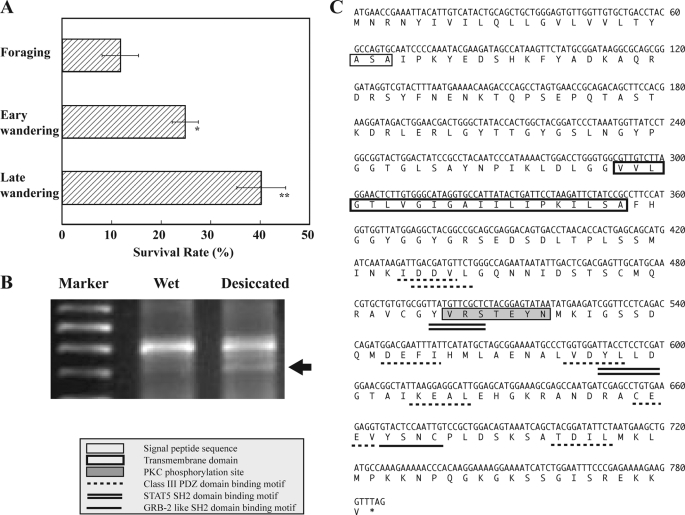
The onset of wandering changes the habitat of Drosophila larvae from wet to dry environments for normal metamorphosis. This behavioral change was interpreted to imply that the resistance of larvae to dry conditions is increased during the transition from foraging to wandering stages. To verify this possibility, we examined the survival rates of larvae that were moved to a dry condition at different developmental stages from foraging to late stage wandering. When larvae were placed under 0% RH for 5 h, the survival rates were progressively increased with larval growth; the survival rate of late stage wandering larvae was considerably greater than those of foraging and early stage wandering larvae (Fig. 1A). These results suggested that foraging and early stage wandering larvae were much more sensitive to dry conditions than late stage wandering larvae.
FIGURE 1.
Survival rates of Drosophila larvae under desiccation and identification of CG14686. A, survival rates of Drosophila larvae at different developmental stages, foraging, early wandering stage (soon after onset of wandering), and late wandering stage (∼4 h after onset of wandering). The test larvae were placed under 0% RH for 5 h and thereafter put on water absorbed cotton in a Petri dish. Survival rates were checked soon after this transfer. Data are given as means ± S.D. for seven separate measurements using 30 larvae each. * and **, significantly different from the value of foraging larvae (*, p < 0.05; **, p < 0.01, Tukey's honestly significant difference (HSD)). B, differential display RT-PCR on total RNAs prepared from whole bodies of control (nonstressed) and stressed larvae under desiccation at 15% RH for 5 h. A DNA band indicated by an arrow coded CG14686. C, nucleotide and deduced amino acid sequences of cloned cDNA for Desi. The putative signal peptide and transmembrane domain are boxed. Src family and STAT5 SH2 domain-binding motifs are underlined and double underlined, respectively. Class III PDZ domain-binding motifs and the protein kinase A phosphorylation site are dotted and underlined and shaded, respectively. These motifs were searched against the Eukaryotic Linear Motif Database.
Identification of Desi (CG14686)
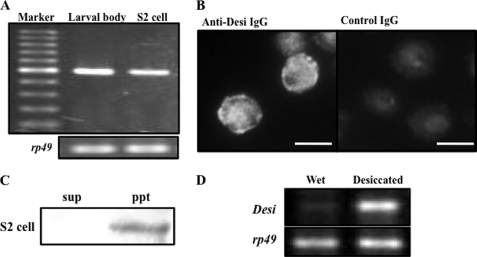
To identify genes whose expression is enhanced by desiccation, we performed differential display RT-PCR on total RNAs prepared from control early third instar (foraging) Drosophila larvae and desiccation-stressed foraging larvae using a set of 120 different primers (annealing control primers 1–120, Seegene, Inc.). Of these, one primer (annealing control primer 94) revealed desiccation-dependent transcriptional enhancement of one DNA band encoding a fragment of CG14686, whose molecular function is unknown (Fig. 1B). CG14686 encoded a 261-amino acid protein with a putative signal peptide sequence in the N terminus and a single-pass transmembrane domain at positions 72 to 95 (Fig. 1C). Because RT-PCR showed CG14686 expression in S2 cells, its immunocytochemical localization in the culture cells was examined to confirm that this gene product is present as a transmembrane protein (Fig. 2A). Although immunoreaction signals were detected in the cytoplasm as well as in the plasma membrane (Fig. 2B), the former signals were expected to be derived from CG14686 in the membranes of organelles. This expectation was confirmed by the discovery of immunoreactive proteins only in the insoluble fraction of the cell extract (Fig. 2C).
FIGURE 2.
CG14686 (Desiccate (Desi)) expression in S2 cells and its desiccation-dependent expression in Drosophila larvae. A, RT-PCR analysis of CG14686 expression in Drosophila larval body and S2 cells. Sequence analysis of the amplified DNA fragments showed that these bands encoded an identical part of CG14686. B, Immunocytochemical analysis of CG14686 product in S2 cells. CG14686 was visualized with anti-CG14686 IgG and an Alex 488-conjugated secondary antibody. Signals indicate nuclei containing DNA conjugated with DAPI. Nonimmunized rabbit IgG was used as control IgG. Scale bar, 10 μm. C, distribution of CG14686 in cell fractions extracted from S2 cells. sup, soluble cytoplasmic fraction; ppt, insoluble membrane fractions of S2 cells. D, RT-PCR analysis of Desi expression in whole bodies of control larvae on water-absorbed cotton in Petri dish and stressed larvae were put in a desiccated Petri dish (15 RH) for 5 h.
Sequence analysis showed that the cytoplasmic tail of CG14686 cDNA contained several conserved motifs, such as SH2 domain-binding motifs, a cAMP-dependent protein kinase (protein kinase A) phosphorylation site, and a PDZ domain-binding motif, implying the contribution to intracellular signal transduction (Fig. 1C). Most of these motifs were found to be conserved in orthologous genes identified in a broad range of insect species such as mosquitoes, bees, and beetles (supplemental Fig. 1). Because the desiccation-induced elevation of this gene expression was confirmed by RT-PCR with specific primers (Fig. 2D), we designated it Desiccate (Desi).
Spatiotemporal Expression Pattern of Desi
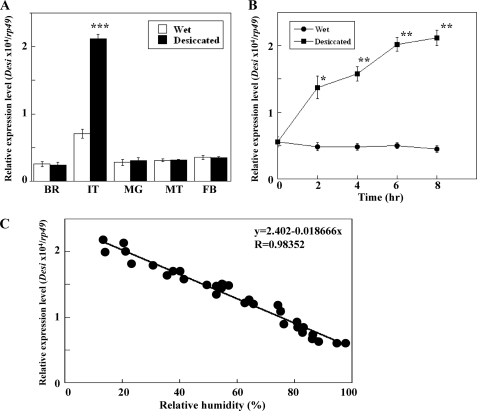
Quantitative real-time PCR was carried out to measure Desi expression levels in various tissues of Drosophila larvae during the foraging stage with and without desiccation stress (15% RH). Although Desi expression was detected in all of the tissues tested, it was highest in the integument and its desiccation-dependent enhancement was also observed only in the integument (Fig. 3A). To confirm the humidity-dependent expression of Desi in the integument of same-aged foraging larvae, the expression levels were measured at various time points after the initiation of desiccation. The results showed that Desi expression increased continuously and that 8-h desiccation elevated Desi expression by 4-fold (Fig. 3B). Furthermore, we demonstrated that Desi expression levels were approximately quadrupled as RH decreased from 100% to 15% (Fig. 3C). In contrast, desiccation-induced elevation of Desi expression was not observed in Drosophila adults (supplemental Fig. 2).
FIGURE 3.
Desi expression in the integuments of Drosophila larvae. A, real-time quantitative RT-PCR analysis of Desi expression in various tissues with and without desiccation stress. Data are given as means for six separate measurements using 15 larvae each. BR, brain; IT, integument; MG, midgut; MT, Malpighian tubules; FB, fat body. ***, significantly different from the value of larvae under wet condition (p < 0.001, Tukey's HSD). B, elevation of Desi expression in the larval integuments during desiccation stress. Foraging second instar larvae were transferred from moist food to desiccated condition (15% RH) at 0 h. Data are given as means for six separate measurements using 20 larvae each. * and **, significantly different from the value of foraging larvae at 0 h (*, p < 0.05; **, p < 0.01, Tukey's HSD). C, humidity-dependent changes in Desi expression in the larval integuments. After foraging second instar larvae were placed at indicated humidities for 6 h, Desi expression in the larval integuments was measured. Each point represents a single determination.
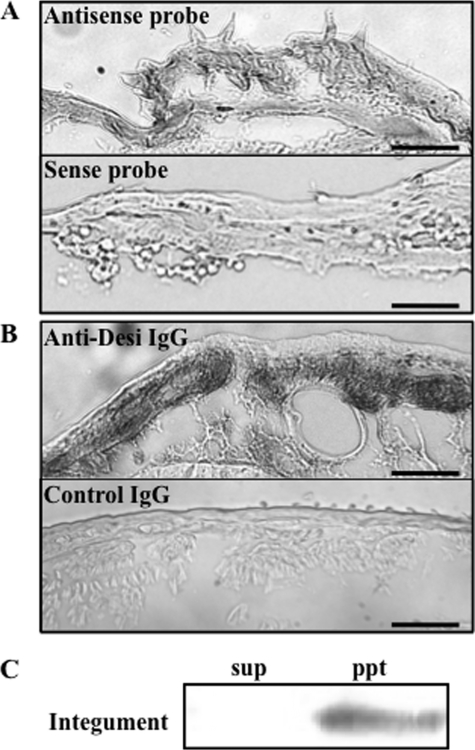
In situ hybridization showed that Desi mRNA is expressed in the epidermal cells (Fig. 4A). Furthermore, immunocytochemistry using anti-Desi IgG showed that Desi immunoreactivities were detected in both the cytoplasm and plasma membrane (Fig. 4B). Like the immunocytochemical observation of Desi in S2 cells, immunoreaction signals in the cytoplasm were expected to be derived from Desi in the membranes of organelles. This interpretation was supported by localization of immunoreactive proteins only in the insoluble fraction of the integument extract (Fig. 4C).
FIGURE 4.
Desi mRNA and protein expression in the integuments of Drosophila larvae. A, in situ hybridization of Desi mRNA probed with antisense or sense probe of Desi RNA. Tissue sections were prepared from larvae during wandering stage. Note that hybridization signals are detected in the epidermal cell layer. Scale bars, 30 μm. B, immunocytochemical staining of Desi protein with anti-Desi IgG. Nonimmunized rabbit IgG was used as control IgG. Bars, 30 μm. C, distribution of Desi in cell fractions extracted from the larval integument. sup, soluble cytoplasmic fraction; ppt, insoluble membrane fractions of the integument.
Stage-dependent Expression Pattern of Desi
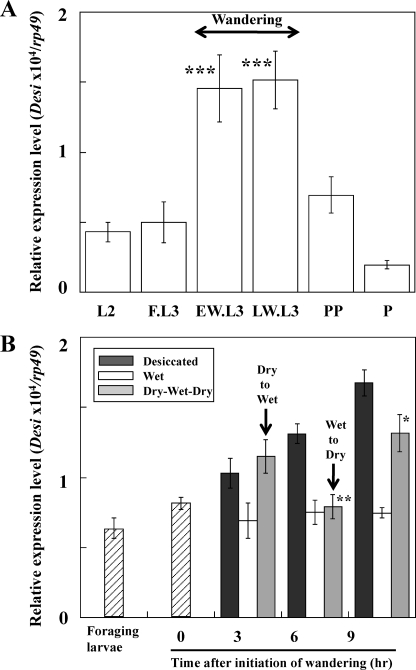
When Drosophila larvae enter the wandering stage after the foraging stage, they leave moist food to search for dry pupation sites. We investigated whether alteration of Desi expression levels depended on this stage-specific behavioral change. The results showed that the Desi expression level was elevated >3-fold around the time of the initiation of wandering and that the high expression level was maintained throughout the wandering stage (Fig. 5A). We then examined whether this elevation of the Desi expression level was due to desiccated condition. After the larvae in a dry place were returned to a wet place (on water-absorbed cotton), Desi expression levels in the larvae reversibly declined within a few hours (Fig. 5B). Furthermore, the decline in Desi expression was reversed by transferring larvae to a dry condition, indicating that the elevation of Desi expression observed in the wandering stage larvae was mainly due to the environmental change from a wet to a dry condition. This interpretation was supported by the observation that the onset of wandering was not advanced by the overexpression of Desi under the direction of the hs-Gal4 driver in the second instar larvae (supplemental Fig. 3).
FIGURE 5.
Desi expression in the integuments of Drosophila larvae during development. A, stage-dependent changes in Desi expression in the larval integuments. Shown are early wandering (EW.L3), within 1 h after initiation of wandering, and late wandering (LW.L3), ∼8 h after initiation of wandering. L2, second instar larvae; FL3, foraging 3rd instar larvae; PP, pupariated larvae; P, pupae. Data are given as means for six separate measurements using 16 animals each. ***, significantly different from the value of second instar larvae (p < 0.001, Tukey's HSD). B, environment-dependent changes in Desi expression in the larval integuments. Wandering larvae were continuously placed in a dry (15% RH) Petri dish (filled bars) or wet condition (on water absorbed cotton in Petri dish) (open bars). Some wandering larvae were transferred to the wet condition at the early wandering stage and thereafter returned to the dry condition (gray bar). Desi expression levels in wet condition were not changed with or without nutrition. Desi expression in the integuments of the larvae was measured at indicated time. * and **, significantly different from the value of wandering larvae (*, p < 0.05; **, p < 0.01, Tukey's HSD). Other explanations are as described in A.
Effects of Manipulation of Desi Expression Levels on Drosophila Larvae
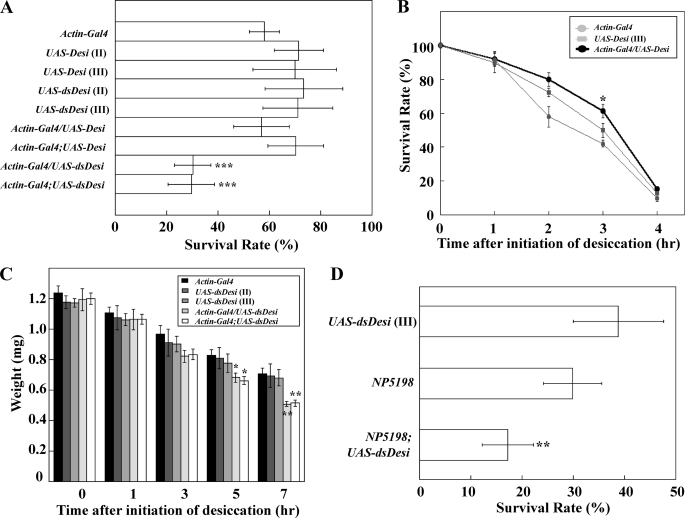
To determine whether Desi contributes to the protection of Drosophila larvae from desiccation, we established transgenic Desi overexpression and knockdown lines under the direction of an Actin-Gal4 driver (supplemental Fig. 4). When both transgenic third instar larvae were put in a dry environment (15% RH) during the foraging stage, the survival rates of the Desi knockdown larvae significantly decreased after 8 h of desiccation, whereas those of Desi overexpression larvae did not change (Fig. 6A). However, second instar larvae overexpressing Desi showed a slight but significant increase in their survival rates when they were put in the dry environment (Fig. 6B). Neither manipulation of Desi expression affected the mortalities of test transgenic larvae during the foraging stage under a wet condition at all (supplemental Fig. 5). The reduction of resistance to desiccation in Desi knockdown larvae was found to be mainly due to an excessive loss of internal water because the desiccation-induced decreases in body weight of the knockdown larvae were significantly greater than those in control larvae, including UAS lines and the Actin-Gal4 line (Fig. 6C). This interpretation was confirmed by the results showing that the weight losses of the Desi-overexpressing second instar larvae under the dry condition (15% RH) were significantly smaller than those of the control transgenic larvae (supplemental Fig. 6). Desiccation-induced mortality was also observed in Desi knockdown larvae in which Desi dsRNA was preferentially expressed in the epidermal cells using the NP5198 driver (Fig. 6D).
FIGURE 6.
Effects of Desi overexpression and knockdown on survival rates and weights of Drosophila larvae. A, survival rates of Desi overexpression and knockdown third instar larvae under desiccated conditions. Transgenic lines carrying UAS-Desi or UAS-dsDesi on second chromosome (II) or third chromosome (III) were used for Desi overexpression and knockdown. Every mutant foraging larva was placed under desiccated condition (15% RH) for 8 h and then transferred to normal diet medium. Survival rates were checked 24 h after this transfer. Data are given as means for seven separate measurements using 50 larvae each. ***, significantly different from the value of control line larvae (p < 0.001, Tukey's HSD). B, survival rates of Desi overexpression second instar larvae. Control and Desi overexpression larvae were put under desiccated condition for indicated periods and then transferred on normal diet medium, and survival rates were checked 1 h after this transfer. *, significantly different from the values of control line larvae (*, p < 0.05, Tukey's HSD). Other explanations are as described in A. C, changes in Desi knockdown larval weights under desiccated condition for indicated periods. Note that weights of Desi knockdown larvae were significantly lower than those of control line larvae after a 5-h desiccation. * and **, significantly different from the value of control line larvae (*, p < 0.05; **, p < 0.01, Tukey's HSD). Other explanations are as described in A. D, survival rates of Desi knockdown larvae, in which Desi dsRNA was preferentially expressed in the epidermal cells using NP5198 driver, under desiccated condition (15% RH) for 10 h. **, significantly different from the value of control larvae UAS-dsDesi (p < 0.01, Tukey's HSD). Other explanations are as described in A.
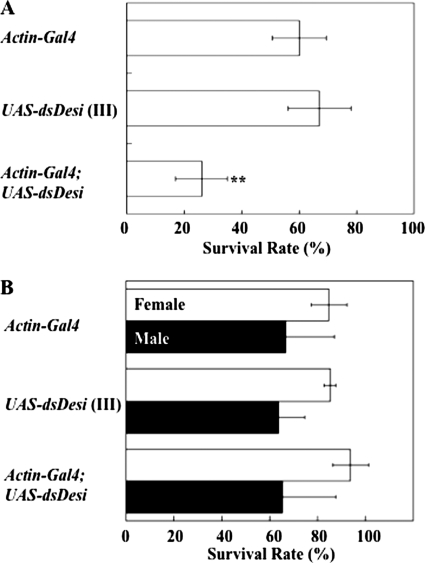
When the Desi knockdown wandering larvae were put in a 35% RH environment, which is lower than the normal humidity of ∼60% RH in which they habitually pupariate, they were able to pupariate, but their mortality rates were significantly higher than those of the control insects before eclosion (Fig. 7A). In contrast, the survival rates of the Desi RNAi adults were not changed under a dry condition (Fig. 7B), indicating that Desi preferentially functions in larvae and pupae for protection from desiccation stress before eclosion.
FIGURE 7.
Effects of Desi knockdown on survival rates of Drosophila pupae and adults. A, survival rates of Desi knockdown pupae under desiccated condition (35% RH) for 6 h. Transgenic line carrying UAS-dsDesi on third chromosome (III) was used for Desi knockdown. Data are given as means for seven separate measurements using 50 larvae each. **, significantly different from the value of control pupae Actin-Gal4 (p < 0.01, Tukey's HSD). B, survival rates of Desi knockdown adults under desiccated condition (15% RH) for 18 h. Note that Desi RNAi did not change survival rates of test adults. Other explanations are as described in A.
Biological Role of Desi
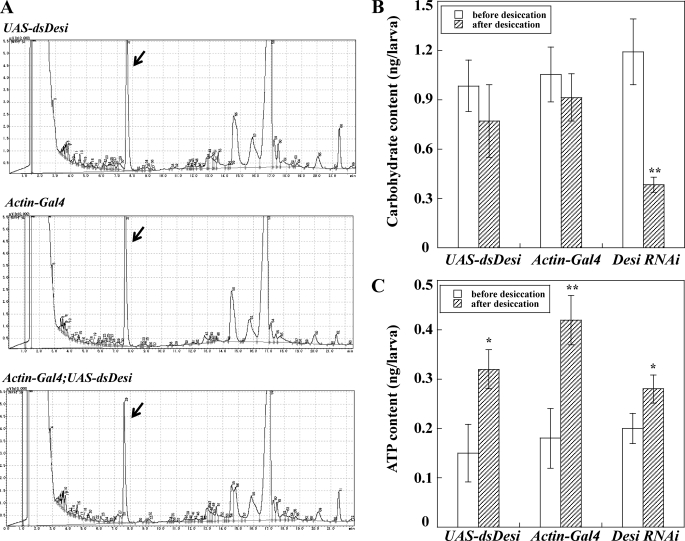
We examined whether the Desi knockdown-induced increase in loss of internal water was due to changes in hydrocarbon components of the cuticular wax layer. Analysis of epicuticular hydrocarbons of control and Desi RNAi larvae did not show any significant difference between the larvae (Fig. 8A). Furthermore, preliminary weight measurements of epicuticle hydrocarbons extracted from control and the RNAi larvae showed no significant difference between both larvae (data not shown), indicating that Desi does not contribute to regulating hydrocarbon synthesis.
FIGURE 8.
Analysis of biological functions of Desi in Drosophila larvae. A, comparison of epicuticular hydrocarbon components between control (UAS-dsDesi and Actin-Gal4) and Desi RNAi larvae. Epicuticular wax was extracted from 100 whole larval bodies of each line. Heneicosane (250 ng) was added as an internal standard as indicated by an arrow. B, carbohydrate contents in control (UAS-dsDesi and Actin-Gal4) and Desi RNAi larvae. Whole larval bodies of each line were homogenized in 1% trichloroacetic acid and the supernatant after centrifugation at 20,000 × g for 10 min at 4 °C was directly used for measurement of carbohydrates. Glucose was used as a standard for calculating carbohydrate concentrations. Data are given as means for five separate measurements using 30 larvae each. **, significantly different from the value of larvae before desiccation (**, p < 0.01, Tukey's HSD). C, ATP contents in control (UAS-dsDesi and Actin-Gal4) and Desi RNAi larvae. * and **, significantly different from the value of larvae before desiccation (*, p < 0.05; **, p < 0.01, Tukey's HSD). Other explanations are as described in B.
Although the excessive loss of internal water of the Desi RNAi larvae is thought to be one of the main causes of their early death, we further investigated changes of metabolites such as carbohydrates and ATP in the Desi RNAi larvae under a dry environment. Quantitative analyses showed that the concentrations of carbohydrates were significantly decreased in the RNAi larvae compared with those of control larvae after desiccation (Fig. 8B). In contrast, ATP concentrations in both the RNAi and control larvae were increased under a dry condition, implying that larvae exposed to the desiccation stress elevated the metabolic rate. However, no significant differences in ATP concentrations were determined between RNAi and control larvae either before or after desiccation (Fig. 8C), suggesting that Desi is not involved in ATP production in response to desiccation.
DISCUSSION
Water conservation is especially essential for the survival of holometabolous insect larvae because they have less lipidic cuticles than pupae and adults. Despite this, Drosophila larvae change their habitat from wet to arid environments during the midthird instar for normal metamorphosis. Although it was recently reported that the sensory neuron of the degenerin/epithelial sodium channel subunit, Pickpocket1, contributes to regulation of the behavioral transition of Drosophila larvae from foraging to wandering stages (18), we do not know how they adapt to the rapid change of habitat from wet to arid conditions. To elucidate the key physiological mechanisms underlying the desiccation resistance that they must acquire around the time of initiation of the wandering stage, we conducted differential display RT-PCR to seek a gene whose expression is enhanced by desiccation. One gene, Desiccate (Desi), that we identified in this study encoded a 261-amino acid single transmembrane protein. The cytoplasmic tail of Desi contains several notable motifs such as SH2 (Src homology 2) domain-binding motifs, a cAMP-dependent protein kinase (protein kinase A) phosphorylation site, and PDZ domain-binding motifs. One of the SH2 domain-binding motifs is a GRB2-like SH2 domain-binding motif (19) and the other is a STAT5-family SH2 domain-binding motif (20). The PDZ domain-binding motifs in Desi are categorized as a class III PDZ domain-binding motif (21). Both binding motifs for SH2 domains and PDZ domains are found in signaling molecules through binding with other component proteins of signal transductions. Furthermore, it has been reported that protein kinase A-induced phosphorylation regulates activities of some receptors, including acetylcholine receptors (22) and N-methyl-d-aspartate receptors (23). Therefore, the presence of the conserved motifs in the Desi cytoplasmic tail suggested the possibility that Desi transmits signals of extracellular desiccation stress to the intracellular signaling pathway. It is worth emphasizing the fact that most of these motifs are conserved in the orthologous genes identified in the EST data bases of mosquitoes, bees, and beetles.
To demonstrate the biological significance of this gene, we established transgenic Drosophila larvae whose Desi expression was either up-regulated by expressing a functional Desi cDNA (UAS-Desi) or down-regulated by expressing Desi dsRNA (UAS-dsDesi). Although a significantly positive effect on desiccation-resistance by overexpression of Desi during the third larval instar was not apparent, the same treatment slightly but significantly increased the survival rates of the second instar larvae under the desiccated condition. This stage-dependent difference in the effect on stress resistance must be due to the difference in the endogenous expression levels of Desi; Desi expression is basically higher in third instar larvae than second instar larvae. Furthermore, it is often observed that larvae repeatedly crawl up and down the food especially at the end of the foraging third instar. Because elevation of Desi expression seemed to have begun already during this stage, it was difficult to detect the positive effect on desiccation-resistance by Desi overexpression in the third instar larvae. Therefore, the overexpression-dependent increase in the survival rates under the desiccated condition occurred only in second instar larvae. Desi knockdown, in contrast, significantly decreased desiccation resistance even during the third larval instar, indicating the contribution of Desi to the protection of Drosophila larvae from desiccation stress. The high mortality rate in Desi RNAi larvae was likely due to increased water loss because the RNAi larvae were the only ones that lost significantly more weight than control larvae after 5 h of desiccation treatment (Fig. 5C). Sørensen et al. (24) reported changes in gene expression in D. melanogaster selected for ecologically relevant environmental stress resistance traits including desiccation, starvation, and cold resistance. However, CG14686 (Desi) was not on the list of genes whose expression was changed in such stress-resistant flies. Furthermore, Telonis-Scott and Hoffmann (25) have reported a desiccation-resistant mutant of D. melanogaster that showed 2-fold higher resistance than control flies when flies were placed under desiccation stress. They had a higher water concentration after desiccation treatment due to decreased rates of water loss but showed no change in body mass, glycogen concentration, hemolymph volume, or the water concentration at death from desiccation. Although the mutant allele is mapped to chromosome 2, specific gene(s) attributed to desiccation resistance have not been identified yet. Although we cannot deny certain functional similarities between the gene(s) mutated in this line and Desi, we recognize a clear difference between the two mutant lines. Although the desiccation resistance of this mutant line was demonstrated using adults, the repressed resistance of Desi transgenic lines to desiccation stresses was demonstrated only in its RNAi larvae. Because there was no significant difference in desiccation resistance between the control and Desi RNAi adults, it is reasonable to expect that the prompt adaptation to desiccation stress in the integument via Desi is more important in larvae than adults.
The proximal causes of mortality from desiccation stress are not well understood. It has been suggested that hemolymph volume and hemolymph solute concentrations are important for the enhancement of desiccation resistance in D. melanogaster (26). Furthermore, Marron et al. (27) reported that the rates of lipid and protein metabolism in Drosophila flies were similar during starvation and desiccation but that carbohydrate metabolism was several times higher during desiccation. This observation is consistent with the previous report that concluded that lower overall rates of water loss in the flies are achieved by reduction of respiratory losses (28). The present study indicated a remarkable decrease in internal water and carbohydrates in the Desi RNAi larvae under a dry condition compared with control larvae. In contrast, ATP concentrations in both the RNAi and control larvae were increased under a dry condition, although no significant differences in ATP concentrations were determined between RNAi and control larvae either before or after desiccation. It is possible to interpret these results to mean that the Desi RNAi larvae lost excess amounts of the internal water needed to enhance the metabolic rate by producing ATP molecules that are used by these animals to maintain homeostasis. To evaluate the desiccation resistance of Drosophila, the importance of recovery processes from desiccation stress was also pointed out. Folk and Bradley (29) showed that the greatest desiccation resistance in a Drosophila population with enhanced desiccation resistance is associated with the restoration of all tested somatic components, whole-body water, dry mass, and sodium concentrations, suggesting the importance of nutrition during rehydration in determining recovery time. Sinclair et al. (6) indicated that up-regulated expression of Frost, a Drosophila stress-responsive gene, was more obvious during recovery from desiccation than during desiccation. These results indicate that temporal factors are involved in desiccation resistance. Although the functional role of Frost was characterized during the adult stage, Desi contributed to enhancing desiccation resistance during the larval stage. To clarify the mechanism underlying the desiccation resistance of insects, it is important to consider the different life stages.
Analyses aimed at revealing the molecular function of Desi will lead to an enhanced understanding of desiccation resistance in adults as well as larvae of Drosophila. The outcome of this type of study can develop a better knowledge of the effects of desiccation stress not only on insects but also on vertebrates.
Supplementary Material
Acknowledgments
We thank T. Tanimura (Kyushu University) for providing the Actin-Gal4 strain and the Bloomington Indiana Stock Center for supplying some of the basic stocks for this study.
This work was supported by a grant-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to Y. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- RH
- relative humidity.
REFERENCES
- 1.Feder M. E., Hofmann G. E. (1999) Annu. Rev. Physiol. 61, 243–282 [DOI] [PubMed] [Google Scholar]
- 2.Sun Y., MacRae T. H. (2005) FEBS J. 272, 2613–2627 [DOI] [PubMed] [Google Scholar]
- 3.Parsell D. A., Lindquist S. (1993) Annu. Rev. Genet. 27, 437–496 [DOI] [PubMed] [Google Scholar]
- 4.Tammariello S. P., Rinehart J. P., Denlinger D. L. (1999) J. Insect Physiol. 45, 933–938 [DOI] [PubMed] [Google Scholar]
- 5.Benoit J. B., Lopez-Martinez G., Phillips Z. P., Patrick K. R., Denlinger D. L. (2010) J. Insect Physiol. 56, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinclair B. J., Gibbs A. G., Roberts S. P. (2007) Insect Mol. Biol. 16, 435–443 [DOI] [PubMed] [Google Scholar]
- 7.Riddiford L. M. (1993) in The Development of Drosophila melanogaster (Bate M., Martinez-Arias A. eds), pp. 899–939, Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 8.Thummel C. S. (2001) Dev. Cell 1, 453–465 [DOI] [PubMed] [Google Scholar]
- 9.Rubin G. M., Spradling A. C. (1982) Science 218, 348–353 [DOI] [PubMed] [Google Scholar]
- 10.Ryuda M., Nakayama H., Hayakawa Y. (2008) Appl. Entomol. Zool. 43, 563–568 [Google Scholar]
- 11.Matsumoto Y., Oda Y., Uryu M., Hayakawa Y. (2003) J. Biol. Chem. 278, 38579–38585 [DOI] [PubMed] [Google Scholar]
- 12.Pfaffl M. W. (2001) Nucleic Acids Res. 29, 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayakawa Y., Ohnishi A., Mizoguchi A., Yamashika C. (2000) Cell Tissue Res. 300, 459–464 [DOI] [PubMed] [Google Scholar]
- 14.Oda Y., Matsumoto H., Kurakake M., Ochiai M., Ohnishi A., Hayakawa Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15862–15867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marican C., Duportets L., Birman S., Jallon J. M. (2004) Insect Biochem. Mol. Biol. 34, 823–830 [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa Y., Chino H. (1982) Insect Biochem. 12, 361–366 [Google Scholar]
- 17.Stanley P. E., William S. G. (1969) Anal. Biochem. 29, 381–392 [DOI] [PubMed] [Google Scholar]
- 18.Ainsley J. A., Kim M. J., Wegman L. J., Pettus J. M., Johnson W. A. (2008) Dev. Biol. 322, 46–55 [DOI] [PubMed] [Google Scholar]
- 19.Chardin P., Cussac D., Maignan S., Ducruix A. (1995) FEBS Lett. 369, 47–51 [DOI] [PubMed] [Google Scholar]
- 20.Beisenherz-Huss C., Mundt M., Herrala A., Vihko P., Schubert A., Groner B. (2001) Mol. Cell Endocrinol. 183, 101–112 [DOI] [PubMed] [Google Scholar]
- 21.Fanning A. S., Anderson J. M. (1996) Curr. Biol. 6, 1385–1388 [DOI] [PubMed] [Google Scholar]
- 22.Nishizaki T., Sumikawa K. (1998) Brain Res. 812, 242–245 [DOI] [PubMed] [Google Scholar]
- 23.Crump F. T., Dillman K. S., Craig A. M. (2001) J. Neurosci. 21, 5079–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sørensen J. G., Nielsen M. M., Loeschcke V. (2007) J. Evol. Biol. 20, 1624–1636 [DOI] [PubMed] [Google Scholar]
- 25.Telonis-Scott M., Hoffmann A. A. (2003) J. Insect Physiol. 49, 1013–1020 [DOI] [PubMed] [Google Scholar]
- 26.Folk D. G., Han C., Bradley T. J. (2001) J. Exp. Biol. 204, 3323–3331 [DOI] [PubMed] [Google Scholar]
- 27.Marron M. T., Markow T. A., Kain K. J., Gibbs A. G. (2003) J. Insect Physiol. 49, 261–270 [DOI] [PubMed] [Google Scholar]
- 28.Gibbs A. G., Fukuzato F., Matzkin L. M. (2003) J. Exp. Biol. 206, 1183–1192 [DOI] [PubMed] [Google Scholar]
- 29.Folk D. G., Bradley T. J. (2004) J. Exp. Biol. 207, 2671–2678 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.