Abstract
Chemical genetics is an emerging approach to investigate the biology of host-pathogen interactions. We screened several inhibitors of ATP-dependent DNA motors and detected the gyrase B inhibitor coumermycin A1 (C-A1) as a potent antiretroviral. C-A1 inhibited HIV-1 integration and gene expression from acutely infected cell, but the two activities mapped to distinct targets. Target discovery identified Hsp90 as the C-A1 target affecting viral gene expression. Chromatin immunoprecipitation revealed that Hsp90 associates with the viral promoter and may directly regulate gene expression. Molecular docking suggested that C-A1 binds to two novel pockets at the C terminal domain of Hsp90. C-A1 inhibited Hsp90 dimer formation, suggesting that it impairs viral gene expression by preventing Hsp90 dimerization at the C terminus. The inhibition of HIV-1 integration imposed by C-A1 was independent of Hsp90 and mapped to the capsid protein, and a point mutation at residue 105 made the virus resistant to this block. HIV-1 susceptibility to the integration block mediated by C-A1 was influenced by cyclophilin A. Our chemical genetic approach revealed an unexpected function of capsid in HIV-1 integration and provided evidence for a role of Hsp90 in regulating gene expression in mammalian cells. Both activities were amenable to inhibition by small molecules and represent novel antiretroviral drug targets.
Keywords: DNA Gyrase, Drug Action, Heat Shock Protein, HIV, Protein Targeting, Coumermycin A1, Hsp90, Integration, Capsid, Chemical Genetics
Introduction
The AIDS epidemic is a major global health problem with an estimated 33.2 million people infected with HIV, mainly in sub-Saharan Africa (1). No effective vaccine is currently available. Highly active antiretroviral therapy is very effective (2) at controlling HIV-1 replication; however, multiple drug resistance and long term persistence of drug-resistant viral reservoirs have been reported (3, 4), making the emergence of secondary epidemics of multidrug-resistant HIV-1 a real long term threat (3–5).
Drugs that target cellular factors necessary for virus replication might be less vulnerable to virus mutation than current antiviral drugs (6). Moreover, most approved drugs already target cellular factors, and if some of these factors turn out to be relevant for HIV-1 replication, it might be possible to exploit existing patented and generic drugs as antiretrovirals with considerable cost savings and clear advantages for resource-poor countries.
This approach depends on the identification of cellular factors involved in HIV-1 replication. Indeed, several studies have recently used high throughput genetic approaches to discover HIV-1 host dependence factors (7). Critically, however, host dependence factors must also be susceptible to inhibition by small molecules to become drug targets. Chemical genetics is an approach whereby small molecules are first screened to find “hits” with the desired phenotype, and the hit molecule is then used as a tool to identify the target (8). Any host factor identified by this approach is, by definition, a drug target. In addition to the more applicable side of this type of screening, chemical genetics may reveal the involvement of unexpected host components and pathways in HIV-1 replication leading to more fundamental discoveries on the biology of mammalian cells.
Here, we used a chemical genetic approach and screened several well characterized small compounds targeting DNA-dependent motor proteins with the rationale that they might inhibit HIV-1 nuclear trafficking or integration, steps that require rearrangement of viral and/or chromosomal DNA (9, 10). We have found that the coumarin antibiotic C-A1,3 a gyrase B (gyrB) inhibitor, impairs both HIV-1 integration and gene expression and that the two activities map to distinct targets. Notably, we found that Hsp90 is the target for C-A1-mediated inhibition of gene expression and that the virus susceptibility to the C-A1-mediated integration block maps to the capsid protein. Our results revealed novel functions of capsid in HIV-1 integration and of Hsp90 in viral gene expression.
EXPERIMENTAL PROCEDURES
Reagents, Viruses, and Cells
Blood was obtained from healthy volunteers after written informed consent according to the approved protocol of the University College London Ethics Committee (reference 0335/001) or from buffy coats obtained from the National Health Service National Blood Service according to governmental ethics regulations. C-A1, novobiocin, etoposide, daunorubicin, doxorubicin, β-lapachone, and GA were purchased from Sigma; 17-AAG was from Enzo Life Sciences. HeLa, 293T, HT1080, and mTOP cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 2 mm glutamine at 37 °C in 5% CO2. Jurkat, SupT1, and C8166 cells were grown in RPMI 1640 medium supplemented with 10% FCS at 37 °C in 10% CO2. The HIV-1 vector (11) was made and purified as described previously (12). HIV primary isolates SF-162, YU-2, and BaL were produced by FuGENE transfection into 293T cells, and supernatant containing viral particles was collected 48 h post-transfection. Reverse transcriptase (RT) activity was measured by the Lenti-RTTM activity assay (Cavidi Tech) following the manufacturer's instructions. PBMCs were isolated from buffy coats from healthy donors by standard Ficoll-Paque density centrifugation and transferred into RPMI 1640 medium containing 10% FCS. PBMCs were activated with 2 μg/ml phytohemagglutinin for 48 h; media were changed, and cells were infected and grown for 3 days in the presence of 10 units/ml IL-2. Macrophages were isolated from buffy coats from healthy donors by standard Ficoll-Paque density centrifugation. Cells were incubated at a density of 5 × 104/well in a volume of 100 μl in 96-well plates for 72 h in the presence of 20 ng/ml M-CSF before media change and infection at day 5.
Hydrolysis of C-A1
To C-A1 (10 mg, 9.92 μmol) in tetrahydrofuran (0.5 ml) was added a solution of lithium hydroxide (50 μl of a 100 mg/ml solution in water). The reaction was stirred overnight at room temperature. Acetic acid (100 μl) was added to stop the reaction. The product was isolated by preparative HPLC (C18, gradient acetonitrile/water, 0.1% formic acid). The monoester product was characterized by LC-MS (ESI, -ve) m/z 1001 (monoester), purity >97%, yield 8.05 mg, 8.5 μmol at 85%.
Infection Assays
Approximately 4 × 105 adherent cells or 0.5 × 106 lymphocytic cells were plated in 24-well plates in 500 μl of media, incubated for 6 h with the compounds, and infected at a m.o.i. ranging from 0.02 to 0.08 using a VSV-G pseudotyped HIV-1 vector or HIV-1 LAIΔenv purified in a sucrose step gradient. Infected cells were incubated with the compounds for 24 h, washed, and analyzed by FACS. Total DNA was extracted from an aliquot of infected cells and analyzed by TaqMan qPCR. In some experiments, 1 aliquot of infected cells was analyzed 24 h post-infection and another aliquot 2 weeks later. For infection with wild type HIV-1, 5 × 105 lymphocytic cells were plated into 24-well plates in 500 μl of medium and cultured in the presence of the compounds for 6 h. The culture was transferred to 96 U-shaped well plates in 100-μl aliquots and infected at an m.o.i. of 0.2. For experiments with C-A1, media were replaced without the compound 24 h later; for experiments with GA in primary macrophages, media were replaced without GA 24 h post-infection; and in PBMCs, media containing GA were replaced daily for 2 days. Cells were grown for 48–72 h, washed once in serum-free medium, fixed in 50% methanol, 50% acetone for 2 min at −20 °C, and immunostained as described previously (13) using anti HIV-1 p24 Ab (38:96 K and EF7 at 1:1 ratio, AIDS Reagent Programme), and secondary anti-mouse Abs were conjugated to β-galactosidase (Southern Biotechnology Associates, Inc.) diluted 1:400. Alternatively, HIV-infected cells were fixed for 20 min at room temperature in 100 μl of 4% paraformaldehyde in PBS, washed and permeabilized in 100 μl of cytofix/cytoperm solution (BD Biosciences) for 30 min at 4 °C. The same primary anti-p24 Ab was detected by an anti-mouse immunoglobulin FITC conjugated and diluted 1:200, and cells were analyzed by FACS. Macrophages immunostained for p24 were counted using an MRX TC Revelation microplate reader (Dynex Technologies).
Hsp90 Knockdown
The following siRNA sequences were used: Hsp90 A2, TCC CGA CGA TAT TAC TAA TGA; Hsp90 A3, AAC ATA TCC CGT GAG ATG TTG TT; Hsp90 B1, GGA GAT TTT CCT TCG GGA GTT. Target and control scramble siRNAs were from Dharmacon. 40 μl/well of Opti-MEM medium containing 50 nm each siRNA (final concentration) were added to 2 μl of Oligofectamine (Invitrogen) previously mixed with 5.5 μl of Opti-MEM, and the mixture was incubated for 15–20 min at room temperature. The siRNA-containing mixture was transferred to 24-well plates, and 2 × 105 HeLa cells/well were subsequently added to a final volume of 500 μl/well. Cells were analyzed by Western blotting to confirm Hsp90 KD.
Docking Studies
For the docking studies, the following software programs were used: FRED (Fast Rigid Exhaustive Docking) version 2.2.4 (2008); FRED_receptor version 2.2 (2006); Omega 2.3.2 (2008) (all from Open Eye Scientific Software Inc., Santa Fe, NM 87507) and MOE version 2008.10 (2008) (Chemical Computing Group, Montreal, Quebec, Canada). For details on the procedure please see supplemental methods.
Site-directed Mutagenesis
Point mutations were introduced into the original NL4.3 HIV-1 backbone by site-directed mutagenesis using the QuikChange II XL kit (Stratagene) following the manufacturer's instructions with primers D31-GAG (CCA AGG GGA AGT GAC ATA GCA GGA ACT ACT AGT AC) and D31-NEF (TCA TTG GTC TTA AAG GTA CGT GAG GTG TGA CTG GAA AAC). Four clones for each mutant were sequenced and tested in infection assays.
Western Blot
Rabbit polyclonal antibody H-114 against Hsp90 α/β was purchased from Santa Cruz Biotechnology and used at 1:200 dilution. mAb TOP2A11-A against Topo2 α was purchased from Alpha Diagnostics and used at 1:100 dilution. mAb A300-949A against Topo2 β was purchased from Bethyl Laboratories and used at 1:2000 dilution. mAb 2011-1 against Topo2 α/β was purchased from Topogen and used at 1:2500 dilution. Secondary anti-mouse and anti-rabbit IgG HRP-conjugated antibodies were purchased from DAKO. After SDS-PAGE, the proteins were transferred to PVDF membranes (Bio-Rad) by semi-dry transfer and probed with the primary antibodies. HRP-conjugated secondary antibodies were used diluted to 1:5000 in PBS containing 10% nonfat milk. Chemiluminescence (ECL, Amersham Biosciences) was used to develop the blots as described by the manufacturer. Autoradiography films were exposed for different periods of time to ensure linearity of the signal.
TaqMan qPCR and RT-qPCR
Approximately 1 × 106 cells were washed twice in PBS, and total DNA was extracted with the QIAamp DNA minikit (Qiagen, UK). Quantitative PCRs were carried out as described previously (14–16) in a 25-μl volume containing 100–300 ng of total DNA using an ABI Prism 7000 sequence detection system. For mRNA quantification, nucleic acids were extracted from 1 × 107 SupT1 cells using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Samples were treated with 2 units/μg ReQ1 DNase in 1× buffer (Promega) for 30 min at 37 °C, and 2 mm EGTA was then added to stop the reaction, and the samples were incubated at 60 °C for 20 min. The Superscript III kit (Invitrogen) was used for first-strand cDNA synthesis by random hexamers following the manufacturer's instructions. Control samples were incubated in parallel without RT. TaqMan quantitative PCR was performed in an ABI Prism 7000 thermocycler with the GFP primers/probe set, and the 18 S ribosomal RNA primers/probe set as follows: 18 S rRNA probe 5′-FAM-AGT CCA CTT TAA ATC CTT-TAMRA-3′; 18 Sr qPCR forward, 5′-TCG AGG CCC TGT AAT TGG AA-3′, and 18 Sr reverse qPCR RC 5′-CCC TCC AAT GGA TCC TCG TT-3′.
Alu-PCR
Analysis of Alu-LTR copies was performed on total cellular DNA using two rounds of amplification (17, 18). The first round was performed in a final volume of 20 μl containing 100 μm of each dNTP, 8 mm MgCl2, 50 nm of Alu-LTR forward primer, 150 nm of Alu-LTR1 reverse primer, and 500 ng of total DNA. Primers sequences for the first round of amplification were as follows: Alu-LTR forward, AAC TAG GGA ACC CAC TGC TTA AG, and LTR1 reverse, TGC TGG GAT TAC AGG CGT GAG (19). The cycle parameters were as follows: 95 °C for 8 min, 95 °C for 10 s, 52 °C for 1 min, and 72 °C for 3 min for 15 cycles followed by 72 °C for 10 min. Samples were then diluted 1:20, and 2 μl were used to quantify the amount of Alu-LTR copies by real time qPCR as described above. Primers sequences were as follows: Alu-LTR forward, AAC TAG GGA ACC CAC TGC TTA AG; LTR2 reverse, TGC TAG AGA TTT TCC ACA CTG ACT; Alu-LTR probe, FAMRA-TAG TGT GTG CCC GTC TGT TGT GTG AC-TAM. The standard curve ranging for 105 to 10 copies was prepared using total DNA extracted from infected cells after quantification of the number of integrants by TaqMan qPCR.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as described previously (20). Briefly, 3 × 107 CEM cells were infected with the VSV-G pseudotyped HIV-1 vector at an m.o.i. of 0.3. Twenty four hours later, cells were chemically cross-linked by the addition of 1/10th volume of fresh 11% formaldehyde solution added directly to cell culture media and incubated for 20 min at room temperature followed by the addition of 1/20th volume of 2.5 m glycine. Cells were then transferred to a 50-ml tube and collected by centrifugation at 4 °C, and the pellet was rinsed twice with PBS and flash-frozen in liquid N2. Cells were resuspended and lysed in lysis buffer 1 (50 mm Hepes-KOH, pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Nonidet P-40, 0.25% Triton X-100), and the nuclei were then washed in 200 mm NaCl, 1 mm EDTA, and lysed in lysis buffer 2 (10 mm Tris, pH 8, 100 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 0.1% sodium deoxycholate, 0.5% N-laurylsarcosine), and sonicated using a Misonix Sonicator 3000, power set to 24 watts, 10 times, 30-s pulses (90 s pause between pulses) at 4 °C. After removal of the insoluble material, the resulting whole-cell extract (WCE) was incubated overnight at 4 °C with 100 μl of Dynal protein-G magnetic beads preincubated with 10 μg of the appropriate antibody for 6–8 h on rotating platform (9 rpm) in a cold room. Three different antibodies were used as follows: mAb against Hsp90 α/β (Axxora) (21), mAb against RNA pol II (8WG16, Abcam), and an unspecific mouse IgG antibody (Santa Cruz Biotechnology). The next day beads were washed six times with RIPA buffer and once with TE containing 50 mm NaCl. Bound complexes were eluted from the beads by heating at 65 °C with occasional vortexing, and cross-linking in the immunoprecipitation and WCE samples was reversed by incubating at 65 °C for 6–7 h. Immunoprecipitation and WCE DNA were then purified by treatment with RNase A, proteinase K and extracted with phenol/chloroform/isoamyl alcohol extractions. ChIP product was quantified by TaqMan qPCR (for details see supplemental Methods).
Sequencing of Parental and Resistant Viruses
The sequences of parental and C-A1-resistant NL4-3 viruses were obtained by PCR amplification of 15 different overlapping segments covering the entire viral genome. Total DNA was extracted from chronically infected cells using the Qiagen DNA extraction kit. Conditions of the PCR and primer sequences were as described previously (16).
RESULTS
We initially hypothesized that ATP-dependent DNA motor proteins might be involved in HIV-1 nuclear import and integration, two steps where structural rearrangements of the viral DNA within the preintegration complex or host chromatin may be required (9, 10). To test this hypothesis, we used a small focused library of relatively well characterized compounds targeting DNA topoisomerase I (Topo1), topoisomerase II (Topo2), and DNA helicases (Fig. 1A) (22–24). Inhibitors of Topo1 and DNA helicases were either too toxic or were weak inhibitors of HIV-1 infection; however, the gyrase B inhibitor coumermycin A1 showed potent antiretroviral activity (Fig. 1A). Novobiocin, which is a coumarin antibiotic structurally related to C-A1, had antiretroviral activity but lower than C-A1 (Fig. 1, A and B).
FIGURE 1.
Antiretroviral activity of C-A1. A, HIV-1 vector and HIV-1 LAI Δenv (VSV-G pseudotyped) virus were tested in SupT1 cells in single cycle assays; HIV-1 NL4.3 was tested in C8166 cells. Infection efficiency was measured by flow cytometry to detect GFP+ cells (HIV-1 vector and HIV-1 LAI Δenv) 24–48 h post-infection or by p24 immunostaining (HIV-1 NL4.3) 72 h post-infection. IC50, dose required to inhibit infection by 50%. CC50, dose inducing death of 50% of the cells. Mean values (n = 2) or mean values ± S.D. (n = 3) are shown. B, chemical structure of novobiocin and C-A1. C, PBMCs infected with HIV-1 SF-162 in the presence of C-A1 and analyzed 72 h post-infection by p24 capsid immunostaining. D, PBMCs infected with three HIV-1 primary isolates in the presence of C-A1 and analyzed 72 h post-infection by p24 CA immunostaining. NI, could not be infected; ND, not done.
C-A1 had a good selectivity index (Fig. 1A), and it modestly increased the proportion of cells in G1 phase of the cell cycle and did not significantly affect growth kinetics (supplemental Fig. S1). Thus, general cell toxicity could not explain its antiviral activity. C-A1 inhibited infection of VSV-G pseudotyped HIV-1 vector (11) and HIV-1 LAIΔenv virus (a near full-length virus with a deletion in the envelope gene) (25) with a similar IC50 but was more potent against wild type HIV-1 NL4.3 (Fig. 1A). C-A1 was tested in human PBMCs infected with three different CCR5-tropic HIV-1 isolates. In four donors C-A1 reduced HIV-1 infection with an IC50 ranging from <50 to 160 nm, whereas in one donor the IC50 ranged from 500 to 2000 nm (Fig. 1, C and D) with no loss of cell viability as detected by trypan blue staining. The lower sensitivity of donor 3 might be due to faster metabolism or poor uptake of C-A1 by this donor's PBMCs. Polymorphisms in genes involved in drug metabolism are well known to influence drug activity (26).
C-A1 Inhibits Two Steps of the HIV-1 Life Cycle
C-A1 was previously shown to have antiretroviral activity, but little is known about its mechanisms of action (27, 28). We performed time of addition (ToA) experiments to examine which step of the replicative cycle was blocked by C-A1. The rationale behind this experiment is that the compound cannot block a step of the viral life cycle if added after the step has been completed. Inhibitors of specific steps (entry, reverse transcription, and integration) were used in parallel to define the temporal dynamics of each step (29). To test if C-A1 blocked virus entry, cells were infected with wild type HIV-1 NL4.3 in the presence of C-A1 or the entry inhibitor AMD3100 (30). AMD3100 potently blocked HIV-1 infection only when added 2 h before or at the time of infection, consistent with its action as an entry inhibitor. In contrast, C-A1 blocked HIV-1 infection even if added at later time points, demonstrating that it interfered with a post-entry step (Fig. 2A). The reverse transcription inhibitor azidothymidine (AZT) lost most of its activity when added 6 h post-infection. The integration inhibitor Raltegravir (31) lost activity when added 8 h post-infection. In contrast, C-A1 was still fully active 8 h post-infection, suggesting that it blocked a step after integration (Fig. 2B).
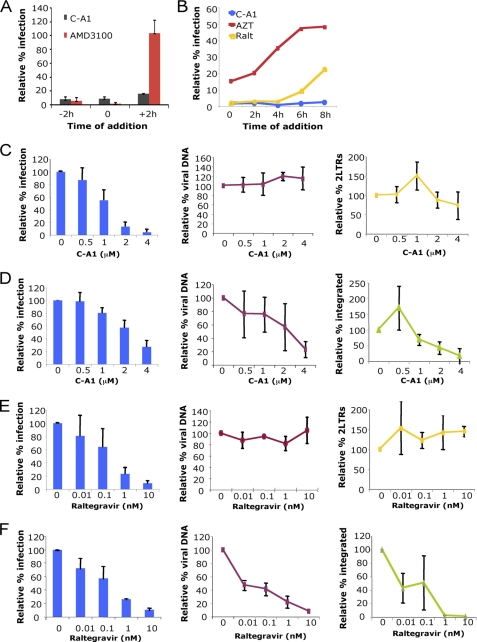
FIGURE 2.
C-A1 blocks HIV-1 integration. A, C8166 cells incubated with C-A1 (2 μm) or AMD3100 (10 nm), added at the indicated time points, infected with HIV-1 NL4.3, and analyzed by FACS 48 h post-infection after intracellular p24 CA immunostaining. Mean values ± S.D. are shown, n = 3. B, SupT1 cells infected with a VSV-G pseudotyped HIV-1 vector in the presence of 8 μm azidothymidine, 100 nm Raltegravir, or 4 μm C-A1; drugs were added at the indicated time post-infection and cells analyzed by FACS 24 h later. Data are representative of two independent experiments. C, SupT1 cells infected with a VSV-G pseudotyped HIV-1 vector and analyzed 24 h post-infection by FACS (left panel) and TaqMan qPCR to detect total viral DNA (middle panel) or two LTR circular viral DNAs (right panel). Mean values ± S.D. are shown, n = 3. Heat-inactivated virus gave no detectable signal. D, same cells shown in C were re-analyzed 2 weeks later by FACS (left panel), TaqMan qPCR to measure total viral DNA (middle panel), and Alu-LTR qPCR (right panel) to measure integrated proviruses. Mean values ± S.D. are shown, n = 3. E and F, same experiments as in C and D, but cells were treated with the indicated doses of Raltegravir for 24 h. Mean values ± S.D. are shown, n = 3.
We sought to confirm these results by directly measuring reverse transcription and integration. C-A1 did not significantly inhibit viral DNA synthesis or accumulation of two LTR viral DNAs, a hallmark of nuclear entry (32), as detected by TaqMan qPCR 24 h post-infection (Fig. 2C). To examine integration, the same cells were re-analyzed 2 weeks later by FACS, TaqMan qPCR to measure viral DNA copies, and Alu-LTR qPCR to directly measure integrated proviruses (Fig. 2D) (17). Control experiments were performed in parallel with Raltegravir (Fig. 2, E and F). Importantly, these experiments showed that C-A1 inhibited HIV-1 integration in a dose-dependent manner (Fig. 2D). The number of viral DNA copies and Alu-LTR integrated proviral copies matched, indicating that most nonintegrated viral DNA is lost 2 weeks post-infection in dividing cells (Fig. 2D). Therefore, in our experimental conditions, given that reverse transcription and nuclear entry proceeded normally, any difference in viral DNA copy number between 24 h and 2 weeks post-infection represented an integration defect.
However, ToA experiments showed that C-A1 still inhibited HIV-1 infection when Raltegravir was losing activity; hence, the integration block was unlikely to fully account for the antiviral effect of C-A1. We hypothesized that C-A1 might interfere with another step of HIV-1 replication in addition to integration, but untangling the two inhibitory activities posed a challenge. To further investigate this possibility, we analyzed the effect of C-A1 on different cell types. The rationale behind this approach was that C-A1 might induce different responses in different cell types and preferentially block one HIV-1 replicative step over another. Several cell lines were treated with the compound, infected with an HIV-1 vector, and analyzed 24 h and 2 weeks later, as described earlier. C-A1 inhibited infection as measured by FACS in all cell types at 24 h without significantly affecting viral DNA synthesis (Fig. 3A, left panels). However, the compound significantly inhibited integration in Jurkat and SupT1, modestly in HeLa, but not in the other cell types tested (Fig. 3A, right panels). This differential susceptibility indicated that C-A1 did indeed impose two distinct blocks to HIV-1 infection and that the two blocks mapped to separate targets.
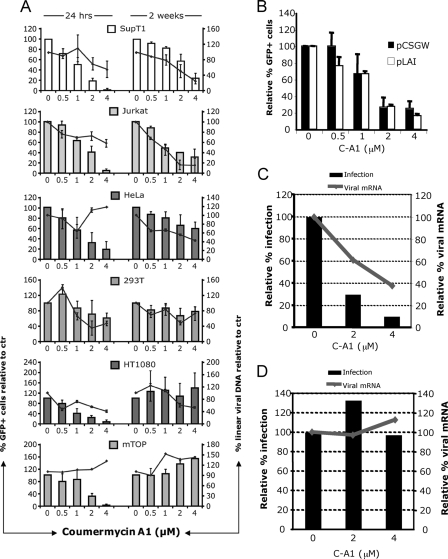
FIGURE 3.
C-A1 exerts an additional block to HIV-1 infection. A, SupT1 (human CD4+ lymphoblastic leukemia), Jurkat (human CD4+ acute lymphoblastic leukemia), HeLa (human epithelial cervix adenocarcinoma), 293T (human embryonic kidney epithelial), HT1080 (human fibrosarcoma), and mTOP (mouse fibroblasts) cells were infected with a VSV-G pseudotyped HIV-1 vector (at m.o.i. of 0.05) and analyzed by FACS (histograms, left y axis) and TaqMan qPCR (line, right y axis) 24 h and then again 2 weeks post-infection. Mean values ± S.D. are shown, n = 3. B, 293T cells transfected with HIV-1 LAIΔenv or the HIV-1 vector plasmids in the presence of the indicated doses of C-A1 and analyzed by FACS 24 h post-transfection. Mean values ± S.D. are shown, n = 3. C, HT1080 cells were infected with VSV-G pseudotyped HIV-1 LAIΔenv and analyzed 24 h later by FACS (bars, left y axis) or by RT-qPCR to measure viral mRNA levels (line, right axis). Viral RNA values were normalized to 18 S RNA copy number. D, SupT1 cells chronically infected with VSV-G pseudotyped HIV-1 LAIΔenv (at m.o.i. of 0.1) were incubated with C-A1 and analyzed 24 h later by FACS (bars, left y axis) or by RT-qPCR to measure viral mRNA levels (line, right axis). Viral RNA values were normalized to 18 S RNA copy number. Control reactions without RT gave undetectable readings. Results are representative of two independent experiments.
To examine if C-A1 impaired viral gene expression, 293T cells were transfected with the HIV-1 LAIΔenv and the HIV-1 vector plasmids in the presence of C-A1 and analyzed by flow cytometry 24 h later. C-A1 significantly reduced gene expression from both plasmids in a dose-dependent way (Fig. 3B). Viral gene expression levels were also examined in HT1080 cells, which were insensitive to the integration block, after acute infection with HIV-1 LAIΔenv. Infection efficiency and viral mRNA levels decreased in a dose-dependent way, consistent with an effect of C-A1 on gene expression (Fig. 3C). We then tested the effect of C-A1 in chronically infected cells, but in this case neither infection nor viral mRNA levels decreased (Fig. 3D), excluding that C-A1 was an unspecific inhibitor of RNA Pol II at this concentration range. Thus, C-A1 impaired both integration and viral gene expression at an early stage post-infection.
Identification of the C-A1 Targets Mediating Inhibition of HIV-1 Infection
The coumarin antibiotics have been shown to bind to the N-terminal ATPase of bacterial gyrB (33). Interestingly, the ATPase of gyrB belongs to the GHKL family, which includes GyrB, Hsp90, histidine kinases, and MutL proteins and their orthologues sharing the so-called Bergerat fold (34). To find candidate targets of C-A1 mediating the blocks to HIV-1 infection, we searched the Uniprot data base for human proteins sharing the GHKL domain and found 12, plus several members of the PMS family (Table 1). The MORC protein was excluded because it is only expressed in germ cells (35). Members of the DNA mismatch repair system (MLH1, PMS1, and PMS-related proteins) were given low priority because several cell lines lacking them support normal HIV-1 replication (36). Topo2, on the other hand, was considered a likely C-A1 target because of its homology to gyrB (37). However, we could not detect any role of Topo2 in HIV-1 infection (supplemental Fig. S2).
TABLE 1.
Proteins having the consensus GHKL ATPase domain in the UniProtKB database (Homo sapiens)
| Target | Activity | Location |
|---|---|---|
| DNA topoisomerase II | Disentangling of chromatin DNA | Nuclear |
| Hsp90 (class A1, A2, and B2) | Heat shock chaperone protein | Cytosolic |
| TRAP1 | Tumor necrosis factor-associated protein (Hsp90 family) | Mitochondrial |
| Gp96 | Heat shock protein | ER |
| MORC | Nuclear protein required for spermatogenesis | Nucleus (germ cells) |
| PDK family | Kinase responsible for the inactivation of the pyruvate dehydrogenase | Mitochondrial matrix |
| BCKDK | Branched chain ketoacid dehydrogenase kinase | Mitochondrial |
| MutL homolog 3 | DNA mismatch repair | Nuclear |
| PMS1 | DNA mismatch repair | Nuclear |
| MLH1 | DNA mismatch repair | Nuclear |
| PMS family | Postmeiotic segregation protein (mismatch repair proteins) | Nuclear |
| Mu-RMS-40.5/.10 | Hypothetical rhabdomyosarcoma antigen (mismatch repair proteins) | Nuclear |
Because the GHKL family includes the Hsp90 ATPase (38) and there is good biochemical evidence showing that the coumarin antibiotics novobiocin and C-A1 are Hsp90 inhibitors (39–41), we tested if Hsp90 was involved in HIV-1 infection. To this end, we performed a parallel chemical screen using GA and 17-AAG, two selective inhibitors of Hsp90 that compete for ATP binding on the N-terminal ATPase (38, 42, 43). A ToA experiment showed that GA and 17-AAG inhibited HIV-1 LAIΔenv infection in single cycle assays even when added 8 h post-infection, suggesting that they imposed a gene expression block (Fig. 4A). GA and 17-AAG inhibited virus infection without any loss of cell viability with an IC50 of 168 ± 98 and 241 ± 67 nm, respectively, but did not impair RT or integration (Fig. 4, B and C). 17-AAG inhibited wild type HIV-1 infection in primary human macrophages and PBMCs more potently than in T-cell lines with no detectable toxicity (Fig. 4D). To understand the basis for the greater antiviral potency of 17-AAG in primary cells, the levels of Hsp90 were examined by Western blotting. Primary cells expressed substantially less Hsp90 compared with T-cell lines (Fig. 4E), which explained their greater sensitivity to 17-AAG.
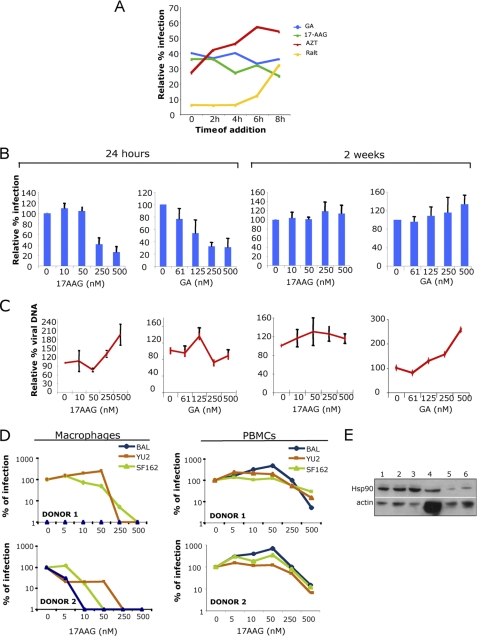
FIGURE 4.
Inhibition of Hsp90 by selective small molecules recapitulates the phenotype of C-A1. A, SupT1 cells infected with a VSV-G pseudotyped HIV-1 vector in the presence of 8 μm azidothymidine, 100 nm Raltegravir, 1 μm GA, or 17-AAG; drugs were added at the indicated time post-infection and cells analyzed by FACS 24 h later. Data are representative of two independent experiments. B, SupT1 cells infected with a VSV-G pseudotyped HIV-1 vector in the presence of the indicated dose of 17-AAG or GA and analyzed by FACS 24 h or 2 weeks post-infection. Mean values ± S.D. are shown, n = 3. C, same cells were analyzed by TaqMan qPCR 24 h and 2 weeks post-infection to detect viral DNA. Mean values ± S.D. are shown, n = 3. D, primary macrophages (left panels) and PBMCs (right panels) were incubated with 17-AAG for 6 h and infected with three HIV-1 primary isolates. Infected cells were immunostained for p24 and automatically counted 72 h post-infection. E, Western blot to detect Hsp90 levels in cell lines and primary cells. Lane 1, Jurkat cells; lane 2, CEM cells; lane 3, SupT1 cells; lane 4, primary macrophages; lane 5, PBMCs; lane 6, 2× PBMCs.
To confirm its role in HIV-1 infection, we transiently knocked down (KD) Hsp90 in HeLa cells by siRNA. Hsp90 was depleted by siRNA to levels that did not induce cell toxicity, and a proportional reduction in HIV-1 infection efficiency was observed in KD cells compared with control cells (Fig. 5, A and B). Moreover, depletion of Hsp90 produced a sensitization to the antiretroviral activity of both C-A1 and GA, supporting the notion that Hsp90 was the target of C-A1 (Fig. 5, B and C) (44). To test if inhibition of Hsp90 impaired viral gene expression, 293T cells were transfected with the HIV-1 LAIΔenv plasmid and the HIV-1 vector plasmid in the presence of GA and analyzed 24 h later by FACS. A dose-dependent inhibition of gene expression was detected (Fig. 5D). SupT1 cells were infected with HIV-1 LAIΔenv in the presence of GA and analyzed 24 h later. Infection efficiency and viral mRNA levels decreased in the presence of the compound, consistent with an effect of GA on gene expression (Fig. 5E). In contrast, chronically infected cells exposed to GA did not show any detectable decrease in HIV-1 infection and viral mRNA levels (Fig. 5F). This phenotype was remarkably similar to the gene expression defect induced by C-A1. To test if Hsp90 could regulate gene expression by associating with the viral promoter, we performed ChIP (Fig. 5, G–J) on cells acutely infected with the HIV-1 vector. Two regions were probed, the promoter in the 5′ LTR and the GFP region, located in proximity of the internal promoter of the viral vector. Antibodies against RNA pol II effectively precipitated both actin DNA, a housekeeping gene, and viral DNA, whereas anti-Hsp90 antibodies only precipitated viral DNA (Fig. 5, H–J), demonstrating the specificity of the procedure. These results indicated that Hsp90 was associated with the viral DNA and supported a role of Hsp90 in regulating viral gene expression.
FIGURE 5.
Hsp90 promotes HIV-1 gene expression. A, Western blot to detect Hsp90 protein levels in HeLa cells following siRNA treatment. B, Si-RNA treated cells were infected with a VSV-G pseudotyped HIV-1 vector with or without C-A1 and GA and analyzed by FACS 24h post-infection. Mean values ± S.D. are shown, n = 4. Paired Student's t test was used to assess statistical significance. Scr, cells treated with scramble siRNA; KD, cells treated with siRNAs targeting Hsp90. C, fold inhibition of HIV-1 vector infection in Hsp90 KD cells relative to control cells in the presence of 2 μm C-A1 or 250 nm GA (n = 4). D, 293T cells transfected with HIV-1 LAIΔenv and the HIV-1 vector (CSGW) plasmids and analyzed by FACS 24 h post-transfection. Mean values ± S.D. are shown, n = 3. E, SupT1 cells were infected with VSV-G pseudotyped HIV-1 LAIΔenv in the presence of the indicated dose of GA and analyzed 24 h later by FACS (bars, left y axis) or by RT-qPCR to measure viral mRNA levels (line, right axis). Viral RNA values were normalized to 18 S RNA copy number. F, SupT1 cells chronically infected with VSV-G pseudotyped HIV-1 LAIΔenv (at m.o.i. of 0.1) were incubated with GA and analyzed 24 h later by FACS (bars, left y axis) or by RT-qPCR to measure viral mRNA levels (line, right axis). Viral RNA values were normalized to 18 S RNA copy number. Control reactions without RT gave undetectable readings. Results are representative of two independent experiments. G, Western blot to detect Hsp90 following ChIP; lane 1, WCE; lane 2, ChIP with nonspecific IgG; lane 3, ChIP with anti-Hsp90 antibody. H, ChIP with anti-pol II, anti-Hsp90, or control IgG antibodies followed by TaqMan qPCR to detect DNA at the actin promoter and in an intergenic control (Upstream) region. Bars represent DNA fold enrichment relative to total DNA concentration. Data are representative of two independent experiments. I, fold enrichment of HIV-1 LTR DNA or GFP DNA (J) in ChIPs for RNA pol II, Hsp90, and control IgG compared with WCE from acutely infected cells measured by TaqMan qPCR. Mean values ± S.D. of triplicate experiments are shown, representative of two independent experiments. K, schematic representation of the viral vector used for ChIP experiments. RRE, rev-responsive element; SFFV, spleen focus-forming virus promoter. The positions of the probes for TaqMan qPCR are indicated by gray bars.
C-A1 Inhibits Hsp90 by Interfering with Its Dimerization
Biochemical studies have shown that C-A1 is an inhibitor of Hsp90 (39–41); however, its mechanism of action is not understood. We have performed molecular docking to identify binding pockets for C-A1 in Hsp90 and to provide further mechanistic insight into the block to HIV-1 infection mediated by this compound. The program FRED-receptor identified one pocket in the N terminus of Hsp90 (p1) and two at the C terminus (p2 and p3) (Fig. 6A). The N-terminal p1 pocket corresponded to the ATPase domain, for which crystal structures are available with selective ligands such as GA, radicicol, and macbecin (45, 46). Using the programs FRED and MOE, it was possible to successfully dock macbecin at the N-terminal ATPase pocket (Fig. 6B), which showed a Chemgauss3 binding score of −60.6 and was fully superimposable with the known macbecin-Hsp90 co-crystal structure. C-A1 could not be docked to the same pocket, even after removing the pyrrole esters groups from either end. Such groups are unstable and are likely to be removed intracellularly by hydrolysis and oxidation; hence, it was important to examine C-A1 in this conformation as well (47). Of the two novel C-terminal pockets, p2 formed only upon dimerization but appeared to be still accessible by small molecules and was delimited at each side by helix 2 and helix 4 of one monomer and by helix 5 of the other monomer (Fig. 6C and supplemental Fig. S3) (48). C-A1, without the pyrrole ester groups at each side, docked to this pocket and made contacts (within a 4.5-Å radius) mainly with residues ERIMKAQA in helix 2, KLLYE in helix 4, and residues SLDEPTSFASRISLNR in helix 5 of the yeast 2cg9 reference structure (supplemental Methods and supplemental Figs. S3 and S4). Pocket p3 was delimited by helix 2 at the top, helix 4 on one side, and helix 3 and the β-strand connecting helix 1 and helix 2 on the other side (48) (supplemental Fig. S3). C-A1 was found to dock to p3 making contacts (within a 4.5 Å radius) with residues EILGDQ in helix 1 and the connected b strand, residues ANMERIMKAQA in helix 2 and AQDKTVKDLTKLLYETALLTSGFSLDEPTSFASR in helix 4, and the connecting domain to helix 5 (supplemental Figs. S3 and S4). The p2 C-A1 docking site sits on the domain shown to be important for Escherichia coli Hsp90 dimerization (supplemental Fig. S4) (48), and several Hsp90 residues making contact with the docked C-A1 molecule were shown to be important for novobiocin binding to Hsp90 in pulldown assays (39, 40). We sought to confirm these results by examining the effect of C-A1 on Hsp90 dimerization. The C-terminal Hsp90 dimerization domain (Hsp90(527–724)) (39) was expressed, purified, and used in dimerization assays in the presence of C-A1 or partially hydrolyzed C-A1, which lacked one of the pyrrolic ester rings. Both forms of the compound inhibited Hsp90 dimer formation; however, the hydrolyzed C-A1 was 50 times more potent than normal C-A1 (Fig. 6, E and F, and supplemental Fig. S5), in agreement with the molecular docking prediction. Together, these results support the notion that C-A1 inhibits Hsp90 function by destabilizing or preventing the formation of the Hsp90 C-terminal dimer (39, 40).
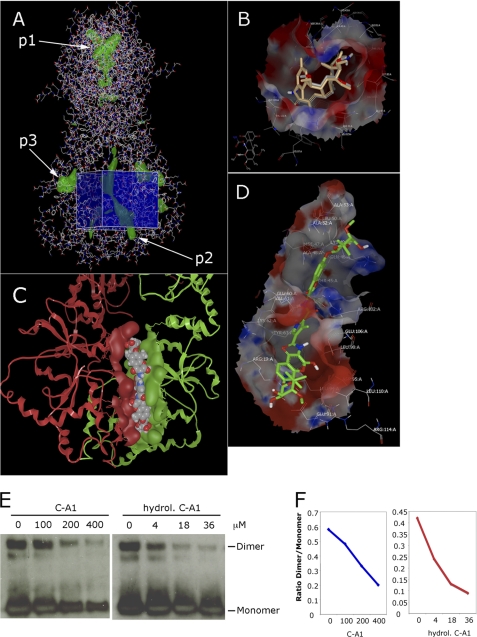
FIGURE 6.
Docking studies reveal two binding pockets for C-A1 in the C-terminal dimerization domain of Hsp90. A, line view in Fred_receptor (OpenEye) of Hsp90 crystal structure Protein Data Bank code 2cg9 from yeast (Sacharomyces cerevisiae). Residues 1–677 (except charged linker) include the N-terminal, middle, and C-terminal domains as a homodimer in the “closed” form. The co-chaperone protein Sba1 has been removed. Green patches represent cavities detected by Fred_receptor; p1 is the N-terminal ATP-binding pocket (in this orientation the two ATP pockets from each monomer are overlaid). P2 is the cleft formed as a result of dimerization, and p3 is found in the C terminus of each monomer. The blue cube represents the volume and location of the docking limits used in FRED. B, VIDA (OpenEye) of the ATP-binding site (p1 pocket) of 2vwc yeast (S. cerevisiae) Hsp90 N-terminal domain (1–219) with bound inhibitor macbecin as found in the crystal structure (gray carbons) and overlaid with docked macbecin (gold carbons). C, C-terminal domains of the homodimer of 2cg9 yeast (S. cerevisiae) Hsp90, secondary structure ribbons colored red and green, respectively. Surface represents the groove resulting from dimerization (p2 pocket) with bound C-A1, represented here as van der Waals spheres in CPK colors. D, stick view of C-A1 (green carbons) in VIDA showing a selected single pose docked into the 1sf8 E. coli C-terminal domain of Hsp90. The binding surface is colored by charge: purple, H-bonding; green, hydrophobic, and blue, mild polar. E, dimerization assay in the presence of C-A1 or hydrolyzed C-A1. 2 μm His-Hsp90β(527–724) was incubated with C-A1 or hydrolyzed (hydrol.) C-A1 at the indicated concentrations prior to chemical cross-linking with bis(sulfosuccinimidyl) suberate, followed by SDS-PAGE and Western blot with anti-Hsp90 antibodies. F, relative intensity of the bands was quantified using ImageJ, and the data are plotted as ratio of Hsp90 dimeric to monomeric forms.
Sensitivity to C-A1 Maps to p24 Capsid
To map the viral determinants of sensitivity to C-A1, wild type NL4.3 virus was serially passaged in culture in the presence of suboptimal concentrations of C-A1 until an escape mutant was detected, which was able to grow in the presence of 1 μm C-A1 (supplemental Fig. S6). The resistant and the parental viruses were sequenced in parallel, and several mutations were detected; however, only three mutations were present in all four clones examined (supplemental Fig. S6). The HIV-1 sequences from the Los Alamos repository were surveyed, and the mutation in the 5′ LTR of the resistant virus was found in several other HIV-1 isolates, suggesting that it might be a nonspecific polymorphism. The two other mutations, A105S in p24 capsid and Q33H in Nef (supplemental Fig. S5B), were specific and were introduced into the wild type NL4.3 backbone by site-directed mutagenesis. Remarkably, the Q33H mutation in Nef did not induce any phenotypic changes (data not shown), but the NL4.3 virus harboring the A105S capsid mutation acquired resistance to C-A1 (Fig. 7B). The IC50 of the mutant virus increased about 5-fold over wild type to 1.63 ± 0.04 μm (Fig. 7C). These data argued that sensitivity to C-A1-mediated inhibition mapped, at least in part, to the capsid protein.
FIGURE 7.
Sensitivity to C-A1-mediated inhibition maps in part to p24 CA. A, A105S mutation in CA detected by sequencing the C-A1-resistant virus. B, A105S mutation was introduced into the parental NL4.3 wild type virus by site-directed mutagenesis. The mutant (D31-GAG) and parental (NL4.3) viruses were then used to infect C8166 cells (at an input of 5pg RT for both viruses) in the presence of the indicated doses of C-A1. RT activity was monitored at regular intervals for 14 days. C, C8166 cells were infected at the same m.o.i. (5 pg RT) with parental and mutant viruses, and the percentage of infected cells was detected by FACS 72 h post-infection after immunostaining for p24 CA. Mean values ± S.D. are shown, n = 3. D, SupT1 cells infected with the VSV-G pseudotyped HIV-1 vector bearing the A105S mutation (D31-GAG) or wild type CA (WT-GAG) at the same m.o.i. in the presence of the indicated dose of GA and analyzed 24 h post-infection by FACS. Mean values ± S.D. are shown, n = 3. E, SupT1 cells infected with D31-GAG or WT-GAG vectors at the same m.o.i. in the presence of the indicated dose of C-A1 and analyzed 24 h post-infection by FACS (left panel) and TaqMan qPCR to detect total viral DNA (middle panel) or two LTR circular viral DNAs (right panel). Mean values ± S.D. are shown, n = 3. F, same infected cells shown in E were re-analyzed 2 weeks later by FACS (left panel), TaqMan qPCR to measure total viral DNA (middle panel) and Alu-LTR qPCR (right panel) to measure integrated proviruses. Mean values ± S.D. are shown, n = 3. Paired Student's t test was used to assess statistical significance.
C-A1 Targets Capsid to Impair HIV-1 Integration
To investigate if the mutant virus escaped the integration or the gene expression block, or both, the A105S capsid mutation was introduced into an HIV-1 vector, and single cycle infection experiments were performed. Parental (WT-GAG) and mutant (D31-GAG) vectors were equally sensitive to GA (Fig. 7D), which only impairs viral gene expression by targeting Hsp90. Cells were then infected with the two vectors and analyzed 24 h and 2 weeks post-infection as described earlier. At the 24-h time point, C-A1 inhibited equally well infection of parental and mutant vectors, and no inhibition of reverse transcription and two LTR viral DNA synthesis was detected (Fig. 7E), in agreement with previous experiments. The same cells were re-analyzed 2 weeks later, and the D31-GAG mutant virus showed resistance to C-A1 (Fig. 7F). To examine integration, we performed TaqMan qPCR to measure viral DNA copies, and Alu-LTR qPCR to directly measure integrated proviruses. The results obtained with the two different approaches matched and demonstrated that the D31-GAG mutant virus was resistant to the integration block imposed by C-A1 (Fig. 7F). Taken together, these results confirmed that C-A1 acted through two different mechanisms and revealed that susceptibility to the C-A1-mediated block to integration maps to capsid.
To explore further the importance of capsid as target of C-A1, we performed a second ToA experiment designed to bypass the anti-Hsp90 activity of the compound. C-A1 was added at different time intervals post-infection and washed out 24 h later. In contrast to the ToA experiment shown in Fig. 2B where cells were analyzed 24 h post-infection, in this experiment cells were analyzed 2 weeks post-infection to detect expression mainly from integrated proviruses. At 2 weeks post-infection, the anti-Hsp90 effect of C-A1 is no longer relevant, as demonstrated previously (Fig. 4). CsA, which prevents binding of cypA to capsid and impairs HIV-1 infectivity (49), was tested in parallel to compare the temporal dynamics of the effect of C-A1 with a drug that targets capsid. Interestingly, in these conditions C-A1 and CsA showed a very similar profile, and both drugs started losing their effect 2–4 h post-infection (Fig. 8A). This result and the results shown in Fig. 2B confirmed that C-A1 acts on two independent steps of the HIV-1 life cycle. More importantly, it was consistent with the possibility that CsA and C-A1 both targeted an early event mediated by the capsid proteins. If this was the case, then one prediction was that C-A1 and CsA might interfere with each other, either synergistically or antagonistically. To test this, cells were infected in the presence of increasing doses of CsA alone or in combination with a fixed amount of C-A1 and analyzed 2 weeks post-infection. The combination of 2 μm CsA with >2 μm C-A1 showed some cytotoxocity; hence, we titrated the two drugs in combination in preliminary experiments to achieve the optimal balance between activity and lack of cytotoxicity. Whereas CsA modestly reduced HIV-1 infection in a dose-dependent way when added alone at the indicated concentrations, it partially but reproducibly rescued infection in the presence of C-A1, indicating that CsA antagonized the effect of C-A1 (Fig. 8B). To further examine this aspect, we tested the effect of C-A1 on an HIV-1 vector bearing the P90A capsid mutation, which has impaired ability to bind cypA (50). Interestingly, the P90A mutant was partially resistant to C-A1 (Fig. 8C), suggesting that cypA is required for optimal antiretroviral activity of C-A1. Taken together, the fact that resistance to C-A1 mapped to capsid, the results of the ToA experiments and the effects of cypA indicated that C-A1 targets capsid and in so doing impairs integration.
FIGURE 8.
C-A1 is influenced by cypA. A, SupT1 cells infected with a VSV-G pseudotyped HIV-1 vector at an m.o.i. of 0.05 in the presence of 4 μm C-A1 or 2 μm CsA; drugs were added at the indicated time post-infection and cells analyzed by FACS 2 weeks post-infection. Data are representative of two independent experiments. B, SupT1 cells were infected with a HIV-1 vector at an m.o.i. of 0.05 in the presence of the indicated concentration of CsA with or without a fixed concentration of C-A1 (2 μm) and analyzed by FACS 2 weeks post-infection. Data are representative of four experiments. C, SupT1 cells were infected at with wild type or a P90A mutant HIV-1 vector at an m.o.i. 0.8 in the presence of the indicated concentrations of C-A1. Samples were analyzed by FACS 2 weeks post-infection. Data are representative of four experiments.
DISCUSSION
The identification of novel host targets is an important approach to develop antimicrobial agents (6), and it may reveal new insight into the biology of virus infection and, more fundamentally, into new cellular processes. By screening small molecule inhibitors of ATP-dependent DNA motors, we detected C-A1, a gyrB inhibitor, as a potent “dual target” antiretroviral drug. C-A1 inhibited viral gene expression and integration independently, via two distinct targets.
The block to viral gene expression mapped to Hsp90. Several lines of evidence support this conclusion. GA and 17-AAG, two specific Hsp90 inhibitors, recapitulated the phenotype induced by C-A1 on viral gene expression, and depletion of Hsp90 at levels that were not toxic to cells inhibited HIV-1 infection to a degree that paralleled the protein KD level. Lower endogenous levels of Hsp90 in primary cells explained their greater sensitivity to the antiviral effect of 17-AAG and C-A1. Moreover, depletion of Hsp90 induced sensitization to the antiretroviral activity of both C-A1 and GA, consistent with the notion that Hsp90 is the actual target of C-A1 (44). Molecular docking studies showed that C-A1 could be docked to two pockets in the C-terminal dimerization domain of Hsp90. A dimerization assay confirmed that C-A1 inhibited Hsp90 dimer formation and that hydrolyzed C-A1, lacking one of the pyrrolic ester rings, was more potent than normal C-A1, in keeping with the molecular docking results and in agreement with previous biochemical analyses aimed at mapping the binding site of C-A1 in Hsp90 (39, 40). Although these results await crystallographic confirmation, they suggested that C-A1 inhibited viral gene expression by interfering with Hsp90 dimerization and are in good agreement with recent evidence showing fast closing and opening dynamics of the Hsp90 C-terminal domain (51).
Both C-A1 and GA prevented gene expression from the HIV-1 LTR as well as a Tat-independent internal promoter present in the HIV-1 vector. Therefore, it is possible that C-A1 impaired viral gene expression by a mechanism that is Tat-independent and precedes assembly of the cdk9-cyclin T1 complex (52). C-A1 and GA suppressed gene expression in acutely infected cells but had little effect in chronically infected cells, suggesting that inhibition of Hsp90 may affect the establishment of an actively transcribing virus in the early phases post-integration. Interestingly, a similar phenotype has been recently described and shown to be caused by inhibition of the sulfonation pathway (53). ChIP experiments showed that Hsp90 was associated with the viral DNA promoter, suggesting that it may be directly involved in regulating gene expression. We propose that Hsp90 promotes chromatin remodeling at the time of integration to induce a favorable environment for viral gene expression. Previous studies have demonstrated chromatin remodeling at the first nucleosome downstream of the transcription start site (nuc-1), which is promoted by Tat and mediated by the SWI/SNF complex (54, 55). Although Hsp90 might participate in this process, our observations with the Tat-independent vector suggest an even earlier and more general mechanism. In support of our interpretation, it has been previously shown that C-A1 inhibits transcriptional initiation from plasmid DNA and integrated phage DNA in E. coli, but it has little effect once steady state levels of transcription have been reached (56, 57). This phenotype in E. coli mapped to gyrB and was linked to DNA supercoiling and chromatin condensation (57). In yeast, Hsp90 has been recently shown to induce transcription by interacting with components of the chromatin remodeling machinery such as The2 proteins and Rvb1 and Rvb2 helicases (58). Hsp90 was also shown to promote rapid nucleosomal removal required for induction of transcription (59). In Drosophila, Hsp90 directly cooperates with Trithorax to maintain the active expression state of target genes (60). Our observations on the effects of C-A1 and GA on HIV-1 gene expression argue for a conserved role of Hsp90 in promoting chromatin remodeling and the early steps of gene expression in human cells.
Interestingly, Hsp90 inhibition did not affect HIV-1 integration. In fact, the block to HIV-1 integration mediated by C-A1 mapped to capsid. When the A105S capsid mutation was introduced into a wild type HIV-1 backbone, the resulting virus was partially resistant to C-A1, and single cycle infection experiments showed that the A105S mutation conferred resistance to the integration block. Further support for the role of capsid came from ToA experiments analyzed 2 weeks post-infection, in which C-A1 and CsA were both found to interfere with an early step of the HIV-1 life cycle. In addition, CsA partially but reproducibly antagonized the antiviral effect of C-A1, and the P90A capsid mutant virus was less sensitive to C-A1 than wild type virus, suggesting that cypA is required for optimal antiretroviral activity. These results further support the notion that capsid is the actual target of C-A1 involved in the block to integration. There is growing evidence that capsid may influence post-nuclear entry events. Specific mutations in capsid impair integration or a step leading to integration (61–64). This phenotype is presumably due to the fact that capsid must be completely shed from the HIV-1 pre-integration complex to prevent steric hindrance, allowing optimal chromatin tethering or binding to host factors such as LEDGF/p75 important for efficient integration (65).
We propose two hypotheses for the mechanism of action of C-A1. First, C-A1 may interfere with the progressive “shedding” of capsid from the viral reverse transcription complex-pre-integration complex or, in other words, disturb uncoating. The target could be the capsid itself or a chaperone, different from Hsp90, binding to capsid. In this regard, the antagonistic effect of CsA suggests that C-A1 may mimic an excess of cypA, which is known to influence infectivity and the stability of the viral core in a cell type-dependent way (49, 66–70). Second, C-A1 may prevent binding to capsid of a different cellular factor important for integration in certain cell types. Our results suggest that this hypothetical host factor must bind to capsid early during infection. Clearly, C-A1 could be used as a tool to further investigate these hypotheses.
C-A1 was developed in the early 1970s by Roche Applied Science and was shown to be well tolerated in toxicological and pharmacokinetics studies in humans but had poor oral bioavailability (71, 72). Because C-A1 blocks two steps of the HIV-1 life cycle by targeting distinct factors, viral resistance is likely to be only partial, in agreement with our findings. Therefore, C-A1 may represent a good starting point for future antiretroviral drug development. Indeed, molecular chaperones have been proposed as targets for antimicrobial therapy (73).
Selective Hsp90 inhibitors derived from GA are currently being tested as anti-cancer agents in phase I to III clinical trials, including trials for patients with Hodgkin and non-Hodgkin lymphomas (clinicaltrials.gov). Our data provide a rationale to further explore the possibility of extending such trials to the increasing number of patients with AIDS-related malignancies, such as non-Hodgkin lymphomas (74). Lowering plasma HIV-1 load as well as delaying viral rebound after chemotherapy has been shown to dramatically improve prognosis in these patients, and because Hsp90 inhibitors should protect macrophages from HIV-1 infection, they are likely to improve the overall treatment outcome when used in association with an appropriately tailored highly active antiretroviral therapy regimen (75).
Supplementary Material
Acknowledgments
We thank Andrew Porter for the HTETOP cells; Caroline Austin for the mTOP cells; Tom Ratajczak for Hsp90 expression plasmids; Elena Sokolskaja for the P90A HIV-1 mutant, and Peter Cherpanov for Raltegravir. We thank Steve Caddick, Paul Kellam, Chris Parry, Laurence Pearl, Deenan Pillay, and Yasu Takeuchi for discussions. We also thank Mahad Noursadeghi for primary macrophages; Anna Forsman, Edward Wright, Greg Towers, and members of his laboratory for reagents.
This work was supported by the Wellcome Trust and University College London Hospital Charities Clinical Research and Development Committee Grant F144 (to A. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Figs. S1–S6, and additional references.
- C-A1
- coumermycin A1
- CsA
- cyclosporine A
- Hsp90
- heat shock protein 90
- cypA
- cyclophilin A
- GA
- geldanamycin
- Topo2
- DNA topoisomerase II
- WCE
- whole-cell extract
- gyrB
- gyrase B
- ToA
- time of addition
- PBMC
- peripheral blood mononuclear cell
- m.o.i.
- multiplicity of infection
- qPCR
- quantitative PCR
- KD
- knockdown
- pol
- polymerase
- VSV
- vesicular stomatitis virus
- 17-AAG
- 17-allylamino-17-demethoxygeldanamycin.
REFERENCES
- 1.UNAIDS (2008) AIDS Epidemic Update 2009, www.UNAIDS.org
- 2.Palella F. J., Jr., Delaney K. M., Moorman A. C., Loveless M. O., Fuhrer J., Satten G. A., Aschman D. J., Holmberg S. D. (1998) N. Engl. J. Med. 338, 853–860 [DOI] [PubMed] [Google Scholar]
- 3.Shafer R. W., Schapiro J. M. (2008) AIDS Rev. 10, 67–84 [PMC free article] [PubMed] [Google Scholar]
- 4.Hué S., Gifford R. J., Dunn D., Fernhill E., Pillay D. (2009) J. Virol. 83, 2645–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith R. J., Okano J. T., Kahn J. S., Bodine E. N., Blower S. (2010) Science 327, 697–701 [DOI] [PubMed] [Google Scholar]
- 6.Kellam P. (2006) Genome Biol. 7, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F. D., Malani N., Fernandes J., D'Orso I., Cagney G., Diamond T. L., Zhou H., Hazuda D. J., Espeseth A. S., König R., Bandyopadhyay S., Ideker T., Goff S. P., Krogan N. J., Frankel A. D., Young J. A., Chanda S. K. (2009) PLoS Pathog. 5, e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stockwell B. R. (2004) Nature 432, 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciuffi A. (2008) Curr. Gene Ther. 8, 419–429 [DOI] [PubMed] [Google Scholar]
- 10.Fassati A. (2006) Retrovirology 3, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demaison C., Parsley K., Brouns G., Scherr M., Battmer K., Kinnon C., Grez M., Thrasher A. J. (2002) Hum. Gene Ther. 13, 803–813 [DOI] [PubMed] [Google Scholar]
- 12.Fassati A., Goff S. P. (2001) J. Virol. 75, 3626–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clapham P. R., McKnight A., Weiss R. A. (1992) J. Virol. 66, 3531–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaitseva L., Cherepanov P., Leyens L., Wilson S. J., Rasaiyaah J., Fassati A. (2009) Retrovirology 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutiño-Moguel T., Fassati A. (2006) Traffic 7, 978–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loh B., Vozzolo L., Mok B. J., Lee C. C., Fitzmaurice R. J., Caddick S., Fassati A. (2010) Chem. Biol. Drug Des. 75, 461–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Doherty U., Swiggard W. J., Jeyakumar D., McGain D., Malim M. H. (2002) J. Virol. 76, 10942–10950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler S. L., Hansen M. S., Bushman F. D. (2001) Nat. Med. 7, 631–634 [DOI] [PubMed] [Google Scholar]
- 19.Mbisa J. L., Delviks-Frankenberry K. A., Thomas J. A., Gorelick R. J., Pathak V. K. (2009) Methods Mol. Biol. 485, 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenner R. G., Townsend M. J., Jackson I., Sun K., Bouwman R. D., Young R. A., Glimcher L. H., Lord G. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17876–17881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim R. H., Kim R., Chen W., Hu S., Shin K. H., Park N. H., Kang M. K. (2008) Carcinogenesis 29, 2425–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachur N. R., Johnson R., Yu F., Hickey R., Applegren N., Malkas L. (1993) Mol. Pharmacol. 44, 1064–1069 [PubMed] [Google Scholar]
- 23.Burden D. A., Osheroff N. (1998) Biochim. Biophys. Acta 1400, 139–154 [DOI] [PubMed] [Google Scholar]
- 24.Pommier Y., Pourquier P., Fan Y., Strumberg D. (1998) Biochim. Biophys. Acta 1400, 83–105 [DOI] [PubMed] [Google Scholar]
- 25.Yamashita M., Emerman M. (2004) J. Virol. 78, 5670–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartkoorn R. C., Kwan W. S., Shallcross V., Chaikan A., Liptrott N., Egan D., Sora E. S., James C. E., Gibbons S., Bray P. G., Back D. J., Khoo S. H., Owen A. (2010) Pharmacogenet. Genomics 20, 112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baba M. (1989) Int. J. Exp. Clin. Chemother. 2, 15–20 [Google Scholar]
- 28.Tachedjian G., Tyssen D., Jardine D., Locarnini S., Birch C. (1992) Antivir. Chem. Chemother. 3, 183–188 [Google Scholar]
- 29.Pannecouque C., Pluymers W., Van Maele B., Tetz V., Cherepanov P., De Clercq E., Witvrouw M., Debyser Z. (2002) Curr. Biol. 12, 1169–1177 [DOI] [PubMed] [Google Scholar]
- 30.De Clercq E. (2003) Nat. Rev. Drug Discov. 2, 581–587 [DOI] [PubMed] [Google Scholar]
- 31.Hazuda D., Iwamoto M., Wenning L. (2009) Annu. Rev. Pharmacol. Toxicol. 49, 377–394 [DOI] [PubMed] [Google Scholar]
- 32.Butler S. L., Johnson E. P., Bushman F. D. (2002) J. Virol. 76, 3739–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis R. J., Singh O. M., Smith C. V., Skarzynski T., Maxwell A., Wonacott A. J., Wigley D. B. (1996) EMBO J. 15, 1412–1420 [PMC free article] [PubMed] [Google Scholar]
- 34.Dutta R., Inouye M. (2000) Trends Biochem. Sci. 25, 24–28 [DOI] [PubMed] [Google Scholar]
- 35.Inoue N., Hess K. D., Moreadith R. W., Richardson L. L., Handel M. A., Watson M. L., Zinn A. R. (1999) Hum. Mol. Genet. 8, 1201–1207 [DOI] [PubMed] [Google Scholar]
- 36.Matheson E. C., Hall A. G. (2003) Carcinogenesis 24, 31–38 [DOI] [PubMed] [Google Scholar]
- 37.Gadelle D., Filée J., Buhler C., Forterre P. (2003) BioEssays 25, 232–242 [DOI] [PubMed] [Google Scholar]
- 38.Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1997) Cell 90, 65–75 [DOI] [PubMed] [Google Scholar]
- 39.Allan R. K., Mok D., Ward B. K., Ratajczak T. (2006) J. Biol. Chem. 281, 7161–7171 [DOI] [PubMed] [Google Scholar]
- 40.Marcu M. G., Chadli A., Bouhouche I., Catelli M., Neckers L. M. (2000) J. Biol. Chem. 275, 37181–37186 [DOI] [PubMed] [Google Scholar]
- 41.Burlison J. A., Blagg B. S. (2006) Org. Lett. 8, 4855–4858 [DOI] [PubMed] [Google Scholar]
- 42.Stebbins C. E., Russo A. A., Schneider C., Rosen N., Hartl F. U., Pavletich N. P. (1997) Cell 89, 239–250 [DOI] [PubMed] [Google Scholar]
- 43.Chiosis G. (2006) Curr. Top. Med. Chem. 6, 1183–1191 [DOI] [PubMed] [Google Scholar]
- 44.Whitehurst A. W., Bodemann B. O., Cardenas J., Ferguson D., Girard L., Peyton M., Minna J. D., Michnoff C., Hao W., Roth M. G., Xie X. J., White M. A. (2007) Nature 446, 815–819 [DOI] [PubMed] [Google Scholar]
- 45.Martin C. J., Gaisser S., Challis I. R., Carletti I., Wilkinson B., Gregory M., Prodromou C., Roe S. M., Pearl L. H., Boyd S. M., Zhang M. Q. (2008) J. Med. Chem. 51, 2853–2857 [DOI] [PubMed] [Google Scholar]
- 46.Roe S. M., Prodromou C., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1999) J. Med. Chem. 42, 260–266 [DOI] [PubMed] [Google Scholar]
- 47.Uetrecht J. P., Trager W. (2007) Drug Metabolism. Chemical and Enzymatic Aspects, pp. 120–130, Informa Healthcare, New York [Google Scholar]
- 48.Harris S. F., Shiau A. K., Agard D. A. (2004) Structure 12, 1087–1097 [DOI] [PubMed] [Google Scholar]
- 49.Thali M., Bukovsky A., Kondo E., Rosenwirth B., Walsh C. T., Sodroski J., Göttlinger H. G. (1994) Nature 372, 363–365 [DOI] [PubMed] [Google Scholar]
- 50.Gitti R. K., Lee B. M., Walker J., Summers M. F., Yoo S., Sundquist W. I. (1996) Science 273, 231–235 [DOI] [PubMed] [Google Scholar]
- 51.Ratzke C., Mickler M., Hellenkamp B., Buchner J., Hugel T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16101–16106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Keeffe B., Fong Y., Chen D., Zhou S., Zhou Q. (2000) J. Biol. Chem. 275, 279–287 [DOI] [PubMed] [Google Scholar]
- 53.Bruce J. W., Ahlquist P., Young J. A. (2008) PLoS Pathog. 4, e1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahmoudi T., Parra M., Vries R. G., Kauder S. E., Verrijzer C. P., Ott M., Verdin E. (2006) J. Biol. Chem. 281, 19960–19968 [DOI] [PubMed] [Google Scholar]
- 55.Tréand C., du Chéné I., Brès V., Kiernan R., Benarous R., Benkirane M., Emiliani S. (2006) EMBO J. 25, 1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith C. L., Kubo M., Imamoto F. (1978) Nature 275, 420–423 [DOI] [PubMed] [Google Scholar]
- 57.Yang H. L., Heller K., Gellert M., Zubay G. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 3304–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao R., Houry W. A. (2005) Biochem. Cell Biol. 83, 703–710 [DOI] [PubMed] [Google Scholar]
- 59.Floer M., Bryant G. O., Ptashne M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tariq M., Nussbaumer U., Chen Y., Beisel C., Paro R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1157–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamashita M., Emerman M. (2009) J. Virol. 83, 9835–9843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dismuke D. J., Aiken C. (2006) J. Virol. 80, 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qi M., Yang R., Aiken C. (2008) J. Virol. 82, 12001–12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamashita M., Perez O., Hope T. J., Emerman M. (2007) PLoS Pathog. 3, 1502–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Engelman A., Cherepanov P. (2008) PLoS Pathog. 4, e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arfi V., Lienard J., Nguyen X. N., Berger G., Rigal D., Darlix J. L., Cimarelli A. (2009) J. Virol. 83, 7524–7535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y., Kar A. K., Sodroski J. (2009) J. Virol. 83, 10951–10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bon Homme M., Carter C., Scarlata S. (2005) Biophys. J. 88, 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yin L., Braaten D., Luban J. (1998) J. Virol. 72, 6430–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grättinger M., Hohenberg H., Thomas D., Wilk T., Müller B., Kräusslich H. G. (1999) Virology 257, 247–260 [DOI] [PubMed] [Google Scholar]
- 71.Kaplan S. A. (1970) J. Pharm. Sci. 59, 309–313 [DOI] [PubMed] [Google Scholar]
- 72.Newmark H. L., Berger J. (1970) J. Pharm. Sci. 59, 1246–1248 [DOI] [PubMed] [Google Scholar]
- 73.Neckers L., Tatu U. (2008) Cell Host Microbe 4, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bonnet F., Chêne G. (2008) Curr. Opin. Oncol. 20, 534–540 [DOI] [PubMed] [Google Scholar]
- 75.Bower M., Stebbing J., Tuthill M., Campbell V., Krell J., Holmes P., Ozzard A., Nelson M., Gazzard B., Powles T. (2008) Blood 111, 3986–3990 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.